In photosynthetic eukaryotes, starch aggregates into insoluble, semicrystalline granules through the action of a debranching enzyme of chlamydial pathogen origin. It is shown that an enzyme of analogous nature to this enzyme but of a different bacterial origin was recruited, by convergent evolution, for the same purpose in single-cell cyanobacteria.
Abstract
Starch, unlike hydrosoluble glycogen particles, aggregates into insoluble, semicrystalline granules. In photosynthetic eukaryotes, the transition to starch accumulation occurred after plastid endosymbiosis from a preexisting cytosolic host glycogen metabolism network. This involved the recruitment of a debranching enzyme of chlamydial pathogen origin. The latter is thought to be responsible for removing misplaced branches that would otherwise yield a water-soluble polysaccharide. We now report the implication of starch debranching enzyme in the aggregation of semicrystalline granules of single-cell cyanobacteria that accumulate both glycogen and starch-like polymers. We show that an enzyme of analogous nature to the plant debranching enzyme but of a different bacterial origin was recruited for the same purpose in these organisms. Remarkably, both the plant and cyanobacterial enzymes have evolved through convergent evolution, showing novel yet identical substrate specificities from a preexisting enzyme that originally displayed the much narrower substrate preferences required for glycogen catabolism.
INTRODUCTION
Bacteria, Archaea, and eukaryotes often store Glc in the form of glycogen. This storage material consists of small hydrosoluble particles composed of α-1,4–linked and α-1,6–branched glucan chains (Shearer and Graham, 2002). Because current structure models envision that each α-1,4–linked chain supports on average two novel branched chains, mathematical modeling predicts that chain density will increase with particle size up to a maximal possible diameter of 42 nm. Such glycogen particles contain up to 55,000 Glc residues with over 36% resting in the outer particle chains (Meléndez et al., 1999). These are thus readily accessible to cell metabolism without the need for polysaccharide debranching. Hence, in glycogen, Glc residues remain rapidly available to cellular enzymes as if they were in the soluble phase but remain much less active osmotically. Archaeplastida (also called Kingdom Plantae) consisting of the Chloroplastida (green algae and all land plants), the Rhodophyceae (red algae), and the Glaucophyta (glaucophytes) store starch granules of potentially unlimited size and no glycogen (Ball et al., 2011). Starch always contains a glycogen-like polymer named amylopectin occasionally mixed with a very moderately branched amylose polysaccharide. Amylopectin aggregates into semicrystalline granules of potentially unlimited diameter. This organization results from an asymmetrical distribution of branches, allowing the formation of double helical structures that align and crystallize into two different allomorphs (the so-called A and B) or a mixture of both (Buléon et al., 1998). This aggregation directly affects the properties of most food sources, including their digestibility as well as the functional properties in all nonfood uses of the polymer.
Starch is osmotically inert, allowing for the accumulation of very large quantities of Glc (60 to 90% of the dry weight) in the storage organs of plants. Until recently, the distribution of starch seemed restricted to photosynthetic eukaryotes, including several secondary endosymbiosis lineages derived from Archaeplastida, such as the cryptophytes (Deschamps et al., 2006), the dinoflagellates (Dauvillée et al., 2009), and some apicomplexa parasites (Coppin et al., 2005). However, more recent studies revealed the existence of starch-like structures in unicellular diazotrophic cyanobacteria belonging to the order Chroococcales (Nakamura et al., 2005; Deschamps et al., 2008; Suzuki et al., 2013). The presence of anomalous glycogen particles had already been identified previously in this clade, while it is only very recently that this material was recognized as starch-like and the term “semi-amylopectin” was coined to describe the major amylopectin-like fraction within these granules (Schneegurt et al., 1994; Suzuki et al., 2013). Four out of the six reported starch-accumulating cyanobacteria strains accumulate only this polysaccharide fraction. However, two different strains also synthesize amylose using an enzyme phylogenetically related to the archaeplastidial granule-bound starch synthase (GBSS), an enzyme known to be selectively responsible for the synthesis of this fraction in plants (Delrue et al., 1992; Deschamps et al., 2008).
Clues as to the nature of the biochemical mechanism distinguishing starch from glycogen synthesis came from the study of glycogen-accumulating mutants of Chlamydomonas and cereals that proved defective for the same GH13-type (for Glycosyl Hydrolase family 13, according to the carbohydrate active enzyme (CAZy) classification) of debranching enzyme (DBE; James et al., 1995; Mouille et al., 1996; Kubo et al., 1999). In the green algae, the substitution of starch by glycogen was complete, thereby hinting that the absence of DBE prevented amylopectin synthesis altogether. This enzyme thus catalyzed an essential previously unrecognized step distinguishing starch from glycogen synthesis. From these observations, several groups proposed that DBE was selectively responsible for removal of misplaced branches, preventing polysaccharide aggregation in an otherwise hydrosoluble precursor (Ball et al., 1996). This model, known as the preamylopectin trimming model, is well sustained by the evidence gathered through mutant analysis in all plant systems examined to date, including Arabidopsis thaliana (Wattebled et al., 2005). However, direct proof of this model will only be obtained by achieving in vitro synthesis of starch granules. The DBEs missing in the glycogen-accumulating plants are known as isoamylases and belong to a family of direct DBEs that apart from Archaeplastida are only distributed in bacteria. In bacteria, such enzymes, generally named GlgX (by reference to the Escherichia coli glycogen metabolism locus coding it), are known to be involved in glycogen catabolism by debranching only those external chains that have been first recessed by the catabolic enzyme glycogen phosphorylase. The E. coli enzyme is thus known to be very selective for chains of 3-4 Glc residues left over by glycogen/starch phosphorylase action on external chains and displays very little residual activity on longer chains (Jeanningros et al., 1976; Dauvillée et al., 2005). This restricted enzyme selectivity prevents futile cycles during polysaccharide synthesis since such activities are unable to debranch directly the products of branching enzyme activity that branches a minimum of six Glc residues on an acceptor chain. In line with their supposed catabolic function, the E. coli GlgX defective mutants overproduce glycogen with short external chains (Dauvillée et al., 2005). Interestingly, analogous results have also been obtained in glycogen-accumulating cyanobacteria (Suzuki et al., 2007).
We now report the existence of both glycogen and starch in a recently axenized marine Cyanobacterium (strain CLg1) related isolate. We show through the isolation of an allelic series of 14 distinct mutants that alteration, decrease, or disappearance of a particular GH13 GlgX type of DBE correlates with the disappearance of the starch fraction and a large increase in the accumulation of glycogen. Remarkably, the wild-type enzyme defective in the mutants has evolved an isoamylase-type of activity very similar to that present in plants. This activity evolved, as was the case for the plant enzyme from preexisting GlgX-like proteins. However, phylogenetic analysis proves that the phylogenetic origin of this gene is independent from the enzyme recruited by the Archaeplastida. These results point to the independent acquisition of starch metabolism in cyanobacteria and plants and suggest that polysaccharide debranching by an isoamylase-like enzyme defines a common mechanism for the synthesis and aggregation of starch polymers from enzymes of a preexisting glycogen metabolism network.
RESULTS
Cyanobacterium sp CLg1 Accumulates Both Glycogen and Starch from a Gene-Rich Suite of Enzymes of Bacterial Glycogen Metabolism
Unlike Archaeplastida, which only contain starch like material, the wild-type axenized strain CLg1 always accumulates two distinct α-1,4–linked and α-1,6–branched polysaccharides fractions in the form of both a major (85%) insoluble and a minor (15%) yet significant water-soluble fraction (Table 1). We confirm that the structure of the insoluble polysaccharide fraction (Figures 1A and 1B) was composed of starch-like granules containing both semicrystalline amylopectin and amylose with chain-length distribution consistent with the presence of starch as previously described (Deschamps et al., 2008). Interestingly, in addition to this major starch-like fraction, a small hydrosoluble fraction representing 15% of the total glucan amount under nitrogen starvation conditions was also observed. The chain-length distribution (Figure 2B) and the negative staining transmission electronic microscopy (TEM) observations (Figure 2A) suggest that this material is a highly branched soluble polysaccharide capable of excluding the uranyl acetate molecule in a fashion similar to rabbit liver glycogen particles (Figure 2G). In addition, TEM images of wild-type CLg1 strain ultrathin sections, in which polysaccharides were stained by the periodic acid thio-carbohydrazide-silver proteinate method, confirm the presence not only of the very obvious large-size starch granules but also of the smaller size glycogen-like particles (Figure 3A). To get a better idea of the nature of the biochemical pathways explaining the presence of these two fractions and to better characterize the nature of the CLg1 strain, we examined the recently reported sequence of CLg1 strain genome (Suzuki et al., 2013). The 16 rRNA and NifH phylogenetic analysis revealed a close relationship to the genus Cyanobacterium (Falcon et al., 2004; Suzuki et al., 2013). This strain, although containing the genes required for nitrogen fixation, remains unable to grow without reduced nitrogen under laboratory conditions.
Table 1. Polysaccharide Composition in Wild-Type and Class A Mutant Strains.
| Starch Granules |
WSP |
% of Each Polysaccharide |
||||
|---|---|---|---|---|---|---|
| Strain | mg/mg of Protein | % | mg/mg of Protein | % | Starch | WSP |
| Wild type | 2.80 ± 0.36 | 100 | 0.50 ± 0.03 | 100 | 84.7 | 15.3 |
| 174H3 | 0.08 ± 0.02 | 3 | 1.89 ± 0.57 | 375 | 4.3 | 95.7 |
| 107D8 | 0.04 ± 0.01 | 1.5 | 0.87 ± 0.23 | 173 | 4.6 | 95.4 |
| 91G1 | 0.09 ± 0.04 | 3.3 | 1.57 ± 0.4 | 312 | 5.5 | 94.5 |
| 153H12 | 0.22 ± 0.20 | 7.7 | 1.47 ± 0.38 | 292 | 12.8 | 87.2 |
| 123B3 | 0.15 ± 0.05 | 5.5 | 1.38 ± 0.14 | 273 | 10.1 | 89.9 |
| 99A7 | 0.09 ± 0.03 | 3.3 | 2.69 ± 1.10 | 533 | 3.3 | 96.7 |
| 21’C6 | 0.07 ± 0.02 | 2.6 | 2.04 ± 0.42 | 403 | 3.4 | 96.6 |
| 118H9 | 0.05 ± 0.03 | 1.7 | 1.64 ± 0.83 | 324 | 2.9 | 97.1 |
| 91D6 | 0.07 ± 0.03 | 2.45 | 1.99 ± 0.35 | 395 | 3.3 | 96.7 |
| 175E12 | 0.12 ± 0.06 | 4.2 | 2.19 ± 0.72 | 435 | 5 | 95.0 |
| 174B6 | 0.17 ± 0.1 | 6 | 1.64 ± 0.51 | 326 | 9.3 | 90.7 |
| 134A4 | 0.06 ± 0.03 | 2.1 | 1.80 ± 0.70 | 354 | 3.2 | 96.8 |
| 81G3 | 0.07 ± 0.02 | 2.6 | 2.37 ± 0.1 | 469 | 3 | 97.0 |
| 80D5 | 0.07 ± 0.04 | 2.5 | 2.13 ± 0.2 | 422 | 3.2 | 96.8 |
The measurements are means ± sd of three independent extractions.
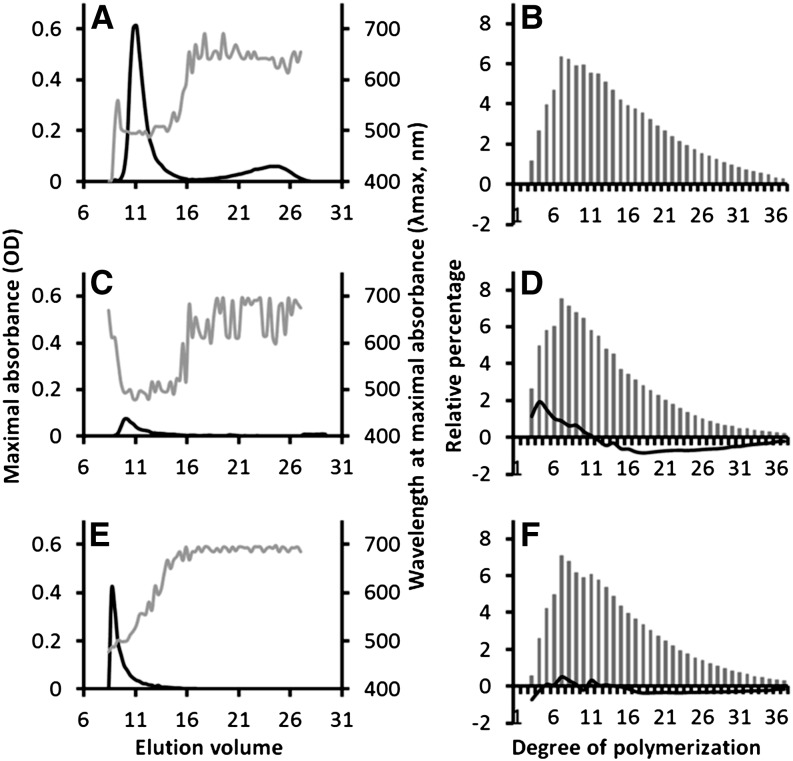
Figure 1.
Structural Analyses of Starch-Like Granules Accumulated by the Wild-Type Strain and Class A Mutants.
Insoluble water polysaccharides purified from the wild type (A). A severe mutant 99A7 (C) and an intermediate mutant 153H12 (E) were subjected to size-exclusion chromatography analysis (CL2B sepharose). The wavelength (λmax, gray lines) at the maximal absorbance (black lines) of the iodine-polysaccharide complex was determined for each fraction. As previously described, starch-like granules of the wild type are composed of semiamylopectin (fractions 9 to 16) and amylose-like polysaccharides (fractions 17 to 27). Both starch-like granules of class A mutants are composed exclusively of abnormal semiamylopectin fraction (λmax values ranging between 490 and 500 nm instead of 507 nm for the wild-type semiamylopectin). Fractions containing semiamylopectin material were pooled and subjected to chain-length distribution analysis. After complete digestion with commercial isoamylase, chains of Glc were separated according to their DP by HPAEC-PAD. The relative abundance for each DP (gray bars) was determined for the wild type (B), the 99A7 mutant (D), and the 153H12 mutant (F) from three independent extractions. Subtractive analyses (percentage of each DP in the mutant’s semiamylopectin minus percentage of each DP in the wild type’s semiamylopectin), depicted as black lines in (D) and (F), reveal an increase of short chains (DP 3 to 10) and a decrease in long-chain content (DP 12 to 35) in the severe mutant 99A7 (in [D]) and no significant difference in the intermediate 153H12 mutant (in [F]).
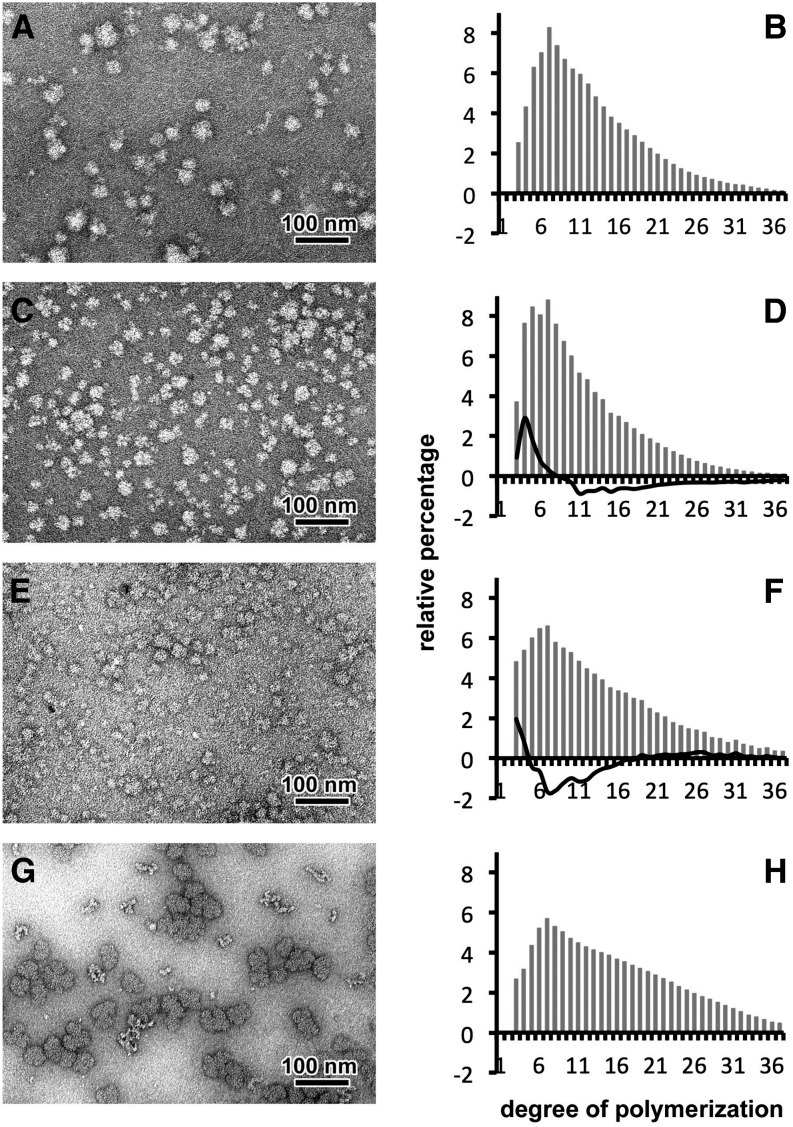
Figure 2.
Characterization of WSPs Accumulated in the Wild-Type Strain and Class A Mutants of Cyanobacterium sp CLg1.
Negative staining following TEM observations suggest that WSP of the wild type (A), the 99A7 mutant (C), and the 153H12 mutant (E) are highly branched polysaccharides with a diameter below 50 nm similar to glycogen particles of rabbit liver (G). After purification and complete digestion with a commercial isoamylase, chains of Glc were separated according to their DP by HPAEC-PAD. From three independent extractions, the relative abundance for each DP (gray bars) was determined for the wild-type strain (B), the severe mutant 99A7 (D), the intermediate 153H12 mutant (F), and glycogen from rabbit liver (H). Subtractive analyses (percentage of each DP of mutant minus percentage of each DP in the wild type), depicted as black lines in (D) and (F), reveal an increase of short chains (DP 3 to 7) and a defect in long-chain content (DP 12 to 35) in the WSPs accumulated in the mutant 99A7 (in [D]) and in the intermediate mutant 153H12 (in [F]). WSP appears to contain fewer chains with a DP ranging between 4 and 16.
Figure 3.
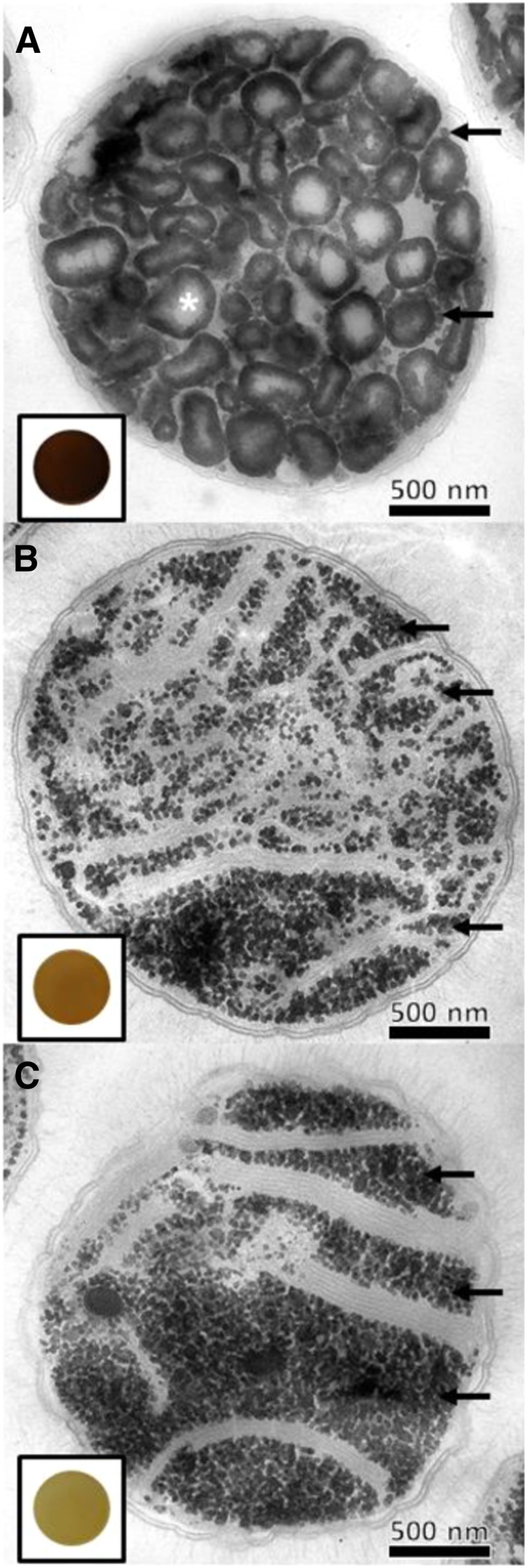
TEM Observations of ultrathin sections (60 nm) of Wild-Type and Class A Mutant Strains.
Polysaccharide contents of the wild-type CLg1 strain (A), and the 99A7 (B) and 175E12 (C) mutant strains were observed after periodic acid thio-carbohydrazide-silver proteinate staining. Both starch-like granules (white star) and glycogen particles (dots indicated with black arrows) are witnessed in the wild-type strain. Starch granules are missing and substituted by a large amount of glycogen particles in class A mutants. The dark-blue iodine stain from a cell patch of the wild-type strain (framed in [A]) is displayed. By contrast, the absence of starch granules in class A mutants is correlated by a yellow-orange stain of cell patches after spraying iodine vapors (framed in [B] and [C]).
Surprisingly, the network evidenced is a rather complex set of bacterial and archean distributed enzymes of glycogen metabolism containing, among others, three glycogen/starch synthases, four glycogen/starch branching enzymes, two GlgX-like DBE sequences, two amylopullulanases, and one indirect DBE–like α-1,6 glucosidase domain (Colleoni and Suzuki, 2012). With two exceptions, no traces of enzymes of eukaryotic affiliation involved in glycogen or starch metabolism were found in this genome. The important exceptions consist of GBSSI and ADP-Glc pyrophosphorylase, which were probably donated to the Archaeplastida through endosymbiotic gene transfer after plastid endosymbiosis. The affiliation of one of the soluble glycogen/starch synthases to the SSIII-IV group of enzymes of Archaeplastida is indirect, and this enzyme groups with a number of other proteobacteria and chlamydia sequences. In this case, either the proteobacteria, or more probably the chlamydia, can be considered as the direct source of the archaeplastidal enzymes (Ball et al., 2013).
Selection and Characterization of the Polysaccharide Structure from Glycogen-Accumulating Mutants of Cyanobacterium sp CLg1
Because of the very slow growth rate of the CLg1 strain and the absence of established transgenesis procedures for this organism, we chose a forward genetic approach (UV mutagenesis) to dissect the genetic determinants of starch metabolism in cyanobacteria (for details, see Methods). Five phenotypic classes of mutants were found after 3 years of screening, segregation, and purification (see Supplemental Figure 1 online). We benefited from the presence of amylose, which yielded a very strong and sensitive iodine stain for the screening of 2.104 cell patches. Of relevance to this study was the finding of a class of mutants (class A), which lacked interaction with iodine and yielded low starch and abundant water-soluble polysaccharides (WSPs). Figure 3 shows that 14 mutants of this class accumulated abundant small size granules resembling the minor glycogen fraction of the wild-type reference. Table 1 shows that 11 out of 14 mutants of this class witness a 94 to 97% decrease of insoluble granules and a replacement by a significant amount of hydrosoluble polysaccharide. However, three additional mutant strains displayed a phenotype intermediate between standard class A mutants and the wild type. The latter accumulated up to 9 to 12% of the wild-type amount of starch and overaccumulated glycogen (Table 1). This will be referred to as the incompletely defective class A mutants. These mutants also gave a clear mutant iodine-stain phenotype of cell patches (see Supplemental Figure 1 online). The substitution of starch by glycogen restricted the total amount of stored glucans in all mutants from two- to threefold. Purification of the water-soluble fraction followed by enzymatic debranching and analysis of the chain-length distribution of the resulting glucans yielded a chain length that mimicked those of glycogen and clearly differed from that of amylopectin-like polymers purified from the wild-type CLg1 reference (Figure 2). Although similar to the chain-length distribution of the minor glycogen fraction from the wild-type reference, the mutant WSP was selectively enriched in very small chains (degree of polymerization [DP] 3 to 6) when compared with the wild type. A very minor amount (2 to 6% of the wild-type mount) of high molecular mass material could still be purified from the glycogen accumulating mutants. This material resembled the polysaccharide accumulating in the single incompletely defective class A mutant 153H12 (12%). Interestingly, the amylopectin-like polysaccharide was equally enriched in very small glucans when compared with the wild type.
The Glycogen-Accumulating Cyanobacteria Are Defective for a Cation Requiring DBE Activity
To get a better understanding of the underlying biochemical cause of the glycogen-accumulating phenotype, we undertook a series of zymogram and enzyme assays for all possible enzymes of bacterial glycogen/starch metabolism. We were unable to find any significant difference in our crude extract assays. A notable increase of one starch phosphorylase isoform was noted through zymograms of the mutants (see Supplemental Figure 2 online). However, this increase was noted in all mutant types that displayed a decrease in total polysaccharide amount and not only in Class A mutants. It thus seems to define a universal secondary effect of altered starch metabolism not only in the Cyanobacterium sp CLg1 but also in other cyanobacteria species (Fu and Xu, 2006). Interestingly, the presence of white-staining glycogen-degrading bands that selectively disappeared from the class A mutants was evidenced from the glycogen/starch synthase gels but was not mirrored by the starch amylopectin- or glycogen-containing gels designed to emphasize starch modifying enzymes (e.g., amylases, branching enzymes, and α-1,4 glucanotransferase) (Figure 4). Because the procedures differed between the hydrolase and synthase zymograms with respect to pH and incubation buffers, we were able to test and narrow down the differences responsible for the identification of this activity selectively in the glycogen/starch synthase activity gels. We found that the missing glucan hydrolase required high levels of cation (see Supplemental Figure 3 online) that were supplied in the glycogen/starch synthase zymograms (i.e., Mg 2+) but not in our other procedures. Figure 3 shows that when starch- or amylopectin-containing gels were supplied with 10 mM MgSO4, a blue staining glucan hydrolase band that disappeared in all class A mutants was evidenced. Interestingly, the incompletely defective class A mutant displays a slower migrating fainter activity. Because such a stain was indicative of the presence of a starch DBE, we purified the activity from wild-type cyanobacteria to near homogeneity. We then subjected amylopectin to the action of the purified enzyme and examined the proton nuclear magnetic resonance (NMR) spectra of the substrate before and after incubation with the glucan hydrolase. Results displayed in Figure 5 demonstrate that amylopectin was completely debranched and that the amount of branch hydrolysis corresponded quantitatively to the appearance of reducing-end resonance signals. These results allow us to identify the purified enzyme as a direct DBE. In addition, we were able to size the activity during our purification procedure as a polypeptide of 82 kD under denaturing conditions and with an apparent molecular mass of 247 kD under nondenaturing conditions (see Supplemental Figure 4 online), which suggests that the native protein eluted as a homotrimeric or as a homodimeric enzyme.
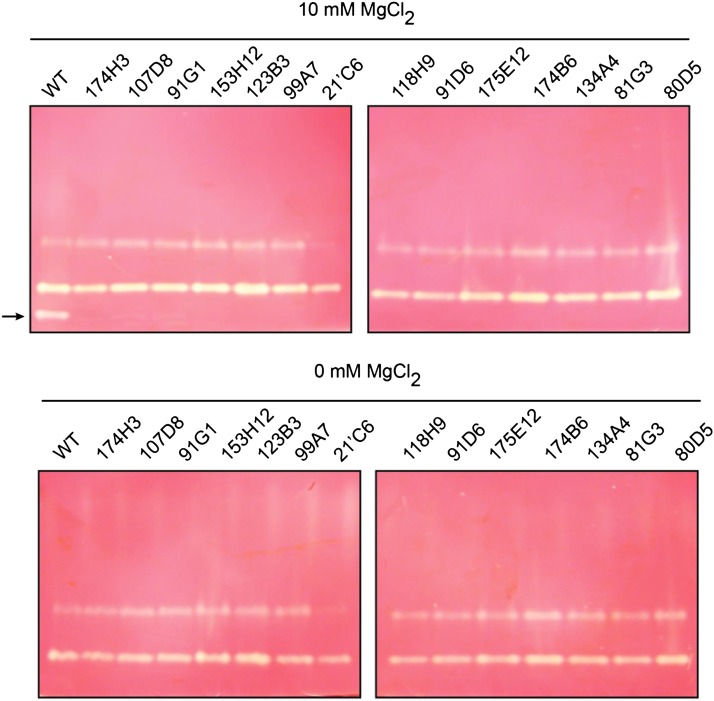
Figure 4.
Zymogram Analysis of Starch-Metabolizing Enzymes from Wild-Type and Class A Mutants.
Total protein of wild-type (WT) and class A mutant strains (21’C6, 80D5, 81G3, 91D6, 91G1, 99A7, 107D8, 118H9, 123B3, 134A4, 153H12, 174B6, 174H3, and 175E12) were separated by native PAGE, followed by transfer of proteins to a native PAGE containing 0.6% (p/v) amylopectin. The native gels were then incubated with (A) or without (B) 10 mM MgCl2. A cation-dependent activity enzyme is witnessed after iodine staining in the wild type’s crude extract and disappears in all class A mutants (black arrow).
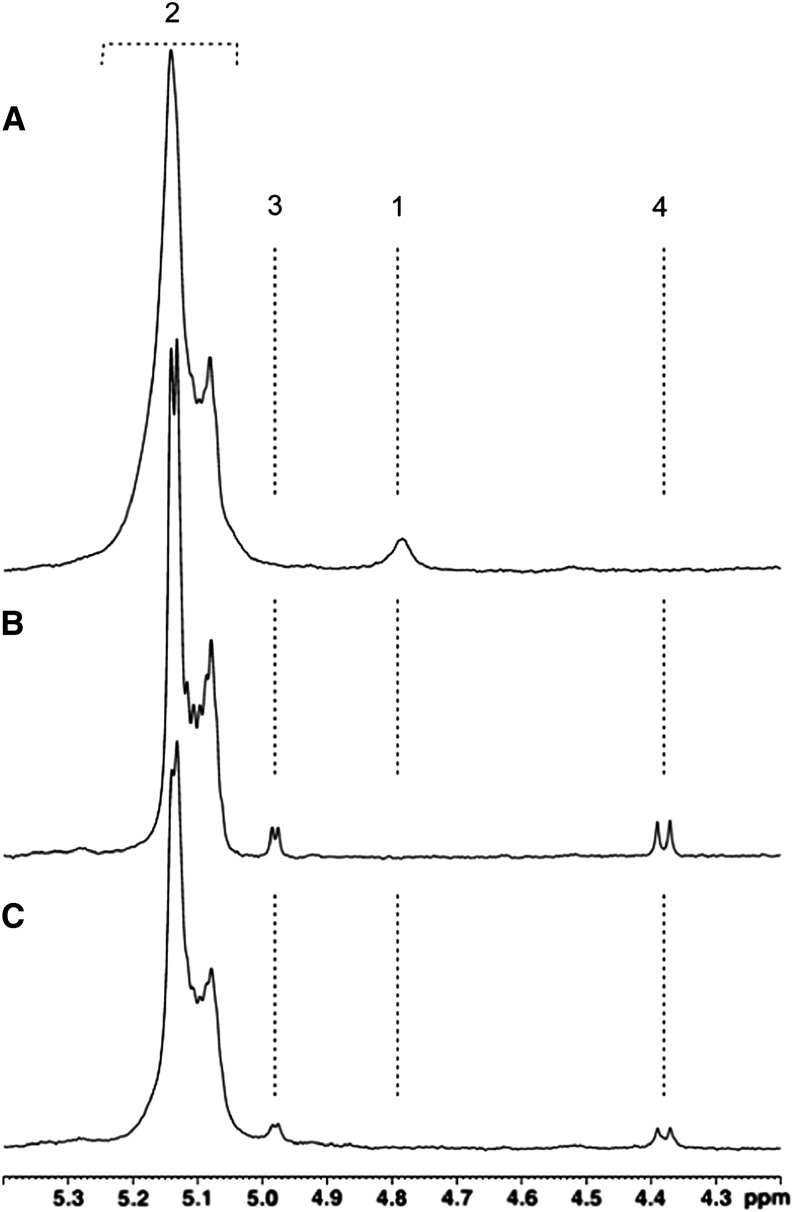
Figure 5.
Part of the 1H-NMR Spectra of Amylopectin in Dimethyl-Sulfoxide.
NMR analysis was performed on amylopectin (A) and amylopectin samples incubated overnight with a commercial isoamylase (B) or with active cation-dependent enzyme purified from wild-type strain (C). Peak #2 (5.2 to 5.08 ppm) and peak #1 (4.79 ppm) represent the signals of 1H engaged in α-1,4 and α-1,6 linkages, respectively. Both incubation experiments with amylopectin ([B] and [C]) result in the release of reducing ends and the apparition of α- (peak #3) and β-anomeric protons (peak #4) at 4.98 and 4.38 ppm, respectively. The presence of peaks #3 and #4 is correlated with a disappearance of the 1H signal engaged in α-1,6 linkages (peak #1). The NMR spectrometry analysis suggests that cation-dependent activity specifically hydrolyzes α-1,6 linkages.
Glycogen-Accumulating Mutants Carry Mutations in a GlgX-Like DBE Activity
The presence of pure enzyme preparations enabled us to identify a dozen of oligopeptide sequences that matched the protein sequences deduced from the genome (see Supplemental Figure 5 online). We were thus able to identify peptides that corresponded to GlgX2, one of the two GlgX-like genes present on the Cyanobacterium sp Clg1 genome sequence. Highly pure enzyme preparations enabled us to size the protein and ascertain the possible presence of distinct enzyme subunits. No bands corresponding to the expected size of GlgX1 and no trypsic fragments matching this enzyme sequence could be retrieved. We therefore conclude that this enzyme is a homomultimer (dimer or trimer). We then sequenced all starch metabolism genes from the 11 class A and three incompletely defective class A mutants and systematically found mutations only in the GlgX2 sequence, altering the amino acid sequence of the protein, while no mutations could be found in any of the other starch metabolism genes. These mutations are summarized in Figure 6. Among these mutations, we found four non-sense mutations resulting in three premature stop codons, one of which was preceded by a frame shift; the other 10 mutants defined different missense alleles. Three of these missense alleles were responsible for the incompletely defective phenotype of the 153H12, 123B3, and 176B6 strains. Because we hereby define an allelic series of 11 (plus three incompletely defective) independent mutants, none of which carried additional mutations in starch metabolism genes (see Supplemental Table 1 online), we can safely conclude that all defective phenotypes recorded in relation to polysaccharide accumulation result directly from the GlgX2 defect and not from other causes, obviating the need for formal complementation of the mutant gene through transgenesis with the wild-type sequence. Successful transgenesis has indeed not been reported in this subgroup of cyanobacteria, and all our attempts were unsuccessful.
Figure 6.
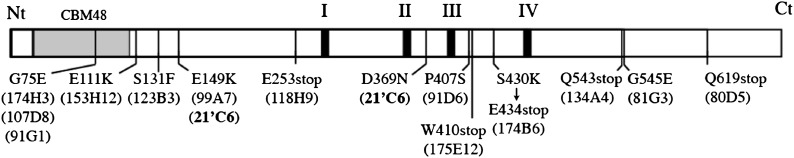
Various Allelic Mutations in the Cyanobacterium sp CLg1 glgX2 Gene.
Forward and reverse primers were designed in the untranslated region of the glgX2 gene. PCR reactions were performed on genomic DNA for each class A mutants. PCR products were cloned and DNA was sequenced on both strands. Punctual mutations (vertical black lines) found in each mutant (name in parentheses) are shown on the glgX2 segment. A mutant strain, 21’C6, harbors two point mutations (bold name). Regions I, II, III, and IV represent highly conserved sequences in the α-amylase family that includes acid residues involved in the catalytic site (Suzuki et al., 2007). The Carbohydrate Binding Module 48 (CBM 48; gray box) is observed at the N terminus (Nt) (Janeček et al., 2011).
The glgX-Like DBE Defines an Isoamylase Type of Activity
In bacteria and cyanobacteria, GlgX-type enzymes display a marked substrate preference for glycogen whose outer chains have been recessed with starch phosphorylase and little activity with either amylopectin or glycogen. The pure GlgX2 enzyme displayed little activity with such substrates or even with glycogen (Figure 7). However, unlike GlgX and in a fashion reminiscent of the plant isoamylase (Hussain et al., 2003), it displayed a marked preference for amylopectin, which it debranched to completion (Figure 7). Hence, the GlgX2 enzyme of Cyanobacterium more closely resembles the potato (Solanum tuberosum) isoamylase substrate specificity than that of the reference Pseudomonas sp enzyme. It certainly deviated from the bacterial GlgX type of activity classically involved in glycogen catabolism.
Figure 7.
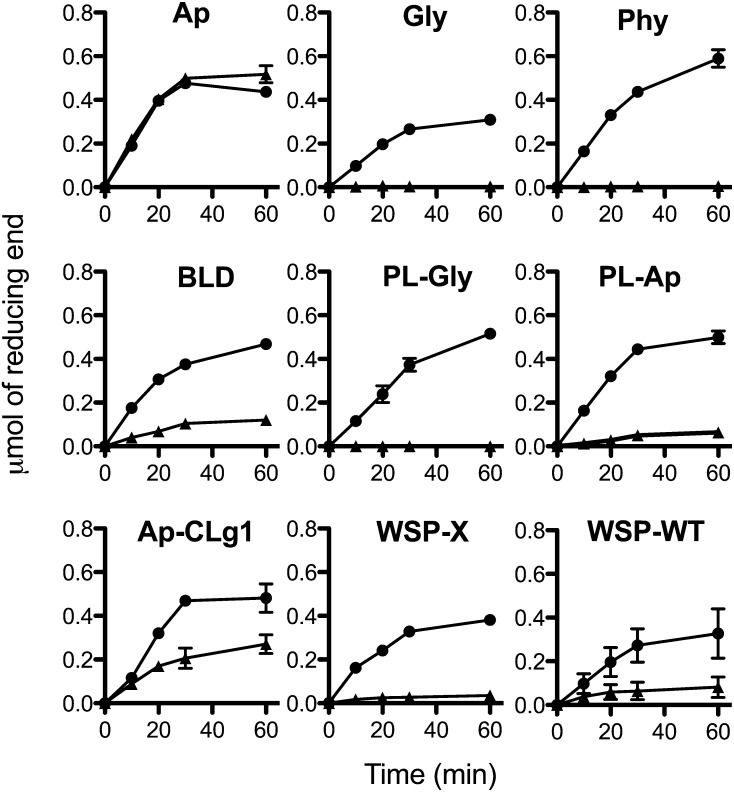
Specificity of the Cation-Dependent Debranching Activity toward Different Polysaccharides.
Both purified DBE activity (black triangle) and commercial isoamylase produced by Pseudomonas sp (megazyme; black circle) were incubated with 0.5% of amylopectin (Ap), glycogen (Gly), phytoglycogen (Phy), β-limit dextrin of amylopectin (BLD), phosphorylase-limit dextrin of glycogen (PL-gly), phosphorylase-limit dextrin of amylopectin (PL-Ap), semiamylopectin of the wild-type CLg1 strain (Ap-CLg1), water-soluble polysaccharide of class A mutant (WSP-X), and water polysaccharide of wild-type CLg1 strain (WSP-WT). The release of reducing ends was determined at 10, 20, 30, and 60 min of incubation using the 3,5-dinitro-2-hydroxybenzoic acid method. In order to compare both DBE activities, the specific activity of commercial isoamylase was adjusted at 0.5 μmol of reducing ends min−1 mL−1 using amylopectin as substrate. Data are the means ± sd of triplicates of two independent purification of cation-dependent debranching activity.
GlgX2 Defines the Enzyme Responsible for Starch Accumulation in Cyanobacterium Only
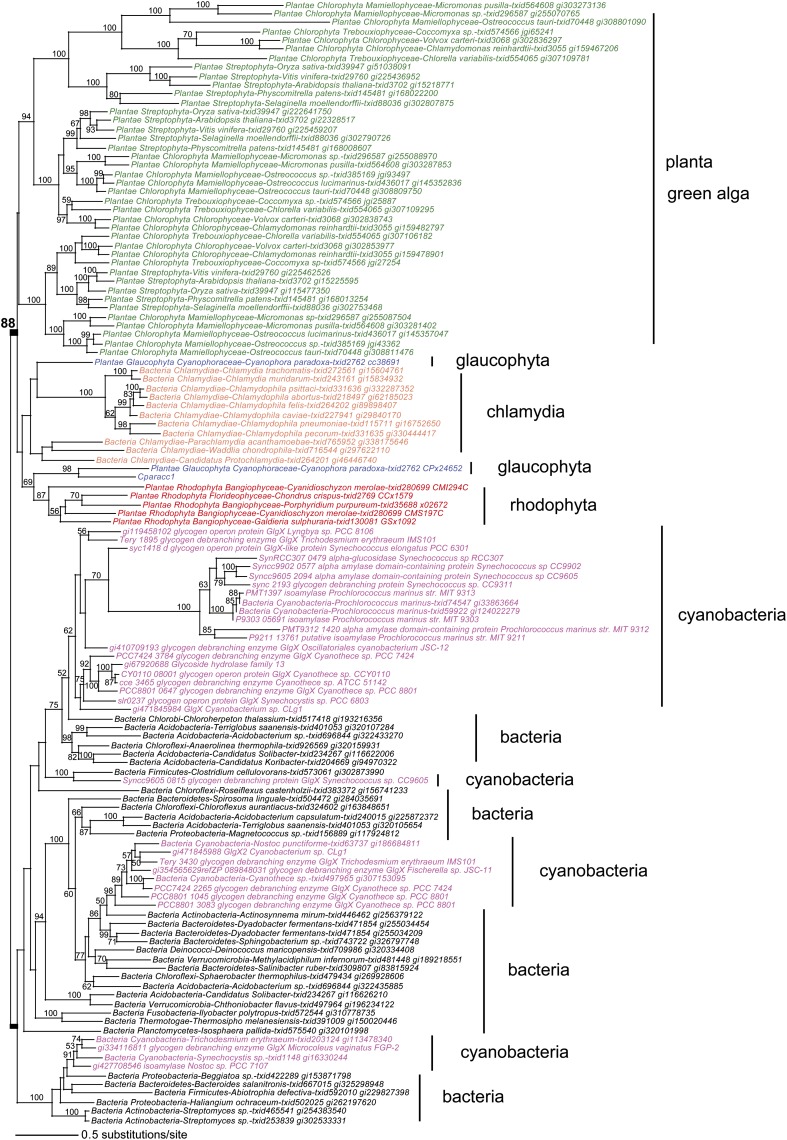
The mutants defined in this work establish that the Cyanobacterium sp CLg1 aggregates starch from a hydrosoluble precursor in a fashion very similar to starch in green algae and land plants. Since in both cases an enzyme from the same CAZy GH 13 subfamily 11 has been recruited to splice out those branches that prevent polysaccharide aggregation during synthesis, it is of interest to examine the phylogeny of these enzyme sequences and correlate the presence of selective enzyme forms to those of starch-like polymers. The phylogeny displayed in Figure 8 (see alignments in Supplemental Data Set 1 online) confirms that the source of the archaeplastidal DBEs cannot be traced to cyanobacteria. The strong monophyletic grouping of the archaeplastida enzymes with those of chlamydiales pathogens establishes the latter as the source of direct DBE, a bacteria-specific activity, in photosynthetic eukaryotes (Ball et al., 2013). In addition, the presence of the particular GlgX2-type enzyme does not correlate with the presence of starch in cyanobacteria. In Figure 8, all other documented starch-accumulating cyanobacteria lack this particular form of the GlgX enzyme, while the closest relatives to GlgX2 are reported to accumulate glycogen. We can therefore conclude that while acquisition of starch in Chloroplastida can still be suggested to be monophyletic (Cenci et al., 2013), transition of glycogen to starch metabolism in cyanobacteria cannot and defines distinctive polyphyletic events.
Figure 8.
Phylogenetic Analysis of Cyanobacterial, Bacterial, and Archaeplastidal Glycogen DBEs.
Starch DBEs in Chloroplastida play an important role in polysaccharide synthesis and in starch degradation. The maximum likelihood phylogeny of these enzymes shows that the three isoforms of isoamylase in Chloroplastida (green text) and the DBE found in Glaucophyta (blue text) and Rhodophyta (red text) do not share the same phylogenetic origin as the cyanobacterial GlgX (magenta text), particularly for Cyanobacterium sp CLg1. Rather, GlgX enzymes in Archaeplastida are sister to Chlamydiae (orange text; bootstrap support value is 90%), strongly suggesting a chlamydial origin of the gene in this eukaryotic supergroup (Ball et al., 2013). Alignments are given in Supplemental Data Set 1 online.
Substrate Preferences of Recombinant GlgX and Isoamylases from Cyanobacteria and Plants
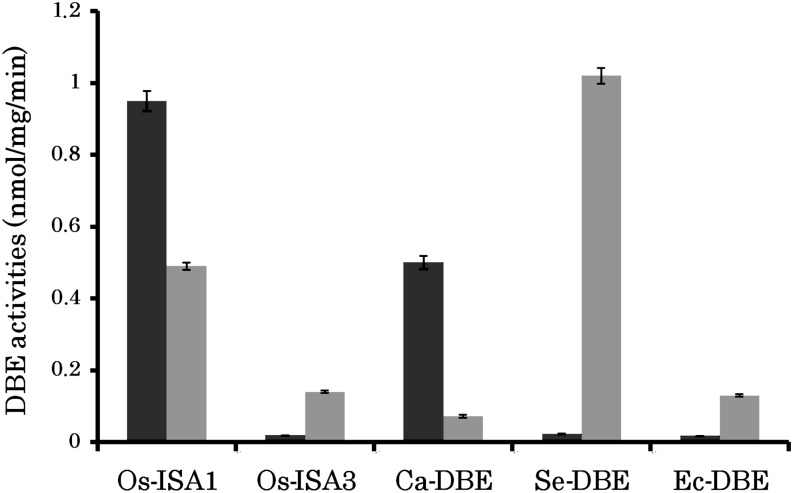
As detailed above, the GlgX2 enzyme responsible for amylopectin synthesis of Cyanobacterium sp CLg1 has converged toward a substrate preference similar to that of the potato ISOAMYLASE1 (ISA1) enzyme. This result would suggest that this enzyme preference may be one of the important features required for the enzyme involved in amylopectin biosynthesis. In order to strengthen this conclusion, we correlated these results to the characterization of recombinant DBEs (rDBEs) produced from organisms well characterized with respect to the structure of the storage polysaccharide they produce. We thus chose to study five distinct rDBEs from four different species: two starch (rice [Oryza sativa] and Cyanothece ATCC51142) and two glycogen accumulators (E. coli and Synechococcus elongatus PCC7942). In rice, it is well established that the ISA1 enzyme is involved in the aggregation of starch granules, while ISA3 is devoted to hydrolysis of external branches present in soluble oligosaccharides produced by either phosphorylases or amylases during starch degradation. In this respect, the function performed by ISA3 is very similar to that of the E. coli GlgX enzyme. Figure 9 summarizes the substrate preferences of the purified recombinant enzyme, while the purity of the preparations is displayed in Supplemental Figure 6 online. Quite evidently, all enzymes suspected (Cyanothece ATCC51142) or proven (rice ISA1) to be involved in amylopectin crystallization display a clear preference for amylopectin debranching, while those involved in glycogen (E. coli and S. elongatus PCC7942) or starch (rice ISA3) breakdown show dramatically reversed preferences. These results underline the importance of the substrate preferences of direct DBEs for either glycogen or starch accumulation in a given organism.
Figure 9.
Activities of Isoamylase-Type DBEs from Various Sources toward Amylopectin and Phytoglycogen.
Enzymatic activities were determined by measuring the amounts of malto-oligosaccharides liberated after the enzymatic reactions with amylopectin (blue bars) or phytoglycogen (brown bars) as glucan substrate, as described in the Methods. The enzymes measured were Os-ISA1, rice ISA1; Os-ISA3, rice ISA3; Ce-ISA, Cyanothece ATCC51142 DBE; Se-DBE, S. elongatus PCC7942 DBE; and Ec-GlgX, E. coli GlgX. Vertical axis presents the debranching activity as expressed by nmol equivalents liberated/μg protein/min, except that the activities of Os-ISA1 were reduced to 0.025 as a result of to their much higher activities compared with other enzymes. Each value represents the mean ± sd of three separate measurements.
DISCUSSION
Cyanobacterium sp CLg1 Accumulates Both Glycogen and Cyanophycean Starch
The significant accumulation of both glycogen and starch-like material in wild-type axenized Cyanobacterium sp CLg1 is a distinctive feature of these organisms when compared with Archaeplastida. In the latter, with the exception of the red alga Porphyridium sordidum (Shimonaga et al., 2008), only starch is known to accumulate with just trace amounts of WSPs and oligosaccharides being sometimes detectable. In this study, however, under conditions of maximal polysaccharide synthesis, 10% of the total storage polysaccharide pool always accumulates in the form of glycogen. We deem this amount to be physiologically significant. The presence of both types of storage polysaccharides may in this case be required to optimize cyanobacterial physiology by making a significant pool of Glc available through a more dynamic and readily accessible form of storage. Indeed, in glycogen, the Glc of the outer chains may be easier to mobilize following fluctuations of physiology both in light and darkness. The crystalline starch-like granules, in turn, will offer a larger pool of osmotically inert carbohydrate stores for delayed use by nitrogenase or cell division at night (Schneegurt et al., 1994). It will be of interest to check for the presence of an analogous fraction in other starch-storing cyanobacteria. We confirm the presence of starch-like granules in Cyanobacterium sp CLg1 composed of both an amylopectin-like high mass fraction and a smaller amylose fraction. The chain-length distribution of the high mass polysaccharide complies to the definition given for semiamylopectin, as it contains fewer of those chains exceeding a DP of 40 (Nakamura et al., 2005). Because the granules also contain a significant amount of amylose (5%), we propose to call this material cyanobacterial starch.
Evolution of Direct DBEs Involved in Amylopectin Maturation May Suffice to Explain the Transition from Glycogen to Starch Metabolism in All Living Cells
Convergent evolution in cyanobacteria and Archaeplastida underlines that polysaccharide debranching may define a common requirement for evolution of aggregated semicrystalline polysaccharides from glycogen metabolism (Cenci et al., 2013). The experiments reported here suggest that Cyanobacterium sp CLg1 use a multimeric DBE to aggregate polysaccharides into insoluble granules. In both cyanobacteria and plants, a glycohydrolase of a similar CAZy GH13 subfamily (subfamily 11 also known as TreX-GlgX) has been recruited. GlgX has been documented in proteobacteria and cyanobacteria to be selectively involved in glycogen degradation through both genetic and biochemical characterizations. In E. coli, GlgX was proven to be restrictive with respect to its substrate preference by debranching (only or preferably) those chains that have been recessed by glycogen phosphorylase during glycogen degradation (Dauvillée et al., 2005). This restrictive specificity was shown through the determination of the three-dimensional structure of the E. coli enzyme to be due to the shorter size of the inferred substrate binding groove (Song et al., 2010). The potato isoamylase was demonstrated to display a preference that accommodates amylopectin with comparatively little activity on glycogen or on either glycogen or amylopectin predigested by glycogen phosphorylase (Hussain et al., 2003). Here, the cyanobacterial enzyme GlgX2 is demonstrated to display the very same substrate preference as that reported for the potato enzyme (Hussain et al., 2003).
To strengthen these conclusions, we characterized rDBEs from two additional cyanobacterial species accumulating either glycogen or semiamylopectin. Both species, unlike Cyanobacterium sp CLg1, contain only one candidate CAZy GH13 subfamily 11 type of enzyme. Because the preference toward amylopectin debranching seems to distinguish GlgX2 from its GlgX progenitor relatives, we focused on this property by comparison to the enzyme activity in the presence of glycogen (in this case phytoglycogen). We further compared these preferences to those of the plant ISA1 enzyme and of the GlgX-like ISA3 plant enzyme. The results obtained so far build a convincing case for the evolution of isoamylase from a GlgX limit dextrinase type (GlgX or ISA3) of DBE as a major step to achieve aggregation of branched glucans into insoluble semicrystalline granules.
Indeed, the switch to an isoamylase specificity, together with a particular type of multimeric organization suggested by a plant study (Kubo et al., 2010; Utsumi et al., 2011), might define the two major determinants determining starch versus glycogen accumulation.
Although the convergent evolution reported in this work seems to imply that debranching of a soluble amylopectin precursor is mandatory to obtain aggregation of starch, we believe that this statement should nevertheless be taken with some caution. First, Streb et al. (2008) reported that in a particular mutant background lacking starch DBE, the presence of an additional mutation in a particular plastidial α-amylase restored starch synthesis in Arabidopsis leaves. This may imply that starch granule aggregation could dispense with the postulated debranching mechanism, although the crystallinity of this residual material still needs to be ascertained. Second, some of the secondary endosymbiosis lineages derived from Archaeplastida, as well as some little studied semiamylopectin accumulating cyanobacteria, were reported to lack candidate-direct DBE genes (Deschamps et al., 2006; Ball et al., 2011; Colleoni and Suzuki, 2012; Curtis et al., 2012). This observation would imply a nonessential function of polysaccharide debranching in amylopectin synthesis. Nevertheless, at least in one case, bona fide DBE activities were reported in biochemical assays, hinting that other glucan hydrolases may have evolved to ensure this function (Deschamps et al., 2006).
In any case, the work detailed in this article proves that at least in Cyanobacterium sp CLg1, a direct DBE has been recruited and evolved an isoamylase specificity and also possibly a multimeric organization that enabled these cyanobacteria to switch from glycogen synthesis to starch metabolism. Because both the chlamydial GlgX recruited by the Archaeplastida ancestors and the GlgX enzyme from cyanobacteria went through a similar convergent path to generate enzymes with similar properties, we propose that polysaccharide debranching could define an essential mechanism that enables conversion of glycogen metabolism into starch in all living cells. Such an evolution might have been favored each time a particular clade would have required a larger sized osmotically-inert storage polysaccharide pool.
METHODS
Media and Culture Conditions
Cyanobacterium sp CLg1 was grown in Artificial Sea Water medium in the absence (AS0 medium) or in the presence of 0.88 mM NaNO3 (ASNIII medium) (Rippka et al., 1979). The axenic strain was grown at 22°C and subjected to light/dark cycles (12 h/12 h) under 0.12 µmol m−2 s−1. Wild-type and mutant strains were maintained on solid ASNIII medium (0.8% noble agar) and transferred onto new plates every 3 months.
Cyanobacterium sp CLg1 Mutagenesis and Iodine Screening
The mutagenesis was performed on log phase Cyanobacterium sp CLg1 cultures plated on solid ASNIII medium (0.9% noble agar) as described by Deschamps et al. (2008). Cells (>105 cells/plate) were subjected to UV irradiation at 0.5, 3.0, or 6.0 cm from the source (Trans-illuminator TS-15 with a peak intensity of 7 mW cm−2 at 254 nm; Ultra-violet Products) for various exposure times (15, 30, 60, and 120 s) and immediately transferred to darkness during 24 h to avoid activation of the photodamage reparation system (Golden, 1988). After 1 month of growth in day-night cycles (12 h/12 h), all surviving colonies belonging to one mutagenesis condition (e.g., 30 s, 1 cm) were gently scraped after adding ASNIII medium over the agar slants and transfer in a 1-liter flask containing 300 mL of ASNIII. Mutagenized cells were diluted and plated on solid ASNIII medium. After 1 month, single colonies were transferred to 96-well plates containing 300 μL of liquid ASNIII medium and incubated for another month. A total of 20,000 cell patches were inoculated in duplicate by loading 40 μL of cell suspension on two solid ASNIII medium plates. After 1 month of growth, mutants impaired in starch metabolism were screened by spraying with iodine crystal vapors. In contrast with the dark-blue staining of wild-type cell patches, cell mutants harboring a distinct iodine staining were picked on sister Petri dishes and transferred to liquid ASNIII medium. The segregation of genome copies was verified for each individual mutant by iodine vapor screening of 48 colonies. Of a total of 20,000 cell patches screened, 88 mutants impaired in starch metabolism were isolated. Among them, 14 mutants harbored a yellow iodine phenotype, indicating that starch was replaced by glycogen synthesis.
Transmission Electron Microscopy Observation
Wild-type and mutant strains were cultivated in 50 mL of liquid AS0 medium and harvested by centrifugation (5 min at 4000g at 4°C) after 2 weeks of growth. The pellets were suspended and incubated during 1 h in fixing buffer containing 2.5% glutaraldehyde and 0.1 M cacodylate buffer at pH 7.2. The fixed cells were centrifuged at 16,000g during 15 min. Cell pellets were washed three times with 0.1 M cacodylate buffer, pH 7.2. The samples were dehydrated by incubating 20 min in increasing ethanol percentages (25, 50, 70, 95, and 100% three times). The impregnation step was conducted in EPON (epoxy resins) resin/ethanol mixtures at different ratios (1:2, 1:1, and 2:1) for 60 min, in EPON 100% for 30 min twice, and finally in EPON 100% overnight at 4°C. Pellets were then transferred to fresh EPON 100% and incubated for 48 h at 60°C for polymerization. Ultrathin sections were cut using a Leica UC6 ultramicrotome and then treated with 1% periodic acid for 30 min and washed six times in water. The sections were immersed in 20% acetic acid containing 1% thiosemicarbazide for 1 h, and a washing series was performed as follows: 20, 10, 5, and 2% acetic acid for 5 min each. The sections were washed six times in water, stained with 1% silver proteinate for 30 min in the dark, and finally washed six times in water.
As described in the following section, WSPs were purified from wild-type and mutant strains subjected to nitrogen starvation. Rabbit liver glycogen was used as a reference. Droplets of diluted WSP suspension were laid onto glow-discharged carbon-coated copper grids. The excess liquid was blotted with filter paper and a droplet of 2% uranyl acetate was added prior to drying.
Both sections and negatively stained preparations were observed using a Philips CM200 microscope operating at 80 kV. The images were recorded on Kodak SO163 film.
Quantification of Water-Soluble and -Insoluble (Starch-Like) Material Produced in Wild-Type and Mutant Strains
Wild-type and mutant strains were grown for 12 d in liquid AS0 medium and harvested at the middle of the light period by centrifugation at 3600g for 15 min at 4°C. Cells were washed three times with extraction buffer (50 mM HEPES, pH 8, 5 mM DTT, 1 mM EDTA, and 0.025% Triton), and the cell suspension (10 mL) was disrupted through a French press. WSP and insoluble polysaccharide (starch-like granules) were separated by spinning the lysate at 16,000g for 15 min at 4°C. The pellet was washed three times with sterile water at 4°C and then solubilized in 100% DMSO at 90°C for 10 min. The WSP (supernatant) and the starch-like granules (in the pellet) were quantified with an amyloglucosidase assay (R Biopharm Starch/amidon). The total protein concentration in the supernatant was determined using the Bradford method (Bio-Rad).
Structural Analysis of Soluble Polysaccharide
WSPs were purified using anion-exchange chromatography (Roth DOWEX 50:8) followed by size exclusion chromatography preequilibrated in 10% DMSO (Toyopearl TSK HW 50) as described (Colleoni et al., 1999). Polysaccharide was quantified in each 1-mL fraction by the phenol-sulfuric acid method (Fox and Robyt, 1991). Polysaccharide fractions were pooled and then subjected to second size exclusion chromatography (Toyopearl TSK HW 55S). Polysaccharide detected in the exclusion volume was totally debranched by Pseudomonas sp isoamylase (Megazyme) in 55 mM sodium acetate buffer, pH 3.5. The chain length distribution of glucan chains was then analyzed by high performance anion exchange chromatography-pulse amperometric detection (HPAEC-PAD) as described (Colleoni et al., 1999).
Structural Analysis of the Insoluble Polysaccharide Fraction
Insoluble polysaccharides were precipitated in 70% ethanol, centrifuged at 6000g for 10 min, and dried at room temperature. The pellet was suspended in 10 mM NaOH and subjected to CL-2B gel permeation chromatography. Each 300-μL fraction was checked for the presence of glucans through their interaction with iodine. Eighty microliters of fraction was incubated with 20 μL of iodine solution (1% KI and 0.1% I2), and the absorbance spectra were monitored to record the wavelength at maximum OD (λmax). The amylopectin, found in the exclusion volume, was then collected and totally debranched as described above for soluble polysaccharides and subjected to HPAEC-PAD.
Purification and Identification of the Cation-Dependent DBE
After 12 d of growth in ASNIII medium, cells were harvested at the middle of the light period by centrifugation at 3000g for 15 min at 4°C and washed three times with Tris-acetate buffer at 4°C (25 mM Tris-acetate, pH 7.5, and 10 mM DTT). Cells were disrupted by sonication or by a French press at 1250 p.s.i. The lysate was centrifuged at 16,000g for 15 min at 4°C. The supernatant (20 mL) was loaded on preparative anion-exchange chromatography column (HitrapQ sepharose Fast Flow, 5-mL column volume; GE Healthcare) preequilibrated in buffer A (25 mM Tris-acetate, pH 7.5, and 10 mM DTT). The proteins were eluted at 4 mL min−1 using a stepwise gradient of 40, 75, and 100% 1 M NaCl. Cation-dependent DBE activity was monitored for each fraction (2 mL) by zymogram analysis. The proteins were separated by nondenaturing PAGE containing 0.15% of β-limit dextrin (Megazyme). After electrophoresis, gels were incubated overnight at room temperature in buffer A with 10 mM MgCl2. The cation dependent activity was then visualized as a white band in the presence of β-limit dextrin of amylopectin or a blue band in the presence of starch polysaccharides after iodine staining. Fractions containing cation-dependent activity were pooled, and ammonium sulfate was added in order to reach a final concentration of 1 M. After filtration, the pooled fraction was loaded on a hydrophobic exchange chromatography column (Hitrap Butyl sepharose Fast Flow, 5-mL column volume; GE Healthcare) preequilibrated in buffer C [25 mM Tris-acetate, pH 7.5, 10 mM DTT, and 1 M (NH4)2SO4]. The proteins were eluted at 4 mL min−1 with a linear gradient of buffer A (0 to 100% in 25 mL). DBE activity of each fraction (2 mL) was monitored by zymogram analysis as described above. Fractions containing DBE were further purified using an anion-exchange chromatography column (MonoQ 5/50 GL, 1-mL column volume; GE Healthcare) preequilibrated in buffer A (25 mM Tris-acetate, pH 7.5, and 10 mM DTT). The proteins were eluted at 1 mL min−1 with a gradient of buffer A containing 1 M NaCl: 0 to 40% in 5 mL, 40 to 60% in 20 mL, 60 to 80% in 10 mL, 80 to 100% in 5 mL. The enzyme activities of each fraction (1 mL) were monitored by zymogram analysis. Fractions containing debranching activity were further concentrated with Amicon Ultra 0.5-mL centrifugal filters (Merck Millipore). Concentrated samples with activity were stored at −80°C in 20% glycerol.
Biochemical Characterization of Cation-Dependent Activity
Purified samples displaying cation-dependent activity (0.526 μmol of reducing end min−1 mL−1) were incubated in 120 mM Tris, 30 mM imidazole, and 30 mM acetic acid, pH 7.5, in presence of 0.5% (w/v) of polysaccharide (amylopectin, glycogen, β-limit dextrin, and phosphorylase-limit dextrin) and 10 mM MgCl2. The release of glucan chains was followed by measuring the increase in reducing ends using the dinitrosalicylic acid method. After 10 min at 99°C, the OD was measured at 540 nm and compared with a standard curve (0 to 100 μg) measured with 1 mg mL−1 of Glc. Glucan chains released during the incubation were analyzed by HPAEC-PAD as described below. The cation-dependent activity was compared with that of isoamylase of Pseudomonas sp diluted 1/100 and incubated with 0.5% polysaccharide and 55 mM sodium acetate, pH 3.5.
Identification of the 80-kD Polypeptide by Nano–Liquid Chromatography–Tandem Mass Spectrometry
Samples with cation-dependent activity were further purified by an additional size-exclusion chromatography preequilibrated with 10 mM Tris/acetate buffer, pH 7.5, 10 mM DTT, and 150 mM NaCl (GE Healthcare Superdex 200; D = 0.8 centimorgans; H = 30 centimorgans). Each fraction (300 μL) was analyzed using zymogram procedures. Fractions containing the cation-dependent activity were then precipitated with 20% TCA (60 μL 20% TCA per 1 mL of sample with 25 μL of 2% sodium deoxycholate) and incubated on ice for 10 to 15 min. After centrifugation for 5 min at 16,100g, the pellets were suspended in 10 μL of buffer A (0.1 M Na2CO3 and 0.1 M DTT). The samples were boiled at 95°C for 5 min in the presence of 5 μL of buffer B (5% SDS, 30% saccharose, and 0.1% blue of bromophenol). Proteins were separated on 7.5% polyacrylamide gels with 0.1% SDS. SDS-PAGE gels were then stained for one night using Roti Blue (Roth) and washed with 25% methanol. The polypeptide was cut off and placed in 60% acetonitrile in bicarbonate ammonium before nano–liquid chromatography–tandem mass spectrometry analysis as described by Gurcel et al. (2008).
Gene Cloning and Sequencing
Starch metabolism genes (see Supplemental Table 2 online), DBE genes (glgX2 and glgX1), branching enzyme genes (be3), and starch/glycogen synthase genes (gbss) were amplified from genomic DNA of wild-type and mutant strains using the following oligonucleotide primers: glgX2F (forward), 5′-ATGTTAATGGGAGATGAATCTATGA-3′; glgX2R (reverse), 5′-TAATTAGTGGTTTTTAGTACTACTAACG-3′; glgX1F, 5′-ATGAACCATAAAACGTTACCTG-3′; glgX1R, 5′-CTATTTTGCCATTAATAAAATGCAAC-3′; gbss, 5′-TCCTCATGAATTGGTGACATAGTATGTT-3′; gbssR, 5′-CAGATACAGGTGAAAATCGTAACGC-3′; be3F, 5′-AGTGAATAGCCAAAAATCAACGAT-3′; and be3R, 5′-TGACCATCCCATTTGGCTCCTA-3′.
PCR (Analytik Jena Flexcycler) was conducted as follows: 95°C for 5 min, 30 cycles of denaturation at 98°C for 30 s, annealing 30 s at 59.6°C for GlgX2, GBSS, and BE3 and at 59.1°C for GlgX1, and extension for 2 min and 30 s at 72°C, and a final elongation step at 72°C for 5 min. The PCR products were cloned into pCR-BluntII-TOPO vector (Invitrogen), transferred into the chemically competent Escherichia coli TOP10 Mach1-TR, and grown on Luria-Bertani agar with kanamycin and X-Gal. Purified plasmids were sequenced by the GATC Biotech Company according to Sanger methods. Each gene was sequenced on both strands using additional primers when required. The presence of a mutation was identified by alignment with the wild-type gene (BLASTn).
Phylogenetic Analysis
Homologs of GlgX were identified in GenBank or other sources using BLASTp and aligned with multiple sequence comparison by log expectation (MUSCLE) (http://www.ebi.ac.uk/Tools/msa/muscle/). The alignment was manually refined using SeAl (http://tree.bio.ed.ac.uk/software/seal/), and blocks of missing data in some taxa or regions of low identity were manually removed (final alignment of 595 amino acids available from Supplemental Data Set 1 online). This reduced alignment was analyzed under maximum likelihood. The best-fitting amino-acid substitution model was selected according to the Akaike informational criterion with ProtTest using the default values (Abascal et al., 2005). The Le Gascuel (Le and Gascuel, 2008) model with heterogeneous gamma rate distribution across sites (+Γ) was selected by ProtTest for this protein data set (see Supplemental Methods 1 online). The Le Gascuel-model parameter values were used under randomized accelerated maximum likelihood (RAxML) v.7.2.8 (Stamatakis 2006) for the maximum likelihood tree searches. The stability of monophyletic groups was assessed using RAxML with 1000 bootstrap replicates. The phylogenetic tree is rooted on the branch uniting the Archaeplastida sequences. This was done to reflect current understanding that supports Archaeplastida monophyly.
Preparation of α-Glucans for rDBE Characterization
The rice (Oryza sativa) amylopectin was prepared from starch in rice endosperm from a GBSSI-deficient waxy mutant line, EM21, derived from the japonica-type rice cultivar ‘Kinmaze’. The starch granules from EM21 were extracted from polished rice seeds using the cold-alkali method as described (Yamamoto et al., 1981). Ten grams of the starch granules was dispersed in 500 mL of dimethylsulfoxide at 80°C under N2 gas. The dispersed starch was precipitated by adding 1500 mL of ethanol and centrifuged at 10,000g for 20 min. The resulting precipitate was washed with acetone and diethyl ether, dried, and stored at −30°C until use as amylopectin.
The rice phytoglycogen was prepared from an isa1-deficient sugary1 mutant line, EM914, derived from the japonica-type rice cultivar ‘Taichung-65’ (Nakamura et al., 1997). The phytoglycogen from polished seeds was extracted with 0.1% (w/v) NaOH solution at 4°C. The extract was filtrated through the 100-µm sieve. The filtrate was collected and phytoglycogen was precipitated with 3 volumes of ethanol. The precipitate was collected by centrifugation at 10,000g for 20 min. To remove the low molecular mass sugars and dextrins, the precipitate was rinsed 10 times with 50% (v/v) ethanol. The resulting precipitate was washed with acetone and diethyl ether, dried, and stored at −30°C until use as phytoglycogen.
Cloning of the Regions Encoding Glucan DBEs
Total RNA was isolated from the developing endosperm of the japonica-type rice cultivar ‘Nipponbare’ according to the manufacturer’s protocol using an RNeasy plant mini kit (Qiagen). The cDNA was synthesized by Superscript III reverse transcriptase (Invitrogen).
The expression vectors for genes encoding the predicted rice mature ISA1 and Pullulanase proteins in E. coli were prepared as described previously (Fujita et al., 2009; Utsumi et al., 2011).
The region encoding the predicted rice mature ISA3 in Nipponbare was amplified by PCR with pGEM-T easy containing the Os-ISA3 gene using the following primer pairs: forward primer (5′-CACGTGTAGCACCACGGCGAGAG-3′) containing BbrPI, and reverse primer (5′-GTCGACCTAAGGCTTTGCCTTGAGC-3′) containing SalI. The DNA fragment was subcloned into pGEM-T easy vector. The sequence encoding the mature Os-ISA3 protein in the pGEM-T easy vector was excised by restriction enzymes BbrPI and SalI. The DNA fragment was ligated with the pET32b (Novagen) expression vector treated with EcoRV and SalI.
The region including the Synechococcus DBE gene was amplified by PCR with genomic DNA from Synechococcus PCC7942 with the following primer pairs: forward primer (5′-CATATGACTGTTTCATCCCGTCGC-3′) containing NdeI and reverse primer (5′-GTCGACCTGCAGGCGGCCGCGAATTC-3′) containing SalI. The DNA fragment was subcloned into pGEM-T easy vector. The Synechococcus DBE in pGEM-T easy was digested with the restriction enzymes NdeI and SalI. The DNA fragment was ligated with the pCold TF (Takara) expression vector treated with NdeI and SalI.
The DNA fragment encoding the Cyanothece ATCC51142 DBE was isolated from genomic DNA of Cyanothece ATCC51142 DBE by PCR using the following primer pairs: forward primer (5′-CATATGAGCTCAAAACCCTTC-3′) containing NdeI and reverse primer (5′-GTCGACTTATGAATTAGACTTTGCC-3′) containing SalI. The DNA fragment was subcloned into pGEM-T easy vector. The Cyanothece DBE in pGEM-T easy was isolated using restriction enzymes NdeI and SalI. The DNA fragment was ligated with the pCold TF expression vector treated with NdeI and SalI.
The DNA fragment encoding the E. coli GlgX was amplified by PCR using the genomic DNA from E. coli K12 strain as template and the following primer pairs: forward primer (5′-CATATGACACAACTCGCCATTG-3′), containing NdeI, and reverse primer (5′-GTCGACTCATCTCTGGAACACACAC-3′) containing SalI. The DNA fragment was subcloned into pGEM-T easy vector. The E. coli GlgX in pGEM-T easy was prepared by hydrolysis with restriction enzymes NdeI and SalI. The treated DNA fragment was ligated in the pCold TF expression vector treated by NdeI and SalI.
E. coli Overexpression of DBE
All of the constructed vectors including DBE genes were introduced into the expression host E. coli BL21 (DE3) star strain (Invitrogen) containing pTf16 chaperone plasmid (Takara). The transformed cells were grown at 37°C in 10 mL of Terrific Broth medium containing 100 µg/mL of carbenicillin and 34 µg/mL of chloramphenicol overnight. The precultured cells were subsequently grown at 37°C with 1.0 liters of Terrific Broth medium containing 100 µg/mL of carbenicillin, 34 µg/mL of chloramphenicol, and 1 mg/mL of l-arabinose. The l-arabinose was used for induction of chaperon protein Tf16. When the OD at 600 nm of the culture medium reached 0.4 to 0.5, the culture was preincubated at 15°C for 30 min. Then, the culture medium was supplemented with isopropylthio-β-d-galactoside to a final concentration of 1 mM, and the rDBE was induced by further incubation at 15°C for 20 h. The cells were collected by centrifugation and stored at –80°C until use.
Purification of Recombinant DBE
The frozen transformed E. coli cells were thawed on ice and suspended in extraction buffer (50 mM imidazol-HCl, pH 7.4, 8 mM MgCl2, 12.5% glycerol, and 50 mM 2-mercaptoethanol). The cells were disrupted by sonication on ice and centrifuged at 10,000g for 30 min at 0°C. The resulting supernatant was applied to the HiTrap HP anion-exchange column (5 mL; GE Healthcare) equilibrated with a buffer A (50 mM imidazol-HCl, pH 7.4, 8 mM MgCl2, and 50 mM 2-mercaptoethanol). After washing the column with the buffer A, the proteins were eluted with a linear gradient of 0.0 to 0.5 M NaCl at a flow rate of 2 mL/min. The peak fractions containing the abundant rDBE protein were collected and were added to one-third volume of 50 mM Na-phosphate, pH7.0, containing 3 M ammonium sulfate. The rDBE protein was applied to the TSKgel Ether-5PW hydrophobic column (7.5 mm in diameter × 75 mm in length; Tosoh Corporation) equilibrated with buffer B (50 mM Na-phosphate, pH 7.0, 50 mM 2-mercaptoethanol, and 1.0 M ammonium sulfate). After washing the column with buffer B, the protein was eluted with a linear gradient of 1.0 to 0 M ammonium sulfate in a buffer solution consisting of 50 mM Na-phosphate, pH 7.0, and 50 mM 2-mercaptoethanol at a flow rate of 1 mL/min. The peak fraction containing the major rDBE protein was collected and concentrated using centrifugal filter units (Ultracel-3K; Millipore). Each DBE preparation was purified to be a near homogeneity using these procedures. The purified DBE preparation was suspended in 50 mM imidazol-HCl, pH 7.4, 8 mM MgCl2, 1 mM DTT, 0.5 M NaCl, and 12.5% glycerol and kept at −80°C until use.
Recombinant ISA Enzymatic Reaction
The reaction mixture routinely contained 10 mM HEPES-NaOH buffer, pH 7.4, 2 mg of glucan, and DBE in a total volume of 400 μL. The reaction was run at 30°C. The amounts of DBE used were 0.13 or 0.13 μg for rOsISA1, 32 or 9.54 μg for rOsISA3, 32 or 32 μg for rCaDBE, 30.3 or 30.3 μg for rSeDBE, 32 or 32 μg for rEcDBE, and 0.11 or 0.11 μg for PaISA when the enzyme was incubated with phytoglycogen or amylopectin, respectively. At appropriate time intervals, the enzymatic reaction was terminated by heating the reaction mixture in a boiling water bath for 5 min.
Assay of DBE Activity
The DBE activity was determined by measuring the amount of reducing ends in the reaction products according to the method of Utsumi et al. (2009). Solution A consisted of 97.1 mg of disodium 2,2’-bicinchoninate, 3.2 g of sodium carbonate monohydrate, and 1.2 g of sodium bicarbonate in a total volume of 50 mL. Solution B consisted of 62 mg of copper sulfate pentahydrate and 63 mg of l-Ser in a total volume of 50 mL. The working reagent was freshly prepared by mixing the equal volume of Solutions A and B (Fox and Robyt, 1991). An amount of 225 μL of the sample was added to 225 μL of the working reagent in a tube, and the mixture was incubated at 80°C for 40 min in a water bath. After incubation, the assay mixture was then cooled to room temperature and incubated for 10 min. An aliquot (150 µL) of the disodium 2,2’-bicinchoninate–treated sample was taken, and its absorbance at 560 nm was measured using a microplate spectrophotometer (Bio-Rad). The absorbance at 560 nm was found to be proportional to the concentration of maltose or Glc in the range of 0 to at least 25 µM of the assay mixture.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: GlgX1 (KC422239.1), GlgX2 (KC422240.1), GlgB1 (JX074061.1), GlgB2 (JX074062.1), GlgB3 (JX074063.1), Apu57 (KF717069), BE57 (KF717075), GlgA1 (KF717072), GlgA2 (KF717073), Apu13 (KF717068), GBSS (KF717071), GlgP (KF717074), MalQ (KF717076), and indirect DBE (KF717070).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cell Patches of the Five Classes of Mutants of Cyanobacterium sp CLg1 Stained with Iodine Vapors.
Supplemental Figure 2. Zymogram of Phosphorylase Activity.
Supplemental Figure 3. Effects of Different Cations on Debranching Enzyme Activity.
Supplemental Figure 4. Purification of Cation-Dependent Debranching Enzyme Activity.
Supplemental Figure 5. Identification of Debranching Enzyme Activity.
Supplemental Figure 6. SDS-PAGE of Purified DBE Preparations from Rice, Cyanobacteria, and E. coli.
Supplemental Table 1. Summary Table of Starch Metabolizing Genes Checked for Mutations.
Supplemental Table 2. List of Primers Used for Gene Cloning and Sequencing Involved in the Storage Polysaccharide Metabolism of Cyanobacterium sp CLg1.
Supplemental Methods 1. Parameter Values for Phylogenetic Analysis.
Supplemental Data Set 1. GlgX and Isoamylase Alignments.
Supplementary Material
Acknowledgments
S.G.B. and C.C. were supported by the Centre National de la Recherche Scientifique, the Université des Sciences et Technologies de Lille, the Région Nord Pas de Calais, and the Agence Nationale pour la Recherche (ANR) grants from Starchevol.
AUTHOR CONTRIBUTIONS
C.C. and S.G.B. designed the research and wrote the article. U.C., M.C., M.D., J.N.-R., and C.T. carried out the mutagenesis campaign and the screening process. Y.U., D.K., S.S., E.S., Y.N. expressed recombinant debranching enzymes and sequenced the CLg1 genome. J.-L.P. and A.D.-T. performed TEM observations. X.R. and E.M. were responsible for HPAEC-PAD and proton NMR analysis, respectively. D.B. and M.-C.A. performed phylogenetic analysis. A.-S.V.-E. carried out mass spectrometry analysis. M.P. and L.S. performed molecular modeling of CLg1-GlgX2.
Glossary
- TEM
transmission electronic microscopy
- DBE
debranching enzyme
- DP
degree of polymerization
- WSP
water-soluble polysaccharide
- rDBE
recombinant DBE
References
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Ball S., Colleoni C., Cenci U., Raj J.N., Tirtiaux C. (2011). The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J. Exp. Bot. 62: 1775–1801 [DOI] [PubMed] [Google Scholar]
- Ball S., Guan H.P., James M., Myers A., Keeling P., Mouille G., Buléon A., Colonna P., Preiss J. (1996). From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell 86: 349–352 [DOI] [PubMed] [Google Scholar]
- Ball S.G., Subtil A., Bhattacharya D., Moustafa A., Weber A.P.M., Gehre L., Colleoni C., Arias M.C., Cenci U., Dauvillée D. (2013). Metabolic effectors secreted by bacterial pathogens: Essential facilitators of plastid endosymbiosis? Plant Cell 25: 7–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buléon A., Colonna P., Planchot V., Ball S. (1998). Starch granules: Structure and biosynthesis. Int. J. Biol. Macromol. 23: 85–112 [DOI] [PubMed] [Google Scholar]
- Cenci U., Nitschke F., Steup M., Minassian B.A., Colleoni C., Ball S. (2013). Transition from glycogen to starch metabolism in Archaeplastida. Trends Plant Sci. (In Press). [DOI] [PubMed] [Google Scholar]
- Colleoni C., et al. (1999). Genetic and biochemical evidence for the involvement of alpha-1,4 glucanotransferases in amylopectin synthesis. Plant Physiol. 120: 993–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni, C., and Suzuki, E. (2012). Storage polysaccharide metabolism in Cyanobacteria. In Essential Reviews in Experimental Biology: Starch: Origins, Structure and Metabolism, Vol. 5, I. Tetlow, ed (London: The Society for Experimental Biology), pp. 213–253. [Google Scholar]
- Coppin A., Varré J.S., Lienard L., Dauvillée D., Guérardel Y., Soyer-Gobillard M.O., Buléon A., Ball S., Tomavo S. (2005). Evolution of plant-like crystalline storage polysaccharide in the protozoan parasite Toxoplasma gondii argues for a red alga ancestry. J. Mol. Evol. 60: 257–267 [DOI] [PubMed] [Google Scholar]
- Curtis B.A., et al. (2012). Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492: 59–65 [DOI] [PubMed] [Google Scholar]
- Dauvillée D., Deschamps P., Ral J.P., Plancke C., Putaux J.L., Devassine J., Durand-Terrasson A., Devin A., Ball S.G. (2009). Genetic dissection of floridean starch synthesis in the cytosol of the model dinoflagellate Crypthecodinium cohnii. Proc. Natl. Acad. Sci. USA 106: 21126–21130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D., Kinderf I.S., Li Z., Kosar-Hashemi B., Samuel M.S., Rampling L., Ball S., Morell M.K. (2005). Role of the Escherichia coli glgX gene in glycogen metabolism. J. Bacteriol. 187: 1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue B., Fontaine T., Routier F., Decq A., Wieruszeski J.M., Van Den Koornhuyse N., Maddelein M.L., Fournet B., Ball S. (1992). Waxy Chlamydomonas reinhardtii: Monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J. Bacteriol. 174: 3612–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps P., et al. (2008). Metabolic symbiosis and the birth of the plant kingdom. Mol. Biol. Evol. 25: 536–548 [DOI] [PubMed] [Google Scholar]
- Deschamps P., et al. (2006). Nature of the periplastidial pathway of starch synthesis in the cryptophyte Guillardia theta. Eukaryot. Cell 5: 954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon L.I., Lindvall S., Bauer K., Bergman B., Carpenter E.J. (2004). Ultrastructure of unicellular N2 fixing cyanobacteria from the tropical north Atlantic and subtropical north Pacific oceans. J. Phycol. 40: 1074–1078 [Google Scholar]
- Fox J.D., Robyt J.F. (1991). Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal. Biochem. 195: 93–96 [DOI] [PubMed] [Google Scholar]
- Fu J., Xu X. (2006). The functional divergence of two glgP homologues in Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 260: 201–209 [DOI] [PubMed] [Google Scholar]
- Fujita N., et al. (2009). Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. J. Exp. Bot. 60: 1009–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden S.S. (1988). Mutagenesis of cyanobacteria by classical and gene-transfer-based methods. Methods Enzymol. 167: 714–727 [DOI] [PubMed] [Google Scholar]
- Gurcel C., Vercoutter-Edouart A.S., Fonbonne C., Mortuaire M., Salvador A., Michalski J.C., Lemoine J. (2008). Identification of new O-GlcNAc modified proteins using a click-chemistry-based tagging. Anal. Bioanal. Chem. 390: 2089–2097 [DOI] [PubMed] [Google Scholar]
- Hussain H., Mant A., Seale R., Zeeman S., Hinchliffe E., Edwards A., Hylton C., Bornemann S., Smith A.M., Martin C., Bustos R. (2003). Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell 15: 133–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James M.G., Robertson D.S., Myers A.M. (1995). Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeček S., Svensson B., MacGregor E.A. (2011). Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enzyme Microb. Technol. 49: 429–440 [DOI] [PubMed] [Google Scholar]
- Jeanningros R., Creuzet-Sigal N., Frixon C., Cattaneo J. (1976). Purification and properties of a debranching enzyme from Escherichia coli. Biochim. Biophys. Acta 438: 186–199 [DOI] [PubMed] [Google Scholar]
- Kubo A., Colleoni C., Dinges J.R., Lin Q., Lappe R.R., Rivenbark J.G., Meyer A.J., Ball S.G., James M.G., Hennen-Bierwagen T.A., Myers A.M. (2010). Functions of heteromeric and homomeric isoamylase-type starch-debranching enzymes in developing maize endosperm. Plant Physiol. 153: 956–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Fujita N., Harada K., Matsuda T., Satoh H., Nakamura Y. (1999). The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 121: 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.Q., Gascuel O.L.G. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Meléndez R., Meléndez-Hevia E., Canela E.I. (1999). The fractal structure of glycogen: A clever solution to optimize cell metabolism. Biophys. J. 77: 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G., Maddelein M.L., Libessart N., Talaga P., Decq A., Delrue B., Ball S. (1996). Preamylopectin processing: A mandatory step for starch biosynthesis in plants. Plant Cell 8: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Kubo A., Shimamune T., Matsuda T., Harada K., Satoh H. (1997). Correlation between activities of starch debranching enzyme and α-polyglucan structure in endosperms of sugary-1 mutants of rice. Plant J. 12: 143–153 [Google Scholar]
- Nakamura Y., et al. (2005). Some Cyanobacteria synthesize semi-amylopectin type alpha-polyglucans instead of glycogen. Plant Cell Physiol. 46: 539–545 [DOI] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111: 1–61 [Google Scholar]
- Schneegurt M.A., Sherman D.M., Nayar S., Sherman L.A. (1994). Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 176: 1586–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer J., Graham T.E. (2002). New perspectives on the storage and organization of muscle glycogen. Can. J. Appl. Physiol. 27: 179–203 [DOI] [PubMed] [Google Scholar]
- Shimonaga T., et al. (2008). Variation in storage alpha-glucans of the Porphyridiales (Rhodophyta). Plant Cell Physiol. 49: 103–116 [DOI] [PubMed] [Google Scholar]
- Song H.N., Jung T.Y., Park J.T., Park B.C., Myung P.K., Boos W., Woo E.J., Park K.H. (2010). Structural rationale for the short branched substrate specificity of the glycogen debranching enzyme GlgX. Proteins 78: 1847–1855 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Streb S., Delatte T., Umhang M., Eicke S., Schorderet M., Reinhardt D., Zeeman S.C. (2008). Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell 20: 3448–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki E., Onoda M., Colleoni C., Ball S., Fujita N., Nakamura Y. (2013). Physicochemical variation of cyanobacterial starch, the insoluble α-glucans in cyanobacteria. Plant Cell Physiol. 54: 465–473 [DOI] [PubMed] [Google Scholar]
- Suzuki E., Umeda K., Nihei S., Moriya K., Ohkawa H., Fujiwara S., Tsuzuki M., Nakamura Y. (2007). Role of the GlgX protein in glycogen metabolism of the cyanobacterium, Synechococcus elongatus PCC 7942. Biochim. Biophys. Acta 1770: 763–773 [DOI] [PubMed] [Google Scholar]
- Utsumi Y., Utsumi C., Sawada T., Fujita N., Nakamura Y. (2011). Functional diversity of isoamylase oligomers: The ISA1 homo-oligomer is essential for amylopectin biosynthesis in rice endosperm. Plant Physiol. 156: 61–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi Y., Yoshida M., Francisco P.B., Jr, Sawada T., Kitamura S., Nakamura Y. (2009). Quantitative assay method for starch branching enzyme with bicinchoninic acid by measuring the reducing terminals of glucans. J. Appl. Glycosci. 56: 215–222 [Google Scholar]
- Wattebled F., Dong Y., Dumez S., Delvallé D., Planchot V., Berbezy P., Vyas D., Colonna P., Chatterjee M., Ball S., D’Hulst C. (2005). Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 138: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Sawada S., Onogaki I. (1981). Effects of quality and quantity of alkali solution on the properties of rice starch. Denpun Kagaku 28: 241–244 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.