Alternative precursor mRNA splicing represents a major mechanism to increase transcriptome diversity in higher eukaryotes. This work demonstrates that a substantial fraction of the alternative splicing variants from Arabidopsis thaliana is subject to turn over via the RNA quality-control mechanism nonsense-mediated decay, having important implications for the regulation of gene expression.
Abstract
The nonsense-mediated decay (NMD) surveillance pathway can recognize erroneous transcripts and physiological mRNAs, such as precursor mRNA alternative splicing (AS) variants. Currently, information on the global extent of coupled AS and NMD remains scarce and even absent for any plant species. To address this, we conducted transcriptome-wide splicing studies using Arabidopsis thaliana mutants in the NMD factor homologs UP FRAMESHIFT1 (UPF1) and UPF3 as well as wild-type samples treated with the translation inhibitor cycloheximide. Our analyses revealed that at least 17.4% of all multi-exon, protein-coding genes produce splicing variants that are targeted by NMD. Moreover, we provide evidence that UPF1 and UPF3 act in a translation-independent mRNA decay pathway. Importantly, 92.3% of the NMD-responsive mRNAs exhibit classical NMD-eliciting features, supporting their authenticity as direct targets. Genes generating NMD-sensitive AS variants function in diverse biological processes, including signaling and protein modification, for which NaCl stress–modulated AS-NMD was found. Besides mRNAs, numerous noncoding RNAs and transcripts derived from intergenic regions were shown to be NMD responsive. In summary, we provide evidence for a major function of AS-coupled NMD in shaping the Arabidopsis transcriptome, having fundamental implications in gene regulation and quality control of transcript processing.
INTRODUCTION
The steady state transcriptome is the output of concurrent transcriptional activity and RNA degradation, both of which are modulated via complex regulatory networks integrating internal and external cues. The accuracy of gene expression is assured by surveillance mechanisms, such as the eukaryotic nonsense-mediated decay (NMD) pathway, which can recognize aberrant transcripts and target them for degradation (Chang et al., 2007; Isken and Maquat, 2008; Nicholson et al., 2010). It was first assumed that NMD functions primarily in the clearance of erroneous transcripts originating, for example, from gene mutations and mistakes during transcription or posttranscriptional processing, but it is now becoming evident that NMD plays a more fundamental role in gene regulation. On the one hand, NMD alters the transcriptome by targeting transcripts derived from pseudogenes (Mitrovich and Anderson, 2005) and ancient transposons (Mendell et al., 2004) as well as mRNA-like noncoding RNAs (ncRNAs; Kurihara et al., 2009). On the other hand, NMD can trigger the degradation of physiological transcript variants, such as products of alternative splicing (AS), thereby providing enormous gene regulatory potential (Lareau et al., 2007a).
The core NMD machinery includes the UP FRAMESHIFT (UPF) proteins UPF1, UPF2, and UPF3 in all eukaryotes investigated so far, but other factors, such as the SUPPRESSOR OF MORPHOLOGICAL DEFECTS ON GENITALIA (SMG) proteins, have not been found in all NMD-containing species (Isken and Maquat, 2008; Nicholson et al., 2010). Evidence for alternative routes of NMD linked to distinct sets of factors exists, but NMD generally requires the RNA helicase UPF1 (Isken and Maquat, 2008; Nicholson et al., 2010). Several NMD factors function not only in NMD but also contribute to other RNA and DNA surveillance processes, including histone mRNA degradation, Staufen-mediated mRNA decay, DNA repair, genome stability control, and telomere maintenance (Lew et al., 1998; Reichenbach et al., 2003; Brumbaugh et al., 2004; Kaygun and Marzluff, 2005; Kim et al., 2005; Azzalin and Lingner, 2006). Previous studies have established the molecular mechanisms underlying NMD as well as the features of target transcripts (Isken and Maquat, 2008; Nicholson et al., 2010). One major class of targets includes mRNAs with premature termination codons (PTCs), which, if not degraded, can lead to the translation of truncated, potentially harmful proteins. Furthermore, NMD targets also include transcripts with upstream open reading frames (uORFs), long or intron-containing 3′ untranslated regions (UTRs), and selenocysteine triplets. The translation-dependent recruitment of UPF1 has been described as a universal feature of NMD target recognition (Nicholson et al., 2010), and competition between 3′ UTR–associated factors distinguishes a premature from a regular translation termination codon. NMD target recognition in mammals was first described to be restricted to a so-called pioneer round of translation (Ishigaki et al., 2001; Chiu et al., 2004); however, this view has been challenged by two recent reports (Durand and Lykke-Andersen, 2013; Rufener and Mühlemann, 2013). Besides PTCs, also the positioning of an exon junction complex downstream of the termination codon can elicit NMD (Le Hir et al., 2001). Interestingly, some transcripts escape NMD despite the presence of the aforementioned features (Eberle et al., 2008; Nicholson et al., 2010). Upon NMD recognition, transcripts are fed into the general mRNA turnover pathways, involving initiation either by endonucleolytic cleavage or deadenylation and decapping (Nicholson and Mühlemann, 2010).
Homologs of the NMD factors UPF1, UPF2, UPF3, and SMG7 have been identified in plants and were functionally linked to the degradation of transcripts with NMD features (Hori and Watanabe, 2005; Arciga-Reyes et al., 2006; Yoine et al., 2006b; Wu et al., 2007; Kerényi et al., 2008; Riehs et al., 2008). Using transient expression assays in Nicotiana benthamiana, phosphorylation of a plant UPF1 homolog as well as the domain requirements of plant UPF1 and SMG7 homologs in the early and late steps of plant NMD have been analyzed (Mérai et al., 2012). Based on our current knowledge, plant NMD is triggered by the same transcript features as observed for NMD in other eukaryotes, namely the presence of PTCs, 3′ UTR–positioned introns, long 3′ UTRs, and uORFs (Kertész et al., 2006; Hori and Watanabe, 2007; Wu et al., 2007; Nyikó et al., 2009, 2013; Saul et al., 2009; Kalyna et al., 2012; Rayson et al., 2012a, 2012b). Interestingly, a recent work studying AS-NMD of a subset of 270 genes from Arabidopsis thaliana identified several transcript variants with intron-retention events that display classical NMD features yet are not NMD sensitive (Kalyna et al., 2012).
While numerous studies on the mechanistic aspects of NMD have greatly advanced our understanding of the eliciting features as well as the functioning of this surveillance pathway, it is still unclear to what extent NMD alters the transcriptome in different species, especially with respect to AS variants. Surveys of deposited data indicated that substantial fractions of all mRNAs are putative NMD targets (Lewis et al., 2003; Hillman et al., 2004; Baek and Green, 2005; Wang and Brendel, 2006). Experimental approaches toward transcriptome-wide identification of NMD targets were mainly based on microarray studies upon NMD suppression, revealing that the abundance of ∼3 to 10% of all analyzed mRNAs is NMD dependent (Rehwinkel et al., 2006). Furthermore, splicing-sensitive microarrays were used to measure exon inclusion rates upon NMD suppression in mouse cells (Ni et al., 2007). The advent of high-throughput RNA sequencing (RNA-seq) techniques revolutionized transcriptome studies and allowed an unprecedented in-depth description of the transcript diversity generated by processes such as AS (Pan et al., 2008; Filichkin et al., 2010; Marquez et al., 2012). Studies using RNA-seq to compare transcriptome profiles between the wild type and single mutants in NMD factors in Caenorhabditis elegans (Ramani et al., 2009) and mammals (McIlwain et al., 2010; Weischenfeldt et al., 2012) indicate that 20 to 30% of all genes produce isoforms that are directly or indirectly targeted by NMD (Ramani et al., 2009; Weischenfeldt et al., 2012).
In this study, we used RNA-seq to define the global alterations in AS patterns between Arabidopsis wild-type seedlings and several types of NMD impairment, providing a comprehensive analysis of AS-derived NMD targets. Our analyses identified 41,491 AS events, of which 3361 and 3238 were significantly altered in upf1upf3 and cycloheximide (CHX)-treated seedlings, respectively. In addition to mRNA targets, we found that NMD regulates the expression of numerous ncRNAs and transcripts derived from intergenic regions, highlighting the prominent role of this surveillance pathway in shaping the transcriptome of a higher eukaryote.
RESULTS
Double Mutant in UPF1 and UPF3 Strongly Accumulates NMD Targets
Previous studies in Arabidopsis (Stauffer et al., 2010; Kalyna et al., 2012) showed that the steady state levels of many NMD targets are relatively low, being a major limitation for their transcriptome-wide identification. To achieve high accumulation of NMD target mRNAs, a double mutant in the NMD factors UPF1 and UPF3 was generated by crossing the previously characterized single mutant lines low-beta-amylase1 (lba1; Yoine et al., 2006b) and upf3-1 (Hori and Watanabe, 2005). In contrast to lba1, homozygous mutants in another upf1 allele (upf1-3) show a growth arrest after radicle emergence from the seed coat (Yoine et al., 2006a). Therefore, it can be assumed that at least the lba1 allele used here is hypomorphic. Genotyping of the following generations did not yield adult, homozygous double mutant plants, and a phenotypic segregation of the progeny from self-pollinated lba1−/−upf3-1+/− plants was observed (Figure 1A). Approximately 0.25 of the descendants of lba1−/−upf3-1+/− and lba1+/−upf3-1−/− plants were arrested early in seedling development, and PCR-based genotyping revealed that those seedlings were homozygous for both mutant alleles (Figure 1B). Quantitative analysis of known NMD targets, derived from AS of the precursor mRNAs expressed from loci At5g53180 (Stauffer et al., 2010) and At4g36960 (Kalyna et al., 2012), showed their increased levels in homozygous double mutants compared with single mutants (Figures 1C and 1D; see Supplemental Figure 1 online). Furthermore, ratios of the splicing variants were analyzed in samples treated with CHX, which triggers the accumulation of NMD targets (Carter et al., 1995). CHX caused pronounced accumulation of the splicing variant II (SII) from At5g53180, harboring a PTC as a result of inclusion of a facultative, so-called cassette exon (Figure 1C). In contrast, SII derived from At4g36960, containing an uORF upon retention of an intron within the 5′ UTR, was unchanged in response to CHX treatment (Figure 1D). Based on these findings, we decided to analyze transcriptome-wide AS patterns of the homozygous single and double NMD factor mutants and CHX-treated plants.
Figure 1.
Double Knockdown of NMD Factors UPF1 and UPF3 Results in a Severe Phenotype and Strong Accumulation of NMD Target RNAs.
(A) Phenotypes of the progeny of the indicated Arabidopsis genotypes from seedlings grown for 11 d. Approximately 0.25 of the progeny of lba1−/− upf3-1+/− plants are arrested early in development (circle).
(B) PCR-based genotyping of the lines shown in (A). Cleaved-amplified polymorphic sequence analysis (1) allowed distinguishing between the wild type (closed circles) and mutant allele (open circles) for UPF1. For UPF3 (3), two different primer combinations specific for the wild type (WT) and mutant allele (m) were used. M indicates a ladder consisting of DNA fragments in 100-bp increments.
(C) Partial gene models and AS variants SI and SII derived from At5g53180, with boxes and lines representing exons and introns, respectively (top). Black boxes show coding regions (according to the representative gene model annotated at TAIR10; SI), the asterisk indicates a translation termination codon, and arrowheads illustrate binding positions of the primers used in RT-PCR (all mutants homozygous). At bottom is a quantitation of the ratio SII:SI using RT-qPCR, normalized to wild-type or control treatment (mock; mean values + sd, n = 3). Bar = 100 nucleotides.
(D) Analysis of At4g36960 splicing variants analogous to the description in (C). The coding sequence of a uORF is indicated. The bracket marks additional minor splicing variants.
Alterations in Transcriptome-Wide Splicing Variant Profiles upon NMD Impairment
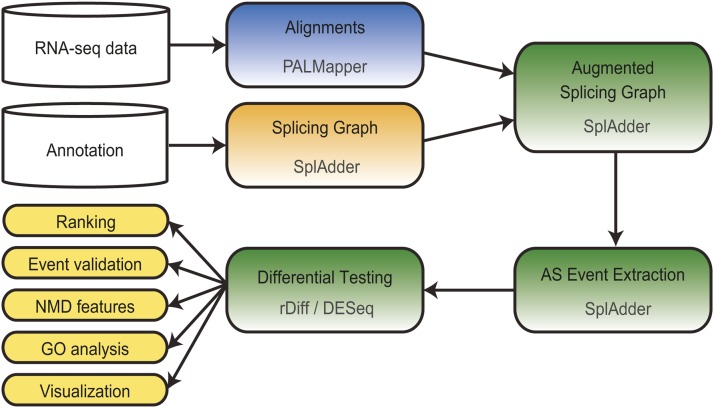
Seedling samples derived from wild-type and NMD mutant plants as well as from mock- and CHX-treated plants were used for the construction of cDNA libraries and RNA-seq analyses. We developed a universal computational pipeline to analyze gene expression and AS patterns in the comparison of control and NMD-impaired samples (Figure 2). Quality filtering and alignment of the ∼100-bp, single-end reads to the Arabidopsis genome (The Arabidopsis Information Resource 10 [TAIR10] version) revealed that 32.9 to 48.1 × 106 and 9.4 to 15.7 × 106 total and spliced reads covering exon–exon borders, respectively, per sample and replicate could be used to deduce gene expression and splicing variant information (see Supplemental Data Set 1A online). Read alignments were used to augment splicing graphs generated from the TAIR10 annotation and to extract 41,491 AS events with sufficient RNA-seq evidence (see Supplemental Data Sets 1B–1G online), which were associated with 11,711 genes from a total of 25,147 expressed genes (definition of expressed based on a Poisson test, false discovery rate [FDR] ≤ 0.05; Gan et al., 2011; see Supplemental Methods 1 online). By exploiting our RNA-seq data for deciphering the full AS diversity, we ensured that the downstream analyses also included NMD substrates that might show low abundance under control conditions and, therefore, might be absent from the available annotation. Subsequently, differential testing of splicing variant ratios on an AS events basis (http://www.bioweb.me/rdiff; Drewe et al., 2013) between the wild type and mutants as well as mock- versus CHX-treated seedlings was performed (see Supplemental Data Sets 1B–1G online). Each individual test of an AS event was assigned a P value, and FDR values for the complete lists were determined. To experimentally validate the FDR values, 10 AS events from the lba1 upf3-1 list in an FDR range of up to 0.3 were randomly selected. For 9 out of 10 candidates, both splicing forms could be detected, and the predicted relative increase of one variant in lba1 upf3-1 compared with the wild type was verified (see Supplemental Figure 2 online). Analysis of the same AS events in the single mutants and CHX-treated plants further substantiated the identity of the respective splicing variants as NMD targets (see Supplemental Figure 2 online). Furthermore, comparison of quantitative changes in the single and double NMD factor mutants revealed synergistic and not only additive effects of the single alleles on the accumulation of most tested NMD targets. Interestingly, in the case of the AS event within At4g19100, both of the analyzed AS variants contained a PTC and were targeted by NMD, as revealed by their increase relative to a PTC-free splicing variant derived from the same gene upon NMD impairment (see Supplemental Figure 2 online). However, one of the two NMD variants displayed a more pronounced accumulation in the mutants compared with the wild type, suggesting either different extents of NMD targeting of those two transcripts and/or an altered AS output.
Figure 2.
Universal Computational Pipeline for RNA-seq Data Analysis.
Comparative analysis of gene expression, isoform expression, and AS patterns based on RNA-seq data from control and NMD-impaired samples.
[See online article for color version of this figure.]
Significantly altered AS events were defined by an FDR threshold of 0.1, resulting in 3361 and 3238 AS events in lba1 upf3-1 and CHX-treated plants, respectively, with an overlap of 635 events (Figure 3A; see Supplemental Table 1 online). As some NMD targets do not accumulate in response to CHX treatment, and to make our candidate selection more stringent, we also considered the overlap between double mutant (FDR ≤ 0.1) and single mutant (uncorrected P ≤ 0.1) lists. We found an overlap between the NMD -factor single and double mutants of 1194 events, and a further 387 events were also changed upon CHX treatment, with the overlaps being significantly larger than expected by chance (permutation test: P < 2 × 10−5). For the single mutants, we chose a less stringent significance threshold value to account for the generally less pronounced accumulation of NMD targets, as observed from our RNA-seq data and by testing individual candidates (Figures 1 and 3). Furthermore, data for the two single mutants were combined by considering the minimum P value for each AS event. However, to be considered as an NMD target, the same isoform was required to accumulate in at least two tests, thereby ensuring a high stringency of the NMD target definition and excluding indirect effects caused by the mutant phenotypes or translation suppression. The validity of our strategy was further corroborated by comparing the overlaps between the various samples using different stringency cutoffs (see Supplemental Figure 3 online). First, with all settings, a major percentage (66.1 to 79.4%) of the events changed upon CHX treatment was not detected in the NMD factor mutants, suggesting that CHX treatment leads to many AS changes in an NMD inhibition–independent manner (see below). Second, the highest fraction of overlap for the double mutant with the other types of NMD impairment was found with the settings shown in Figure 3A. Third, using the lower stringency for the single mutants decreased the fraction of overlap with the other NMD impairments from 84.1 to 48.1% but increased the total number more than sixfold (1772 versus 280 events). Most of the remaining events detected only in the single mutants are probably caused by splicing variant changes that are not directly linked to NMD deficiency but show more pronounced alterations than many NMD targets, which display only a moderate increase in the single mutants compared with lba1 upf3-1. Furthermore, several genes contained more than one AS event significantly altered upon NMD impairment (Figure 3A). Considering the significantly altered AS events in the double mutant and CHX-treated plants, 3004 genes contained AS events that responded under at least one condition. Most of these genes (2915) are annotated as protein coding, representing 13.3% of the 21,842 protein-coding, multiple-exon genes from Arabidopsis (352 of these genes are annotated as single-exon genes in TAIR10 but have AS support from our data).
Figure 3.
Transcriptome-Wide Comparison of Splicing Variant Patterns between Control and NMD-Impaired Arabidopsis Seedlings.
(A) Size-proportional Venn diagram of significantly altered AS events (with numbers of corresponding genes in parentheses) comparing lba1 upf3-1 and CHX treatment (FDR ≤ 0.1) and lba1 upf3-1 with the two single mutants (double mutant FDR ≤ 0.1; single mutant P ≤ 0.1). Asterisks provide information on statistical significance, and contradictory events with significant changes in opposite directions have been excluded (for numbers, see Supplemental Table 1 online).
(B) Relative frequencies of AS types in all detected events as well as for the indicated combinations of NMD impairments: lba1 upf3-1 + CHX (1), changed under both conditions; lba1 upf3-1 + SM – CHX (2), changed in lba1 upf3-1 and in at least one of the single mutants but not upon CHX treatment; all NMD events (3), combination of the two previous sets. alt 5′/3′ ss, Alternative 5′/3′ splice sites.
(C) Direction of splicing change for subsets 1, 2, and 3 defined in (B). Direction of change for alternative 5′/3′ splice sites refers to usage of the downstream and upstream splice sites, respectively.
(D) Gene models of alternatively spliced regions (top), representative coverage plots (relatively scaled to the same height in all figures) with the altered region in black (middle), and quantitative analysis of splicing variants (bottom) for selected AS events identified by RNA-seq. Splicing variant ratios were determined by Bioanalyzer quantitation (mean values + sd, n = 3). Boxes and lines in gene models represent exons and introns, respectively, with black boxes depicting coding regions (according to the representative gene model annotated at TAIR10; SI). Asterisks indicate translation termination codons, and arrowheads illustrate the binding positions of primers used in RT-PCR. WT, Wild type. Bars = 100 nucleotides.
We next defined a stringent subset of NMD-linked AS events, which were altered in the double mutant and, in the same direction, in at least one of the single mutants and/or in response to CHX (Figure 3B, all NMD events). This category encompassed 1872 AS events, derived from 1126 genes, and was used for the downstream analysis of NMD target features (see below). The frequencies of the different AS types for all events and those significantly altered in one or several NMD-impaired systems were determined (Figure 3B). Out of all events detected in the various samples, 43.4% accounted for alternative 3′ splice sites, 24.4 and 21.6% for intron retention and alternative 5′ splice sites, respectively, and only 10.6% for cassette exons. Comparable fractions of AS types were found for events represented in the wild type or single types of NMD impairment, with most unique events detected in lba1 upf3-1 and CHX-treated seedlings (see Supplemental Table 2 online). When considering only AS events significantly altered, a similar distribution was found for lba1 upf3-1 alone and its combination with the single mutants as for all detected events (Figure 3B). In contrast, CHX-responsive AS events were enriched for cassette exon and intron retentions, while the fractions of both alternative 5′ and 3′ splice sites were reduced. Subsequently, the direction of change for the subsets combining altered AS events in the mutants and upon CHX treatment was compared (Figure 3C). For all combinations, most intron retention events showed a relative increase of the spliced variant upon NMD impairment, suggesting the frequent occurrence of so-called cryptic introns (i.e., erroneous removal of exonic regions by the spliceosome). Considering their relative increase upon NMD impairment, the clearance of these incorrect splicing products under normal conditions, most probably triggered by the presence of NMD features, can be assumed. Interestingly, cassette exons were almost exclusively upregulated for the combination of lba1 upf3-1 and CHX treatment, revealing for this subset a prevalence of so-called poison exons, which led to termination of the ORF and, in consequence, triggered NMD upon inclusion. In contrast, CHX-independent cassette exon events found to be altered only in the NMD factor mutants did not show any trend but a similar proportion of inclusion and skipping for the NMD-sensitive variant. Considering all events, however, the more frequent occurrence of poison exons versus the skipping of cassette exons as an NMD trigger became apparent. For alternative 5′ splice site events, splicing variants derived from usage of the downstream 5′ splice sites were overrepresented upon NMD impairment, whereas no clear trend was visible for alternative 3′ splice sites.
Examples for different AS types were selected from the subset of NMD-dependent events for experimental verification (Figure 3D; see Supplemental Figures 2 and 4A–4D online). The corresponding coverage plots were consistent with an independent experimental validation, both of which further substantiated NMD targeting of one of the splicing variants. In line with our findings for At4g36960 (Figure 1D) and At4g17370 (see Supplemental Figure 2H online), the intron-retention variant derived from At5g17550 was elevated in the NMD mutants but not in response to CHX. Analysis of previously characterized AS events that lead to two protein-coding splicing variants, which therefore are both not expected to be NMD targets, revealed no ratio changes in the NMD mutant samples (see Supplemental Figures 4E and 4F online), indicating that the observed splicing pattern changes indeed reflect the accumulation of NMD targets and not a general perturbation of AS. In contrast, CHX treatment resulted in altered AS ratios for both examples, highlighting the occurrence of side effects upon treatment with this translation inhibitor (see below). Finally, we compared our data set with AS-NMD events reported previously by Kalyna et al. (2012). For most of these events, ratio changes were detected in our RNA-seq data as well, although relatively high P values were found in some cases (see Supplemental Table 3 online). Furthermore, Kalyna et al. (2012) described a subset of AS events not altered in the upf1-5 and upf3-1 mutants and/or upon CHX treatment. Concordantly, these events were found to be unchanged in the double mutant described here.
The Majority of NMD-Sensitive Transcripts Show Classical NMD Features and Are Derived from Genes Linked to Diverse Processes
To analyze the presence of previously described NMD-triggering features, the locations of the detected AS events relative to the ORF of the representative gene model from TAIR10 were determined (Figure 4A; see Supplemental Data Sets 2A and 2B online). Only 5.3% of the events altered upon NMD suppression were confined to the 5′ and 3′ UTR, indicating an underrepresentation of NMD-linked events compared with all AS events within these regions. For 5′ UTR–positioned events, the occurrence of uORFs that are either relatively long (>35 amino acids) or that overlap with the start of the main ORF was determined (see Supplemental Data Set 2C online). Both features were present at higher rates (1.5- to 2-fold) in the NMD-responsive transcript variants. AS events within the 3′ UTR can elicit NMD by introducing splice junctions more than 50 nucleotides downstream of the stop codon and by generating long 3′ UTRs. A threshold for long 3′ UTRs was defined based on the annotated 3′ UTRs of TAIR10 representative transcript models, 90% of which have a 3′ UTR length equal to or smaller than 347 nucleotides. Accordingly, all 3′ UTRs above this value were considered to be exceptionally long, agreeing with previous studies (Kertész et al., 2006; Wachter et al., 2007; Kalyna et al., 2012). Analysis of these 3′ UTR–linked features revealed identical fractions of long 3′ UTRs for the transcripts relatively upregulated and downregulated upon NMD impairment, whereas more downstream splice junctions were found for the NMD-associated transcripts (see Supplemental Data Set 2C online). Taken together, only a minor fraction of the NMD-responsive AS events identified in this study are confined to the 5′ and 3′ UTR. These events show an overrepresentation of uORFs and splice junctions more than 50 nucleotides downstream of the stop codon for the splicing variants relatively increased upon NMD impairment.
Figure 4.
NMD-Responsive Transcripts Are Highly Enriched in Premature Termination Codons.
(A) Fractions of positions of AS events as defined by mapping to the corresponding representative TAIR10 model. NA, Not mapable due to the absence of a coding sequence. Analyzed AS event sets are as follows: lba1 upf3-1 + CHX, changed under both conditions; lba1 upf3-1 + SM – CHX, changed in lba1 upf3-1 and in at least one of the single mutants but not upon CHX treatment; all NMD, combination of two previous sets.
(B) Presence (+, black bars) or absence (−, white bars) of PTCs or splice junctions more than 50 nucleotides (nts) downstream of the stop codon in transcript variants downregulated or upregulated in NMD-impaired samples (derived from the All NMD set).
(C) Analysis of the two splicing variants for the presence/absence of a PTC for all coding sequence–linked AS events altered upon NMD impairment.
(D) Median 3′ UTR length of the indicated transcript sets. For the altered events, upregulated and downregulated transcript variants were separately analyzed.
(E) Distribution of mean-smoothed 3′ UTR lengths relative to the test P values for lba1 upf3-1 and wild-type transcript isoforms, which were assigned according to their relative increase. Dashed line, FDR = 0.1.
(F) Fraction of NMD targets within the top-most protein-coding multiple-exon (PCME) genes relative to coverage for lba1 upf3-1. Under the top 7139 genes with reads per kilobase per million mapped reads (RPKM) ≥ 10, 17.5% are likely NMD targets (dashed line).
We observed striking differences in the occurrence of NMD features for the AS events positioned within the coding sequence. Here, most of the NMD-regulated splicing variants contained PTCs as well as downstream splice junctions (Figure 4B; see Supplemental Data Sets 2C and 2D online). More than 94% of the NMD-sensitive splicing variants contained a PTC, which was absent in most of the corresponding alternative splice forms (Figure 4C). As a substantial fraction of the genes with NMD-responsive AS events harbored more than one significantly altered event (31% for set “all NMD”), we next tested if integrating multiple events has an effect on PTC frequency (see Supplemental Methods 1 online). For only 4 out of 1485 single integration events giving rise to a PTC, possible event combinations could be integrated that resulted in a PTC loss (see Supplemental Data Set 2G online). Thus, even though it is unknown how these individual AS events are combined in natural transcripts, we can exclude a considerable effect on the presence of PTCs. Furthermore, multiple AS events associated with one gene can be either independent or coregulated at the splicing level, although the frequency of coregulated AS in plants is currently unknown. Thus, some of the AS events detected as NMD responsive in our analysis might actually not trigger NMD but might be coupled on the level of regulation to another NMD-triggering AS event within the same transcript. To account for this possibility, we next restricted our analysis to genes containing only one significantly altered AS event (see Supplemental Data Set 3 online). We found similar fractions of NMD feature occurrence as in our previous analysis; for example, 92.3 and 95.1% of the splicing variants relatively upregulated upon NMD impairment for all and single-event genes, respectively, contained classical NMD features. Thus, we conclude that most of the events detected by our analysis pipeline can directly trigger NMD.
Given that most of the NMD-suppressed splicing variants contained PTCs and thus resulted in long 3′ UTRs, we thought that the parameter 3′ UTR length might be used to analyze the enrichment of NMD target features for the single types of NMD impairment compared with our more stringent subset. For the all-NMD set, the median 3′ UTR length was considerably increased in the splicing variants upregulated upon NMD impairment compared with the corresponding downregulated isoforms (Figure 4D; see Supplemental Data Set 2E online), which was expected due to the frequent occurrence of PTCs in the respective splicing variants (see above). A similar difference was observed when comparing 3′ UTR lengths between the NMD-suppressed transcripts and the data sets of TAIR10 representative gene models as well as all detected AS variants from this work. Interestingly, the lba1 upf3-1 subset showed similar median 3′ UTR length patterns to the more stringent all-NMD fraction combined from different types of NMD impairments. To test for a correlation of the 3′ UTR length of splicing variants and their induction in the double mutant, we plotted the 3′ UTR length against the P value from the test of the wild type versus lba1 upf3-1 (Figure 4E; see Supplemental Figure 5 online). Interestingly, a negative correlation was found between the P value and the 3′ UTR length of the splicing variant showing a relative increase in the double mutant (−0.23, P < 10−15; Figure 4E; see Supplemental Figure 5 online). This observation revealed that longer 3′ UTRs cause more pronounced accumulation of the respective splicing variants in lba1 upf3-1. This correlation was also seen for P values above the stringent cutoff used in our analysis (Figure 4E, dashed line), suggesting the existence of an even higher number of bona fide NMD targets than determined above. Importantly, our approach to NMD target identification depends on the statistical power, which decreases for lowly expressed genes represented by small numbers of sequencing reads. Thus, we might underestimate the fraction of NMD-linked AS events for genes with relatively low expression levels. In line with this, we found a smaller fraction of NMD targets with decreasing levels of gene expression (Figure 4F). To correct for this bias, we used the 7275 genes with ≥10 reads per kilobase per million mapped reads as a more stable basis for the estimation of the fraction of NMD targets (Figure 4F, dashed line). Accordingly, we estimate that 17.4% of the 21,842 multiple-exon, protein-coding genes produce NMD targets. Furthermore, to determine the proportion of mRNA being degraded via NMD, we computationally quantified the individual isoforms in the wild-type and lba1 upf3-1 samples with rQuant (Bohnert et al., 2009). To do so, the expression of each gene in wild-type and lba1 upf3-1 samples was normalized to the level of the isoform that was most abundant in the wild type and, therefore, unlikely to be targeted by NMD. Considering all annotated genes and those containing significantly altered AS events in a comparison of wild-type and lba1 upf3-1 seedlings, 6.8 and 17.5% of the total transcript mass, respectively, were subjected to NMD, representing a considerable proportion of the transcriptional output. In this context, the mass of a single transcript is the number of its nucleotides; thus, the total mass of an isoform can be computed from its length multiplied by the occurrences of this isoform. Between wild-type replicates, no considerable difference of transcript mass could be observed (0.01 and 2.4% for all genes and those with NMD-dependent AS events, respectively).
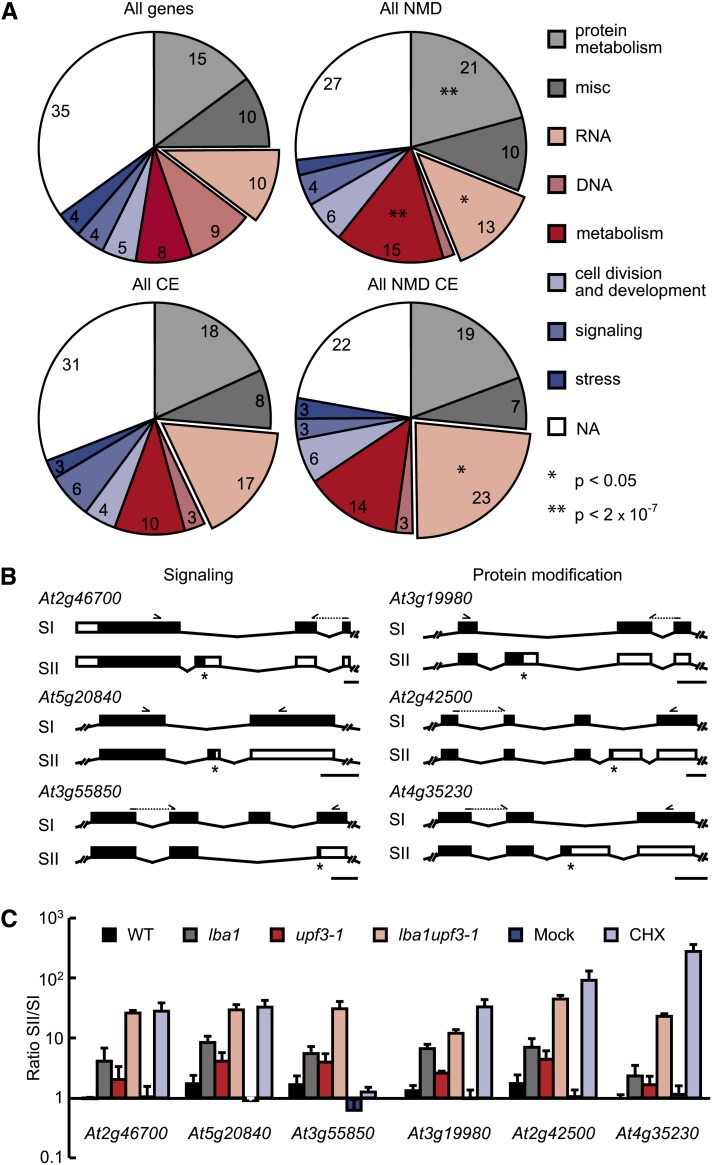
To test if coupled AS-NMD is overrepresented among particular gene functions, we analyzed the distribution of all genes and those generating NMD-regulated splicing variants in functional groups, combining Gene Ontology (GO) terms using the MapMan software (http://mapman.gabipd.org; Thimm et al., 2004; see Supplemental Data Sets 4A and 4B online). Our analysis highlighted the widespread role of coupled AS-NMD in diverse biological processes (Figure 5A; see Supplemental Data Sets 4A and 4B online), and comparing the set of NMD-associated genes with all annotated genes, we found an enrichment of the functional groups protein metabolism, metabolism, and, to lesser extent, RNA (from a hypergeometrical test after Bonferroni correction, P < 0.05). We also found a pronounced reduction of the category DNA, as the majority of all genes within this group correspond to transposons and related functions. Considering the NMD-regulated cassette exons related to all cassette exons with experimental support from our wild-type RNA-seq data, an overrepresentation of the category RNA was found (from a hypergeometrical test after Bonferroni correction, P < 0.05), in line with previous studies showing poison exon-mediated regulation for many RNA binding proteins (Lareau et al., 2007a; Filichkin et al., 2010; Palusa and Reddy, 2010). Interestingly, NMD-responsive cassette exons were also detected for genes belonging to other functional groups, including signaling and posttranslational protein modification. Experimental validation of six selected candidates belonging to these two categories confirmed the presence of NMD-responsive cassette exons, either the skipping or inclusion of which caused PTC generation (Figures 5B and 5C; see Supplemental Figure 6 online). Specifically, cassette exon inclusion introduces a PTC in the case of a calcium-dependent protein kinase (At2g46700) and a phosphoinositide phosphatase family protein (At5g20840), whereas for the putative PHYTOCHROME A (PHYA) signaling component At3g55850, NMD targeting results from exon skipping. The tested candidates encoding posttranslational protein modification components all generated NMD targets by poison exon inclusion and comprised phytochrome-associated protein phosphatase 3 (At3g19980), a protein phosphatase 2A catalytic subunit (At2g42500), and brassinosteroid signaling kinase 1 (At4g35230). Comparison of the annotated full-length proteins and the predicted translation products derived from the PTC-containing splicing variants revealed that splicing to the NMD-targeted mRNA results in a loss or truncation of most of the annotated protein domains (see Supplemental Table 4 online). At this point, however, it is unknown if the NMD-regulated splicing variants result in any substantial protein accumulation.
Figure 5.
NMD Targets Are Derived from Genes Linked to Diverse Biological Processes.
(A) Distribution of functional groups for all genes (top left), those associated with NMD regulation (top right), all genes with cassette exon (CE) events having experimental support from our wild-type RNA-seq data (bottom left), and genes with NMD-linked cassette exons (bottom right). The two segments without numbers correspond to fractions of 2% each. Significant changes based on a hypergeometrical, Bonferroni-corrected test in comparisons of juxtaposed sets are indicated. NA, not assigned.
(B) Gene models of alternatively spliced regions for selected genes of the GO terms signaling (left) and posttranslational protein modification (right). RNA-seq data analysis revealed the presence of NMD-responsive cassette exons in all instances. Boxes and lines represent exons and introns, respectively, with black boxes depicting coding regions (according to the representative gene model annotated at TAIR10; SI). Asterisks indicate translation termination codons, and arrowheads illustrate binding positions of primers used in RT-PCR. Bars = 100 nucleotides.
(C) Quantitative analysis of ratios of splicing variants depicted in (B) using Bioanalyzer assays (mean values + sd, n = 3) for control (wild-type [WT] and mock) and NMD-impaired seedlings.
The detection of coupled AS-NMD for genes with functions in signaling and posttranslational protein modification raised the question of whether the accumulation of these respective NMD-targeted splicing variants is a constitutive or regulated process, with possible implications for the expression control of these genes. To address this question, we analyzed splicing variant ratios for the genes At2g46700, At3g55850, At3g19980, and At2g42500 after NaCl treatment, which was previously shown to alter AS in Arabidopsis (Filichkin et al., 2010). Interestingly, relative levels of the NMD target splicing variant were elevated upon NaCl treatment for all of the tested examples (Figure 6), indicating that AS and/or NMD of the corresponding transcripts is regulated in an abiotic stress–dependent manner. In contrast, application of mannitol with equal osmolarity did not result in significant splicing variant ratio changes for those candidates (see Supplemental Figure 7 online), indicating that the observed effects are not caused by the osmotic effect of NaCl. In summary, our findings suggest an expanded scope of coupled AS and NMD in gene regulation, with implications for fundamental biological processes such as signaling and posttranslational protein modification as well as in the response to NaCl stress.
Figure 6.
NaCl Treatment Alters Splicing Variant Ratios for Genes with Coupled AS-NMD.
(A) RT-PCR analysis of splicing variant ratios for the depicted genes from Arabidopsis seedlings treated with mock (−) or NaCl (+) solutions for the indicated durations. Product bands corresponding to the major splicing variants SI and SII (for details, see Figure 5B) are indicated.
(B) Quantitative analysis of AS ratios depicted in (A) using Bioanalyzer assays (mean values + sd, n = 3), each normalized to the ratio SII:SI of the control sample at the 0.5-h time point.
A previous analysis of global splicing pattern changes upon UPF2 depletion in mouse tissues also identified a substantial fraction of PTC-free transcripts that might not be direct NMD targets (Weischenfeldt et al., 2012). Alternatively, changes in AS caused by NMD suppression might result in altered levels of these transcripts, as also supported by the finding that 10 out of 19 inspected splicing factors showed deregulated expression (Weischenfeldt et al., 2012). Given that most of the NMD-sensitive splicing variants identified in our study contained classical NMD features, however, we did not expect that a major fraction of these targets would be derived from the perturbed expression of splicing factors upon knockdown of the NMD factors UPF1 and UPF3 in Arabidopsis. To test this, gene expression levels of known Arabidopsis splicing factors were differentially tested using DEseq (Anders and Huber, 2010), revealing that 20.6% were significantly altered in lba1 upf3-1 compared with the wild type (all upregulated; P < 0.01; see Supplemental Data Set 4C online). This is a similar fraction to that observed for all 33,602 annotated Arabidopsis genes, of which 22.7% were altered in their expression in lba1 upf3-1 relative to the wild type (10 and 12.7% upregulated and downregulated, respectively; P < 0.01). All splicing factors with altered expression in the double mutant were upregulated. This might be explained by the stabilization of NMD-regulated splicing variants, the occurrence of which has been described for several representatives from this group of proteins. Furthermore, analyzing the expression of the representative isoforms with rQuant (Bohnert et al., 2009) revealed a lower fraction of deregulated splicing factors in Arabidopsis lba1 upf3-1 (see Supplemental Data Set 4C online) compared with a previous study of UPF2-depleted mouse tissues (Weischenfeldt et al., 2012). This might contribute to the observed differences between our analysis and the previous work by Weischenfeldt et al. (2012) in the proportions of identified targets containing classical, NMD-eliciting features.
NMD Regulates the Expression of Numerous ncRNAs and Transcripts Derived from Intergenic Regions
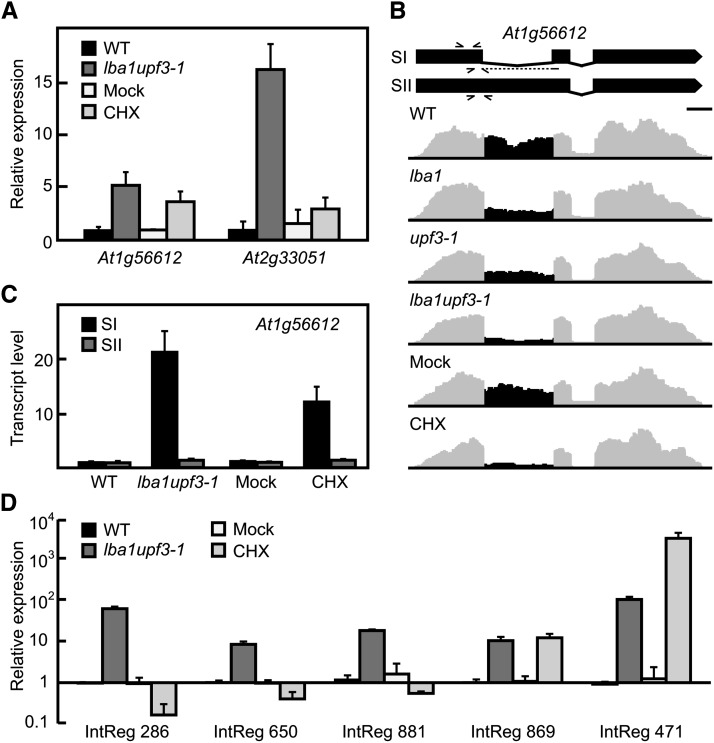
Previous work found 20 mRNA-like ncRNAs to be upregulated in Arabidopsis NMD mutants relative to the wild type (Kurihara et al., 2009). Interestingly, analysis of the gene expression of all annotated ncRNAs for lba1 upf3-1 samples revealed that out of 121 ncRNAs with altered expression (P < 0.01), 79% were upregulated, while for the single mutants, all 22 differentially expressed ncRNAs showed increased levels (see Supplemental Data Set 5A online). In CHX-treated samples, only 49 ncRNAs exhibited differential expression (67% upregulated), suggesting that many ncRNAs are regulated by the NMD factors in a translation-independent manner. Using RT–quantitative PCR (qPCR), we confirmed the upregulation of ncRNAs derived from At1g56612 and At2g33051 (Figure 7A). Interestingly, separate quantitation of the two splicing variants expressed from At1g56612 revealed that only the intron-spliced variant was an NMD target (Figures 7B and 7C). Total levels of both tested ncRNAs increased in the double mutant, whereas CHX treatment resulted in a substantial increase only for At1g56612. Furthermore, we speculated that pseudogenes might frequently generate NMD targets due to the absence of a positive selection for a functional ORF. In line with this hypothesis, out of 74 differentially expressed pseudogenes (P < 0.01), 66 were upregulated in double mutant compared with wild-type seedlings (see Supplemental Data Set 5B online).
Figure 7.
NMD Targeting of ncRNAs and Transcripts from Intergenic Regions.
(A) RT-qPCR analysis of annotated ncRNAs in control and NMD-impaired seedlings (mean values + sd, n = 3 for At2g33051, n = 4 for At1g56612). WT, Wild type.
(B) Gene model and representative coverage plots (altered region in black); arrowheads show binding positions of primers used in (A), at top, or (C), below individual transcript models.
(C) RT-qPCR analysis of individual splicing variants for At1g56612 (mean values + sd, n = 4).
(D) RT-qPCR analysis of total transcript levels for newly identified transcripts in samples from control and NMD-suppressed plants (mean values + sd, n = 3).
Based on the large number of annotated ncRNAs that showed an increased levels upon NMD impairment, we reasoned that additional, so far not identified NMD-dependent transcriptional units might exist. Searching for read clusters mapping to unannotated genomic locations identified 200 intergenic regions (IntRegs; see Supplemental Data Set 5C online). Furthermore, transcript recognition using mixed integer transcript identification (MITIE) (Behr et al., 2013) provided splicing evidence for 50 of them. Using rapid amplification of cDNA ends (RACE), the 5′ and 3′ ends of two candidates were experimentally determined (see Supplemental Figure 8 online). The obtained results were in good agreement with the transcript predictions and also confirmed the presence of a 5′ cap structure as well as polyadenylation of those transcripts. Out of 108 substantially expressed regions, most (62 up, 12 down) were more than twofold upregulated in the double mutant. Five candidates were selected for validation, confirming their increased levels in the double mutant (Figure 7D). Interestingly, two of them also accumulated upon CHX treatment, which might be explained by translation of the respective RNAs. Sequence inspection of the two candidates that were analyzed by RACE revealed in both instances the presence of potential ORFs (see Supplemental Figure 8 online); however, only IntReg 869, but not IntReg 881, accumulated in response to CHX treatment.
DISCUSSION
Our study provides a comprehensive analysis of NMD-targeted AS variants in Arabidopsis, revealing a major role of this RNA surveillance mechanism in defining the steady state transcriptome. Considering AS changes found in at least one type of NMD impairment, 13.3% of all intron-containing, protein-coding genes were affected, which is in agreement with an extrapolation of 13% from a recent study analyzing 270 selected genes in Arabidopsis (Kalyna et al., 2012). However, our analysis still represents an underestimation of the extent of NMD of transcript variants. First, our study was limited to seedling samples and, therefore, did not account for the full AS diversity, which also includes splicing variants confined to certain tissues, developmental stages, and stress conditions. Second, we did not consider potential NMD targets that are produced in an AS-independent manner, such as differential transcription initiation or alternative 3′ end processing. Third, low levels of gene expression or high decay rates limit the detection of significant splicing ratio changes. Taking the bias of low steady state transcript levels into account, 17.4% of the multi-exon, protein-coding genes are predicted to generate NMD-targeted splicing variants. Importantly, in contrast to most previous transcriptome-wide studies of NMD targets (Rehwinkel et al., 2006), the approach taken here relies on changes in AS ratios, thereby excluding indirect targets displaying total expression level changes. While it cannot be ruled out that the various types of NMD impairment also lead to AS changes, as reported in an earlier study of UPF2-depleted mouse tissues (Weischenfeldt et al., 2012), the presence of classical NMD features in 92.3% of the NMD-sensitive splicing variants clearly supports their identity as bona fide NMD targets. The higher proportion of likely direct NMD targets found in this study compared with the analysis by Weischenfeldt et al. (2012) can be explained by using mutants in other NMD factors, the combination of different types of NMD impairment, as well as species-specific differences. Furthermore, in contrast to UPF2 depletion in mouse, knockdown of UPF1 and UPF3 in Arabidopsis leads to less pronounced changes in the expression levels of known splicing factors and, therefore, is not expected to generally perturb AS. The latter aspect has also been experimentally confirmed by analyzing known, NMD-independent AS events.
Comparison of AS pattern changes between the mutants and CHX-treated samples revealed only a partial overlap, which on the one hand can be explained by the occurrence of splicing artifacts attributable to the CHX-mediated translational block. On the other hand, a major fraction of NMD targets with retained introns accumulated only in the NMD factor mutants, but not upon CHX treatment. Given that CHX treatment resulted in strong NMD target accumulation for the other types of splicing variants, we suggest a translation-independent decay mechanism for intron-retaining transcripts. Translation of these mRNAs might be prevented as a result of their recognition as being incompletely spliced, which can cause nuclear retention of mRNAs (Chang and Sharp, 1989; Legrain and Rosbash, 1989). Interestingly, in Arabidopsis, an enrichment of aberrant transcripts and the localization of UPF2 and UPF3 within the nucleolus have been described (Kim et al., 2009), suggesting a role of this compartment in the recognition of substrates for NMD factor–mediated decay. While a translation-independent route of NMD has not been documented so far (Nicholson et al., 2010), the involvement of UPF1 in additional processes such as the mammalian Staufen-mediated mRNA decay pathway (Park and Maquat, 2013) has been well established. Given that Staufen-mediated mRNA decay is also restricted to translated mRNAs, our findings might extend UPF1/UPF3 functions to translation-independent mRNA decay of incompletely spliced mRNAs in plants. Furthermore, our study and a previous analysis (Kalyna et al., 2012) revealed a strong underrepresentation of intron-retention variants among the identified NMD targets, suggesting the possible involvement of further NMD-independent pathways for the clearance of these transcripts. While future studies need to address the degradation of intron-retention mRNAs with NMD features and the impact of translation on this process in more detail, our results also strongly suggest that CHX responsiveness alone should not be used to judge the NMD targeting of a particular transcript variant.
From the NMD-eliciting features previously investigated in plants (Kertész et al., 2006; Kerényi et al., 2008; Nyikó et al., 2009), the presence of a PTC seems to be most prevalent among NMD targets, as it has also been found in a study of 270 selected Arabidopsis genes (Kalyna et al., 2012). However, as RNA-seq data lead to a low read coverage at transcript ends, UTR-associated events might be underrepresented. Furthermore, NMD features within UTRs can also be introduced by AS-independent processes that have not been considered in this study. Interestingly, 7.7% of the NMD-responsive AS events were not associated with classical target features. While some of these events might be altered due to secondary effects or their coregulation with another NMD-triggering event within the same transcript, novel types of NMD-triggering features or translation of an ORF differing from the annotation also could explain their NMD responsiveness.
Our study revealed that NMD alters the expression of genes linked to diverse cellular functions, which is consistent with previous findings in other eukaryotes (He et al., 2003; Mendell et al., 2004; Rehwinkel et al., 2006). Among NMD-regulated cassette exons, we found an enrichment of genes associated with RNA metabolic functions. This observation is in line with the well-characterized role of coupled AS-NMD in regulating the expression of RNA-binding proteins (Lareau et al., 2007b; Isken and Maquat, 2008; Schöning et al., 2008; Palusa and Reddy, 2010; Kalyna et al., 2012). For example, polypyrimidine tract binding proteins from mammals (Wollerton et al., 2004) and plants (Stauffer et al., 2010) are subject to auto-regulatory and cross-regulatory circuits, in which elevated protein levels stimulate splicing to an NMD-targeted variant. Interestingly, NMD-triggering poison exons were also detected in genes with functions in cellular signaling and posttranslational protein modification, and a relative increase of the NMD-targeted splicing variants in response to NaCl stress was observed. While, at this point, we cannot distinguish if the observed changes are caused by an altered splicing output or diminished NMD activity, our findings revealed that coupled AS and NMD for these and probably many other genes represents a stress-regulated process, with potentially far-reaching biological implications. For example, some of the identified NMD targets might be stabilized under certain conditions and translated into a protein as a consequence of repressed NMD activity, thereby having a biological function beyond being the side product of a gene regulatory mechanism. Precedence for such a mechanism has recently been provided in the context of NMD-mediated axonal pathfinding (Colak et al., 2013). Specifically, NMD controls axon guidance involving the local accumulation and translation of the Robo3.2 mRNA in NMD factor–deficient commissural neurons (Colak et al., 2013). A wider role of NMD in the fine-adjustment of gene expression, in addition to its function in the efficient decay of transcript variants, seems likely and is also supported by the finding that the expression of NMD factors is regulated in feedback loops by NMD in animal and plant species (Kerényi et al., 2008; Saul et al., 2009; Huang et al., 2011; Nyikó et al., 2013). Besides triggering the degradation of mRNA targets, NMD is also involved in the decay of ncRNAs (Kurihara et al., 2009). Here, we identified many additional instances of NMD-sensitive ncRNAs and intergenic regions. Future studies will need to address whether the accumulation of representatives from this class of NMD targets is regulated during plant development (e.g., by altered NMD activity) and identify the physiological functions of these RNAs.
Based on their low abundance and low evolutionary conservation, it was suggested that many NMD targets are not functionally relevant (Pan et al., 2006; Isken and Maquat, 2008). However, the efficient clearance of nonfunctional AS variants might be a prerequisite for the suppression of negative side effects arising from highly variable splicing outputs, thereby providing the basis for an enormous evolutionary flexibility. In addition, the spliceosome might be prone to alternate base pairing, and an evolutionary pressure to introduce PTCs would suppress the production of nonfunctional or even deleterious proteins from these transcripts (Lareau et al., 2007a). Similarly, the majority of differentially expressed pseudogenes was upregulated upon NMD impairment, suggesting also a role of this surveillance pathway in silencing expression from nonfunctional genes.
The double mutant used in this study showed an arrest during early seedling development, in contrast to only subtle phenotypes of the single mutants. Previous studies in Arabidopsis had also reported seedling lethality for a knockout upf1 allele (upf1-3; Yoine et al., 2006a) as well as for a double mutant in two other upf1 and upf3 alleles (Arciga-Reyes et al., 2006). Given that NMD targets strongly overaccumulate in the double mutant, its NMD defect most likely accounts for the dramatic phenotype, but critical functions in another process cannot be ruled out. Interestingly, analysis of NMD mutants in other eukaryotes suggested that the severity of the phenotype upon NMD knockdown depends on the overall complexity of the organism and its extent of AS. NMD mutants in Saccharomyces cerevisiae (Leeds et al., 1991) and C. elegans (Hodgkin et al., 1989) are viable, but lethality of the corresponding mutants in Drosophila melanogaster (Metzstein and Krasnow, 2006), zebrafish (Wittkopp et al., 2009), and mouse (Medghalchi et al., 2001) was observed. In support of the hypothesis that overaccumulation of NMD targets is largely responsible for the negative effects observed in NMD factor mutants, numerous recessively inherited diseases become dominant negative when NMD fails to target the respective PTC variants (Holbrook et al., 2004).
Interestingly, several recent studies revealed altered defense responses in Arabidopsis NMD mutants (Jeong et al., 2011; Rayson et al., 2012a, 2012b; Riehs-Kearnan et al., 2012; Shi et al., 2012), and the severe seedling-lethal phenotype of the upf1-3 mutant could be partially rescued in the pad4 mutant background with an impaired pathogen signaling pathway (Riehs-Kearnan et al., 2012). Moreover, the upf1-3 mutant was reported to have strongly enhanced PR1 transcript levels (Riehs-Kearnan et al., 2012), which were also suppressed in the pad4 background and might be caused by elevated salicylic acid levels. Thus, the phenotype of the lba1 upf3-1 mutant analyzed in this work could also result from an autoimmune response. In line with this hypothesis, analysis of our RNA-seq data for the total expression of salicylic acid–responsive genes such as PR1-5, PAD4, SAG13, WRKY70, and FRK1 showed increased levels in the lba1 upf3-1 mutant compared with the wild type (see Supplemental Data Set 1I online). Future studies need to address whether the lba1 upf3-1 phenotype is indeed caused by altered salicylic acid signaling. Furthermore, the data presented in this study can be mined to identify target genes potentially linking NMD suppression to the induction of autoimmune responses, with R genes being prime candidates. Taken together, deregulated expression of either signaling components or effectors in pathogen defense might be largely responsible for the early lethality of strong NMD-factor mutants in Arabidopsis.
METHODS
Plant Growth and Treatments
Arabidopsis thaliana seeds were surface-sterilized in 3.75% NaOCl and 0.01% Triton X-100 and plated on one-half-strength Murashige and Skoog (MS) medium supplemented with 2% Suc and 0.8% phytoagar (Duchefa). After stratification, seedlings were cultivated for 11 d under continuous light conditions (20°C, 60% humidity). For CHX treatment, seedlings were transferred to liquid one-half-strength MS medium with 2% Suc containing 10 µg mL−1 CHX or water as mock treatment. Seedlings were incubated for 4 h at room temperature and slight agitation. NaCl treatment was performed as described previously (Filichkin et al., 2010). Briefly, seedlings were grown for 12 d under long-day conditions (16-h days, 8-h nights, 20°C, 60% humidity) on solid one-half-strength MS medium with 2% Suc in Petri dishes as described above. For stress treatment, 20 mL of 0.5 M NaCl was added to the Petri dishes with seedlings, resulting in complete submergence of the plants, and samples were taken at the indicated time points. Mock treatment was performed with water.
Genotyping of the mutants was performed using cleaved-amplified polymorphic sequences as described before (Yoine et al., 2006a) or PCR testing for the presence of a T-DNA insertion in lba1 or upf3-1 mutants, respectively.
RNA Isolation, RT-PCR, and qPCR
RNA isolation, RT, and RT-qPCR were performed as described previously (Stauffer et al., 2010) using the Bio-Rad CFX384 cycler and the MESA Blue qPCR Mastermix (Eurogentec). For quantifying single transcript variants via RT-qPCR, PP2A (AT1G13320) was used as a reference transcript. RT-PCR fragments were analyzed using 2% agarose Tris-acetate (TAE, 40 mM Tris, 20 mM acetate, 1 mM EDTA, pH 8.0) gels and ethidium bromide staining followed by visualization under UV irradiation. For quantitation of transcript isoform ratios, the Agilent 2100 Bioanalyzer was used. Oligonucleotides used for RT-PCR and RT-qPCR analyses are listed in Supplemental Figure 9 online. Numbers of biological replicates are stated in the corresponding figure legends.
Splice variants identified in coamplification assays were subcloned using the StrataClone PCR Cloning Kit (Agilent) and sequenced (see Supplemental Figure 9 online).
Library Preparation and Illumina Sequencing
RNA isolation was performed using the EURx RNA isolation kit (Roboklon) according to the manufacturer’s instructions. Libraries were prepared using the Illumina TruSeq sample preparation kit starting from 2 to 4 µg of total RNA and following the instruction manual (TruSeq RNA Sample Preparation v2 Guide, November 2010). All samples were analyzed at least in biological duplicate. For further details, see Supplemental Methods 1 online.
RNA-seq Data Analysis
All RNA-seq data were aligned using the spliced alignment tool PALMapper (Jean et al., 2010) and postprocessed using the RNA-geeq toolbox (A. Kahles, J. Behr, and G. Rätsch, unpublished data). AS events were extracted from the TAIR10 annotation augmented with additional splicing evidence from the alignments using Splicing Adder (SplAdder, www.bioweb.me/spladder). The differential testing of alternative isoform expression and alternative gene expression based on negative binomial tests taking biological variance into account were made with rDiff (Stegle et al., 2010; Drewe et al., 2013) and DESeq (Anders and Huber, 2010), respectively. For NMD feature evaluation, an analysis pipeline was implemented in MATLAB, Python, and as shell scripts. For further details, see Supplemental Methods 1 online. To estimate the fraction of the total transcript mass that is subject to NMD, we compared the total transcript expression for the wild type and the lba1 upf3-1 mutant. For this, the alignments of wild-type and lba1 upf3-1 replicates were merged, respectively, and in silico quantification was performed using rQuant (Bohnert et al., 2009), leading to a quantification value for all transcript isoforms. For each gene, the gene expression of the wild type was normalized to the mutant level based on the isoform that showed the highest wild-type expression and, therefore, was most likely not targeted by NMD. The cumulative difference of all remaining isoforms was added up over all genes. The fraction of degraded transcript mass results from the ratio of the total difference and the total transcript mass in the mutant. In this context, the mass of a transcript isoform is its length multiplied by the occurrences of this isoform. The same methodology was applied to the two wild-type replicates.
Functional Clustering of NMD-Regulated Genes
Functional clustering was performed using the MapMan software (http://mapman.gabipd.org; Thimm et al., 2004). All analyzed genes were assigned a gene function based on the corresponding GO terms listed in TAIR10. Based on these GO terms, genes were clustered in 35 different functional groups and further combined as depicted in Supplemental Data Sets 4A and 4B online.
Determination of the Ends of Transcripts Derived from Intergenic Regions
RNA ligase–mediated RACE was performed using the GeneRacer kit (Invitrogen) including the SuperScript III RT module. All steps for oligo ligation and cDNA synthesis were executed according to the manufacturer’s instructions using 1 µg of total RNA from lba1 upf3-1 double mutants as starting material. Downstream PCR amplification steps were performed using gene-specific primers as listed in Supplemental Figure 9 online.
Data Access
Supplemental data sets and read alignments can be accessed from http://www.raetschlab.org/suppl/nmd. RNA-seq data visualization is available at http://gbrowse.cbio.mskcc.org/gb/gbrowse/NMD2013/. RNA-seq data have also been deposited in the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE41432.
Accession Numbers
Sequence data from this article can be found in TAIR databases under the following accession numbers: At5g47010 (UPF1, LBA1), At1g33980 (UPF3), At5g53180 (AtPTB2), At4g36960, At1g11650, At5g17550, At1g58080, At2g30260, At4g04350, At4g16990, At3g58760, At1g79600, At3g66654, At5g65720, At1g28660, At4g17370, At4g19100, At2g27720, At3g17609 (HYH), At3g23280 (XBAT35), At2g46700, At5g20840, At3g55850, At3g19980, At2g42500, At4g35230, At2g33051, At1g56612, and At1g13320 (PP2AA3). A list of all further analyzed genes is given in Supplemental Data Set 1 online. Supplemental Data Sets 1 to 5 are deposited in the DRYAD repository (doi:10.5061/dryad.hb7j1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantitation of Coamplified PCR Products and Coverage Plots for NMD Candidate Genes.
Supplemental Figure 2. Gene Models, Coverage Plots, and Splice Ratios for AS Events Chosen for FDR Testing.
Supplemental Figure 3. Venn Diagrams and Numbers of AS Events Changed upon NMD Impairment Using Different Stringency Cutoffs.
Supplemental Figure 4. Gel Images, Gene Models, and Coverage Plots for NMD-Regulated and Control AS Events.
Supplemental Figure 5. Correlation of 3′ UTR Lengths with Predicted Significance of Differential Transcript Expression.
Supplemental Figure 6. Gene Models, Coverage Plots, and Gel Images for Candidates from GO Terms Signaling and Posttranslational Protein Modification.
Supplemental Figure 7. Osmotic Stress Does Not Alter Splicing Variant Ratios for Genes Displaying NaCl-Responsive AS-NMD.
Supplemental Figure 8. Full-Length Sequences of Transcripts from Intergenic Regions.
Supplemental Figure 9. Oligonucleotides for the Detection of Splicing Variants and Corresponding Splicing Isoform Sequences as Revealed by Sequencing.
Supplemental Table 1. Numbers of AS Events and Corresponding Gene Fractions.
Supplemental Table 2. Types of AS Events Detected in Different Samples.
Supplemental Table 3. Comparison of Previously Published Analyses of Coupled AS-NMD with RNA-seq Data.
Supplemental Table 4. Impact of AS on the Presence of Protein Domains for Candidate Genes from GO Terms Signaling and Posttranslational Protein Modification.
Supplemental Methods 1. Sequencing of mRNA Libraries and Computational Methods.
Supplemental Data Set 1. Read Statistics of RNA-seq Data and Computational Analysis of Transcriptome-Wide AS and Gene Expression.
Supplemental Data Set 2. Analysis of NMD Target Features for All Events.
Supplemental Data Set 3. Analysis of NMD Target Features for Genes Containing Single Events.
Supplemental Data Set 4. Categorization of NMD-Regulated and Reference Gene Sets into Functional Subgroups.
Supplemental Data Set 5. Expressed Intergenic Regions and NMD Impairment–Responsive ncRNAs and Pseudogenes.
Supplementary Material
Acknowledgments
We are grateful to Christa Lanz, Jens Riexinger, and the Genome Center (Max Planck Institute) for performing the Illumina sequencing and to Vipin T. Sreedharan for visualization of RNA-seq data in GBrowse. We thank the Nottingham Arabidopsis Stock Centre for providing seeds of the single mutants described in this work. Furthermore, we thank the central facilities of the Center for Plant Molecular Biology (University of Tübingen), the Max Planck Society, and the Sloan-Kettering Institute. This work was supported by an Emmy Noether fellowship (WA 2167/2-1) and the Deutsche Forschungsgemeinschaft (RA1894/2-1).
AUTHOR CONTRIBUTIONS
G.D., A.K., G.R., and A.W. designed the research; G.D., A.K., A.K.K., E.S., J.B., G.R., and A.W. performed the research; P.D. contributed new analytical tools; all authors contributed to data analysis and discussion; and A.W., G.D., and A.K. wrote the article.
Glossary
- NMD
nonsense-mediated decay
- AS
alternative splicing
- ncRNAs
noncoding RNAs
- PTC
premature termination codon
- UTR
untranslated region
- ORF
open reading frame
- uORF
upstream open reading frame
- RNA-seq
RNA sequencing
- CHX
cycloheximide
- SI
splicing variant I
- SII
splicing variant II
- TAIR10
The Arabidopsis Information Resource 10
- FDR
false discovery rate
- GO
Gene Ontology
- qPCR
quantitative PCR
- RACE
rapid amplification of cDNA ends
- MS
Murashige and Skoog
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciga-Reyes L., Wootton L., Kieffer M., Davies B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47: 480–489 [DOI] [PubMed] [Google Scholar]
- Azzalin C.M., Lingner J. (2006). The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 16: 433–439 [DOI] [PubMed] [Google Scholar]
- Baek D., Green P. (2005). Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl. Acad. Sci. USA 102: 12813–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr J., Kahles A., Zhong Y., Sreedharan V.T., Drewe P., Rätsch G. (2013). MITIE: Simultaneous RNA-Seq-based transcript identification and quantification in multiple samples. Bioinformatics 29: 2529–2538 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert R., Behr J., Rätsch G. (2009). Transcript quantification with RNA-Seq data. BMC Bioinformatics 10 (suppl. 13): P5 [Google Scholar]
- Brumbaugh K.M., Otterness D.M., Geisen C., Oliveira V., Brognard J., Li X., Lejeune F., Tibbetts R.S., Maquat L.E., Abraham R.T. (2004). The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell 14: 585–598 [DOI] [PubMed] [Google Scholar]
- Carter M.S., Doskow J., Morris P., Li S., Nhim R.P., Sandstedt S., Wilkinson M.F. (1995). A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270: 28995–29003 [DOI] [PubMed] [Google Scholar]
- Chang D.D., Sharp P.A. (1989). Regulation by HIV Rev depends upon recognition of splice sites. Cell 59: 789–795 [DOI] [PubMed] [Google Scholar]
- Chang Y.F., Imam J.S., Wilkinson M.F. (2007). The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chiu S.Y., Lejeune F., Ranganathan A.C., Maquat L.E. (2004). The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 18: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak D., Ji S.J., Porse B.T., Jaffrey S.R. (2013). Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell 153: 1252–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewe P., Stegle O., Hartmann L., Kahles A., Bohnert R., Wachter A., Borgwardt K., Rätsch G. (2013). Accurate detection of differential RNA processing. Nucleic Acids Res. 41: 5189–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S., Lykke-Andersen J. (2013). Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat. Struct. Mol. Biol. 20: 702–709 [DOI] [PubMed] [Google Scholar]
- Eberle A.B., Stalder L., Mathys H., Orozco R.Z., Mühlemann O. (2008). Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 6: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.K., Mockler T.C. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X., et al. (2011). Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Li X., Spatrick P., Casillo R., Dong S., Jacobson A. (2003). Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- Hillman R.T., Green R.E., Brenner S.E. (2004). An unappreciated role for RNA surveillance. Genome Biol. 5: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Papp A., Pulak R., Ambros V., Anderson P. (1989). A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J.A., Neu-Yilik G., Hentze M.W., Kulozik A.E. (2004). Nonsense-mediated decay approaches the clinic. Nat. Genet. 36: 801–808 [DOI] [PubMed] [Google Scholar]
- Hori K., Watanabe Y. (2005). UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 43: 530–540 [DOI] [PubMed] [Google Scholar]
- Hori K., Watanabe Y. (2007). Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol. 48: 1072–1078 [DOI] [PubMed] [Google Scholar]
- Huang L., Lou C.H., Chan W., Shum E.Y., Shao A., Stone E., Karam R., Song H.W., Wilkinson M.F. (2011). RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell 43: 950–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Li X., Serin G., Maquat L.E. (2001). Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L.E. (2008). The multiple lives of NMD factors: Balancing roles in gene and genome regulation. Nat. Rev. Genet. 9: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean, G., Kahles, A., Sreedharan, V.T., De Bona, F., and Rätsch, G. (2010). RNA-Seq read alignments with PALMapper. Curr Protoc Bioinformatics Chapter 11: Unit 11.6. [DOI] [PubMed] [Google Scholar]
- Jeong H.J., Kim Y.J., Kim S.H., Kim Y.H., Lee I.J., Kim Y.K., Shin J.S. (2011). Nonsense-mediated mRNA decay factors, UPF1 and UPF3, contribute to plant defense. Plant Cell Physiol. 52: 2147–2156 [DOI] [PubMed] [Google Scholar]
- Kalyna M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H., Marzluff W.F. (2005). Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12: 794–800 [DOI] [PubMed] [Google Scholar]
- Kerényi Z., Mérai Z., Hiripi L., Benkovics A., Gyula P., Lacomme C., Barta E., Nagy F., Silhavy D. (2008). Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 27: 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertész S., Kerényi Z., Mérai Z., Bartos I., Pálfy T., Barta E., Silhavy D. (2006). Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 34: 6147–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Koroleva O.A., Lewandowska D., Pendle A.F., Clark G.P., Simpson C.G., Shaw P.J., Brown J.W. (2009). Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 21: 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Furic L., Desgroseillers L., Maquat L.E. (2005). Mammalian Staufen1 recruits Upf1 to specific mRNA 3’UTRs so as to elicit mRNA decay. Cell 120: 195–208 [DOI] [PubMed] [Google Scholar]
- Kurihara Y., et al. (2009). Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 2453–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L.F., Brooks A.N., Soergel D.A., Meng Q., Brenner S.E. (2007a). The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv. Exp. Med. Biol. 623: 190–211 [DOI] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. (2007b). Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929 [DOI] [PubMed] [Google Scholar]
- Leeds P., Peltz S.W., Jacobson A., Culbertson M.R. (1991). The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5: 2303–2314 [DOI] [PubMed] [Google Scholar]
- Legrain P., Rosbash M. (1989). Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583 [DOI] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Izaurralde E., Moore M.J. (2001). The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew J.E., Enomoto S., Berman J. (1998). Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 18: 6121–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Green R.E., Brenner S.E. (2003). Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 100: 189–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y., Brown J.W., Simpson C., Barta A., Kalyna M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22: 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain D.R., Pan Q., Reilly P.T., Elia A.J., McCracken S., Wakeham A.C., Itie-Youten A., Blencowe B.J., Mak T.W. (2010). Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. USA 107: 12186–12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi S.M., Frischmeyer P.A., Mendell J.T., Kelly A.G., Lawler A.M., Dietz H.C. (2001). Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 10: 99–105 [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. (2004). Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Mérai Z., Benkovics A.H., Nyikó T., Debreczeny M., Hiripi L., Kerényi Z., Kondorosi E., Silhavy D. (2012). The late steps of plant nonsense-mediated mRNA decay. Plant J. 73: 50–62 [DOI] [PubMed] [Google Scholar]
- Metzstein M.M., Krasnow M.A. (2006). Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2: e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovich Q.M., Anderson P. (2005). mRNA surveillance of expressed pseudogenes in C. elegans. Curr. Biol. 15: 963–967 [DOI] [PubMed] [Google Scholar]
- Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Jr (2007). Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 21: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P., Mühlemann O. (2010). Cutting the nonsense: The degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 38: 1615–1620 [DOI] [PubMed] [Google Scholar]
- Nicholson P., Yepiskoposyan H., Metze S., Zamudio Orozco R., Kleinschmidt N., Mühlemann O. (2010). Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyikó T., Kerényi F., Szabadkai L., Benkovics A.H., Major P., Sonkoly B., Mérai Z., Barta E., Niemiec E., Kufel J., Silhavy D. (2013). Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res. 41: 6715–6728 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyikó T., Sonkoly B., Mérai Z., Benkovics A.H., Silhavy D. (2009). Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 71: 367–378 [DOI] [PubMed] [Google Scholar]
- Palusa S.G., Reddy A.S. (2010). Extensive coupling of alternative splicing of pre-mRNAs of serine/arginine (SR) genes with nonsense-mediated decay. New Phytol. 185: 83–89 [DOI] [PubMed] [Google Scholar]
- Pan Q., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J. (2006). Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 20: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Park E., Maquat L.E. (2013). Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA 4: 423–435 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A.K., Nelson A.C., Kapranov P., Bell I., Gingeras T.R., Fraser A.G. (2009). High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol. 10: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayson S., Arciga-Reyes L., Wootton L., De Torres Zabala M., Truman W., Graham N., Grant M., Davies B. (2012a). A role for nonsense-mediated mRNA decay in plants: Pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS ONE 7: e31917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayson S., Ashworth M., de Torres Zabala M., Grant M., Davies B. (2012b). The salicylic acid dependent and independent effects of NMD in plants. Plant Signal. Behav. 7: 1434–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Raes J., Izaurralde E. (2006). Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem. Sci. 31: 639–646 [DOI] [PubMed] [Google Scholar]
- Reichenbach P., Höss M., Azzalin C.M., Nabholz M., Bucher P., Lingner J. (2003). A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 13: 568–574 [DOI] [PubMed] [Google Scholar]
- Riehs N., Akimcheva S., Puizina J., Bulankova P., Idol R.A., Siroky J., Schleiffer A., Schweizer D., Shippen D.E., Riha K. (2008). Arabidopsis SMG7 protein is required for exit from meiosis. J. Cell Sci. 121: 2208–2216 [DOI] [PubMed] [Google Scholar]
- Riehs-Kearnan N., Gloggnitzer J., Dekrout B., Jonak C., Riha K. (2012). Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res. 40: 5615–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufener S.C., Mühlemann O. (2013). eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 20: 710–717 [DOI] [PubMed] [Google Scholar]
- Saul H., et al. (2009). The upstream open reading frame of the Arabidopsis AtMHX gene has a strong impact on transcript accumulation through the nonsense-mediated mRNA decay pathway. Plant J. 60: 1031–1042 [DOI] [PubMed] [Google Scholar]
- Schöning J.C., Streitner C., Meyer I.M., Gao Y., Staiger D. (2008). Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 36: 6977–6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Baldwin I.T., Wu J. (2012). Arabidopsis plants having defects in nonsense-mediated mRNA decay factors UPF1, UPF2, and UPF3 show photoperiod-dependent phenotypes in development and stress responses. J. Integr. Plant Biol. 54: 99–114 [DOI] [PubMed] [Google Scholar]
- Stauffer E., Westermann A., Wagner G., Wachter A. (2010). Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J. 64: 243–255 [DOI] [PubMed] [Google Scholar]
- Stegle O., Drewe P., Bohnert R., Borgwardt K.M., Rätsch G. (2010). Statistical tests for detecting differential RNA-transcript expression from read counts. Nat. Preced.http//dx..org/10.1038/npre.2010.4437.1. [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Wachter A., Tunc-Ozdemir M., Grove B.C., Green P.J., Shintani D.K., Breaker R.R. (2007). Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19: 3437–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.B., Brendel V. (2006). Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 103: 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J., Waage J., Tian G., Zhao J., Damgaard I., Jakobsen J.S., Kristiansen K., Krogh A., Wang J., Porse B.T. (2012). Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 13: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp N., Huntzinger E., Weiler C., Saulière J., Schmidt S., Sonawane M., Izaurralde E. (2009). Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell. Biol. 29: 3517–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton M.C., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W. (2004). Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13: 91–100 [DOI] [PubMed] [Google Scholar]
- Wu J., Kang J.H., Hettenhausen C., Baldwin I.T. (2007). Nonsense-mediated mRNA decay (NMD) silences the accumulation of aberrant trypsin proteinase inhibitor mRNA in Nicotiana attenuata. Plant J. 51: 693–706 [DOI] [PubMed] [Google Scholar]
- Yoine M., Nishii T., Nakamura K. (2006a). Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 47: 572–580 [DOI] [PubMed] [Google Scholar]
- Yoine M., Ohto M.A., Onai K., Mita S., Nakamura K. (2006b). The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 47: 49–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.