Laccases and peroxidases are encoded by large gene families in plants, and both enzymes have been implicated in the polymerization of monolignols during lignification. Loss of function of three LACCASE genes in Arabidopsis essentially eliminates lignification in root and stem tissue, in the absence of reductions in peroxidase transcripts, indicating that laccase is essential for lignification.
Abstract
The evolution of lignin biosynthesis was critical in the transition of plants from an aquatic to an upright terrestrial lifestyle. Lignin is assembled by oxidative polymerization of two major monomers, coniferyl alcohol and sinapyl alcohol. Although two recently discovered laccases, LAC4 and LAC17, have been shown to play a role in lignin polymerization in Arabidopsis thaliana, disruption of both genes only leads to a relatively small change in lignin content and only under continuous illumination. Simultaneous disruption of LAC11 along with LAC4 and LAC17 causes severe plant growth arrest, narrower root diameter, indehiscent anthers, and vascular development arrest with lack of lignification. Genome-wide transcript analysis revealed that all the putative lignin peroxidase genes are expressed at normal levels or even higher in the laccase triple mutant, suggesting that lignin laccase activity is necessary and nonredundant with peroxidase activity for monolignol polymerization during plant vascular development. Interestingly, even though lignin deposition in roots is almost completely abolished in the lac11 lac4 lac17 triple mutant, the Casparian strip, which is lignified through the activity of peroxidase, is still functional. Phylogenetic analysis revealed that lignin laccase genes have no orthologs in lower plant species, suggesting that the monolignol laccase genes diverged after the evolution of seed plants.
INTRODUCTION
Lignin is a phenylpropanoid-derived polymer naturally present in specific cell types of vascular plants, particularly those with secondarily thickened cell walls where it plays critical roles in mechanical support, water transport, and pathogen defense. As a branch of the phenylpropanoid pathway, monolignol synthesis occurs in the cytosol via a series of aromatic hydroxylation and O-methylation reactions as well as successive side-chain reductions. Generally, the lignin polymer is considered to arise from three monomers: p-coumaryl alcohol (H unit, minor unit), coniferyl alcohol (G unit), and sinapyl alcohol (S unit), which differ in their degree of methoxylation. However, other units can also be incorporated into lignin polymers. For example, it has recently been reported that the lignin in the seed coat of the monocotyledonous angiosperm Vanilla plainfolia is a homopolymer of caffeyl alcohol (C unit) (Chen et al., 2012). After their synthesis in the cytosol, monolignols are transported across the plasma membrane by yet-to-be-fully-defined mechanisms, but likely including ATP binding cassette–like transporters (Miao and Liu, 2010), for free-radical–based oxidative polymerization in the cell wall (Miao and Liu, 2010; Alejandro et al., 2012).
It was first suggested over 20 years ago that laccases are involved in oxidative lignin polymerization in plant species (Sterjiades et al., 1992; Bao et al., 1993). However, early studies mostly addressed oxidation of lignin precursors in vitro. Recently, study of the Arabidopsis thaliana laccase4 (lac4) and lac17 double knockout mutant provided the first in vivo evidence that these two laccase genes are involved in monolignol polymerization (Berthet et al., 2011). However, the fact that disruption of both laccase genes only leads to a moderate reduction in lignin content and that deposition of S lignin units is barely changed in the mutant, indicates that there must be additional lignin laccase genes, or alternate polymerization enzymes, in Arabidopsis. Of the 17 putative laccase genes identified in the Arabidopsis genome (McCaig et al., 2005; Hoegger et al., 2006), eight have significant transcript expression in the highly lignified inflorescence stem (Berthet et al., 2011).
Here, we identify LAC11 as a further laccase involved in monolignol polymerization. LAC11 is expressed in the Arabidopsis stem and is under control of SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 (SND1), a master regulator of secondary cell wall biosynthesis. The lac11 single knockout mutant shows normal lignin deposition, and the lac11 lac4 and lac11 lac17 double knockout mutants only show mild lignin reduction. However, simultaneous disruption of LAC4, LAC11, and LAC17 almost completely abolishes lignin deposition, causing severe plant growth arrest. Our data imply that peroxidase does not function in a redundant route for lignin polymerization, at least in the vascular tissue of Arabidopsis.
RESULTS
Identification of Arabidopsis Laccases Regulated by Secondary Cell Wall Transcriptional Regulators
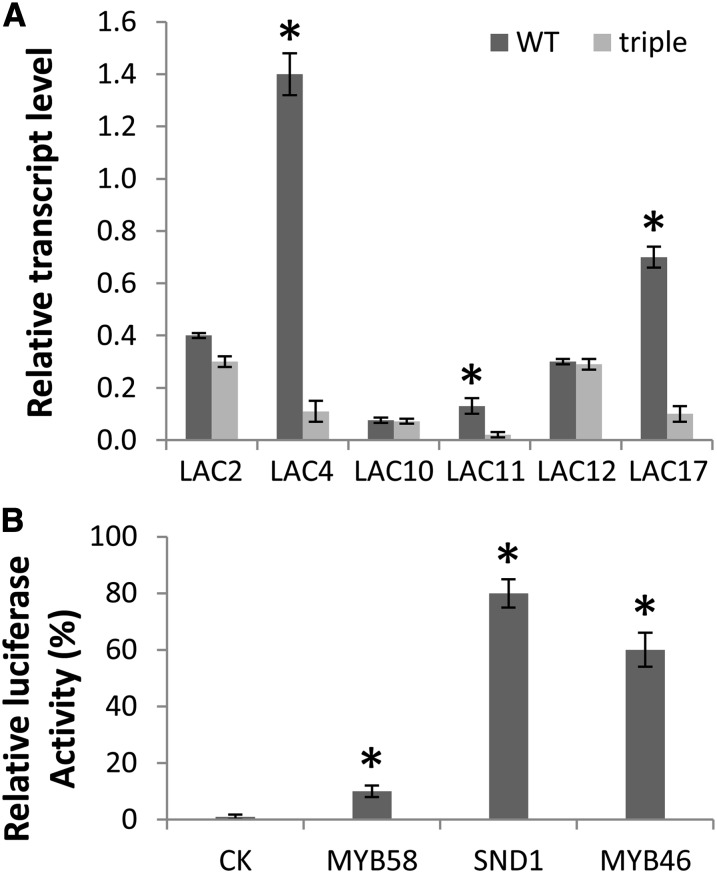
To identify novel monolignol laccases, annotated laccases with relatively high transcript expression in lignifying stem tissue (Berthet et al., 2011; Turlapati et al., 2011) were selected as candidates. Because monolignol polymerization is part of the secondary cell wall biosynthesis program, monolignol laccases should theoretically be under the control of the NAC SECONDARY WALL THICKENING PROMOTING FACTOR (NST) transcription factor family (Mitsuda et al., 2005, 2007). Therefore, transcript levels of putative monolignol laccases were compared in the Arabidopsis nst1 nst2 nst3 triple mutant background along with the wild-type control by quantitative real-time PCR (Figure 1A). lac4 and lac17 were previously identified as monolignol laccases (Berthet et al., 2011), and these two genes are significantly downregulated in the nst1 nst2 nst3 triple mutant, indicating that this is a reasonable criterion for screening for additional monolignol laccases. In addition to LAC4 and LAC17, the expression level of LAC11 was also significantly reduced in the triple mutant (Figure 1A).
Figure 1.
Arabidopsis LAC11 Is Regulated by a Cell Wall Transcriptional Regulator.
(A) Levels of putative lignin laccase transcripts in the wild type (WT) and nst1 nst2 nst3 triple mutant as determined by quantitative real-time PCR. Expression of genes is normalized to actin. Error bars represent se of three biological replicates. Asterisks indicate values that were determined by the Student’s t test to be significantly different from their equivalent control (P < 0.05)
(B) Transactivation of the LAC11 promoter by cell wall regulatory transcription factors. Each transcription factor was coexpressed in Arabidopsis protoplasts with the firefly luciferase reporter gene driven by the LAC11 promoter. The induction of the reporter gene was measured by assaying firefly luciferase activity. The control, which has no transcription factor coexpressed, is set to 1.0. A 35S promoter–driven Renilla luciferase was coexpressed to standardize the transfection efficiency. Error bars represent se of three biological replicates. CK, control check.
To further investigate how LAC11 is regulated by secondary cell wall transcription factors, a protoplast-based in vitro transfection assay was used to test the effects of coexpression of the transcription factors SND1, myeloblastoma family (MYB) 46, and MYB58 on trans-activation of a reporter gene, firefly luciferase, driven by the promoter of LAC11. SND1 (also named NST3) and MYB46 are master transcription switches for the entire secondary cell wall biosynthesis pathway, whereas MYB58 is a lignin-specific transcription factor (Mitsuda et al., 2007; Zhong et al., 2007; Zhou et al., 2009). The firefly luciferase gene construct was generated using 1 kb of DNA upstream of the start codon of LAC11, and all transcription factors were driven by the cauliflower mosaic virus 35S promoter. Coexpression of SND1 or MYB46 significantly activated the expression of the reporter gene by around 80- and 60-fold, respectively, whereas MYB58 activated the LAC11 promoter less efficiently (Figure 1B).
Simultaneous Disruption of LAC4, LAC11, and LAC17 Causes a Severe Plant Growth Defect
To investigate the function of LAC11 in planta, a LAC11 T-DNA insertion line (SALK_063746, lac11-1) was identified in the Salk Institute (SALK) collection. Sequencing of the T-DNA insertion product indicated that the lac11-1 mutant has a T-DNA insertion in the fifth exon of the coding region (Figure 2A). RT-PCR on total RNA extracted from stem tissues confirmed that this T-DNA insertion line was null for LAC11 expression (Figure 2B).
Figure 2.
Identification of the lac11 Mutant.
(A) Schematic diagram of the structure of LAC11 and T-DNA insertion in lac11-1. Numbers indicate nucleotide positions from the site of initiation of translation. Boxes represent exons. Black lines represent introns.
(B) RT-PCR analysis of LAC11 transcripts. Actin is used as a positive control. WT, the wild type.
(C) Three-week-old wild-type and mutant plants grown in the growth chamber. Top left panel is a magnified view of the lac4-2 lac11-1 lac17-1 triple mutant.
[See online article for color version of this figure.]
The lac11-1 single mutant exhibited neither an altered overall growth phenotype nor reduction in lignin level (see Supplemental Figure 1 and Supplemental Table 1 online). Therefore, double mutants were generated by crossing lac11-1 with either lac4-2 or lac17-1 (Berthet et al., 2011), and the double mutants were further crossed to generate the triple mutant. All three double knockout mutants, namely, lac4-2 lac11-1, lac4-2 lac17-1, and lac11-1 lac17-1 showed similar growth to the wild type. By contrast, the triple mutant exhibited a severe growth defect and stopped growing in soil after development of the first two pairs of leaves (Figure 2C). To rule out the possibility that the phenotype of the triple mutant is caused by an unknown insertion, segregation analysis was performed. One hundred and eight lac4-2 lac11-1 homozygous and lac17-1 heterozygous plants as well as 72 lac4-2 lac17-1 homozygous and lac11-1 heterozygous mutant plants were selfed, seed collected, and the plants grown for phenotyping. Twenty-six out of 108 offspring of lac4-2 lac11-1 homozygous and lac17-1 heterozygous plants were triple mutants, and 16 out of 72 offspring of lac4-2 lac17-1 homozygous and lac11-1 heterozygous plants were triple mutants. All the triple mutants had the same growth defect as shown in Figure 2C. All plants with the other genotypes grew normally.
To investigate the impact of laccase deficiency on lignin biosynthesis, lignin composition of the lac11-1 single mutant and the double knockout mutants lac4-2 lac11-1 and lac11-1 lac17-1 was determined by thioacidolysis using entire mature inflorescence stems of 6-week-old plants (Table 1). The thioacidolysis yield of the lac11-1 single mutant was similar to that of wild-type plants. However, the lac4-2 lac11-1 and lac11-1 lac17-1 mutants released significantly smaller amounts of guaiacyl (G) and syringyl (S) monomers (Table 1). The total lignin contents of the entire mature inflorescence stems of 6-week-old plants were also determined by the acetyl bromide method to further confirm the impact of LAC11 deficiency on lignin biosynthesis. The lac4-2 lac11-1 and lac11-1 lac17-1 double knockout mutants had significantly reduced total lignin content, whereas the acetyl bromide lignin level of the single lac11-1 mutant did not differ from that of the wild-type control (see Supplemental Table 1 online). Phloroglucinol staining of roots showed that all three double mutants possessed similar patterns and levels of lignin staining (see Supplemental Figure 2 online).
Table 1. Lignin Composition of Arabidopsis lac11 Single and Double Mutants as Measured by Thioacidolysis.
| Line | H (µmol/g) | G (µmol/g) | S (µmol/g) | Total (µmol/g) |
|---|---|---|---|---|
| Wild type | 1.44 ± 0.1 | 187 ± 3 | 66 ± 1 | 255 ± 4 |
| lac4-2 | 1.45 ± 0.1 | 172 ± 3* | 63 ± 2 | 236 ± 5* |
| lac11-1 | 1.56 ± 0.2 | 177 ± 6 | 64 ± 2 | 243 ± 9 |
| lac17-1 | 1.45 ± 0.1 | 178 ± 4 | 67 ± 4 | 247 ± 8 |
| lac4-2 lac11-1 | 1.57 ± 0.1 | 165 ± 2* | 56 ± 3* | 222 ± 5* |
| lac11-1 lac17-1 | 1.40 ± 0.1 | 167 ± 8* | 55 ± 1* | 223 ± 10* |
All data are means ± se (n = 6). Asterisks indicate values that were determined by the Student’s t test to be significantly different from their equivalent control (P < 0.05).
Comprehensive Phenotypic Analysis of the lac4 lac11 lac17 Triple Mutant
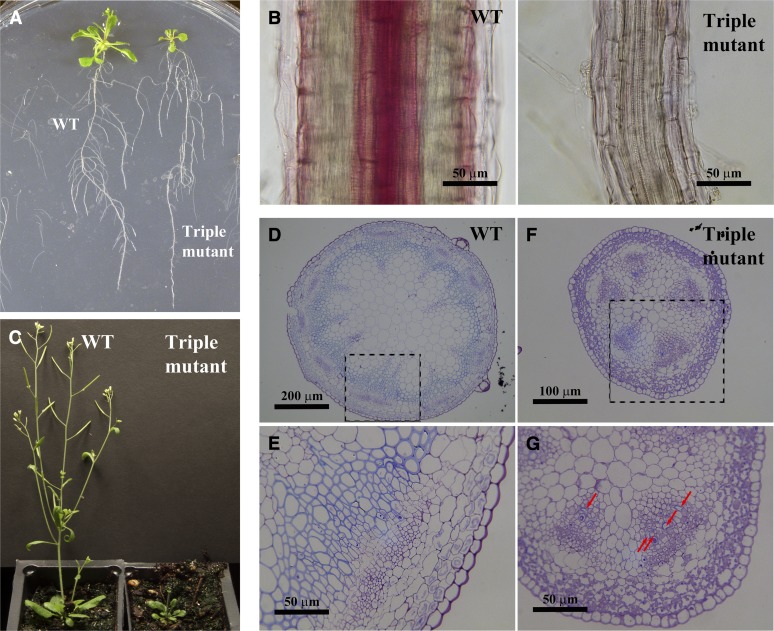
Since the laccase triple mutant is very small, it is difficult to quantify lignin content. Therefore, lignin histochemical staining was used to examine lignin accumulation. At 3 weeks after germination, rosette leaves of the triple mutant are much smaller than those of the wild type, but the overall root growth, including primary root length and lateral root numbers, appears normal (Figure 3A). However, examination under the microscope indicates that the triple mutant has roots with reduced diameter (Figure 3B). Phloroglucinol staining of roots indicated that lignin accumulation is barely detectable in the mutant compared with the wild type; even the normally strongly lignified vascular elements have no obvious lignin staining in the mutant (Figure 3B).
Figure 3.
Phenotypic Analysis of the Laccase Triple Mutant.
(A) Three-week-old seedlings of the wild type (WT) and the laccase triple mutant, which has smaller leaves but relatively normal root development.
(B) Phloroglucinol staining of longitudinal sections of roots of 3-week-old plants. The triple mutant shows little lignification, while the wild-type root has strong lignification in vascular tissues.
(C) Six-week-old plants of the wild type and the laccase triple mutant.
(D) to (G) Stem cross section of 6-week-old plants at two different magnifications. The wild type is shown in (D) and (E) and the triple mutant in (F) and (G). (E) and (G) are higher magnifications of the dotted areas in (D) and (F), respectively. Red arrows show lignified cells in the mutant.
Although the laccase triple mutant is extremely dwarfed, it still develops a small stem-like structure at around 6 weeks, but then never grows longer than about 1 cm. (Figure 3C). To investigate vascular development in the stem, both wild-type and laccase triple mutant stems were embedded in LR White resin, and cross sections from the basal part (just above the soil line) were stained with Toluidine Blue O. The mutant stem was much smaller in diameter; more importantly, the arrangement of the vascular tissues was completely different from that of the wild type (Figures 3D to 3G). The vascular tissues of the mutant resemble those of the juvenile stage of wild-type plants (Jun et al., 2002), even though the tissue would be expected to be mature at 6 weeks after germination. Higher magnification of stem cross sections indicated that very few vascular elements in the mutant are lignified, whereas lignification was observed in most of the xylem tissues in the wild type (Figures 3E and 3G). UV autofluorescence further confirmed that there was no visible phenolic material accumulated in cell walls of the triple mutant, whereas lignin autofluorescence was evident in xylem tissues of the wild type. Additionally, what appear to be chloroplasts (Carmo-Silva et al., 2009) were dominant in the cortical cells of the mutant (see Supplemental Figure 3 online), consistent with the entire stem tissues from the mutant being developmentally comparable to the juvenile stage of the wild type (Figure 3F).
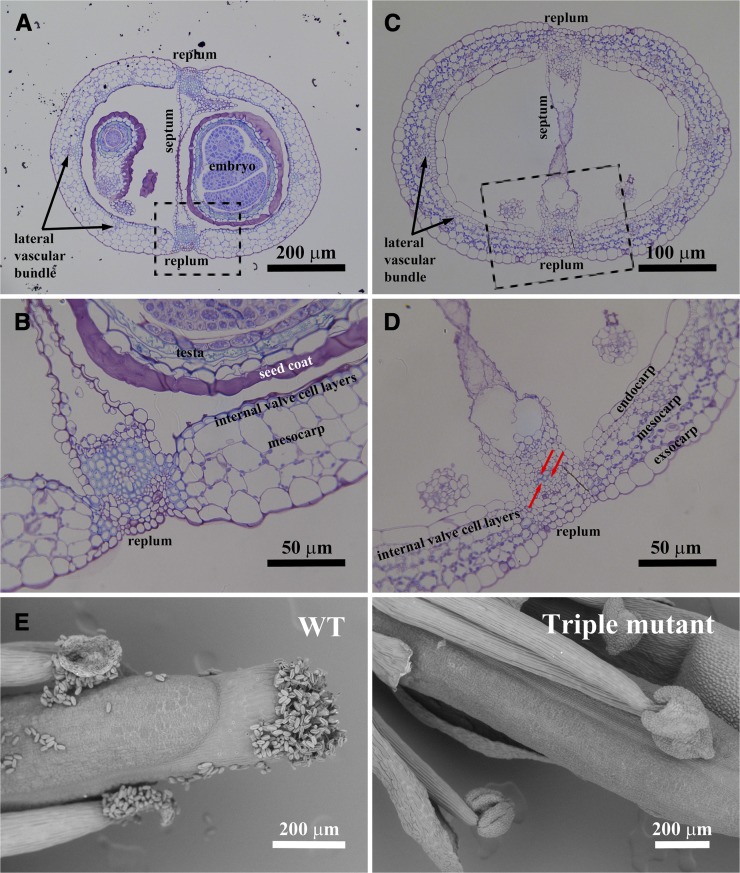
Occasionally, some laccase triple mutants developed siliques, but their size was much smaller than those formed in the wild type (see Supplemental Figure 4 online). Lignification is an important step toward maturation in Arabidopsis siliques (Liljegren et al., 2004). To investigate secondary cell wall development in the triple mutant siliques, cross sections were stained with Toluidine Blue O. Compared with the wild type, the siliques from the laccase triple mutant retain a normal structure, although mature seeds are missing. In the mutant, lignification of vascular bundles located in replums was almost nonexistent, except in the few cells highlighted by the red arrows (Figures 4A and 4C). Furthermore, those vascular elements were of reduced diameter in the triple mutant. Internal valve cell layers (sclerenchyma cells) lost lignin completely in the triple mutant, although the layers themselves were still visible (Figures 4B and 4D).
Figure 4.
Phenotype of the Siliques and Anthers of the Arabidopsis Laccase Triple Mutant.
(A) to (D) Cross sections of siliques of the wild type and laccase triple mutant at two different magnifications. (A) and (B) are the wild type, and (C) and (D) are the triple mutant. (B) and (D) are higher magnifications of the dotted areas in (A) and (C), respectively.
(E) Anther development of the wild type (WT) and laccase triple mutant. Pollen is released from wild-type anthers, whereas the triple mutant has an anther-indehiscent phenotype. Red arrows show lignified cells in the mutant.
The small silique phenotype mimics that of the nst1 nst2 double mutant, which also has disrupted secondary wall thickenings in the anther endothecium, leading to loss of dehiscence (Mitsuda et al., 2005). Therefore, anther development of the triple laccase mutant was also investigated. Anther morphology of the mutant was changed compared with the wild type, and no pollen was released from anthers of the triple mutant (Figure 4E).
Casparian Strip Formation Is Not Affected in the lac4 lac11 lac17 Triple Mutant
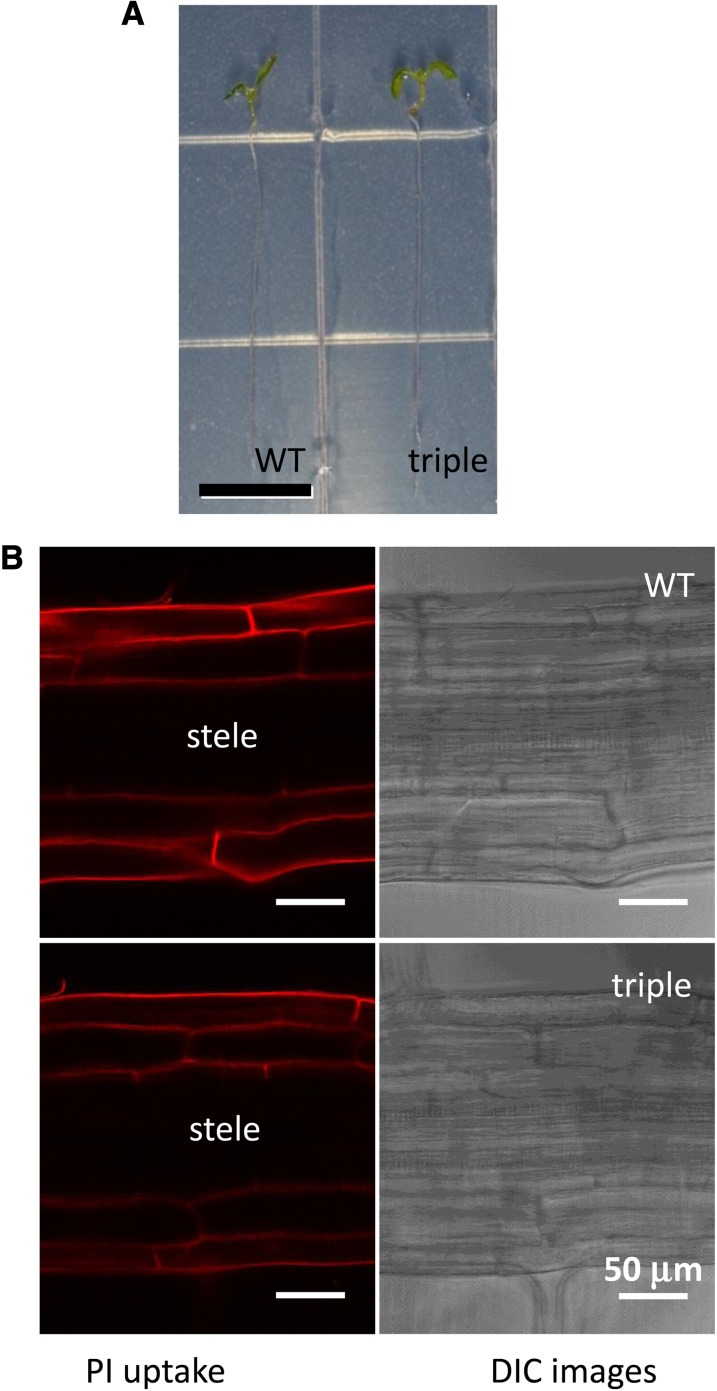
The Casparian strip is a lignin-based ring-like structure in the root endodermis of vascular plants (Naseer et al., 2012). Recently, it has been reported that peroxidase is required for monolignol oxidation in the Casparian strip (Lee et al., 2013). Because lignin deposition is severely disrupted in all of the tested tissues of the laccase triple mutant, we determined whether Casparian strip development is also affected. The diffusion of the fluorescent dye propidium iodide (PI) into the inner cell layers of the stele was used as an assay of Casparian strip function; disruption of the Casparian strip allows PI to diffuse into the stele (Naseer et al., 2012). Six-day-old seedlings were used for PI staining, and the roots of the triple mutant look very similar to those of the wild-type control at this stage (Figure 5A). Even though lignin deposition is almost totally abolished in the roots of 3-week-old triple mutant seedlings (Figure 3B), the diffusion of PI into the stele is completely blocked in both the wild type and triple mutant, indicating that the Casparian strip develops normally in the mutant (Figure 5B).
Figure 5.
Laccase Activity Is Not Required for Casparian Strip Formation.
(A) Six-day-old seedlings of the wild-type (WT) control and laccase triple mutant, showing comparable root length. Bar = 0.5 cm.
(B) The diffusion barrier for PI in the stele is present in both the wild-type control and laccase triple mutant.
Genome-Wide Transcript Profiling in the Laccase Triple Mutant
To explore gene expression profiles in the triple mutant, microarray analysis was performed. Three biological replicates of RNA extracted from the 3-week-old whole seedlings (Figure 3A) of the mutant and two replicates from the wild type were used for analysis using the Affymetrix Arabidopsis gene chip. Genes potentially associated with monolignol biosynthesis or polymerization with different expression levels between the wild-type control and triple mutant (with a fold-change threshold of 2 and statistically significant threshold of Bonferroni corrected P value of 2.19202E-06) were selected and are presented in Table 2 and Supplemental Table 2 online. In total, 975 genes were upregulated and 938 were downregulated in the triple mutant. The complete array data are presented in Supplemental Data Set 1 online.
Table 2. Comparison of the Expression of Lignin Biosynthetic Genes between the Laccase Triple Mutant and the Wild Type.
| Putative Gene | At No. | Gene Description | Mutant/Wild Type | Average Wild Type | sd Wild Type | Average Mutant | sd Mutant |
|---|---|---|---|---|---|---|---|
| PHE AMMONIA-LYASE2 | At3g53260 | Lignin biosynthetic gene | 1.12 | 2382 | 37 | 2676 | 436 |
| CINNAMATE-4-HYDROXYLASE | At2g30490 | Lignin biosynthetic gene | 1.53 | 1583 | 14 | 2423 | 364 |
| 4-COUMARATE:COA LIGASE1 | At1g51680 | Lignin biosynthetic gene | 2.11 | 1073 | 73 | 2255 | 178 |
| HYDROXYCINNAMOYL-COENZYME A SHIKIMATE/QUINATE HYDROXYCINNAMOYLTRANSFERASE | At5g48930 | Lignin biosynthetic gene | 1.43 | 1334 | 124 | 1911 | 84 |
| COUMAROYL SHIKIMATE 3′-HYDROXYLASE | At2g40890 | Lignin biosynthetic gene | 1.08 | 1134 | 198 | 1230 | 203 |
| CAFFEOYL COENZYME A O-METHYLTRANSFERASE1 | At4g34050 | Lignin biosynthetic gene | 1.78 | 4502 | 190 | 8056 | 798 |
| CINNAMOYL COA REDUCTASE | At1g15950 | Lignin biosynthetic gene | 1.11 | 1204 | 176 | 1338 | 45 |
| FERULATE 5-HYDROXYLASE | At4g36220 | Lignin biosynthetic gene | 0.69 | 415 | 76 | 286 | 7.8 |
| CAFFEIC ACID/5-HYDROXYFERULIC ACID O-METHYLTRANSFERASE | At5g54160 | Lignin biosynthetic gene | 1.56 | 2403 | 64 | 3748 | 485 |
| CINNAMYL ALCOHOL DEHYDROGENASE5 | At4g34230 | Lignin biosynthetic gene | 2.30 | 572 | 29 | 1357 | 121 |
| LAC4 | At2g38080 | Lignin laccase | 0.20 | 185 | 21 | 37 | 4.0 |
| LAC11 | At5g03260 | Lignin laccase | 0.14 | 141 | 16 | 19 | 2.0 |
| LAC17 | At5g60020 | Lignin laccase | 0.20 | 171 | 0.23 | 35 | 4.7 |
| LAC2 | At2g29130 | Putative laccase | 2.50 | 75 | 12 | 185 | 28 |
| LAC12 | At5g05390 | Putative laccase | 1.20 | 60 | 16 | 71 | 3.0 |
| Prx72 | At5g66390 | Lignin peroxidase | 3.50 | 239 | 65 | 829 | 158 |
| PER64 | At5g42180 | Lignin peroxidase | 2.10 | 321 | 26 | 676 | 191 |
| PEROXIDASE | At5g51890 | Lignin peroxidase | 2.90 | 338 | 17 | 998 | 108 |
| ABCG29 | At3g16340 | Monolignol transporter | 2.40 | 215 | 24 | 519 | 25 |
| UGT72E2 | At5g66690 | Monolignol glucosyltransferase | 5.20 | 292 | 70 | 1507 | 361 |
| UGT72E3 | At5g26310 | Monolignol glucosyltransferase | 2.00 | 64 | 3.2 | 128 | 38 |
Expression of monolignol biosynthesis genes was not much affected in the triple mutant, with the exception of approximately twofold increases in transcripts encoding 4-COUMARATE:CoA LIGASE1 and CINNAMYL ALCOHOL DEHYDROGENASE5 (Table 2). Thus, although lignin polymerization is blocked in the mutant, there appears to be little or no feedback on the biosynthetic machinery, at least not at the transcript level. The three lignin laccase genes LAC4, LAC11, and LAC17 showed an expected reduction in expression in the triple mutant. By contrast, the other two laccase genes that are expressed highly in stem, LAC2 and LAC12 (Figure 1A), showed either similar or even higher expression in the mutant compared with the control. These genes are therefore not functionally redundant with the lignin laccase genes.
Peroxidase PER64 has been recently reported to play a role in lignin deposition in the endodermis (Lee et al., 2013). PER64 transcript levels were not affected in the triple mutant, consistent with the fact that the Casparian strip develops normally (Figure 5A). AtPrx72 and the Arabidopsis ortholog of ZPO-C (At5g51890) are additional peroxidase genes that have been proposed to be involved in lignin polymerization (Sato et al., 2006; Herrero et al., 2013). However, lignin polymerization is severely disrupted in the triple mutant, although these two peroxidase genes were both upregulated (Table 2).
ABCG29 has been reported as a (H-unit) monolignol transporter in Arabidopsis (Alejandro et al., 2012). Interestingly, this gene was slightly upregulated, by over twofold, in the laccase triple mutant (Table 2), suggesting potential feedback regulation as a result of monolignol overaccumulation due to lack of laccase activity.
The Arabidopsis uridine diphosphate glycosyltransferases UGT72E2 and UGT72E3 have been shown to glucosylate monolignols (Lanot et al., 2006). The microarray data indicated that these two genes were upregulated in the laccase triple mutant by fivefold and twofold, respectively (Table 2). This could represent a mechanism for detoxifying monolignols in the absence of effective polymerization.
Lignans are dimers of coniferyl alcohol linked by the central (C8) carbon atoms and are distributed widely in the plant kingdom (Nakatsubo et al., 2008). In Arabidopsis, At1g32100 and At4g13660 encode pinoresinol reductase1 (PrR1) and PrR2, respectively, both of which redundantly catalyze the conversion of pinoresinol to lariciresinol in lignan biosynthesis (Nakatsubo et al., 2008). Although PrR1 transcripts were not significantly elevated in the laccase triple mutant, PrR2 transcript levels were increased by fourfold (see Supplemental Table 2 online). This suggests that disrupted monolignol polymerization may redirect the monolignol pools to the lignan biosynthesis pathway. At-ligB encodes an extradiol ring cleavage enzyme that is involved in the recently discovered arabidopyrone biosynthesis pathway (Weng et al., 2012). This enzyme uses caffealdehyde as substrate, a compound that is also shared by the monolignol biosynthesis pathway (Weng et al., 2012). In the laccase triple mutant, At-ligB transcript levels were increased by 2.5-fold (see Supplemental Table 2 online).
Transcript levels of genes involved in sinapate ester metabolism were also altered in the triple mutant. Sinapate:UDP-Glc glucosyltransferase catalyzes the synthesis of sinapoylglucose (Fraser et al., 2007). The microarray data suggested that this gene was upregulated by over twofold in the mutant (see Supplemental Table 2 online). Sinapoylglucose:malate sinapoyltransferase catalyzes the conversion of sinapoylglucose to 1,2-disinapoylglucose (Fraser et al., 2007; Fraser and Chapple, 2011). This gene was found to be downregulated by around threefold in the triple mutant (see Supplemental Table 2 online).
Altered Metabolite Profile in the Laccase Triple Mutant
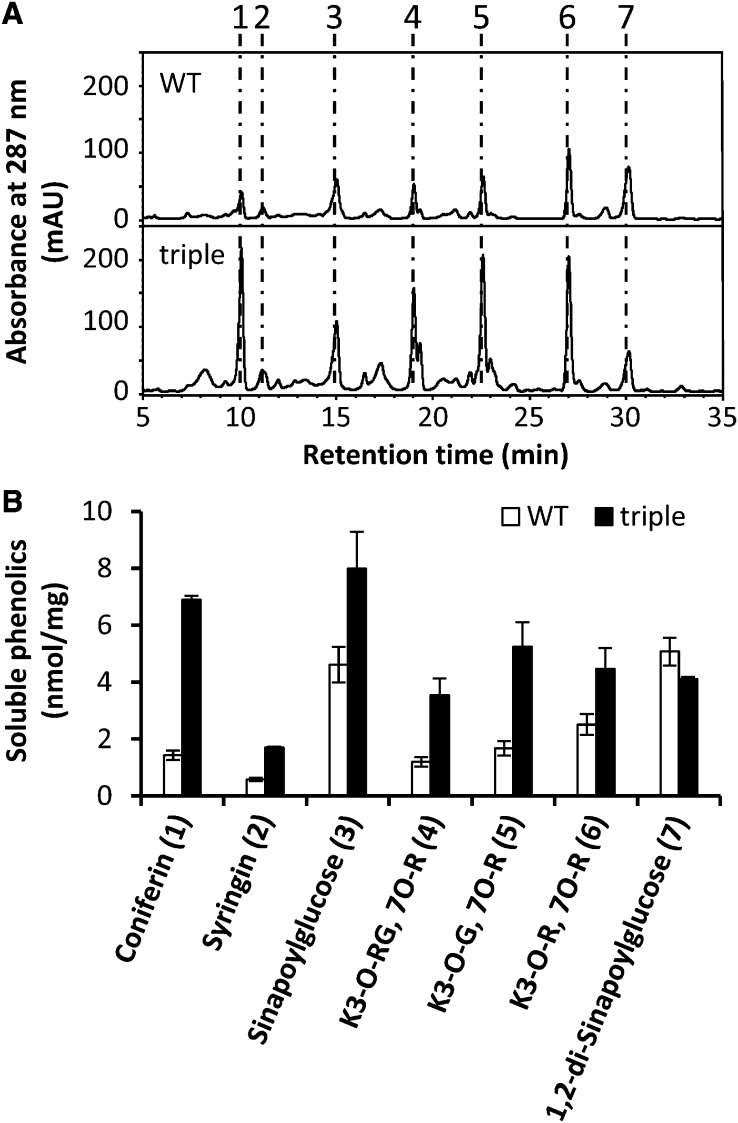
Because expression of monolignol biosynthesis genes is relatively unchanged in the laccase triple mutant in spite of the disruption of lignin polymerization, we speculated that monolignol derivatives might accumulate in the mutant. High performance liquid chromatography analysis was therefore performed to compare profiles of soluble metabolites in the wild type and laccase triple mutant. The profile of soluble phenolics was significantly altered in the laccase triple mutant (Figure 6A). Consistent with the fact that both UGT72E2 and UGT72E3 are significantly upregulated, levels of coniferyl alcohol glucoside (coniferin) and sinapyl alcohol glucoside (syringin) were both strikingly increased in the mutant (Figure 6B). Levels of sinapoylglucose, which is considered to be derived from sinapaldehyde, were also found to be elevated in the triple mutant (Figure 6B), in addition to levels of three flavonoids (kaempferol glycosides) (Figure 6A). Consistent with the fact that the expression of sinapoylglucose:malate sinapoyltransferase is downregulated in the mutant, 1,2-di-sinapoylglucose is slightly reduced in the mutant (Figure 6A).
Figure 6.
HPLC Analysis of Soluble Phenolics Accumulated in the Wild Type and Laccase Triple Mutant.
(A) Soluble leaf phenolics extracted from whole plants in 50% methanol and analyzed by reverse-phase HPLC. Coniferin (1), syringin (2), sinapoylglucose (3), kaempferol 3-O-[6″-O-(rhamnosyl) glucoside] 7-O-rhamnoside (K3-O-RG, 7-O-R) (4), kaempferol 3-O-glucoside 7-O-rhamnoside (K3-O-G, 7-O-R) (5), kaempferol 3-O-rhamnoside 7-O-rhamnoside (K3-O-R, 7-O-R) (6), and 1, 2-disinapoylglucose (7) were found in wild-type (WT) and lac triple mutant extracts. mAU, milliabsorbance units.
(B) Levels of soluble phenolic compounds in the wild type and lac triple mutants. The data are the means ± sd (n = 3).
Phylogenetic Analysis of Arabidopsis Putative Laccases
Because our data suggest that LAC4, LAC11, and LAC17 are most likely the three major lignin laccases in stems and roots of Arabidopsis, we considered their evolutionary origins by conducting phylogenetic analysis of eight fully sequenced plant species, including an alga, liverwort, fern, moss, gymnosperm, one dicot (Arabidopsis), and one monocot (rice [Oryza sativa]), and four other species with EST assemblies (see Supplemental Table 3 online for species names).
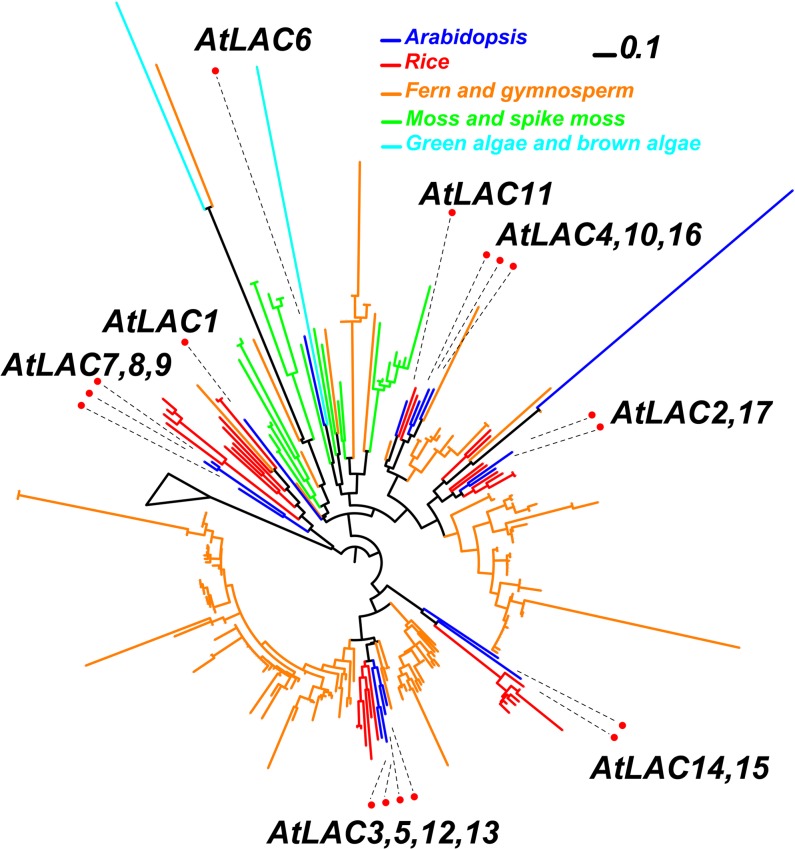
In our phylogenetic analysis, the 17 At-LAC genes form seven clusters with their corresponding putative orthologs in other plants. At-LAC1 and At-LAC6 seem to not have corresponding rice orthologs, and both are clustered with laccases from lower land plants (moss and spike moss), suggesting they might be more ancient than the other At-LAC genes and diverged before mosses appeared. All the other five clusters, except for the one containing At-LAC7, 8, and 9, have gymnosperm homologs within the clusters, suggesting that they diverged before the appearance of gymnosperms. Some green and brown algal homologs are found basal to the land plant laccase homologs, representing the ancient laccase orthologs in the algal species. Interestingly, there is only one copy of laccase in each of the three algae (Chlamydomonas reinhardtii, Ectocarpus siliculosus, and Volvox carteri), suggesting that gene duplication played a major role in the expansion of the lac gene family in land plants.
Although they are closely related, LAC4, LAC11, and LAC17 grouped in two different clusters (Figure 7; see Supplemental Figure 5 and Supplemental Data Set 2 online). Interestingly, the three laccase proteins have gymnosperm homologs but no orthologs from lower plant species. This indicates that LAC4, LAC11, and LAC17 are seed plant specific and diverged after the evolution of seed plants.
Figure 7.
Phylogeny of Laccase Homologous Proteins from Eight Fully Sequenced Plant and Algal Genomes and Four Plants with Assembled ESTs in PlantGDB.
The TIGRFAM (http://www.jcvi.org/cgi-bin/tigrfams/index.cgi) hidden Markov model TIGR03389 was constructed to represent the plant laccase family and used to search against the translated peptides of the 12 selected species (see Supplemental Table 3 online). The bar represents evolutionary distance of 0.1 amino acid substitutions per site.
DISCUSSION
Involvement of Laccases and Peroxidases in Lignification
The lignin polymer is formed through dehydrogenative polymerization of monolignols. Different classes of enzymes have been suggested to be involved in monolignol polymerization, such as peroxidases, laccases, and polyphenol oxidases (Boerjan et al., 2003). Peroxidases use hydrogen peroxide to oxidize their substrate. The peroxidases in plants are encoded by multigene families. In Arabidopsis, 73 peroxidases have been identified and annotated (Tognolli et al., 2002). Early studies on tree lignification suggested that peroxidase but not laccase activity is involved in lignin polymerization (Harkin and Obst, 1973). In addition, overexpression of peroxidase can lead to ectopic lignin deposition (Ostergaard et al., 2000; Quiroga et al., 2000; Barceló et al., 2002), and antisense-mediated downregulation of a peroxidase resulted in up to 50% reduction of lignin content in tobacco (Nicotiana tabacum; Blee et al., 2003). However, despite this early evidence, the involvement of peroxidases in monolignol polymerization is still a matter of some debate.
Laccases are copper-containing glycoproteins that are encoded by multigene families in plants. In contrast with peroxidases, laccases consume oxygen instead of hydrogen peroxide to oxidize their substrates. Many laccases are reported to be expressed in lignifying cells in a variety of plant species (Sterjiades et al., 1992; Bao et al., 1993). Unlike the high number of peroxidase genes in plant genomes, there are only 17 laccase genes annotated in Arabidopsis (Turlapati et al., 2011). The first genetic evidence of laccase involvement in monolignol polymerization came from analysis of the LAC15-deficient Arabidopsis mutant, which showed significant lignin reduction in the seed coat (Liang et al., 2006). However, because the seed coat is not a highly lignified tissue in Arabidopsis, LAC15 has not been considered to be the major lignin laccase. The identification of LAC4 and LAC17 provided the first solid evidence of laccase involvement in lignin polymerization in stem vascular tissue (Berthet et al., 2011). However, the lac4 lac17 double knockout mutant still has high lignin content, and the plants look developmentally normal, suggesting that there may be additional laccases, or other oxidases, contributing to monolignol polymerization in Arabidopsis.
Although disruption of LAC11 did not result in any obvious lignin reduction, simultaneous knock out of LAC11 along with either LAC4 or 17 leads to significant lignin reduction. In fact, the double knockout mutants lac4 lac11 and lac11 lac17 have lower lignin content than any of the three single knockout mutants, indicating that all three genes are lignin laccases. The triple mutant, which appears to completely lack lignin in roots, shows blocked development at the stage of appearance of a few pairs of dark green leaves, and the plant is very small compared with the wild type at the same developmental stage. This phenotype could be attributed to the lack of lignin and secondary cell walls, overaccumulation of toxic free forms of monolignols due to laccase deficiency, spillover to another metabolic pathway due to overaccumulation of monolignols, or loss of a developmental regulator as a result of laccase depletion.
Casparian strips are ring-like cell-wall structures in the root endodermis of vascular plants. They function as an apoplastic barrier to nutrients in plants (Geldner, 2013). There used to be considerable debate regarding the chemical nature of Casparian strips, with both suberin and lignin considered to be major components. Recently, it was demonstrated that, in Arabidopsis, Casparian strips consist of lignin polymer without suberin (Naseer et al., 2012). The subcellular precision of lignin deposition in the endodermis is achieved by localized peroxidase expression and locally restricted production of reactive oxygen species (Lee et al., 2013). In the laccase triple mutant, lignin deposition was nearly totally abolished even in the otherwise mostly lignified vascular elements of the root. However, Casparian strip development does not seem to be affected because penetration of the fluorescent dye PI across the stele was still blocked. This suggests that lignification in the Casparian strip is still present, supported by peroxidases, which are still expressed in the laccase triple mutant. Overall, our data support a model in which lignin polymerization in vascular tissues is driven primarily by laccases, rather than from the combinatorial action of both laccases and peroxidases. In the laccase triple mutant, over 20 peroxidase genes out of the total of 73 annotated peroxidases in Arabidopsis are upregulated by at least twofold, including the peroxidases that have been previously implicated in lignin polymerization (see Supplemental Table 2 online). However, this is still not able to compensate for the loss of function of the three laccase genes. It therefore remains unclear why one-third of the total peroxidases are upregulated in the triple mutant. Our results, while pointing to the essential role of laccases in developmentally regulated vascular lignification, do not rule out a role for peroxidases if, for example, laccase-mediated monolignol polymerization were an essential step for initiation of the lignification process, with peroxidases functioning later in the process.
Metabolic Changes in the Laccase Triple Mutant
The disruption of lignin laccase expression leads to redirection of flux through the phenylpropanoid pathway. The substrates of lignin polymerization would overaccumulate due to lack of laccase activity. The glycosyltransferases UGT72E2 and UGT72E3 that have been shown to glucosylate monolignols (Lanot et al., 2006) are both upregulated in the laccase triple mutant. This results in increased levels of both coniferin and syringin, the monolignol glucosides (Figure 6). Monolignol glucosylation is considered to increase the solubility and decrease the reactivity of monolignols. Given that monolignol glucosides are the vacuolar storage form, the upregulation of UGT72E2 and UGT72E3 in the triple mutant could be seen as a detoxification mechanism (Miao and Liu, 2010). Likewise, sinapoylglucose levels were also increased in the mutant. These could be derived from the otherwise overaccumulated sinapyl alcohol pool, via activated expression of sinapate:UDP-Glc glucosyltransferase, the enzyme responsible for glucosylation of sinapate. However, sinapate metabolism is complex (Fraser and Chapple, 2011), and other pathways could potentially be involved.
The above metabolic changes, associated with potential detoxification, result in no observable increase in free monolignols. Therefore, it is unlikely that the growth defect in the laccase triple mutant is simply the result of monolignol toxicity. It is more likely associated with the almost complete loss of lignin, resulting in severe impairment of vascular function.
Evolution of Lignin Laccases
Although some lower plant species, including red and green algae, have been reported to contain lignin or lignin-like polymers, their lignin content is much lower than that of land plant species (Martone et al., 2009; Sørensen et al., 2011). For instance, angiosperm (Arabidopsis) and gymnosperm (Pinus taeda) cell walls are estimated to contain around 100 times higher lignin content than the red alga Calliarthron cheilosporioides (Martone et al., 2009). Lignin composition is also different among terrestrial plants and marine plants. Gymnosperm walls usually contain no S lignin, but do have significant levels of H lignin in addition to the predominant G lignin; the red alga C. cheilosporioides has almost 40% H units, of which most land plants have only trace amounts; the lycophyte Selaginella has more S units than G units, whereas G units are usually dominant in angiosperm cell walls (Jin et al., 2005; Martone et al., 2009). Some lower plant species have functionally similar lignin biosynthetic genes to those of angiosperms, but these have evolved independently. For example, the lycophyte Selaginella has a ferulic acid/coniferaldehyde/coniferyl alcohol 5-hydroxylase that evolved independently from angiosperm F5Hs (Weng et al., 2008). Therefore, some key genes in the lignin biosynthetic pathway could be unique to seed plants, which might help explain why lignin polymers in seed plants are different from those in lower plant species. The fact that the Arabidopsis lignin laccases LAC4, LAC11, and LAC17 have no direct orthologous proteins in lower plant species indicates that these lignin laccases are seed plant specific and have diverged after the evolution of seed plants.
In conclusion, our identification of lac11 as a functional lignin laccase gene adds a missing piece to the puzzle of lignin polymerization, indicates the critical role of laccases in lignification, and suggests that peroxidase and laccase do not serve redundant functions in lignification at least in the vascular tissues of the stem and root of Arabidopsis. However, it remains unclear whether peroxidase participates in lignification in combination with laccase in other cell types or during certain specific developmental stages.
METHODS
Plant Growth Conditions
Arabidopsis thaliana plants were grown in MetroMix 350 soil under 16-h-light/8-h-dark cycles at 23°C during the day and 21°C during the night, with 70 to 80% RH and 150 μmol m−2 s−1 light intensity. Arabidopsis seedlings were grown aseptically on agar plates in half-strength Murashige and Skoog medium under a 16-h-light/8-h-dark cycle, with 55% RH, at 22°C and a light intensity of 150 μmol photons m−2 s−1.
Plant Materials
The lac4 mutant GabiKat-720G02 was obtained from the GabiKat collection at the University of Bielefeld. The lac11 mutant Salk_063746 and the lac17 mutant Salk_016748 were obtained from the ABRC at Ohio State University.
RNA Extraction and Real-Time PCR
Mature stems from 6-week-old plants were collected and used for RNA extraction with a Qiagen RNeasy plant mini kit. Qiagen DNase was applied to eliminate DNA contamination. Reverse transcription was performed using Superscript Reverse Transcriptase II (Invitrogen). Triplicate cDNA samples were used for quantitative RT-PCR, and the data were analyzed using SDS 2.2.1 software (Applied Biosystems). The PCR efficiency was estimated using LinRegPCR software, and the transcript levels were determined by relative quantification using the Arabidopsis actin gene as a reference.
Determination of Lignin Content and Composition
Mature stems from 6-week-old plants were harvested for lignin analysis using previously published procedures (Zhao et al., 2010a).
Transfection of Leaf Protoplasts for Transactivation Analysis
Leaves from 4-week-old greenhouse-grown Arabidopsis were used as a source of protoplasts. One reporter construct (promoter-luciferase) and one effector construct (35S:transcription factor) were cotransfected into protoplasts as described (Zhao et al., 2010b). A reference construct containing the Renilla luciferase gene driven by the 35S promoter was also cotransfected to determine the transfection efficiency. Luciferase activities were determined using the dual-luciferase reporter assay system (Promega). The firefly luciferase activity was calculated by normalizing against the Renilla luciferase activity in each transfection event. Data are presented as averages ± sd of three biological replicates.
PI Staining
For visualization of the Casparian strip, 6-d-old seedlings were examined with a Leica TCS SP2 AOBS confocal microscope (Leica Microsystems Heidelberg) equipped with a ×63 water immersion objective. Seedlings were incubated in the dark for 10 min in a fresh solution of 10 mM PI and rinsed two times in distilled water. Excitation and detection windows were set according to Naseer et al. (2012) and Lee et al. (2013) with minor modifications. PI was excited with the 514-nm line of the argon/krypton laser and emission detected at 550 to 700 nm.
Microscopy Observations
Six-week-old Arabidopsis stems and siliques from both the wild type (Columbia) and laccase triple mutant were fixed in 2.5% (v/v) glutaraldehyde and 4% paraformaldehyde (Electron Microscopy Sciences) in PBS, pH 7.2, for 12 h at 4°C. Stems from the mature portions (just above the soil line) were dissected and siliques from the nearly fully fertilized stage (still green) were picked. Samples were then washed with PBS, dehydrated in a graded ethanol series, and embedded in LR White resin (London Resin Co.). The resin was polymerized at 55°C for 3 d. Cross sections (0.5 µm) were cut with a diamond knife on a Leica EM UC7 ultramicrotome (Leica Mikrosysteme). For Toluidine Blue O staining, 0.5-µm semithin sections were placed onto glass slides, stained with 1% (w/v) Toluidine Blue O (with 1% [w/v] sodium borate) for 5 min, and observed under a Nikon Optiphot-2. For visualization of total lignin autofluorescence, 0.5-µm semithin sections were immersed in 50% (v/v) glycerol and observed under a Leica TCS SP2 AOBS confocal laser scanning microscope (Leica Microsystems CMS) illuminated with a 405-nm blue diode laser and emission detected at 460 nm. To obtain meaningful comparisons, laser intensity, pinhole, and photomultiplier gain settings were kept constant between samples. Additionally, morphological details of anthers and pollen were examined under a Hitachi TM3000 tabletop scanning electron microscope (Hitachi High-Technologies) operated at 5 kV.
Microarray Analysis
Total RNA was isolated from whole seedlings of 3-week-old plants (Figure 3A) with Tri-reagent according to the manufacturer’s protocol (Invitrogen). RNA was cleaned and concentrated using the RNeasy MinElute cleanup kit (Qiagen), and 500 ng of purified RNA was used for microarray analysis in each of three biological replicates. Probe labeling, hybridization, and scanning were conducted according to the manufacturer’s instructions (Affymetrix). Data were normalized using Robust Multichip Average algorithms. The presence/absence call for each probe set was obtained from DNA chip (Li and Wong, 2001). Genes with significantly different expression levels between the wild-type control and mutants were selected using associative analysis (Dozmorov and Centola, 2003). The type-I family-wise error rate was reduced using a Bonferroni-corrected P value threshold of 0.05/N, where N represents the number of genes present on the chip. The false discovery rate was monitored and controlled by the Q value (false discovery rate), which was calculated using extraction of differential gene expression (Leek et al., 2006).
Metabolite Extraction and Determination
Soluble phenolic compounds were extracted twice from 20.0 ± 0.05 mg of ground lyophilized plant material with 1.0 mL of 50% methanol plus 1.5% acetic acid for 3 h at room temperature. 7-Hydroxy-coumarin (Sigma-Aldrich) was added as an internal standard. Pooled supernatants were vacuum-dried, redissolved in 1.0 mL of 50% methanol, and clarified by centrifugation at 15,300g.
Compounds were identified by comparing their UV spectra and retention times with those of authentic standards and quantified by means of standard curves. HPLC was performed on a Beckman System Gold HPLC device, consisting of a programmable solvent module 126, a System Gold 508 autosampler, and a System Gold 168 diode array detector (Beckman Coulter). A Phenomenex Luna 5μ C18 reverse-phase column (5-μm particle, 250 × 4.6 mm) was used, with solvent A being 0.1% phosphoric acid in water and solvent B acetonitrile. The gradient was as follows: 8% B for 1 min; 8% B to 35% B in 45 min; at 46 min, B from 35% to 100% in 1 min.
Phylogenetic Analysis
The TIGRFAM (http://www.jcvi.org/cgi-bin/tigrfams/index.cgi) hidden Markov model TIGR03389 was constructed to represent the plant laccase family and used to search against the translated peptides of the 12 selected species (see Supplemental Table 3 online). If the genome was sequenced, the automated predicted peptides were used; otherwise, the translated peptides in six frames from the assembled EST UniGenes in PlantGDB were used (Dong et al., 2005). The homologs found in the three searches under the E-value cutoff of < 0.01 were collected respectively and subjected to multiple sequence alignments using the MAFFT v6.717 program (Katoh et al., 2005) with the L-INS-I method. Two phylogenies were built, one with homologs found in the eight fully sequenced plant and algal genomes and the other with homologs from all 12 plants. The phylogenies were further reconstructed using the FastTree v2.1.1 program (Price et al., 2010). The local support values beside the nodes were computed by resampling the site likelihoods 1000 times and performing the Shimodaira Hasegawa test to show the confidence levels with regard to the clustering of relevant proteins into one group. The 17 Arabidopsis laccase genes were labeled in the phylogenies and displayed using the Interactive Tree of Life Web server (Letunic and Bork, 2007).
Accession Numbers
The Arabidopsis Information Resource accession numbers for the genes reported here are as follows: At2g29130 (LAC2), At2g38080 (LAC4), At5g03260 (LAC11), At5g05390 (LAC12), At5g60020 (LAC17), AT2G46770 (NST1), AT3G61910 (NST2), and AT1G32770 (SND1). Additional accession numbers are provided in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Seven-Week-Old Wild-Type and lac11-1 Plants Grown in the Growth Chamber.
Supplemental Figure 2. Phloroglucinol Staining of Roots of 3-Week-Old Plants.
Supplemental Figure 3. UV Autofluorescence of Stem Cross Sections.
Supplemental Figure 4. Siliques of the Wild Type and the Laccase Triple Mutant.
Supplemental Figure 5. Phylogeny of Higher and Lower Plant LACCASE Proteins.
Supplemental Table 1. Total Lignin Content of Arabidopsis lac11 Single and Double Mutants as Determined by the Acetyl Bromide Method.
Supplemental Table 2. Additional Lignin-Associated Genes with Altered Expression in the Laccase Triple Mutant.
Supplemental Table 3. Plant Species Used in the Phylogenetic Analysis.
Supplemental Data Set 1. Complete Microarray Data Set.
Supplemental Data Set 2. Alignment Used to Generate the Phylogeny Presented in Figure 7.
Supplementary Material
Acknowledgments
We thank Chenggang Liu for critical reading of the article and Yuhong Tang for assistance with microarray analysis. This work was supported by the Samuel Roberts Noble Foundation.
AUTHOR CONTRIBUTIONS
Q.Z. and R.A.D. designed the research. Q.Z., J.N., F.C., Y.Y., J.Y., and H.S. performed research. Q.Z., J.N., F.C., Y.Y., C.F., X.W., Z.-Y.W. and R.A.D. analyzed data. R.A.D. and Q.Z. wrote the article.
Glossary
- PI
propidium iodide
References
- Alejandro S., Lee Y., Tohge T., Sudre D., Osorio S., Park J., Bovet L., Lee Y., Geldner N., Fernie A.R., Martinoia E. (2012). AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 22: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Bao W., O’malley D.M., Whetten R., Sederoff R.R. (1993). A laccase associated with lignification in loblolly pine xylem. Science 260: 672–674 [DOI] [PubMed] [Google Scholar]
- Barceló A.R., Pomar F., Ferrer M.A., Martínez P., Ballesta M.C., Pedreño M.A. (2002). In situ characterization of a NO-sensitive peroxidase in the lignifying xylem of Zinnia elegans. Physiol. Plant. 114: 33–40 [DOI] [PubMed] [Google Scholar]
- Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cézard L., Le Bris P., Borrega N., Hervé J., Blondet E., Balzergue S., Lapierre C., Jouanin L. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blee K.A., Choi J.W., O’Connell A.P., Schuch W., Lewis N.G., Bolwell G.P. (2003). A lignin-specific peroxidase in tobacco whose antisense suppression leads to vascular tissue modification. Phytochemistry 64: 163–176 [DOI] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Carmo-Silva A.E., Francisco A., Powers S.J., Keys A.J., Ascensão L., Parry M.A., Arrabaça M.C. (2009). Grasses of different C4 subtypes reveal leaf traits related to drought tolerance in their natural habitats: Changes in structure, water potential, and amino acid content. Am. J. Bot. 96: 1222–1235 [DOI] [PubMed] [Google Scholar]
- Chen F., Tobimatsu Y., Havkin-Frenkel D., Dixon R.A., Ralph J. (2012). A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. USA 109: 1772–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q., Lawrence C.J., Schlueter S.D., Wilkerson M.D., Kurtz S., Lushbough C., Brendel V. (2005). Comparative plant genomics resources at PlantGDB. Plant Physiol. 139: 610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov I., Centola M. (2003). An associative analysis of gene expression array data. Bioinformatics 19: 204–211 [DOI] [PubMed] [Google Scholar]
- Fraser C.M., Chapple C. (2011). The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9: e0152, /10.1199/tab.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.M., Thompson M.G., Shirley A.M., Ralph J., Schoenherr J.A., Sinlapadech T., Hall M.C., Chapple C. (2007). Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol. 144: 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N. (2013). The endodermis. Annu. Rev. Plant Biol. 64: 531–558 [DOI] [PubMed] [Google Scholar]
- Harkin J.M., Obst J.R. (1973). Lignification in trees: Indication of exclusive peroxidase participation. Science 180: 296–298 [DOI] [PubMed] [Google Scholar]
- Herrero J., Fernández-Pérez F., Yebra T., Novo-Uzal E., Pomar F., Pedreño M.A., Cuello J., Guéra A., Esteban-Carrasco A., Zapata J.M. (2013). Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 237: 1599–1612 [DOI] [PubMed] [Google Scholar]
- Hoegger P.J., Kilaru S., James T.Y., Thacker J.R., Kües U. (2006). Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 273: 2308–2326 [DOI] [PubMed] [Google Scholar]
- Jin Z., Matsumoto Y., Tange T., Akiyama T., Higuchi M., Ishii T., Iiyama K. (2005). Proof of the presence of guaiacyl–syringyl lignin in Selaginella tamariscina. J. Wood Sci. 51: 424–426 [Google Scholar]
- Jun J.H., Ha C.M., Nam H.G. (2002). Involvement of the VEP1 gene in vascular strand development in Arabidopsis thaliana. Plant Cell Physiol. 43: 323–330 [DOI] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanot A., Hodge D., Jackson R.G., George G.L., Elias L., Lim E.K., Vaistij F.E., Bowles D.J. (2006). The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 48: 286–295 [DOI] [PubMed] [Google Scholar]
- Lee Y., Rubio M.C., Alassimone J., Geldner N. (2013). A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412 [DOI] [PubMed] [Google Scholar]
- Leek J.T., Monsen E., Dabney A.R., Storey J.D. (2006). EDGE: Extraction and analysis of differential gene expression. Bioinformatics 22: 507–508 [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2007). Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128 [DOI] [PubMed] [Google Scholar]
- Li C., Wong W.H. (2001). Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Davis E., Gardner D., Cai X., Wu Y. (2006). Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Roeder A.H., Kempin S.A., Gremski K., Østergaard L., Guimil S., Reyes D.K., Yanofsky M.F. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853 [DOI] [PubMed] [Google Scholar]
- Martone P.T., Estevez J.M., Lu F., Ruel K., Denny M.W., Somerville C., Ralph J. (2009). Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 19: 169–175 [DOI] [PubMed] [Google Scholar]
- McCaig B.C., Meagher R.B., Dean J.F. (2005). Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221: 619–636 [DOI] [PubMed] [Google Scholar]
- Miao Y.C., Liu C.J. (2010). ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. USA 107: 22728–22733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Iwase A., Yamamoto H., Yoshida M., Seki M., Shinozaki K., Ohme-Takagi M. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsubo T., Mizutani M., Suzuki S., Hattori T., Umezawa T. (2008). Characterization of Arabidopsis thaliana pinoresinol reductase, a new type of enzyme involved in lignan biosynthesis. J. Biol. Chem. 283: 15550–15557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer S., Lee Y., Lapierre C., Franke R., Nawrath C., Geldner N. (2012). Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl. Acad. Sci. USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard L., Teilum K., Mirza O., Mattsson O., Petersen M., Welinder K.G., Mundy J., Gajhede M., Henriksen A. (2000). Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 44: 231–243 [DOI] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. (2010). FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga M., Guerrero C., Botella M.A., Barceló A., Amaya I., Medina M.I., Alonso F.J., de Forchetti S.M., Tigier H., Valpuesta V. (2000). A tomato peroxidase involved in the synthesis of lignin and suberin. Plant Physiol. 122: 1119–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Demura T., Yamawaki K., Inoue Y., Sato S., Sugiyama M., Fukuda H. (2006). Isolation and characterization of a novel peroxidase gene ZPO-C whose expression and function are closely associated with lignification during tracheary element differentiation. Plant Cell Physiol. 47: 493–503 [DOI] [PubMed] [Google Scholar]
- Sørensen I., Pettolino F.A., Bacic A., Ralph J., Lu F., O’Neill M.A., Fei Z., Rose J.K., Domozych D.S., Willats W.G. (2011). The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 68: 201–211 [DOI] [PubMed] [Google Scholar]
- Sterjiades R., Dean J.F., Eriksson K.E. (1992). Laccase from sycamore maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol. 99: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognolli M., Penel C., Greppin H., Simon P. (2002). Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288: 129–138 [DOI] [PubMed] [Google Scholar]
- Turlapati P.V., Kim K.W., Davin L.B., Lewis N.G. (2011). The laccase multigene family in Arabidopsis thaliana: Towards addressing the mystery of their gene function(s). Planta 233: 439–470 [DOI] [PubMed] [Google Scholar]
- Weng J.K., Li Y., Mo H., Chapple C. (2012). Assembly of an evolutionarily new pathway for α-pyrone biosynthesis in Arabidopsis. Science 337: 960–964 [DOI] [PubMed] [Google Scholar]
- Weng J.K., Li X., Stout J., Chapple C. (2008). Independent origins of syringyl lignin in vascular plants. Proc. Natl. Acad. Sci. USA 105: 7887–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Gallego-Giraldo L., Wang H., Zeng Y., Ding S.Y., Chen F., Dixon R.A. (2010a). An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 63: 100–114 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Wang H., Yin Y., Xu Y., Chen F., Dixon R.A. (2010b). Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch. Proc. Natl. Acad. Sci. USA 107: 14496–14501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Richardson E.A., Ye Z.H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Lee C., Zhong R., Ye Z.H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.