Abstract
We identified the first prokaryotic urea carboxylase (UCA) from a member of the alpha subclass of the class Proteobacteria, Oleomonas sagaranensis. This enzyme (O. sagaranensis Uca) was composed of 1,171 amino acids, and its N-terminal region resembled the biotin carboxylase domains of various biotin-dependent carboxylases. The C-terminal region of the enzyme harbored the Met-Lys-Met motif found in biotin carboxyl carrier proteins. The primary structure of the enzyme was 45% identical to that of the urea carboxylase domain of urea amidolyase from Saccharomyces cerevisiae. O. sagaranensis Uca did not harbor the allophanate hydrolase domain found in the yeast enzyme, but a separate gene with structural similarity was found to be adjacent to the uca gene. Purified recombinant O. sagaranensis Uca displayed ATP-dependent carboxylase activity towards urea (Vmax = 21.2 μmol mg−1 min−1) but not towards acetyl coenzyme A (acetyl-CoA) and propionyl-CoA, indicating that the gene encoded a bona fide UCA and not an acetyl-CoA or propionyl-CoA carboxylase. The enzyme also exhibited high levels of activity towards acetamide and formamide. Kinetic parameters of the enzyme reaction were determined with ATP, urea, acetamide, and formamide. O. sagaranensis could grow on urea, acetamide, and formamide as sole nitrogen sources; moreover, ATP-dependent urea-degrading activity was found in cells grown with urea but not in cells grown with ammonia. The results suggest that the UCA of this organism may be involved in the assimilation of these compounds as nitrogen sources. Furthermore, orthologues of the O. sagaranensis uca gene were found to be widely distributed among Bacteria. This implies that there are two systems of urea degradation in Bacteria, a pathway catalyzed by the previously described ureases and the UCA-allophanate hydrolase pathway identified in this study.
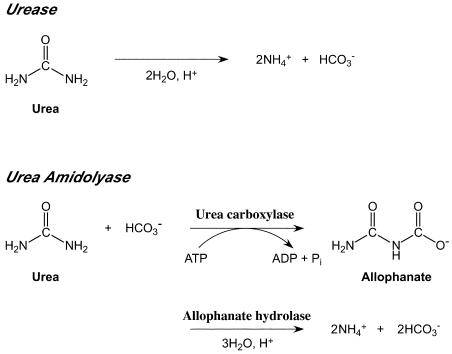
Enzymes involved in the biological transformation of urea have been found in diverse organisms. Two distinct enzymes are known to degrade urea to ammonia and carbon dioxide; these enzymes are urease and urea amidolyase (Fig. 1). Urease has been extensively characterized and catalyzes the ATP-independent cleavage of the amide bonds of urea. This enzyme is nickel dependent, and a number of accessory proteins are necessary for formation of the active enzyme (3). Ureases from plants, fungi, and bacteria have been studied. In plants, urease is known to be responsible for the recycling of nitrogen from urea which is metabolically derived from the degradation of arginine or purines and ureides (21). The ureases of ureolytic bacteria and fungi contribute to assimilation of urea as a nitrogen source (12, 13). Urease also plays important roles in the pathogenesis of a number of bacterial species, including Helicobacter pylori, Proteus mirabilis, and Staphylococcus saprophyticus (5, 14).
FIG. 1.
Reactions catalyzed by urease and urea amidolyase. The two reactions catalyzed by UCA and allophanate hydrolase in the urea amidolyase reaction are shown individually.
Although detailed studies of ureases have been performed, the enzymatic properties and physiological roles of urea amidolyases are not well known. The presence of urea amidolyase or its activity has been reported only for yeast and algae (11, 19). These organisms lack a urease, and it has been supposed that urea amidolyase functionally replaces urease. Urea amidolyase catalyzes the ATP-dependent hydrolysis of urea to ammonia and carbonic acid with allophanate as an intermediate. The overall reaction consists of two independent enzymatic reactions, (i) carboxylation of urea (forming allophanate) and (ii) hydrolysis of allophanate to NH3 and CO2 (27). While in yeast both activities derive from a single polypeptide (7, 15, 22), the activities in algae are the result of two separate enzymes, urea carboxylase (UCA), which catalyzes the first reaction mentioned above, and allophanate hydrolase, which catalyzes the second reaction (Fig. 1) (24).
UCA is a member of the biotin-dependent carboxylase family of enzymes (8, 17). These enzymes catalyze carboxylation of a variety of substrates via carboxylation of a prosthetic biotin group, followed by transcarboxylation of the specific substrates. The yeast UCA, which is fused with allophanate hydrolase on a single peptide, has been purified and biochemically characterized (19, 25, 27). The gene from Candida utilis (Pichia jadinii; accession no. E05855) has been cloned and sequenced (15), and regulation of gene expression has been studied with the enzyme from Saccharomyces cerevisiae (accession no. M64926) (7, 28). Studies of UCA have been limited to these organisms; in particular, the presence of this enzyme has not been reported previously in a prokaryotic organism. Recently, complete genome sequencing has revealed the presence of genes exhibiting similarity to the UCA domains of yeast urea amidolyase, suggesting that UCAs may not be restricted to eukaryotes.
Characterization and classification of a bacterium isolated from an oil field in Sagara, Shizuoka, Japan, have been described recently (9). This bacterium, Oleomonas sagaranensis, is a member of the alpha subclass of the class Proteobacteria (α-proteobacteria) and represents a novel genus. Here, we describe identification and biochemical characterization of a bona fide UCA from this organism (O. sagaranensis Uca) and clearly show that UCAs occur in Bacteria.
MATERIALS AND METHODS
Bacterial strains and plasmids.
O. sagaranensis strain HD-1 was isolated from an oil field in Sagara, Shizuoka, Japan (9). Escherichia coli strain DH5α and pUC118 were used for gene cloning and DNA manipulation. E. coli strain BL21(DE3) (Stratagene, La Jolla, Calif.) was used as a host with the expression vector pET21a (Novagen, Madison, Wis.).
DNA manipulation.
Standard DNA manipulation was carried out by using the methods described by Sambrook and Russell (20). Restriction enzymes and DNA polymerases were purchased from Toyobo (Osaka, Japan), Takara Shuzo (Kyoto, Japan), and New England Biolabs (Beverly, Mass.). Each enzyme was used according to the recommendations of the manufacturer. Genomic DNA, λ phage DNA, and plasmid DNA were isolated by using the QIAGEN Genomic-tip system, a QIAGEN lambda kit, and a QIAGEN plasmid kit (QIAGEN, Hilden, Germany), respectively. DNA ligation was performed by using Ligation high (Toyobo). A GFX PCR DNA and gel band purification kit (Amersham Biosciences, Little Chalfont, Buckinghamshire, United Kingdom) was used to recover DNA fragments from agarose gels.
Growth experiments with various nitrogen sources.
O. sagaranensis cells were cultivated in a modified Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) (20) and shaken (110 rpm) at 30°C. When the cells reached a density of 1.5 mg (wet weight)/ml, they were harvested and washed with modified BS medium without a nitrogen source as described below. Cells were centrifuged and resuspended in the same medium at a cell density of 15 mg/ml. Fifty microliters of this suspension was inoculated into 5 ml of modified BS medium without a nitrogen source containing the following compounds (per liter of deionized water): KH2PO4, 1.4 g; NaHCO3, 0.22 g; Na2SO4, 0.20 g; FeSO4 · 7H2O, 0.2 mg; Na2WO4 · 2H2O, 0.1 mg; Na2SeO4, 0.1 mg; trace mineral solution, 10 ml; and vitamin solution, 10 ml (16). The trace mineral solution contained the following compounds (per liter of deionized water): nitrilotriacetic acid, 1.5 g; MgSO4 · 7H2O, 3.0 g; MnSO4 · 2H2O, 0.5 g; NaCl, 1.0 g; FeSO4 · 7H2O, 0.1 g; CoCl2, 0.1 g; CaCl2 · 2H2O, 0.1 g; ZnSO4, 0.1 g; CuSO4 · 5H2O, 0.01 g; AlK(SO4)2, 0.01 g; H3BO3, 0.01 g; and Na2MoO4 · 2H2O, 0.01 g. As a nitrogen source, 5 mM NH4Cl, 5 mM acetamide, 5 mM formamide, or 2.5 mM urea was added initially and after 24 h. For each culture, 0.1% butanol was added as a carbon source initially and after 24 h. Cultures were shaken at 90 rpm at 30°C for 48 h.
Partial purification of biotinylated proteins from O. sagaranensis.
In order to prepare the cell extract, O. sagaranensis cells were cultivated in a modified LB medium at 30°C. After cultivation for 72 h, the cells were harvested, resuspended in 50 mM potassium phosphate buffer (pH 7.0), disrupted by sonication on ice, and then centrifuged for 20 min at 20,000 × g and 4°C. The biotinylated proteins in the supernatant were partially purified at 4°C by using TetraLink tetrameric avidin resin (Promega, Madison, Wis.) according to the manufacturer's instructions.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described previously (2, 10), and proteins were visualized by Coomassie brilliant blue staining (2).
Detection of biotinylated proteins by using streptavidin-HRP conjugate.
Biotinylated proteins were detected with streptavidin-horseradish peroxidase (HRP) conjugate. After SDS-PAGE, the proteins were electrotransferred to polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). The membranes were blocked by shaking for 1 h in phosphate-buffered saline (PBS) (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl) containing 5% skim milk. After two washes with PBS, the membranes were incubated for 1 h with streptavidin-HRP conjugate (ZYMED Laboratories, San Francisco, Calif.) in PBS containing 1% bovine serum albumin. The membranes were washed three times with buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.01% (vol/vol) Triton X-100, and detection was carried out with 4-chloro-1-naphthol and hydrogen peroxide as described elsewhere (6).
Determination of N-terminal amino acid sequences.
In order to determine the N-terminal amino acid sequence, proteins were electroblotted onto a polyvinylidene difluoride membrane after SDS-PAGE. The membrane was stained with an amido black solution (1.5% amido black, 30% methanol, 10% acetic acid) for 2 min and was destained with destaining solution (30% methanol, 10% acetic acid). N-terminal amino acid sequences were determined with a protein sequencer (491 cLC; Applied Biosystems, Foster City, Calif.).
Isolation, sequencing, and characterization of the O. sagaranensis uca gene.
Construction of a genomic DNA library of O. sagaranensis strain HD-1 has been described previously (9). A partial DNA fragment containing O. sagaranensis uca was amplified from O. sagaranensis genomic DNA by PCR by using two primers. One primer (5′-ATCCTSATCGCSAAYCGSGG-3′, where S = G or C and Y = C or T) was designed from the N-terminal sequence of O. sagaranensis Uca, and the other primer (5′-CTGSAGSCGSGTRTTSACYTC-3′, where R = A or G) corresponded to a highly conserved region in urea amidolyase genes and their orthologues. By using the amplified DNA fragment and a digoxigenin DNA labeling kit (Roche Diagnostics, Mannheim, Germany), probes were constructed, and a phage clone carrying the complete O. sagaranensis uca gene was screened from the genomic library by plaque hybridization. A DNA fragment containing the O. sagaranensis uca gene and its flanking region was subcloned into pUC118. DNA sequencing was performed by using an ABI PRISM BigDye terminator v3.0 cycle sequencing Ready Reaction kit (Applied Biosystems) and a DNA sequencer (ABI 3100; Applied Biosystems). Nucleotide and deduced amino acid sequence analyses, multiple alignment, open reading frame searches, and molecular weight calculations were performed with the DNASIS software (Hitachi Software, Yokohama, Japan). Homology searches were carried out by using the BLAST program (1) at Takara Bio (Ohtsu, Japan).
Expression of the O. sagaranensis uca gene in E. coli and purification of the protein product.
NdeI and BamHI sites were introduced by PCR into the N- and C-terminal regions of the O. sagaranensis uca gene, respectively. The primers used were 5′-AAAAACATATGTTCAAGAAAATCCTGATCGCCAATCGGGGCG-3′ and 5′-AAAAAGGATCCTCAGGCCGCTTCGATCAGGGCGACGATCTGG-3′ (underlining indicates the NdeI and BamHI sites). After the DNA sequence was confirmed, the NdeI-BamHI fragment was inserted into the pET21a expression vector, resulting in pET-uca. E. coli BL21(DE3) carrying pET-uca was grown overnight in LB medium containing 100 μg of ampicillin ml−1 at 37°C. The culture was inoculated (1%) into fresh LB medium containing 100 μg of ampicillin ml−1, and cultivation was continued at 37 or 17°C until the optical density at 660 nm reached 0.4. Then, expression of the O. sagaranensis uca gene was induced by addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) along with 0.2 mM biotin and incubation for 4 h at 37°C or for 24 h at 17°C.
All purification steps were carried out on ice or at 4°C. The cells harvested from the culture incubated at 17°C were washed and suspended in 50 mM Tris-HCl buffer (pH 8.0) (column buffer). The cells were disrupted by sonication on ice and centrifuged for 30 min at 20,000 × g twice; this was followed by ultracentrifugation at 100,000 × g for 1 h to remove the cell debris. The soluble fraction obtained was brought to 30 to 50% saturation with solid (NH4)2SO4. The precipitated proteins were recovered by centrifugation for 15 min at 8,000 × g and resuspended in a minimum volume of column buffer, and this was followed by dialysis against 1 liter of the same buffer twice. The desalted protein solution was subjected to the following chromatographic steps by using an ÄKTA Explorer 10S (Amersham Biosciences). (i) For Resource Q chromatography, the protein solution was applied to a Resource Q column (diameter, 1.6 cm; volume, 6 ml; Amersham Biosciences) which had been equilibrated with column buffer at a flow rate of 1.5 ml · min−1. The column was washed with 3 bed volumes of column buffer and developed with a 15-ml increasing linear gradient of 0 to 0.5 M NaCl. The O. sagaranensis Uca protein eluted at salt concentrations between 260 and 390 mM, and the volume was 3.9 ml (three 1.3-ml fractions). (ii) For Resource PHE chromatography, an (NH4)2SO4 solution (3 M) was added to the combined fractions from Resource Q chromatography to obtain a final concentration of 0.5 M. A Resource PHE column (diameter, 1.6 cm; volume, 6 ml; Amersham Biosciences) was equilibrated with 5 bed volumes of column buffer containing 0.5 M (NH4)2SO4, and then the sample was applied to the column at a flow rate of 2 ml · min−1. After the column was washed with 3 bed volumes of this buffer, it was developed with an 18-ml decreasing linear gradient of 0.5 to 0 M (NH4)2SO4. The O. sagaranensis Uca protein eluted at salt concentrations between 175 and 30.5 mM, and the volume was 5.2 ml (four 1.3-ml fractions). (iii) For gel filtration chromatography, the sample obtained from Resource PHE chromatography was applied to a TSK GEL G4000SW gel filtration column (diameter, 2.15 cm; volume, 100 ml; Tosoh, Tokyo, Japan) which had been equilibrated with 1.5 bed volumes of column buffer containing 0.15 M NaCl. The flow rate was 1.5 ml · min−1. The fractions which contained the O. sagaranensis Uca protein were collected (6.5 ml; five 1.3-ml fractions) and desalted and/or concentrated if necessary.
Estimation of the native molecular mass of O. sagaranensis Uca.
To estimate the native molecular weight of O. sagaranensis Uca, gel filtration chromatography with a Superdex-200 column (diameter, 1.0 cm; volume, 24 ml; Amersham Biosciences) and an ÄKTA Explorer 10S apparatus was performed. An HMW gel filtration calibration kit and an LMW gel filtration calibration kit (Amersham Biosciences) containing thyroglobulin (669 kDa), ferritin (450 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), ovalbumin (45 kDa), chymotrypsinogen (25 kDa), and RNase A (13.7 kDa) were used as molecular mass standards.
Enzyme activity assay.
UCA catalyzes the carboxylation of urea coupled with the hydrolysis of ATP to ADP. The apparent activity of O. sagaranensis Uca was monitored spectrophotometrically by coupling the ADP-forming reaction with the reactions catalyzed by pyruvate kinase and lactate dehydrogenase (18, 19). Unless indicated otherwise, the reaction mixture (1.0 ml) contained 50 mM HEPES buffer (pH 7.5), 50 mM KCl, 8 mM MgSO4, 1 mM ATP, 1.5 mM phosphoenolpyruvate, 8 mM KHCO3, 0.15 mM NADH, 50 mM urea, 10 U of pyruvate kinase, and 25 U of lactate dehydrogenase. The oxidation of NADH linked with ADP generation was determined at 340 nm at 30°C, and 1 U of activity was defined as 1 μmol of NAD formed per min. Changes in absorbance in a control reaction without urea were subtracted.
ATP-dependent and -independent formation of ammonia from urea was examined with the cell extracts of O. sagaranesis grown with urea or ammonia. Cells were grown in LB medium and inoculated into minimal medium as described above. The minimal medium was the medium described above, with slight modifications. The medium contained either 5 mM NH4Cl or 2.5 mM urea as a nitrogen source, or it contained no nitrogen source. Each of the media was prepared with or without 0.2 mg NiCl2 per liter. Biotin was present in all media at a concentration of 0.2 mM. Cultivation in each minimal medium was repeated three times. Urea-dependent ammonia-generating activity in the cell extracts was measured by using a coupled assay with NAD-dependent glutamate dehydrogenase from bovine liver (Sigma, St. Louis, Mo.). The reaction mixtures (1 ml) contained 50 mM HEPES (pH 7.5), 0.24 mM NADH, 0.81 mM 2-oxoglutarate, 8 mM MgSO4, 8 mM KHCO3, 50 mM KCl, 15 U glutamate dehydrogenase, and 60 μg of cell extract. When necessary, 50 mM urea, 1 mM ATP, and various amounts of avidin and acetohydroxamate were added to examine the activity in detail. Reactions were monitored consecutively at 25°C. The ammonia released in the absence of urea was subtracted from all observed data.
Effects of temperature, pH, and metal cations on enzyme activity.
To examine the effect of pH, reactions were carried out in 1.0-ml reaction mixtures containing one of the following buffers at a concentration of 50 mM: morpholineethanesulfonic acid (MES)-NaOH (pH 5.5 to 7.0), HEPES-NaOH (pH 6.5 to 8.5), Tris-HCl (pH 7.5 to 9.0), and CHES-NaOH (pH 8.5 to 9.5). When the effect of temperature was determined, reaction mixtures were incubated at temperatures between 20 and 60°C. To examine the effects of divalent metal ions, reactions were carried out at 25°C with one of the following metal cations at a concentration of 0 to 10 mM: Mg2+, Mn2+, Co2+, Ni2+, and Ca2+.
Nucleotide sequence accession numbers.
The UCA and allophanate hydrolase gene sequences have been deposited in the GenBank/EMBL/DDBJ databases under accession no. AB158433 and AB158434, respectively.
RESULTS
Utilization of various compounds as nitrogen sources in O. sagaranensis.
We previously examined utilization of various compounds as carbon sources in O. sagaranensis. Among the carbon sources, various sugars, including glucose, fructose, maltose, and lactose, did not support growth of strain HD-1. However, alcohols, including ethanol, n-propanol, n-butanol, and n-tetradecanol, supported high levels of growth, as did a few organic acids, such as acetate, propionate, and pyruvate. This strain could also utilize hydrocarbons, including toluene, benzene, phenol, and n-tetradecane (9).
Here we examined whether O. sagaranensis was able to utilize compounds other than ammonium chloride as sole nitrogen sources. n-Butanol was supplied as the carbon source in a modified BS medium. Cells were inoculated at an initial cell density of 0.15 mg/ml. Without any nitrogen source, we observed very little growth, and the cell density was 0.2 mg/ml after 48 h of cultivation. Addition of ammonium chloride to the medium led to a cell density of 0.9 mg/ml after 48 h. We also observed similar levels of growth when urea, acetamide, or formamide was added to the media, and the densities were 0.9, 0.9, and 1.0 mg/ml after 48 h, respectively.
Detection of an ATP-dependent urea-degrading activity in O. sagaranensis induced in the presence of urea.
The ability of O. sagaranensis to assimilate urea, acetamide, or formamide as a sole nitrogen source suggested that the organism contained an enzyme that could degrade these compounds to form ammonia. We therefore examined the ATP-dependent (urea amidolyase) and ATP-independent (urease) formation of ammonia from urea in the cell extracts of O. sagaranensis cells grown on urea or ammonia. Cells were grown with these nitrogen sources in the presence and absence of nickel, which is necessary for urease activity. The results are shown in Table 1. An ATP-independent ammonia-generating activity was observed in all cell extracts, and the highest levels were detected in cells grown with urea and nickel. ATP-dependent activity could be detected in cells grown with urea but not in cells grown with ammonia. The ATP-dependent activity was avidin sensitive (data not shown), indicating that a biotin protein was involved. The ATP-independent activity remaining after avidin treatment was completely eliminated by addition of 0.125 mM acetohydroxamic acid, a urease inhibitor.
TABLE 1.
Urea-dependent ammonia-generating activity of O. sagaranensis
| Culture conditions
|
NH3-generating activity (mU mg−1)
|
||
|---|---|---|---|
| Nitrogen source | Ni2+ | ATP dependent | ATP independent |
| Urea | − | 44 | 150 |
| + | 36 | 176 | |
| Ammonia | − | <1 | 146 |
| + | <1 | 105 | |
Presence of multiple biotinylated proteins in O. sagaranensis.
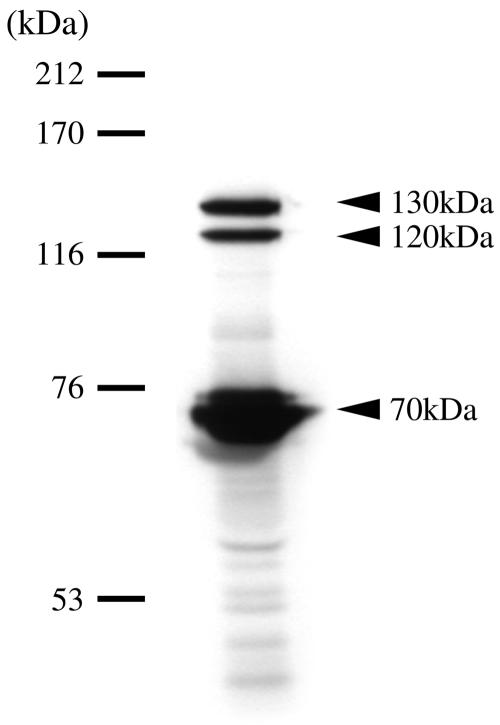
As urea amidolyases have not been identified in prokaryotes, we searched for the presence of such an enzyme in O. sagaranensis. The UCA component of urea amidolyase is a biotinylated protein; we therefore analyzed the biotinylated proteins in cell extracts using a streptavidin-HRP conjugate (see Materials and Methods). We observed four biotinylated proteins in O. sagaranensis. Along with a protein having a molecular weight of approximately 20,000 (data not shown), three high-molecular-weight proteins were also detected (Fig. 2). In particular, two proteins had extremely high molecular weights, approximately 120,000 and 130,000.
FIG. 2.
Presence of multiple biotinylated proteins in O. sagaranesis. Biotinylated proteins in the cell extracts of O. sagaranensis were partially purified with TetraLink tetrameric avidin resin as described in Materials and Methods. The partially purified extract was subjected to SDS-PAGE and detected with streptavidin-HRP conjugate. The arrowheads indicate the positions of three biotinylated proteins with molecular masses of approximately 130, 120, and 70 kDa.
Gene cloning of the 130-kDa biotinylated protein of O. sagaranensis.
The molecular mass of the UCA domain of urea amidolyase from S. cerevisiae is approximately 130 kDa. We therefore analyzed the N-terminal amino acid sequences of the 120- and 130-kDa biotinylated proteins. The sequence of the 120-kDa protein was MFRRILIANRGEIAIRIARA, while that of the 130-kDa protein was MFKKILIANRGEIAARVIK. These sequences exhibited similarity to various biotin carboxylase subunits of bacterial acetyl coenzyme A (acetyl-CoA) and propionyl-CoA carboxylases. Using these sequences and sequences conserved in various urea amidolyase genes and putative uca orthologues in the databases, we were able to amplify a DNA fragment corresponding to a portion of the 130-kDa protein. Using this fragment as a probe, we isolated the entire gene (O. sagaranensis uca) from a genomic DNA library of O. sagaranensis.
Primary structure of the 130-kDa biotinylated protein, O. sagaranensis Uca.
O. sagaranensis uca was composed of 3,513 bp, corresponding to a protein with 1,171 amino acid residues and a molecular weight of 125,211. The primary structure of O. sagaranensis Uca resembled those of the UCA domains of yeast urea amidolyases (45% identical to the domains of both S. cerevisiae and C. utilis). The UCA domains of the two yeast enzymes were 69% identical to each other. O. sagaranensis Uca did not have a region corresponding to the allophanate hydrolase domain found in the yeast enzymes. We instead noticed the presence of a putative gene for allophanate hydrolase adjacent to O. sagaranensis uca, initiating 15 bp downstream. Sequence comparisons and the location of specific motifs revealed the domain structure of O. sagaranensis Uca. The N-terminal portion of O. sagaranensis Uca (residues 1 to 440) exhibited similarity with the biotin carboxylase domains of various biotin-dependent carboxylases, such as acetyl-CoA carboxylase, propionyl-CoA carboxylase, and pyruvate carboxylase. Active site residues of the biotin carboxylase of acetyl-CoA carboxylase from E. coli have been determined from the three-dimensional structure of the protein (8, 23, 26). Among these residues, residues interacting with the phosphate groups of ATP (Lys116, Gly165, and Gly166) were mostly conserved as Lys116, Gly163, and Gly164 in O. sagaranensis Uca, while His236 was replaced by Asn234. The residues involved in binding with the adenine base (201Glu-Lys-Tyr-Leu204) were 199Glu-Arg-Phe-Val202 in O. sagaranensis Uca. In the C-terminal region of O. sagaranensis Uca, we found a Met-Lys-Met motif in which the lysine residue is known to be biotinylated in various biotin carboxyl carrier proteins (BCCP). Therefore, the BCCP domain of O. sagaranensis Uca is most likely to reside in the C-terminal region (residues 1,095 to 1,171) of the protein. The central region of the protein should therefore correspond to the carboxyltransferase domain. This region exhibited similarity to the central portion of the UCA domains of the yeast enzymes, but it did not resemble the carboxyltransferase domains of the multidomain acetyl-CoA carboxylase from yeast (29) or the carboxyltransferase domains of the bacterial acetyl-CoA, propionyl-CoA, and pyruvate carboxylases (Fig. 3). The enzyme also did not exhibit notable similarity to the recently characterized archaeal acyl-CoA carboxylase (4).
FIG. 3.
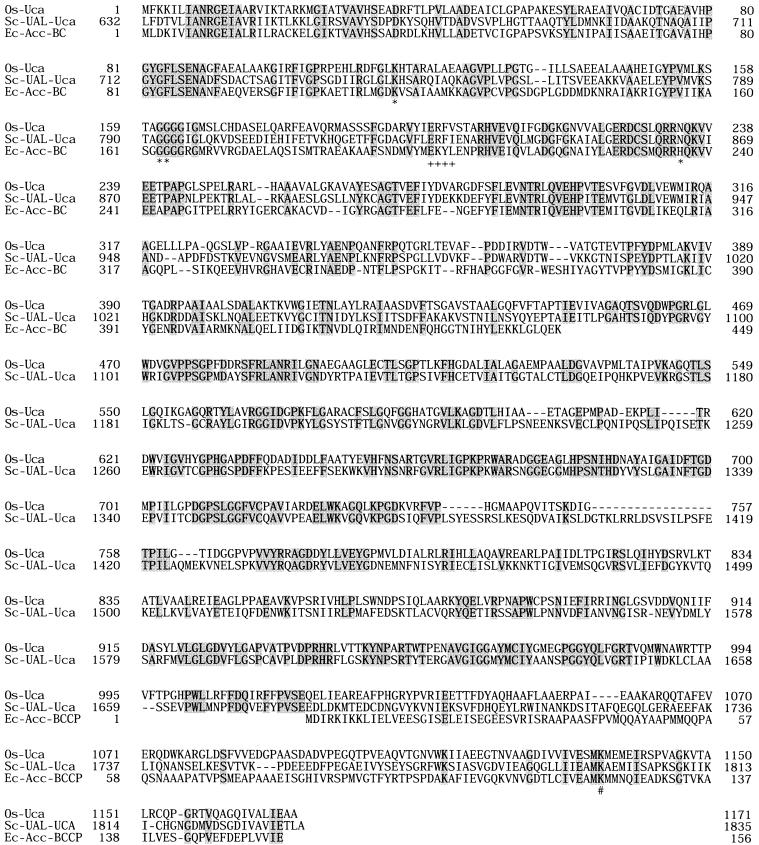
Alignment of the amino acid sequences of various biotin-dependent carboxylases. Os-Uca, O. sagaranensis Uca; Sc-UAL-Uca, UCA domain of urea amidolyase from S. cerevisiae; Ec-Acc-BC, biotin carboxylase of acetyl-CoA carboxylase from E. coli; Ec-Acc-BCCP, BCCP of acetyl-CoA carboxylase from E. coli. The shaded residues are the residues that are the same in all of the sequences. Asterisks indicate residues involved in binding to the phosphate group; plus signs indicate residues involved in binding to the adenine base; and the number sign indicates the lysine residue in the biotin binding motif. Details are described in the text.
Gene expression and purification of recombinant O. sagaranensis Uca.
In order to characterize the protein product of the O. sagaranensis uca gene, we expressed the gene in E. coli BL21(DE3). Biotin was added to the medium at a concentration of 0.2 mM. When we performed a gene expression experiment at 37°C, the majority of the recombinant protein was obtained as aggregated inclusion bodies. We therefore lowered the cultivation temperature to 17°C, and as a result, approximately one-half of the recombinant protein was obtained in a soluble form. The presence of the prosthetic biotin group in the soluble protein was confirmed by blotting and by using the streptavidin-HRP conjugate (data not shown). By using four purification steps, the recombinant protein was purified 37.5-fold with a yield of 4.1% (Table 2). The homogeneity of the purified protein was confirmed by SDS-PAGE (data not shown).
TABLE 2.
Purification of recombinant O. sagaranensis Uca
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U mg−1)a | Yield (%) | Purifi- cation (fold) |
|---|---|---|---|---|---|
| Cell extract | 301 | 150 | 0.50 | 100 | 1.0 |
| (NH4)2SO4 (30-50%) | 89.9 | 75.1 | 0.83 | 50.2 | 1.7 |
| Resource Q | 11.0 | 25.0 | 2.27 | 16.7 | 4.6 |
| Resource PHE | 0.83 | 9.94 | 11.9 | 6.6 | 24.1 |
| TSK GEL G4000SW | 0.33 | 6.13 | 18.6 | 4.1 | 37.5 |
Activity was measured with 50 mM urea and 1 mM ATP.
Oligomeric structure and biotinylation efficiency of recombinant O. sagaranensis Uca.
The molecular mass of purified O. sagaranensis Uca determined by gel filtration chromatography was approximately 130 kDa, indicating that the recombinant protein was a monomer. We also examined the degree of biotinylation of the purified protein. Aliquots of purified O. sagaranensis Uca were subjected to chromatography with a HiTrap streptavidin HP column, and the amount of protein bound to the column was compared to the amount that was present in the flowthrough fraction. We confirmed that the O. sagaranensis Uca in the flowthrough fraction was not biotinylated. In this experiment, we found that 60% of the purified O. sagaranensis Uca was biotinylated.
UCA activity of O. sagaranensis Uca.
The UCA activity of purified O. sagaranensis Uca was examined by enzymatically measuring the production of ADP with pyruvate kinase and lactate dehydrogenase. Significant levels of ATP hydrolysis were observed when the purified recombinant O. sagaranensis Uca was incubated in the presence of urea, ATP, Mg2+, K+, and HCO3− along with the components of the coupling enzymes (Vmax, 21.2 U mg−1) (see below). When urea was omitted from the reaction mixture, we observed only trace levels of ATP hydrolysis corresponding to approximately 0.7 U mg−1, most likely due to spontaneous decarboxylation of the biotin group in the absence of substrate. When we did not add HCO3− to the reaction mixture, we observed transient ATP hydrolysis, which eventually ceased due to depletion of contaminating HCO3− in the mixture. Recovery of ATP hydrolysis was observed in this mixture only after addition of HCO3− to the mixture; it was not observed after addition of the other components. We also compared the amounts of urea before and after the reaction and found that urea was consumed. The level of consumption was lower than that expected from the amount of ATP hydrolysis, probably due to the spontaneous degradation of allophanate to urea. When urea was replaced with acetyl-CoA or propionyl-CoA, we observed ATP hydrolysis at levels equivalent to those observed when no substrate was added. These observations strongly indicate that O. sagaranensis Uca is a UCA and not an acetyl-CoA carboxylase or a propionyl-CoA carboxylase.
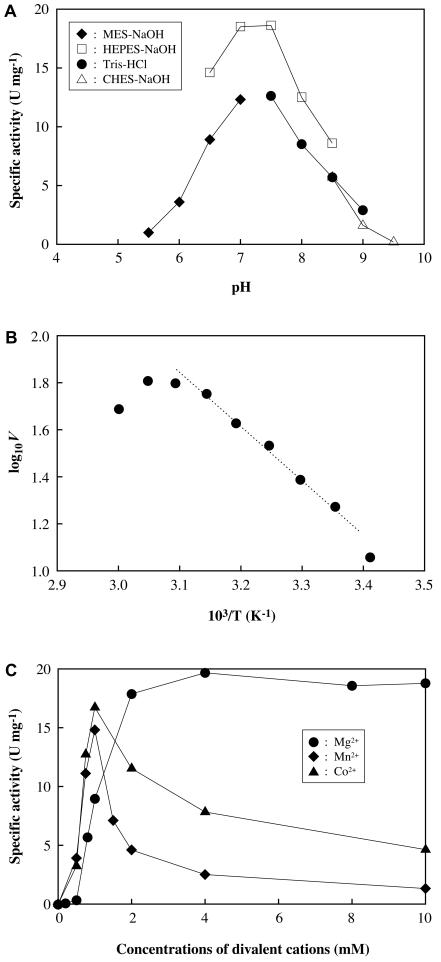
The effects of pH, temperature, and metal cations on the enzyme activity of O. sagaranensis Uca were also examined. With the assay methods used, the optimal pH for O. sagaranensis Uca was between 7.0 and 7.5 (Fig. 4A). Inactivation of O. sagaranensis Uca, along with the coupling enzymes, was not observed in the pH range examined (pH 5.5 to 9.5), as incubation of the enzyme at each pH value, followed by activity measurements at neutral pH, led to trivial or no loss of activity. As biotin-dependent carboxylases are known to utilize HCO3− and not CO2 or H2CO3, the decrease in activity at an acidic pH most likely reflected the low availability of HCO3−. At an alkaline pH, ionization of the carboxylic group on the biotin moiety can be presumed to occur, decreasing the electrophilicity of the carbonyl carbon. The effect of temperature was also examined, and we found that the activation energy of the reaction was 43.6 kJ/mol, based on the Arrhenius plot shown in Fig. 4B. Divalent metal cations were required for enzyme activity; Mg2+, Mn2+, and Co2+ supported activity, while Ca2+ and Ni2+ did not. In the case of Mn2+ and Co2+, concentrations higher than 1 mM led to a decrease in enzyme activity. The presence of Mg2+ led to an increase in activity at concentrations up to 4 mM, and higher concentrations did not inhibit enzyme activity (Fig. 4C). The sigmoidal nature of enzyme activity in the presence of Mg2+ was due to binding to ATP in the reaction mixture. Varying the concentration of ATP in the reaction mixture led to corresponding differences in the concentrations at which the Mg2+-dependent activity abruptly increased (data not shown). This suggests that additional Mg2+ besides the Mg2+ chelated to ATP may be necessary for the enzyme activity of O. sagaranensis Uca.
FIG. 4.
Effects of pH (A), temperature (B), and metal cations (C) on the activity of O. sagaranesis Uca. In the temperature experiments the logarithm of specific activity was plotted against 1,000/T (K−1) (Arrhenius plot), and the reaction mixtures used contained 50 mM HEPES buffer (pH 7.5), 50 mM KCl, 8 mM MgSO4, 1 mM ATP, 1.5 mM phosphoenolpyruvate, 8 mM KHCO3, 0.15 mM NADH, 10 mM urea, 10 U of pyruvate kinase, and 25 U of lactate dehydrogenase. In the pH experiments, the same reaction mixtures were used except that HEPES buffer was replaced with different buffers, as indicated. In the metal cation experiments, the same reaction mixtures were used except that MgSO4 was replaced with the metal cations indicated at various concentrations.
Substrate specificity and kinetic examination of O. sagaranensis Uca.
We further examined the substrate specificity of O. sagaranensis Uca. The substrates used were examined at a concentration of 10 mM and are listed in Table 3. Among these compounds, O. sagaranensis Uca displayed activity with urea, acetamide, and formamide. As O. sagaranensis Uca displayed only trivial activity with the other substrates at a concentration of 10 mM, we performed kinetic analyses of the reactions with urea, acetamide, and formamide along with ATP. Reactions with ATP, urea, acetamide, and formamide all followed Michaelis-Menten kinetics. With urea as the substrate, the Km towards ATP was 42.4 ± 2.5 μM, while the Vmax was 20.7 ± 0.5 μmol min−1 mg of protein−1. The parameters for urea, acetamide, and formamide are shown in Table 4. While the kcat values for urea and acetamide were similar, a much lower Km was observed for urea, indicating that urea was the most preferred substrate with the highest kcat/Km. Acetamide and formamide displayed similar kcat/Km values, which were approximately 5% of the value for urea.
TABLE 3.
Comparison of O. sagaranensis Uca activities on various nitrogen compounds
| Substrate | Activity (U mg−1)a |
|---|---|
| Urea | 12.3 |
| Acetamide | 2.20 |
| Formamide | 1.55 |
| l-Citrulline | 0.03 |
| Arginine succinate | 0.17 |
| l-Arginine | 0.03 |
| l-Ornithine | 0.03 |
| l-Glutamine | 0.02 |
| l-Asparagine | 0.05 |
| Thiourea | 0.08 |
| Carbamyl phosphate | 0.10 |
Activities were measured by monitoring ATP hydrolysis in the presence of 10 mM substrate.
TABLE 4.
Kinetic parameters for ATPase activity of O. sagaranensis Uca
| Substrate | Vmax (μmol mg−1 min−1) | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| Urea | 21.2 ± 0.5 | 7.17 ± 0.41 | 44.2 ± 1.0 | 6.16 |
| Acetamide | 19.3 ± 0.4 | 126 ± 7 | 40.3 ± 0.8 | 0.32 |
| Formamide | 8.06 ± 0.52 | 56.5 ± 7.7 | 16.8 ± 1.1 | 0.30 |
DISCUSSION
In this paper, we report the first identification and characterization of a prokaryotic UCA, the UCA from the α-proteobacterium O. sagaranensis (O. sagaranensis Uca). The results clearly indicate that UCAs are not confined to eukaryotes and are present in Bacteria. In contrast to the yeast UCAs, which are fused to allophanate hydrolases, O. sagaranensis Uca was a monofunctional UCA without the allophanate hydrolase domain (see below). Based on the primary structure of O. sagaranensis Uca, we searched the databases for orthologues of the O. sagaranensis uca gene. Although some open reading frames were annotated as open reading frames for acetyl-CoA carboxylases or propionyl-CoA carboxylases, orthologues of O. sagaranensis uca were detected in the genomes of α-proteobacteria (Caulobacter crescentus [accession no. NC_002696], Rhodopseudomonas palustris [NZ_AAAF01000001]), β-proteobacteria (Burkholderia fungorum [NZ_AAAJ02000045]), γ-proteobacteria (Xanthomonas axonopodis [NC_003919], Pseudomonas fluorescens [NZ_AAAT02000050], Pseudomonas syringae [NC_004578], Microbulbifer degradans [NZ_AABI02000012]), and ɛ-proteobacteria (Wolinella succinogenes [NC_005090]), as well as in the genomes of the high-G+C-content gram-positive organisms Streptomyces avermitilis (NC_003155) and Corynebacterium efficiens (NC_004369). This indicates that the presence of a UCA in O. sagaranensis is not an exceptional case, and the enzyme seems to be widely distributed in the bacteria. A fused allophanate hydrolase domain was not found in any of these orthologues. No orthologues of O. sagaranensis uca were found in the domain Archaea.
When we expressed the gene in E. coli without addition of biotin, we found that only a small portion of the recombinant enzyme was biotinylated. When biotin was added to the medium, we found a large increase in the population of proteins that were biotinylated. Under optimal conditions (0.2 mM biotin), we found that 60% of the purified, recombinant O. sagaranensis Uca was biotinylated. In order to increase this level, it may be necessary to decrease the expression level of O. sagaranensis uca or, alternatively, to coexpress the gene encoding biotin ligase from O. sagaranesis, which attaches the biotin molecule to the lysine residue of the Met-Lys-Met motif.
Although the kinetic parameters of O. sagaranensis Uca have been determined, there are few enzymes to which the values can be compared. The Km towards urea (7.17 mM) was higher than the values observed for the urea amidolyases from S. cerevisiae and C. utilis (0.39 and 0.1 mM, respectively) (19, 27). On the other hand, the Km towards ATP (0.042 mM) was much lower than the values observed for the enzymes from S. cerevisiae (0.25 mM) and C. utilis (0.1 mM).
At present, we cannot determine the physiological role of O. sagaranensis Uca in O. sagaranensis. However, our results strongly suggest that this enzyme contributes to the degradation of urea and possibly acetamide and formamide to generate ammonia, which can be utilized as a nitrogen source. This suggestion is supported by the substrate specificity of the enzyme (Tables 3 and 4) and the strong induction of ATP-dependent urea-degrading activity observed in cells grown in the presence of urea (Table 1). The presence of the protein in the cell extracts of O. sagaranensis cells grown on urea, acetamide, and formamide has also been confirmed (data not shown). Furthermore, adjacent to the O. sagaranensis uca gene, we found a putative gene for allophanate hydrolase, the enzyme necessary to cleave the amide bonds generated by UCA. When we searched the databases, we also found putative allophanate hydrolase genes adjacent to the putative UCA genes in a number of organisms, including C. crescentus, X. axonopodis, and P. syringae. These facts strongly suggest that the bacterial UCAs are involved in urea degradation.
The ATP-independent urea-degrading activity that we observed in O. sagaranensis is intriguing. This activity is comparatively high even in cells grown in the presence of ammonia. Moreover, the activity is found in cells grown without nickel, an essential component of previously described ureases. This activity may reflect the presence of a nickel-independent amidase(s) that can recognize urea as a substrate. It is also possible that along with the nickel-independent urea-degrading activity, the slightly higher ATP-independent activity observed in the urea medium with nickel (176 mU mg−1) than in the medium without nickel (150 mU mg−1) was due to an induced activity of urease.
Along with UCAs, acetyl-CoA carboxylases, propionyl-CoA carboxylases, and pyruvate carboxylases are well-known biotin-dependent carboxylases. O. sagaranensis was found to harbor four biotin-dependent enzymes. Of the four proteins, this study revealed that the largest protein is a UCA. The 20-kDa protein had a molecular mass that was similar to those of the usual BCCPs of acetyl-CoA carboxylases found in many bacteria, implying that it may be a component of the acetyl-CoA carboxylase involved in fatty acid biosynthesis. The functions of the remaining two proteins, the 120- and 70-kDa proteins, are also very interesting, and studies on the recombinant proteins are currently in progress.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Benoit, S., and R. J. Maier. 2003. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J. Bacteriol. 185:4787-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuakrut, S., H. Arai, M. Ishii, and Y. Igarashi. 2003. Characterization of a bifunctional archaeal acyl coenzyme A carboxylase. J. Bacteriol. 185:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, C. M., and S. E. F. D'Orazio. 1993. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol. Microbiol. 9:907-913. [DOI] [PubMed] [Google Scholar]
- 6.Della-Penna, D., R. E. Christoffersen, and A. B. Bennett. 1986. Biotinylated proteins as molecular weight standards on Western blots. Anal. Biochem. 152:329-332. [DOI] [PubMed] [Google Scholar]
- 7.Genbauffe, F. S., and T. G. Cooper. 1991. The urea amidolyase (DUR1,2) gene of Saccharomyces cerevisiae. DNA Seq. 2:19-32. [DOI] [PubMed] [Google Scholar]
- 8.Jitrapakdee, S., and J. C. Wallace. 2003. The biotin enzyme family: conserved structural motifs and domain rearrangements. Curr. Protein Pept. Sci. 4:217-229. [DOI] [PubMed] [Google Scholar]
- 9.Kanamori, T., N. Rashid, M. Morikawa, H. Atomi, and T. Imanaka. 2002. Oleomonas sagaranensis gen. nov., sp. nov., represents a novel genus in the α-Proteobacteria. FEMS Microbiol. Lett. 217:255-261. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Leftley, J. W., and P. J. Syrett. 1973. Urease and ATP:urea amidolyase activity in unicellular algae. J. Gen. Microbiol. 77:109-115. [Google Scholar]
- 12.Mackay, E. M., and J. A. Pateman. 1982. The regulation of urease activity in Aspergillus nidulans. Biochem. Genet. 20:763-776. [DOI] [PubMed] [Google Scholar]
- 13.Mobley, H. L. T., and R. P. Hausinger. 1989. Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53:85-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mobley, H. L. T., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiya, Y., and T. Imanaka. 1993. Cloning and nucleotide sequence of the urea amidolyase gene from Candida utilis. J. Ferment. Bioeng. 75:245-253. [Google Scholar]
- 16.Robb, F. T., and A. R. Place. 1995. Thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Roon, R. J., J. Hampshire, and B. Levenberg. 1972. Urea amidolyase. The involvement of biotin in urea cleavage. J. Biol. Chem. 247:7539-7545. [PubMed] [Google Scholar]
- 18.Roon, R. J., and B. Levenberg. 1970. ATP:urea amidolyase (ADP) (Candida utilis). Methods Enzymol. 17:317-324. [Google Scholar]
- 19.Roon, R. J., and B. Levenberg. 1972. Urea amidolyase. I. Properties of the enzyme from Candida utilis. J. Biol. Chem. 247:4107-4113. [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, 3 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Sirko, A., and R. Brodzik. 2000. Plant ureases: roles and regulation. Acta Biochim. Pol. 47:1189-1195. [PubMed] [Google Scholar]
- 22.Sumrada, R. A., and T. G. Cooper. 1982. Urea carboxylase and allophanate hydrolase are components of a multifunctional protein in yeast. J. Biol. Chem. 257:9119-9127. [PubMed] [Google Scholar]
- 23.Thoden, J. B., C. Z. Blanchard, H. M. Holden, and G. L. Waldrop. 2000. Movement of the biotin carboxylase B-domain as a result of ATP binding. J. Biol. Chem. 275:16183-16190. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, J. F., and A.-M. E. Muenster. 1971. Separation of the Chlorella ATP:urea amido-lyase into two components. Biochem. Biophys. Res. Commun. 43:1049-1055. [DOI] [PubMed] [Google Scholar]
- 25.Waheed, A., and P. A. Castric. 1977. Purification and properties of the urea amidolyase from Candida utilis. J. Biol. Chem. 252:1628-1632. [PubMed] [Google Scholar]
- 26.Waldrop, G. L., I. Rayment, and H. M. Holden. 1994. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochemistry 33:10249-10256. [DOI] [PubMed] [Google Scholar]
- 27.Whitney, P. A., and T. Cooper. 1973. Urea carboxylase from Saccharomyces cerevisiae. Evidence for a minimal two-step reaction sequence. J. Biol. Chem. 248:325-330. [PubMed] [Google Scholar]
- 28.Whitney, P. A., T. G. Cooper, and B. Magasanik. 1973. The induction of urea carboxylase and allophanate hydrolase in Saccharomyces cerevisiae. J. Biol. Chem. 248:6203-6209. [PubMed] [Google Scholar]
- 29.Zhang, H., Z. Yang, Y. Shen, and L. Tong. 2003. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science 299:2064-2067. [DOI] [PubMed] [Google Scholar]