Abstract
The genus Vibrio consists of more than 100 species grouped in 14 clades that are widely distributed in aquatic environments such as estuarine, coastal waters, and sediments. A large number of species of this genus are associated with marine organisms like fish, molluscs and crustaceans, in commensal or pathogenic relations. In the last decade, more than 50 new species have been described in the genus Vibrio, due to the introduction of new molecular techniques in bacterial taxonomy, such as multilocus sequence analysis or fluorescent amplified fragment length polymorphism. On the other hand, the increasing number of environmental studies has contributed to improve the knowledge about the family Vibrionaceae and its phylogeny. Vibrio crassostreae, V. breoganii, V. celticus are some of the new Vibrio species described as forming part of the molluscan microbiota. Some of them have been associated with mortalities of different molluscan species, seriously affecting their culture and causing high losses in hatcheries as well as in natural beds. For other species, ecological importance has been demonstrated being highly abundant in different marine habitats and geographical regions. The present work provides an updated overview of the recently characterized Vibrio species isolated from molluscs. In addition, their pathogenic potential and/or environmental importance is discussed.
Keywords: Vibrionaceae, genus Vibrio, molluscan microbiota, new species, pathogenicity, ecology
INTRODUCTION
Coastal and estuaries environments are growing areas of bivalve molluscs which become an important industry in many countries, due to the increasing importance of these animal as protein for human consumption. Due to their filter-feeding habit, bivalves accumulate a rich and diverse bacterial microbiota, composed of various species belonging to different genera like Vibrio, Pseudomonas, Acinetobacter, Photobacterium, Moraxella, Aeromonas, Micrococcus, and Bacillus (Murchelano and Brown, 1970; Kueh and Chan, 1985; Prieur et al., 1990).
Vibrios are Gram-negative curved rods that occur naturally in marine, estuarine, and freshwater systems worldwide. They occupy habitats ranging from the deep sea to shallow aquatic environments (Reen et al., 2006), being some species important for natural systems, including the carbon cycle and osmorregulation (Johnson, 2013), as free-living inhabitants in the water column or associated to particulate material. On the other hand, vibrios are also associated with a wide variety of poikilotherm and homoiotherm animals, including humans, for some of which are pathogens. Paillard et al. (2004) recognized that the emergence of vibrios as etiological agents of diseases is likely to increase over the coming years due to ocean warming.
The repeated episodes of mortality due to bacterial infections constitute one of the main problems in the culture of bivalve molluscs, since they reduce the production and cause high economical losses. Some members of the genus Vibrio have been described as the main aetiological agents of diseases affecting all life stages of molluscan shellfish (Liu et al., 2000; Allam et al., 2002; Waechter et al., 2002; Lee et al., 2003; Anguiano-Beltrán et al., 2004; Estes et al., 2004; Gay et al., 2004a, b; Paillard et al., 2004; Gómez-León et al., 2005; Prado et al., 2005; Labreuche et al., 2006a, b; Garnier et al., 2007, 2008).
The aim of the present work is to provide an overview on the diversity of Vibrionaceae associated with bivalve molluscs. Special emphasis is made on the species described in the last years in an attemp to clarify, not only their taxonomy, but also their pathogenic or ecological importance.
VIBRIOS AS MICROBIOTA OF BIVALVE MOLLUSCS
In the literature, studies analyzing the diversity, distribution, and density of marine bacteria associated with bivalve molluscs are scarce. These studies date back to the 1960s and in general, results agree in the dominance of Gram negative over Gram positive bacteria in molluscs, as well as in the high abundance of bacteria belonging to the genus Vibrio (Colwell and Liston, 1960; Beenson and Johnson, 1967; Kueh and Chan, 1985).
From 1990s the diversity of Vibrio species associated with bivalves in different geographical areas has been the subject of various studies (Montilla et al., 1994; Hariharan et al., 1995; Arias et al., 1999; Pujalte et al., 1999; Maugeri et al., 2000; Caballo and Stabili, 2002; Castro et al., 2002; Guisande et al., 2004; Beaz-Hidalgo et al., 2008, 2010a; Lafisca et al., 2008). The main common conclusions obtained from these studies were that environmental parameters, such as variations in the water temperature and salinity, can influence the diversity of Vibrio spp. in the environment, as well as the physiological state of the bivalve and its susceptibility to bacterial infections (Arias et al., 1999; Pujalte et al., 1999; Maugeri et al., 2000; Paillard et al., 2004; Garnier et al., 2007).
In most studies, the predominating species associated with bivalves from different geographical locations (Spain, Canada, Italy, or Brazil), all from temperate climates, were either V. splendidus, V. alginolyticus, V. harveyi, or any combination of these species (Montilla et al., 1994; Arias et al., 1999; Pujalte et al., 1999). More recently, Beaz-Hidalgo et al. (2008) analyzed the diversity of Vibrio spp. in cultured Manila clams (Venerupis philippinarum) and carpet-shell clams (Venerupis decusata) by means of phenotypic and genotypic methods. The predominant species that accounted for 66.6% of the total identified strains were Vibrio cyclitrophicus, V. splendidus, and V. alginolyticus. Other species such as V. fluvialis, V. vulnificus, and V. mimicus have also been associated with molluscs (Maugeri et al., 2000; Caballo and Stabili, 2002). In those studies, the identification of Vibrio species was established using only phenotypic methods and, therefore, the real diversity present in bivalves could be underestimate. In fact, a high phenotypic variability was described within the V. splendidus-like and the V. harveyi-like groups which makes impossible to discriminate among several species (Thompson et al., 2005; Le Roux and Austin, 2006; Pascual et al., 2010).
As mentioned, most of the studies described the influence of environmental parameters (i.e., salinity and water temperature) on the diversity and alternance of Vibrio species (Kaspar and Tamplin, 1993; Motes et al., 1998; Arias et al., 1999; Pujalte et al., 1999). For instance, in bivalves from the Mediterranean Sea, V. splendidus has been found to be dominant during winter and spring and V. harveyi during the warmer months (Arias et al., 1999; Pujalte et al., 1999). Another example, in shellfish-growing areas of the US Northern Gulf Coast, the densities of V. vulnificus were high and almost constant at temperatures above 26°C and/or at salinity below 25 ppt, but decreased drastically below this temperature and/or above this salinity (Motes et al., 1998). The latter species together with Vibrio parahaemolyticus and V. cholerae are considered important human pathogens, producing important outbreaks after the consumption of contaminated shellfish (mainly oysters), and therefore have been the subject of many studies (Morris, 2003; Su and Liu, 2007; Jones and Oliver, 2009).
NEW Vibrio SPECIES ASSOCIATED TO MOLLUSCS
The introduction of molecular techniques such as the fluorescent amplified fragment length polymorphism (FAFLP) and multilocus sequence analysis (MLSA) has allowed a more precise identification of Vibrio species which were previously masked under other taxa (Thompson et al., 2001, 2005; Beaz-Hidalgo et al., 2008, 2010a; Pascual et al., 2010).
In this sense, molecular studies have demonstrated the genetic diversity and the polyphyletic nature of V. splendidus (Thompson et al., 2001, 2005; Le Roux et al., 2002) and have enabled many new species to be described, such as Vibrio kanaloae, V. pomeroyi, V. chagasii, or V. gallaecicus (Thompson et al., 2003c; Beaz-Hidalgo et al., 2009b). Furthermore, phenotypically identified V. harveyi strains were re-classified as V. campbellii by FAFLP, DNA–DNA hybridization (DDH), and MLSA (Gomez-Gil et al., 2004a; Thompson et al., 2007). Pascual et al. (2010) investigated the usefulness of an MLSA approach with six housekeeping genes to discriminate six tightly related species with DDH values close to 70%, namely V. harveyi, V. campbellii, V. rotiferianus, V. parahaemolyticus, V. alginolyticus, and V. natriegens. They recognized the genes toxR (cholera toxin transcriptional activator) and rpoD (Sigma factor σ70) as the most reliable for species identification, and proposed a scheme for species definition on the basis of the similarities of the concatenated sequences of the most resolving genes.
In the last decade, more than 50 new species have been described in the genus Vibrio, many of them associated to marine environments and aquatic eukaryotic organisms. To mention some examples, among the new species described as free-living seawater bacteria are V. agarivorans (Macián et al., 2001b), V. ruber (Shieh et al., 2003), V. aestivus and V. quintilis (Lucena et al., 2012), V. azureus (Yoshizawa et al., 2009), or V. sagamiensis (Yoshizawa et al., 2010). Associated to different marine organisms have been described, among others, V. caribbeanicus from sponges (Hoffmann et al., 2012), V. hemicentroti from sea urchin (Kim et al., 2013), V. corallilyticus, V. maritimus, V. shiloi, V. stylophorae, and V. variabilis from corals (Kushmaro et al., 2001; Ben-Haim et al., 2003; Chimetto et al., 2011; Sheu et al., 2011), V. rotiferianus from rotifers (Gomez-Gil et al., 2003a), V. comitans, V. gallicus, V. inusitatus, V. neonates, V. rarus, and V. superstes from abalones (Hayashi et al., 2003; Sawabe et al., 2004a, b, 2007a), V. atypicus, V. hispanicus, V. jasicida, V. owensii, V. pacinii, V. zhanjiangensis, and V. zhuhaiensis from crustaceans (Gomez-Gil et al., 2003b, 2004b; Cano-Gómez et al., 2010; Wang et al., 2010; Jin et al., 2012, 2013; Yoshizawa et al., 2012), V. hippocampi from sea horses (Bálcazar et al., 2010), and V. alfacsensis, V. sinaloensis, and V. tasmaniensis from fish (Thompson et al., 2003d; Gomez-Gil et al., 2008, 2012).
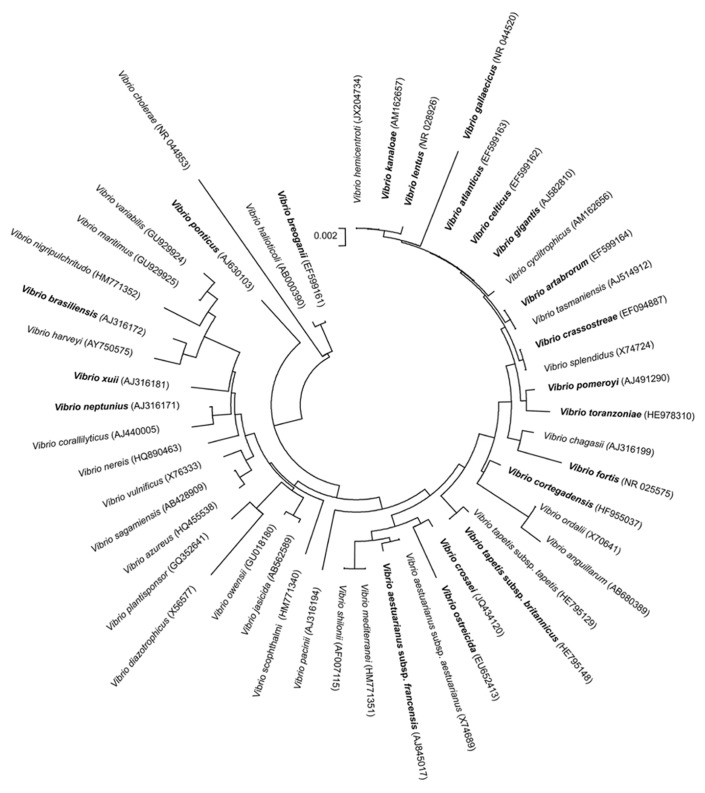
Regarding the vibrios described as associated with bivalve molluscs, and beside the well known species V. alginolyticus, V. harveyi, V. mytili, V. parahaemolyticus, V. pectenicida, or V. vulnificus (Pujalte et al., 1993; Lambert et al., 1998; Arias et al., 1999; Pujalte et al., 1999; Maugeri et al., 2000; Caballo and Stabili, 2002;Paillard et al., 2004; Beaz-Hidalgo et al., 2010a; Romalde et al., 2013), since the turn of the century 19 new species and 2 new subspecies have been described within the genus Vibrio (Figure 1). These new species and subspecies are listed below, in alphabetical order, including their key features, as well as their pathogenic potential and/or ecological relevance. It is noteworthythat about 50% of the new species belong to only one clade, Splendidus, comprising taxa phylogenetically closely related.
FIGURE 1.
Phylogenetic tree based on partial 16S rRNA gene sequences obtained by the neighbor joining method, including the Vibrio species associated with molluscs together with other representative species of the genus. In bold are represented 19 new species and 2 new subspecies described since the turn of the century. GenBank sequence accession numbers of the correspondent type strains are given in parentheses. Bar, no. of substitutions per nucleotide position.
Vibrio aestuarianus SUBSP. Francensis (Garnier et al., 2008)
The species Vibrio aestuarianus was described back by Tison and Seidler (1983) on the basis of a group of isolates obtained from oysters, clams and seawater in Oregon and Washington coasts (USA). Few years later, it was described as sharing close similarity with Vibrio anguillarum, and V. pelagius (Pillidge et al., 1987). This finding was confirmed 20 years later when Sawabe et al. (2007b) on the basis of MLSA of all known Vibrio species included V. aestuarianus within the Anguillarum clade of the genus. It has been described as a ubiquitous species in different geographic areas from the Baltic Sea (Eiler et al., 2006) to Hong Kong (Wang et al., 2006).
In the last years, it was associated with the syndrome known as “summer mortality” of the oyster (Crassostrea gigas) in the French coasts (Gay et al., 2004b; Garnier et al., 2007). The syndrome is thought to be multifactorial involving physiological and environmental factors as well as pathogens. Labreuche et al. (2006a, b) demonstrated the pathogenic potential of V. aestuarianus in experimental oyster challenges. The characterization of a group of isolates obtained from diseased oysters in France led to the description of a new subspecies, named V. aestuarianus subsp. francensis, on the basis of DDH values close to the boundary limit for species definition (70%) and several phenotypic differences with the American isolates.
Vibrio artabrorum (Diéguez et al., 2011)
In a previous study (Beaz-Hidalgo et al., 2008), a collection of isolates obtained from Manila and carpet-shell clams and tentatively allocated to the genus Vibrio based on their phenotypic features were analyzed by FAFLP. One of the groups established, cluster 70, consisted of 8 isolates that could not be assigned to any of the known species of the genus Vibrio. Analysis of the 16S rRNA gene sequences allocated the isolates within the Splendidus clade forming a tight group. MLSA of five housekeeping genes, atpA (α-subunit of bacterial ATP synthase), pyrH [uridine monophosphate (UMP) kinase], recA (RecA protein), rpoA (RNA polymerase α-chain), and rpoD, supported their inclusion in that clade forming a well differentiated group with respect to the rest of species, being its closest neighbors V. pomeroyi and V. kanaloae. DNA–DNA reassociation values confirmed its status of new species within the genus Vibrio. It is considered as an environmental species without pathogenic potential for clams.
Vibrio atlanticus (Diéguez et al., 2011)
From the same study of Beaz-Hidalgo et al. (2008) another group of five strains, designated as cluster 5, was likely to be also a new Vibrio species, being further characterized using the same approach employed for the description of V. artabrorum. The phenotypical characterization, chemotaxonomy, MLSA, and DDH techniqeus confirmed the hypothesis that the clam isolates constituted a new species, related with V. tasmaniensis, V. kanaloae, and V. cyclitrophicus within the Splendidus clade. As V. artabrorum, and since until now no pathogenic activity can be proved for V. atlanticus, it seems to be part of the normal environmental and clam microbiota.
Vibrio brasiliensis (Thompson et al., 2003a)
Six isolates obtained from lion’s paw scallop (Nodipecten nodosus) larvae were identified as a tight group during a wide study on vibrios by FAFLP (Thompson et al., 2001). Further characterization of those isolates on the basis of phenotypic features, 16S rRNA gene sequencing, G + C content and DDH, allowed the description of the new species V. brasiliensis within the Orientalis clade. Its pathogenic potential was demonstrated in experimental challenges using rainbow trout (Oncorhynchus mykiss) and Artemia nauplii as animal models (Austin et al., 2005). The extracellular products (ECP) of the strain tested were also harmful to the animals.
Vibrio breoganii (Beaz-Hidalgo et al., 2009a)
A group of seven non-motile, facultative anaerobic alginolytic marine bacteria isolated from cultured Manila and carpet-shell clams in Galicia (NW Spain) were characterized employing a polyphasic approach, including the sequence analysis of the 16S rRNA gene and three housekeeping (atpA, recA, and rpoA) genes, FAFLP fingerprinting, G + C content, DDH, and phenotypic features. Phylogenetic analysis based on 16s rRNA gene sequences showed that the isolates were closely related to the species V. comitans, V. rarus, and V. inusitatus, with sequence similarities of approximately 99%. However, phylogenies based on the sequences of the housekeeping genes grouped the isolates together and allocated them within the Halioticoli clade, although they can be differentiated from the other species in the clade by their FAFLP profiles. DDH experiments confirmed that they represented a new Vibrio species, that was named V. breoganii.
Some years later, this species was consistently detected in a meta-analysis of three separated studies aimed to identify the ecological population structure of Vibrionaceae in the Plum Island Sound Estuary (Ipswich, MA, USA), mainly associated to large particles and zooplankton (Preheim et al., 2011). Interestingly, population of V. breoganii showed essentially identical results with respect to genetic breadth in all studies, regardless the season or the sampling method. This species constitutes a good example of how genotypic clusters established by MLSA can serve as a reasonable initial definition of cohesive unit from an ecological point of view, as well as of the ubiquity of Vibrio species in the marine environment.
In 2012, a strain of this species was included in one of the first studies examining the complete methylation pattern of a bacterial genome (Murray et al., 2012). The knowledge of the methylome could be of great interest due to the recognized importance of methylation for understanding fundamental microbiological processes, microbe adaptability, and disease pathogenicity.
Vibrio celticus (Beaz-Hidalgo et al., 2010b)
A group of four motile facultative anaerobic marine isolates obtained from cultured pullet carpet-shell (Venerupis pullastra) and Manila clams during 2004 and 2005 in Galicia (NW Spain) were studied using a polyphasic approach. It was found that they formed a tight phylogenetic group based on sequences of the 16S rRNA gene and four housekeeping (atpA, recA, rpoA, and rpoD) genes, indicating that the four isolates represented a novel species in the Splendidus clade of the genus Vibrio, for which the name V. celticus was proposed. In addition, the strains showed potential pathogenic activity for adult clams in virulence assays.
Recently, a study on the diversity of Vibrio spp. in the Eastern English Channel by means of sequencing of the housekeeping gene pyrH (Tall et al., 2013), revealed that V. celticus was the predominant species among other 20 Vibrio species isolated at ambient environmental temperature.
Vibrio cortegadensis (Lasa et al., 2013b)
It was described as a results of the polyphasic characterization of a group of four marine strains isolated from carpet-shell and Manila clams in Galicia (NW Spain). The study of the phenotypic characteristics, the analysis of chemotaxonomic features, the sequencing of the 16S rRNA and five housekeeping (atpA, pyrH, recA, rpoA, and rpoD) genes, as well as DDH, allowed the identification of the isolates within the genus Vibrio, being their closest neighbors V. tapetis, V. pomeroyi, and V. crassostreae (97.9%). The phylogenetic analysis of the five concatenated genes indicated the allocation of these strains in between the Splendidus and Anguillarum clades.
Vibrio crassostreae (Faury et al., 2004)
Described in 2004 on the basis of five strains obtained from oyster haemolimph, and originally identified as V. splendidus-like isolates. The authors employed a polyphasic approach including besides biochemical tests, fatty-acid methyl ester (FAME) analysis, 16S rRNA, and gyrB (DNA gyrase subunit B) genes sequencing, FAFLP fingerprinting, and DDH. Although all the genetic studies supported that the five strains constituted a novel Vibrio species within the Splendidus clade, their differentiation of the closest relatives was not possible on the basis of 17 phenotypic characters. However, the presence of fatty acids 16:0 iso and 14:0 iso allowed the differentiation of the new species from other V. splendidus-like species. It was described as a species with pathogenic potential for the oyster C. gigas (Gay et al., 2004a).
Vibrio crosaei (González-Castillo et al., 2014)
The description of this new Vibrio species was based on the study and characterization of one isolate obtained from cultured oyster (C. gigas) in Sonora (Mexico). The phenotypic characteristics and the 16S rRNA gene sequence of the isolate clearly placed it within the genus Vibrio, with V. orientalis and V. rotiferianus as closest relatives. Curiously, these both species belong to different clades, as proposed by Sawabe et al. (2007b), the Orientalis clade and the Harveyi clade, respectively. MLSA technique clarified the definitive allocation of the isolate within the Orientalis clade, and the DNA relatedness measures by DDH experiments confirmed that it constituted a new Vibrio species. The proposed name, Vibrio crosaei, was chosen to honor Prof. Dr. Jorge Crosa, microbiologist and specialist in vibrios.
Vibrio fortis (Thompson et al., 2003b)
The species was defined on the basis of 10 isolates obtained between 1994 and 1999 from different hosts, including healthy and diseased lion’s paw scallop larvae, diseased C. gigas larvae, shrimp (Litopenaeus vannamei) larvae, Atlantic salmon (Salmo salar), as well as sea water. The geographical origins included different North and South American Countries, Tasmania, and UK. Genotypic analysis such as 16S rRNA gene sequencing and DDH confirmed the deliniation of this new species, differentiating it from the closets neighbors V. pelagius or V. mytili. Austin et al. (2005) confirmed the pathogenic potential of the species using rainbow trout and A. nauplii as animal models.
Vibrio fortis was further isolated from spotted rose snapper (Lutjanus guttatus) in Mexico (Gomez-Gil et al., 2007) and from crown-of-thorns starfish (Acanthasther planci) in Australia and Guam (Rivera-Posada et al., 2011). It was also identified as one of the predominant Vibrio species in the Cariaco Basin, Venezuela (García-Amado et al., 2011).
The extracellular polymeric substances from this biofilm forming Vibrio species were characterized (Kavita et al., 2013), showing potential for industrial applications.
Vibrio gallaecicus (Beaz-Hidalgo et al., 2009b)
This species within the Splendidus clade was described on the basis of the characterization of three strains isolated from Manila clams in Galicia (NW Spain). Phylogenetic analysis of the 16S rRNA gene and four housekeeping (atpA, pyrH, recA, and rpoA) genes, indicated that these strains were closely related to the Splendidus clade, being its closest relatives V. splendidus, V. gigantis, and V. pomeroyi. The FAFLP fingerprints and DDH values supported the MLSA results. It is considered as an environmental species without proved pathogenic potential.
Vibrio gigantis (Le Roux et al., 2005)
The polyphasic characterization of four isolates obtained from C. gigas haemolymph allowed the description of this new Vibrio species within the Splendidus clade. Although 16S rRNA gene sequence analysis did not permit a clear differentiation of V. gigantis from other phenotypically related species, other techniques including FAFLP, DDH, and sequencing of four housekeeping [gyrB, rctB (replication origin-binding protein), rpoD, and toxR] genes demonstrated that the isolates formed a tight genomic group, clearly differentiated from the neighboring species. The authors suggested that, as other Vibrio species present in the shellfish haemolymph, V. gigantis may play a role in the health of the host.
Vibrio kanaloae (Thompson et al., 2003a)
It was described on the basis of five isolates with different origins, including diseased oyster (Ostrea edulis) larvae from France, shrimp (Penaeus chinensis) from China and sea water from Hawaii (USA). Therefore, it has been described as an ubiquitous species in the aquatic environment. The five strains were originally detected in a wide FAFLP study (Thompson et al., 2001) as a separate cluster, showing a pattern clearly different from other Vibrio species, with which share the main phenotypic traits of the genus. Further DDH experiments confirmed that they were in fact a new species within the Splendidus clade.
Later studies on the virulence of other related strains were performed on the basis of experimental infections of C. gigas. After injection of strains, bacteria were localized at the periphery of the muscle and induced extensive lesions of the translucent part of the adductor muscle. Unfortunately, although using a polyphasic approach these strains were confirmed to be V. splendidus-related, no clear discrimination between V. kanaloae and V. pomeroyi was possible with the techniques employed. (Gay et al., 2004b). Austin et al. (2005) confirmed its pathogenic potential for aquatic animals including fish and crustaceans.
Vibrio lentus (Macián et al., 2001a)
The study of 12 marine bacteria by means of cultural and physiological characterization, ribotyping, G+C content, DDH, and phylogenetic analysis on the 16S and 23S rRNA genes allowed the description of this new Vibrio species in 2001. All the strains had been isolated from Mediterranean oysters in Spain, and were phenotypically similar to V. splendidus. The name V. lentus was proposed since the strains showed a slow growth on Marine Agar. Thus, colonies of some of the isolates were not larger than 0.2 mm diameter after 3 days of incubation. Some years later V. lentus was isolated from diseased wild octopus (Octopus vulgaris) and from turbot (Scophthalmus maximus) also in Spain (Farto et al., 2003; Montes et al., 2003, 2006). In the case of octopus, experimental infections by bath challenge demonstrated that V. lentus was able to reproduce the skin lesions, colonize the internal organs, and induce mortality in healthy octopuses (Farto et al., 2003).
The presence of a lethal extracellular 39-kDa protease, similar to that of Vibrio pelagius, was detected in 15% of the ECP assayed belonging to strains of the Vibrio splendidus–V. lentus related group by Farto et al. (2006), which suggested their potential risk for the health of reared aquatic organisms.
Vibrio neptunius (Thompson et al., 2003c)
Described during a polyphasic study of 21 isolates with diverse origins, like healthy and diseased lion’s paw scallop larvae, rotifers, and turbot larvae. The results clearly indicated that this group od strains constituted a new species within the Corallilyticus clade of the genus. Vibrio neptunius was further identified as aetiological agent of a mortality episode of oyster (O. edulis) larvae occurred in a Galician hatchery (Prado et al., 2005). Pathogenicity was confirmed in experimental tests where it shown to cause high mortalities (ranging from 98.5 to 100%) in 72 to 96 h after inoculation of larval cultures. The work of Prado and co-workers constituted the first description of V. neptunius as a molluscan pathogen. Later studies with New Zealand Greenshell mussel (Perna canaliculus) larvae confirmed the pathogenic potential for other molluscan species (Kesarcodi-Watson et al., 2009a,b).
Vibrio neptunius was also found in environmental studies as a predominant bacteria in the anoxic zone (García-Amado et al., 2011). In addition, during a study searching for novel antimicrobials in marine Vibrionaceae (Wietz et al., 2010), V. neptunius has been identified as a potential resource of antibacterial compounds with future applicability.
Vibrio ostreicida (Prado et al., 2014)
The species description relies on three strains isolated from a flat oyster (O. edulis) hatchery in Spain after episodes of high mortality (Prado et al., 2005). Pathogenicity was confirmed in experimental tests where the strains were able to cause high larval mortalities. The results of the phenotypic and genotipic analysis revealed that this group of strains constituted a new Vibrio species, closely related to V. pectenicida.
Vibrio pomeroyi (Thompson et al., 2003a)
As in the case of V. kanaloae, V. pomeroyi was originally detected as a group of four isolates showing a characteristic FFAFLP pattern during a study on the genomic diversity amongst Vibrio isolates from different sources (Thompson et al., 2001). Two strains had been isolated from healthy bivalve larvae (N. nodosus) in Brasil and two from turbot in Spain. They were confirmed as a new Vibrio species within the Splendidus clade by means of DDH, phenotypic characterization, and FAME analysis. The studies of Gay et al. (2004a) and Austin et al. (2005) mentioned before demonstrated either non- or low virulence of V. pomeroyi in animal models.
Vibrio ponticus (Macián et al., 2004)
It has been described to accommodate four marine bacteria isolated from mussels, fish, and seawater at the Mediterranean coast of Spain. Phylogenetic analysis locate these strains in the vicinity of the Fluvialis–Furnissii clade, sharing with these species similarities slightly higher tan 97% in their 16S rRNA gene sequences. Since one of the isolates were isolated after direct plating of a kidney sample from a diseased gilthead seabream (Sparus aurata), the pathogenic potential of the species cannot be discarded.
Vibrio tapetis subsp. britannicus (Balboa and Romalde, 2013)
Vibrio tapetis, described by Borrego et al. (1996), is the causative Vibriotapetis, describedby Borrego et al. (1996), isthecausativeagent of an epizootic infection described in adult clams called Brown Ring Disease (BRD) constituting a major limiting factor for the culture of Manila clams. This pathogen was considered for years as a highly homogeneous taxon on the basis of its phenotypical features, but the isolation of new strains from different hosts revealed some variability both at serological and genetic level, allowing the description of three major groups related to the host origin of the isolates (Castro et al., 1997; Romalde et al., 2002; Rodríguez et al., 2006). Balboa and Romalde (2013) performed for the first time a phylogenetic study for this pathogen, where V. tapetis strains appeared clearly separated in two main robust clusters, one containing the isolates from the British Isles and other one containing the isolates from all other geographic origins. The two clusters, that showed values of DDH between 65.05 and 79.8%, were easily distinguishable for their capacity to produce acid from mannitol and arabinose and for the use of citrate. On the basis of these results, not only an emended description was provided for V. tapetis, but also the new subspecies V. tapetis subsp. britannicus was proposed.
Vibrio toranzoniae (Lasa et al., 2013a)
It was recently described in a polyphasic study of four strains isolated from cultured carpet-shell and Manila clams in the Northwest of Spain. The techniques utilized included phylogenetic analysis based on sequences of 16S rRNA and MLSA of five housekeeping genes (atpA, recA, pyrH, rpoA, and rpoD), DDH, FAME analysis and more than 100 phenotypic traits. All the closest relatives were Vibrio species included in the Splendidus clade, such as V. kanaloae, V. artabrorum, V. gigantis, or V. celticus, from which it can be easily differentiated by several phenotypic characteristics. Current studies of some Chilean Vibrio strains isolated from fish seem to indicate that the geographical and host distribution of this species could be wider than expected.
Vibrio xuii (Thompson et al., 2003c)
Three isolates obtained from bivalve and shrimp systems were identified as a tight group during a wide study on vibrios by FAFLP (Thompson et al., 2001). Further characterization of those isolates on the basis of phenotypic features, 16S rRNA gene sequencing, G + C content, and DDH, allowed the description of the new species V. xuii within the Nereis clade. Considered as an environmental species, V. xuii demonstrated either non- or low virulence in the animal models (Austin et al., 2005).
MOLLUSC AND OTHER Vibrionaceae
Other Vibrionaceae described in the last years also associated to molluscan shellfish are representatives of the genera Aliivibrio and Photobacterium.
The genus Aliivibrio was established by Urbanczyk et al. (2007) to accommodate the species V. fischeri, V. logei, V. salmonicida, and V. wodanis, on the basis of a study based on 16S rRNA gene sequencing and MLSA which results indicated that the four species represented a lineage within the Vibrionaceae distinct from other genera. Therefore, the authors proposed the reclassification of the species as Aliivibrio fischeri (the type species), A. logei, A. salmonicida and A. wodanis, respectively. The genus includes symbiotic (A. fischeri) and pathogenic (A. salmonicida) species for marine organisms (Urbanczyk et al., 2007).
Photobacterium was one of the oldest established genus in the family Vibrionaceae. The type species is Photobacterium phosphoreum, which had been described by Cohn in 1878 as “Micrococcus phosphoreus” (Gomez-Gil et al., 2011). At the time of writing, the genus Photobacterium contained 23 species (http://www.vibriobiology.net). Although most species have no described pathogenic activity and are common inhabitants of marine environment, some species, i.e., both subspecies of Photobacterium damselae, are pathogenic for aquatic animals, mainly for fish.
Aliivibrio finisterrensis (Beaz-Hidalgo et al., 2010c)
This species was described after the phenotypic and genotypic characterization of four strains isolated from cultured Manila clam in the north-western coast of Spain. Phylogenetic analyses based on the 16S rRNA gene sequences indicated that these bacteria were closely related to A. wodanis, A. salmonicida, A. fischeri, and A. logei with sequence similarities between 98.1 and 96.0%. Phylogenetic analysis based on MLSA of four housekeeping genes and FAFLP experiments clearly showed that these novel isolates form a tight genomic group different from any currently known Aliivibrio species.
Photobacterium swingsii (Gomez-Gil et al., 2011)
The characterization of six Gram-negative coccobacilli, isolated from Pacific oysters (C. gigas) from Mexico and haemolymph of spider crabs (Maja brachydactyla) from Spain, allowed the description of this species within one of the oldest established genera in the family Vibrionaceae. Repetitive palindromic PCR (REP-PCR) analysis revealed a high degree of genomic homogeneity among the isolates. Several phenotypic traits differentiated the isolates from the type strains of species of the genus Photobacterium, including its closest relatives P. aplysiae and P. frigidiphilum.
FUTURE PERSPECTIVES
Although classification of bacteria into a natural system has been hampered by the lack of a generally applicable species concept, the introduction of MLSA has provided much higher resolution for microbial identification and taxonomy (Gevers et al., 2005). The groups or species defined by means of MLSA are of particular interest for microbial ecology, since some theories predict that they correspond to ecologically cohesive populations (Fraser et al., 2009; Preheim et al., 2011). Some examples have been mentioned in this review, such as V. breoganii or V. celticus which, some years after their description as species, have been identified in ecological studies in different geographical areas as predominant populations. It is likely expected that in the near future more efforts will be made to identify ecological populations using these or other approaches, including single-cell amplification of multilocus genes or single-cell genomics (Stepanauskas and Sieracki, 2007; Rodrigue et al., 2009). As indicated by Preheim et al. (2011), to establish reproducible associations between bacterial species and environmental categories may be helpful to predict their occurrence and to get a deeper knowledge on the ecological factors driving their evolution.
It has been indicated that a phylogenetic hypothesis based on complete genomes is desired for Vibrionaceae (Dikow and Smith, 2013), and the new pyrosequencing and bioinformatic tools available would be very helpful to obtain such goal. Comparative genome analyses have already revealed a variety of genomic events, including mutations, chromosomal rearrangements, loss of genes by decay or deletion, and gene acquisitions through duplication or horizontal transfer (e.g., in the acquisition of bacteriophages, pathogenicity islands, and super-integrons), that are probably important driving forces in the evolution and speciation of vibrios (Hazen et al., 2010; Morrison et al., 2012; Daccord et al., 2013; Dikow and Smith, 2013).
On the other hand, a better knowledge of the in situ or real-time function of vibrios is needed, both in the environment or within the microbiota of aquatic animals (Frias-Lopez et al., 2008; Jones et al., 2008). Metatranscriptomics would be a valuable method, not only to reveal “near instantaneous” responses to environmental changes, but also to determine the real role of vibrios in different habitats or hosts.
CONCLUDING REMARKS
The present study overviewed the diversity of Vibrio species associated with bivalve molluscs. Ongoing studies on the disease and pathogenicity of bivalves primarily relies on the use of phenotypic and molecular methods for an exact species identification. It remains to be investigated to what extent some of the recently discovered species are commensal, opportunistic or pathogenic organisms. Knowledge of the infection mechanisms used by classical and emerging Vibrio spp. to develop disease in bivalve molluscs will help to establish adequate preventive measures to control the transmission of these pathogens in hatcheries and in coastal growing areas.
Finally, and as pointed out by Grimes et al. (2009), future information on completed genomes, metagenomics, and metatranscriptomics will increase the understanding on the biology and ecology of vibrios, providing new insights and solutions to problems with disease, nutrient cycling in the ocean, and opportunities in marine biotechnology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The studies of the University of Santiago reviewed here were supported in part by grants AGL2003-09307-C02-01, AGL2006-13208-C02-01, and AGL2010-18438 from the Ministerio de Ciencia y Tecnología (Spain).
REFERENCES
- Allam B., Paillard C., Ford S. E. (2002). Pathogenicity of Vibrio tapetis, the etiological agent of brown ring disease in clams. Dis. Aquat. Org. 48 221–231 10.3354/dao048221 [DOI] [PubMed] [Google Scholar]
- Anguiano-Beltrán C., Lizárraga-Partida M. L., Searcy-Bernal R. (2004). Effect of Vibrio alginolyticus on larval survival of the blue mussel Mytilus galloprovincialis. Dis. Aquat. Org. 59 119–123 10.3354/dao059119 [DOI] [PubMed] [Google Scholar]
- Arias C. R., Macián M. C., Aznar R., Garay E., Pujalte M. J. (1999). Low incidence of Vibrio vulnificus among Vibrio isolates from sea water and shellfish of the western Mediterranean coast. J. Appl. Microbiol. 86 125–134 10.1046/j.1365-2672.1999.00641.x [DOI] [PubMed] [Google Scholar]
- Austin B., Austin D., Sutherland R., Thompson F., Swings J. (2005). Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii. Environ. Microbiol. 7 1488–1495 10.1111/j.1462-2920.2005.00847.x [DOI] [PubMed] [Google Scholar]
- Balboa S., Romalde J. L. (2013). Multilocus sequence analysis of Vibrio tapetis, the causative agent of Brown Ring Disease: description of Vibrio tapetis subsp. britannicus subsp. nov. Syst. Appl. Microbiol. 36 183–187 10.1016/j.syapm.2012.12.004 [DOI] [PubMed] [Google Scholar]
- Bálcazar J. L., Pintado J., Planas M. (2010). Vibrio hippocampi sp. nov., a new species isolated from wild seahorses (Hippocampus guttulatus). FEMS Microbiol. Lett. 307 30–34 10.1111/j.1574-6968.2010.01955.x [DOI] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Cleenwerck I., Balboa S., De Watcher M., Thompson F. L., Swings J., et al. (2008). Diversity of vibrios with reared clams in Galicia (NW Spain). Syst. Appl. Microbiol. 31 215–222 10.1016/j.syapm.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Cleenwerck I., Balboa S., Prado S., de Vos P., Romalde J. L. (2009a). Vibrio breoganii sp. nov., a non motile alginolytic marine bacterium within the Vibrio halioticoli clade. Int. J. Syst. Evol. Microbiol. 59 1589–1594 10.1099/ijs.0.003434-0 [DOI] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Doce A., Pascual J., Toranzo A. E., Romalde J. L. (2009b). Vibrio gallaecicus sp. nov., isolated from cultured clams in north-western Spain. Syst. Appl. Microbiol. 32 111–117 10.1016/j.syapm.2008.12.002 [DOI] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Balboa S., Romalde J. L., Figueras M. J. (2010a). Diversity and pathogenicity of Vibrio species in cultured bivalve molluscs. Environ. Microbiol. Rep. 2 34–43 10.1111/j.1758-2229.2010.00135.x [DOI] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Diéguezguez A. L., Cleenwerck I., Balboa S., Doce A., de Vos P., et al. (2010b). Vibrio celticus sp. nov., a new Vibrio species belonging to the Splendidus clade with pathogenic potential for clams. Syst. Appl. Microbiol. 33 311–315 10.1016/j.syapm.2010.06.007 [DOI] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Doce A., Balboa S., Barja J. L., Romalde J. L. (2010c). Aliivibrio finisterrensis sp. nov., isolated from Manila clams, Ruditapes philippinarum and emended description of the genus Aliivibrio. Int. J. Syst. Evol. Microbiol. 60 223–228 10.1099/ijs.0.010710-0 [DOI] [PubMed] [Google Scholar]
- Beenson R. J., Johnson P. T. (1967). Natural bacterial flora of the bean clam, Donax gouldi. J. Invertebr. Pathol. 9 104–110 10.1016/0022-2011(67)90049-3 [DOI] [Google Scholar]
- Ben-Haim Y., Thompson F. L., Thompson C. C., Cnockaert M. C., Hoste B., Swings J., et al. (2003). Vibrio corallilyticus sp. nov., a temperatura-dependent pathogen of the coral Pocillopora damicordis. Int. J. Syst. Evol. Microbiol. 53 309–315 10.1099/ijs.0.02402-0 [DOI] [PubMed] [Google Scholar]
- Borrego J. J., Castro D., Luque A., Paillard C., Maes P., García M., et al. (1996). Vibrio tapetis sp. nov., the causative agent of the brown ring disease affecting cultured clams. Int. J. Syst. Bacteriol. 46 480–484 10.1099/00207713-46-2-480 [DOI] [Google Scholar]
- Caballo R. A., Stabili L. (2002). Presence of vibrios in seawater and Mytillus galloprovincialis (Lam.)from the Mar Piccolo of Taranto(Ionian Sea). Water Res. 36 3719–3729 10.1016/S0043-1354(02)00107-0 [DOI] [PubMed] [Google Scholar]
- Castro D., Pujalte M. J., Lopez-Cortes L., Garay E., Borrego J. J. (2002). Vibrios isolated from the cultured manila clam (Ruditapes philippinarum): numerical taxonomy and antibacterial activities. J. Appl. Microbiol. 93 438–447 10.1046/j.1365-2672.2002.01709.x [DOI] [PubMed] [Google Scholar]
- Castro D., Romalde J. L., Vila J., Magariñosos B., Luque A., Borrego J. J. (1997). Intraspecific characterization of Vibrio tapetis strains by use of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling. Appl. Environ. Microbiol. 63 1449–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gómez A., Goulden E. F., Owens L, Høj L. (2010). Vibrio owensii sp. nov., isolated from cultured crustaceans in Australia. FEMS Microbiol. Lett. 302 175–181 10.1111/j.1574-6968.2009.01850.x [DOI] [PubMed] [Google Scholar]
- Su Y. C., Liu C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24 549–558 10.1016/j.fm.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Chimetto L. A., Cleenwerck I., Moreira A. P., Brocchi M., Willems A., de Vos P., et al. (2011). Vibrio variabilis sp. nov. and V. maritimus sp. nov., isolated from Palythoa caribaeorum. Int. J. Syst. Evol. Microbiol. 61 3009–3305 10.1099/ijs.0.026997-0 [DOI] [PubMed] [Google Scholar]
- Colwell R. R., Liston J. (1960). Microbiology of shellfish. Bacteriological study of the natural flora of Pacific oysters (Crassostraea gigas). Appl. Microbiol. 8 104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord A., Ceccarelli D., Rodrigue S., Burrus V. (2013). Comparative analysis of mobilizable genomic islands. J. Bacteriol. 195 606–614 10.1128/JB.01985-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diéguez A. L., Beaz-Hidalgo R., Cleenwerck I., Balboa S., De Vos P., Romalde J. L. (2011). Vibrio atlanticus sp. nov., and Vibrio artabrorum sp. nov. isolated from clam (Ruditapes decussatus and R. philippinarum). Int. J. Syst. Evol. Microbiol. 61 2406–2411 10.1099/ijs.0.025320-0 [DOI] [PubMed] [Google Scholar]
- Dikow R. B., Smith W. L. (2013). Genomelevel homology and phylogeny of Vibrionaceae (Gammaproteobacteria: Vibrionales) with three new complete genome sequences. BMC Microbiol. 13:80 10.1186/1471-2180-13-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiler A., Johansson M., Bertilsson S. (2006). Environmental influences on Vibrio populations in northern temperate and boreal coastal waters (Baltic and Skagerrak Seas). Appl. Environ. Microbiol. 72 6004–6011 10.1128/AEM.00917-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes R. M., Friedman C. S., Elston R. A., Herwig R P. (2004). Pathogenicity testing of shellfish hatchery bacterial isolates on Pacific oyster Crassostrea gigas larvae. Dis. Aquat. Org. 58 223–230 10.3354/dao058223 [DOI] [PubMed] [Google Scholar]
- Farto R., Armada S. P., Montes M., Guisande J. A., Pérez M. J., Nieto T. P. (2003). Vibrio lentus associated with diseased wild octopus (Octopus vulgaris). J. Invertebr. Pathol. 83 149–156 10.1016/S0022-2011(03)00067-3 [DOI] [PubMed] [Google Scholar]
- Farto R., Armada S. P., Montes M., Perez M. J., Nieto T. P. (2006). Presence of a lethal protease in the extracellular products of Vibrio splendidus-Vibrio lentus related strains. J. Fish Dis. 29 701–707 10.1111/j.1365-2761.2006.00746.x [DOI] [PubMed] [Google Scholar]
- Faury N., Saulnier D., Thompson F. L., Gay M., Swings J, Le Roux F. (2004). Vibrio crassostreae sp. nov., isolated from the haemolymph of oysters (Crassostrea gigas). Int. J. Syst. Evol. Microbiol. 54 2137–2140 10.1099/ijs.0.63232-0 [DOI] [PubMed] [Google Scholar]
- Fraser C., Alm E. J., Polz M. F., Spratt B. G., Hanage W. P. (2009). The bacterial species challenge: making sense of genetic and ecological diversity. Science 323 741–746 10.1126/science.1159388 [DOI] [PubMed] [Google Scholar]
- Frias-Lopez J., Shi Y., Tyson G. W., Coleman M. L., Schuster S. C., Chisholm S. W., et al. (2008). Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. U.S.A. 105 3805–3810 10.1073/pnas.0708897105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Amado M. A., Bozo-Hurtado L., Astor Y., Suárez P., Chistoserdov A. (2011). Denaturing gradient gel electrophoresis analyses of the vertical distribution and diversity of Vibrio spp. populations in the Cariaco Basin. FEMS Microbiol. Ecol. 77 347–356 10.1111/j.1574-6941.2011.01116.x [DOI] [PubMed] [Google Scholar]
- Garnier M., Labreuche Y., García C., Robert M., Nicolas J. L. (2007). Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacific oyster Crassostrea gigas. Microb. Ecol. 53 187–196 10.1007/s00248-006-9061-9 [DOI] [PubMed] [Google Scholar]
- Garnier M., Laubreche Y., Nicolas J. L. (2008). Molecular and phenotypic characterization of Vibrio aestuarianus subsp. francensis subsp. nov., a pathogen of the oyster Crassostrea gigas. Syst. Appl. Microbiol. 31 358–365 10.1016/j.syapm.2008.06.003 [DOI] [PubMed] [Google Scholar]
- Gay M., Berthe F. C. J, Le Roux F. (2004a). Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas. Dis. Aquat. Org. 59 49–56 10.3354/dao059049 [DOI] [PubMed] [Google Scholar]
- Gay M., Renault T., Pons A. M, Le Roux F. (2004b). Two Vibrio splendidus related strains collaborate to kill Crassostrea gigas: taxonomy and host alterations. Dis. Aquat. Org. 62 65–74 10.3354/dao062065 [DOI] [PubMed] [Google Scholar]
- Gevers D., Cohan F. M., Lawrence J. G., Spratt B. G., Coenye T., Feil E. J., et al. (2005). Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3 733–739 10.1038/nrmicro1236 [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Fajer-Avila E, García-Vargas F. (2007). Vibrios of the spotted rose snapper Lutjanus guttatus Steindachner, 1869 from northwestern Mexico. J. Appl. Microbiol. 102 1518–1526 10.1111/j.1365-2672.2006.03199.x [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Fajer-Avila E., Pascual J., Macián M. C., Pujalte M. J., Garay E., et al. (2008). Vibrio sinaloensis sp. nov., isolated from the spotted rose snapper, Lutjanus guttatus Steindachner, 1869. Int. J. Syst. Evol. Microbiol. 58 1621–1624 10.1099/ijs.0.65719-0 [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Roque A., Chimetto L., Moreira A. P., Lang E., Thompson F. L. (2012). Vibrio alfacsensis sp. nov., isolated from marine organisms. Int. J. Syst. Evol. Microbiol. 62 2955–2961 10.1099/ijs.0.033191-0 [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Roque A., Rotllant G., Peinado L., Romalde J. L., Doce A., et al. (2011). Photobacterium swingsii sp. nov., isolated from marine organisms. Int. J. Syst. Evol. Microbiol. 61 315–319 10.1099/ijs.0.019687-0 [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Soto-Rodríguez S., García-Gasca A., Roque A., Vázquez-Juárez A., Swings J. (2004a). Molecular identification of V. harveyi-related isolates associated with diseased aquatic organisms. Microbiology 150 1769–1777 10.1099/mic.0.26797-0 [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Thompson F. L., Thompson C. C., Garcia-Gasca A., Roque A., Swings J. (2004b). Vibrio hispanicus sp. nov., isolated from Artemia sp. and sea water in Spain. Int. J. Syst. Evol. Microbiol. 54 261–265 10.1099/ijs.0.02775-0 [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Thompson F. L., Thompson C. C., Swings J. (2003a). Vibrio rotiferianus sp. nov., isolated from cultures of the rotifer Brachionus plicatilis. Int. J. Syst. Evol. Microbiol. 53 239–243 10.1099/ijs.0.02430-0 [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Thompson F. L., Thompson C. C., Swings J. (2003b). Vibrio pacinii sp. nov., from cultured aquatic organisms. Int. J. Syst. Evol. Microbiol. 53 1569–1573 10.1099/ijs.0.02670-0 [DOI] [PubMed] [Google Scholar]
- Góomez-León J., Villamil L., Lemos M. L., Novoa B. (2005). Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl. Environ. Microbiol. 71 98–104 10.1128/AEM.71.1.98-104.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Castillo A., Balboa S., Romalde J. L., Gomez-Gil B. (2014). Vibrio crosaei sp. nov., isolated from a cultured oyster Crassostrea gigas. Int. J. Syst. Evol. Microbiol. (in press). [DOI] [PubMed] [Google Scholar]
- Grimes D. J., Johnson C. N., Dillon K. S., Flowers A. R., Noriea N. F., III, Berutti T. (2009). What genomic sequence information has revealed about Vibrio ecology in the ocean – a review. Microb. Ecol. 58 447–460 10.1007/s00248-009-9578-9 [DOI] [PubMed] [Google Scholar]
- Guisande J. A., Montes M., Farto R., Armada S. P., Pérez M. J., Nieto T. P. (2004). A set of tests for the phenotypic identification of culturable bacteria associated with Galician bivalve mollusk production. J. Shellfish Res. 23 599–610 [Google Scholar]
- Hariharan H., Giles J. S., Heaney S. B., Arsenault G., McNair N., Rainnie D. J. (1995). Bacteriological studies on mussels and oysters from six river systems in Prince Edward Island, Canada. J. Shellfish Res. 14 527–532 [Google Scholar]
- Hayashi K., Moriwaki J., Sawabe T., Thompson F. L., Swings J., Gudkovs N., et al. (2003). Vibrio superstes sp. nov., isolated from the gut of Australian abalones Haliotis laevigata and Haliotis rubra. Int. J. Syst. Evol. Microbiol. 53 1813–1817 10.1099/ijs.0.02625-0 [DOI] [PubMed] [Google Scholar]
- Hazen T. H., Pan L., Gu J.-D., Sobecky P. A. (2010). The contribution of mobile genetic elements to the evolution and ecology of vibrios. FEMS Microbiol. Ecol. 74 485–499 10.1111/j.1574-6941.2010.00937.x [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Monday S. R., Allard M. W., Strain E. A., Whittaker P., Naum M., et al. (2012). Vibrio caribbeanicus sp. nov., isolated fron the marine sponge Scleritoderma cyanea. Int. J. Syst. Evol. Microbiol. 62 1736–1743 10.1099/ijs.0.032375-0 [DOI] [PubMed] [Google Scholar]
- Jin C., Luo P., Zuo H., Chen J., Chen M., Wang W. (2012). Vibrio zhanjiangensis sp. nov., isolated from sea water of shrimp farming pond. Antonie Van Leeuwenhoek 101 743–751 10.1007/s10482-011-9688-5 [DOI] [PubMed] [Google Scholar]
- Jin C., Luo P., Zuo H., Chen J., Chen M., Wang W. (2013). Vibrio zhuhaiensis sp. nov., isolated from a Japanese prawn (Marsupenaeus japonicus). Antonie Van Leeuwenhoek 103 989–996 10.1007/s10482-013-9878-4 [DOI] [PubMed] [Google Scholar]
- Johnson C. N. (2013). Fitness factors in vibrios: a mini-review. Microb. Ecol. 65 826–851 10.1007/s00248-012-0168-x [DOI] [PubMed] [Google Scholar]
- Jones M. K., Oliver J. D. (2009). Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77 1723–1733 10.1128/IAI.01046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. K., Warner F., Oliver J. D. (2008). Survival of and in situ gene expression by Vibrio vulnificus at varying salinities in estuarine environments. Appl. Environ. Microbiol. 74 182–187 10.1128/AEM.02436-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar C. W., Tamplin M. L. (1993). Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl. Environ. Microbiol. 59 2425–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavita K., Mishra A., Jha B. (2013). Extracellular polymeric substances from two biofilm forming Vibrio species: characterization and applications. Carbohydr. Polym. 94 882–888 10.1016/j.carbpol.2013.02.010 [DOI] [PubMed] [Google Scholar]
- Kesarcodi-Watson A., Kaspar H., Lategan M. J., Gibson L. (2009a). Two pathogens of Greenshell mussel larvae, Perna canaliculus: Vibrio splendidus and a V. coralliilyticus/neptunius-like isolate. J. Fish Dis. 32 499–507 10.1111/j.1365-2761.2009.01006.x [DOI] [PubMed] [Google Scholar]
- Kesarcodi-Watson A., Kaspar H., Lategan M. J., Gibson L. F. (2009b). Challenge of New Zealand Greenshell mussel Perna canaliculus larvae using two Vibrio pathogens: a hatchery study. Dis. Aquat. Org. 86 15–20 10.3354/dao02100 [DOI] [PubMed] [Google Scholar]
- Kim D., Baik K. S., Hwang Y. S., Choi J. S., Kwon J., Seong C. N. (2013). Vibrio hemicentroti sp. nov., an alginate lyase producing bacterium, isolated from the gut microflora of sea urchin (Hemicentrotus pulcherrimus). Int. J. Syst. Evol. Microbiol. 63 3697–3703 10.1099/ijs.0.047951-0 [DOI] [PubMed] [Google Scholar]
- Kueh C. S. W., Chan K. Y. (1985). Bacteria in bivalve shellfish with special reference to the oyster. J. Appl. Bacteriol. 59 41–47 10.1111/j.1365-2672.1985.tb01773.x [DOI] [PubMed] [Google Scholar]
- Kushmaro A., Banin E., Loya Y., Stackebarndt E., Rosenberg E. (2001). Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int. J. Syst. Evol. Microbiol. 51 1383–1388 [DOI] [PubMed] [Google Scholar]
- Labreuche Y., Lambert C., Soudant P., Boulo V., Huvet A., Nicolas J. L. (2006a). Cellular and molecular hemocyte responses of the Pacific oyster, Crassostrea gigas, following bacterial infection with Vibrio aestuarianus strain 01/32. Microb. Infect. 8 2715–2724 10.1016/j.micinf.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Labreuche Y., Soudant P., Goncalves M., Lambert C., Nicolas J. L. (2006b). Effects of extracellular products from the pathogenic Vibrio aestuarianus strain 01/32 on lethality and cellular immune responses of the oyster Crassostrea gigas. Dev. Comp. Immunol. 30 367–379 10.1016/j.dci.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Lafisca A., Pereira C. S., Giaccone V, Rodriguez D. D. P. (2008). Enzymatic characterization of Vibrio alginolyticus strains isolated from bivalves harvested at Venice lagoon (Italy) and Guanabara Bay (Brazil). Rev. Inst. Med. Trop. S. Paulo 50 199–202 10.1590/S0036-46652008000400002 [DOI] [PubMed] [Google Scholar]
- Lambert C., Nicolas J. L., Cilia V., Corre S. (1998). Vibrio pectenicida sp. nov., a pathogen of scallop (Pecten maximus) larvae. Int. J. Syst. Bacteriol. 48 481–487 10.1099/00207713-48-2-481 [DOI] [PubMed] [Google Scholar]
- Lasa A., Diéguez A. L., Romalde J. L. (2013a). Vibrio toranzoniae sp. nov., a new member of the Splendidus clade in the genus Vibrio. Syst. Appl. Microbiol. 36 96–100 10.1016/j.syapm.2012.11.005 [DOI] [PubMed] [Google Scholar]
- Lasa A., Diéguez A. L., Romalde J. L. (2013b). Vibrio cortegadensis sp. nov., isolated from clams. Antonie van Leeuwenhoek. 10.1007/s10482-013-0078-z [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Lee K. K., Ping-Chung L., Huang C. Y. (2003). Vibrio parahaemolyticus infections for both humans and edible mollusc abalone. Microb. Infect. 5 481–485 10.1016/S1286-4579(03)00065-0 [DOI] [PubMed] [Google Scholar]
- Le Roux F., Austin B. (2006). “Vibrio splendidus,” in The Biology of Vibrios eds Thompson. F.L., Austin B., Swings J. (Washington: ASM Press; ) 285–296 [Google Scholar]
- Le Roux F., Gay M., Lambert C., Waechter M., Poubalanne S., Chollet B., et al. (2002). Comparative analysis of Vibrio splendidus related strains isolated during Crassostrea gigas mortality events. Aquat. Liv. Resour. 15 251–258 10.1016/S0990-7440(02)01176-2 [DOI] [Google Scholar]
- Le Roux F., Goubet A., Thompson F. L., Faury N., Gay M., Swings J., et al. (2005). Vibrio gigantis sp. nov., isolated from the haemolymph of cultured oysters (Crassostrea gigas). Int. J. Syst. Evol. Microbiol. 55 2251–2255 10.1099/ijs.0.63666-0 [DOI] [PubMed] [Google Scholar]
- Liu P. C., Chen C. Y., Huang C. Y., Lee K. K. (2000). Virulence of Vibrio parahaemolyticus isolated from cultured small abalone, Haliotis diversicolor supertexta, with withering syndrome. Lett. Appl. Microbiol. 31 433–437 10.1046/j.1365-2672.2000.00843.x [DOI] [PubMed] [Google Scholar]
- Lucena T., Ruvira M. A., Arahal D. R., Macián M. C., Pujalte M. J. (2012). Vibrio aestivus sp. nov. and Vibrio quintilis sp. nov., related to Marisflavi and Gazogenes clades, respectively. Syst. Appl. Microbiol. 35 427–431 10.1016/j.syapm.2012.08.002 [DOI] [PubMed] [Google Scholar]
- Macián M. C., Garay E., Grimont P. A. D., Pujalte M. J. (2004). Vibrio ponticus sp. nov., a neighbour of V. fluvialis-V. furnissii clade, isolated from gilthead seabream, mussels and seawater. Syst. Appl. Microbiol. 27 535–540 10.1078/0723202041748127 [DOI] [PubMed] [Google Scholar]
- Macián M. C., Ludwig W., Aznar R., Grimont P. A. D., Schleifer K. H., Garay E., et al. (2001a). Vibrio lentus sp. nov., isolated from Mediterranean oysters. Int. J. Syst. Evol. Microbiol. 51 1449–1456 [DOI] [PubMed] [Google Scholar]
- Macián M. C., Ludwig W., Schleifer K. H., Pujalte M. J., Garay E. (2001b). Vibrio agarivorans sp. nov. a novel agarolytic bacterium. Int. J. Syst. Evol. Microbiol. 51 2031–2036 10.1099/00207713-51-6-2031 [DOI] [PubMed] [Google Scholar]
- Maugeri T. L., Caccamo D., Gugliandolo C. (2000). Potentially pathogenic vibrios in brackish waters and mussels. J. Appl. Microbiol. 89 261–266 10.1046/j.1365-2672.2000.01096.x [DOI] [PubMed] [Google Scholar]
- Montes M., Farto R., Pérez M. J., Nieto T. P., Larsen J. L., Christensen H. (2003). Characterization of Vibrio strains isolated from turbot (Scophthalmus maximus) culture by phenotypic analysis, ribotyping and 16S rRNA gene sequence comparison. J. Appl. Microbiol. 95 693–703 10.1046/j.1365-2672.2003.02028.x [DOI] [PubMed] [Google Scholar]
- Montes M., Farto R., Pérez M. J., Armada S. P., Nieto T. P. (2006). Genotypic diversity of Vibrio isolates associated with turbot (Scophthalmus maximus) culture. Res. Microbiol. 157 487–495 10.1016/j.resmic.2005.11.008 [DOI] [PubMed] [Google Scholar]
- Montilla R., Palomar J., Santmarti M., Fuste C, Viñas M. (1994). Isolation and characterization of halophilic Vibrio from bivalves bred in nurseries at the Ebre Delta. J. Invertebr. Pathol. 63 178–181 10.1006/jipa.1994.1033 [DOI] [Google Scholar]
- Morris J. G. (2003). Cholera and other types of Vibriosis: a story of human pandemics and oysters on the half shell. Food Safety 37 272–280 10.1086/375600 [DOI] [PubMed] [Google Scholar]
- Morrison S. S., Williams T., Cain A., Froelich B., Taylor C., Baker-Austin C., et al. (2012). Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates. PLoS ONE 7:e37553 10.1371/journal.pone.0037553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motes M. L., De Paola A., Cook D. W., Veazey J. E., Hunsucker J. C., Garthright W. E., et al. (1998). Influence of water temperature and salinity on Vibrio vulnificus in Northen Gulf and Atlantic Coast Oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64 1459–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchelano R. A., Brown C. (1970). Heterotrophic bacteria in Long Island Sound. Mar. Biol. 7 1–6 10.1007/BF00346801 [DOI] [Google Scholar]
- Murray I. A., Clark T. A., Morgan R. D., Boitano M., Anton B. P., Luong K., et al. (2012). The methylomes of six bacteria. Nucleic Acids Res. 40 11450–11462 10.1093/nar/gks891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard C., Le Roux F., Borrego J. J. (2004). Bacterial disease in marine bivalves, a review of recent studies: trends and evolution. Aquat. Living Resour. 17 477–498 10.1051/alr:2004054 [DOI] [Google Scholar]
- Pascual J., Macián M. C., Arahal D. R., Garay E., Pujalte M. J. (2010). Multilocus sequence analysis of the central clade of the genus Vibrio using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Evol. Microbiol. 60 1844–1848 10.1099/ijs.0.010702-0 [DOI] [PubMed] [Google Scholar]
- Pillidge C. J., MacDonell M. T., Colwell R. R. (1987). Nucleotide sequence of the 5S rRNA from Listonella (Vibrio) aestuarianus ATCC 35048. Nucleic Acids Res. 15 1879 10.1093/nar/15.4.1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado S., Romalde J. L., Barja J. L., Toranzo A. E. (2014). Vibrio ostreicida sp. nov., a new pathogen for bivalve larvae. Int. J. Syst. Evol. Microbiol. (in press) [DOI] [PubMed] [Google Scholar]
- Prado S., Romalde J. L., Montes J., Barja J. L. (2005). Pathogenic bacteria isolated from disease outbreaks in shellfish hatcheries. First description of Vibrio neptunius as an oyster pathogen. Dis. Aquat. Org. 67 209–215 10.3354/dao067209 [DOI] [PubMed] [Google Scholar]
- Preheim S. P., Timberlake S., Polz M. F. (2011). Merging taxonomy with ecological population prediction in a case study of Vibrionaceae. Appl. Environ. Microbiol. 77 7195–7206 10.1128/AEM.00665-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieur D., Mével G., Nicolas J. L., Plusquellec A., Vigneulle M. (1990). Interactions between bivalve molluscs and bacteria in the marine environment. Oceanogr. Mar. Biol. Annu. Rev. 28 277–352 [Google Scholar]
- Pujalte M. J., Ortigosa M., Maciá>n M. C., Garay E. (1999). Aerobic and facultative anaerobic heterotrophic bacteria associated to Mediterranean oysters and seawater. Int. Microbiol. 2 259–266 [PubMed] [Google Scholar]
- Pujalte M. J., Ortigosa M., Urdaci M. C., Garay E, Grimont P. A. D. (1993). Vibrio mytili sp. nov., from mussels. Int. J. Syst. Bacteriol. 43 358–362 10.1099/00207713-43-2-358 [DOI] [Google Scholar]
- Reen F. J., Almagro-Moreno S., Ussery D., Boyd E. F. (2006). The genomic code inferring Vibrionaceae niche specialization. Nat. Rev. Microbiol. 9 697–704 10.1038/nrmicro1476 [DOI] [PubMed] [Google Scholar]
- Rivera-Posada J. A., Pratchett M., Cano-Gomez A., Arango-Gomez J. D., Owens L. (2011). Refined identification of Vibrio bacterial flora from Acanthasther planci based on biochemical profiling and analysis of housekeeping genes. Dis. Aquat. Org. 96 113–123 10.3354/dao02389 [DOI] [PubMed] [Google Scholar]
- Rodrigue S., Malmstrom R. R., Berlin A. M., Birren B. W., Henn M. R., Chisholm S. W. (2009). Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE 4:e6864 10.1371/journal.pone.0006864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez J. M., López-Romalde S., Beaz R., Alonso C., Castro D., Romalde J. L. (2006). Molecular fingerprinting of Vibrio tapetis strains using three PCR-based methods: ERIC-PCR, REP-PCR and RAPD. Dis. Aquat. Org. 69 175–183 10.3354/dao069175 [DOI] [PubMed] [Google Scholar]
- Romalde J. L., Castro D., Magariños B., López-Cortés L., Borrego J. J. (2002). Comparison of ribotyping, randomly amplified polymorphic DNA, and pulsed-field gel electrophoresis for molecular typing of Vibrio tapetis. Syst. Appl. Microbiol. 25 544–550 10.1078/07232020260517689 [DOI] [PubMed] [Google Scholar]
- Romalde J. L., Diéguez A. L., Doce A., Lasa A., Balboa S., López C., et al. (2013). “Advances in the knowledge of the microbiota associated with clams from natural beds”, in Clam Fisheries and Aquaculture ed. Da Costa F. (New York: Nova Science Publishers; ) 163–190 [Google Scholar]
- Sawabe T., Fujimura Y., Niwa K., Aono H. (2007a). Vibrio comitans sp. nov., Vibrio rarus sp. nov. and Vibrio inusitatus sp. nov., from the gut of the abalones Haliotis discus discus, H. gigantea, H. madaka and H. rufescens. Int. J. Syst. Evol. Microbiol. 57 916–922 10.1099/ijs.0.64789-0 [DOI] [PubMed] [Google Scholar]
- Sawabe T., Kita-Tsukamoto K., Thompson F. L. (2007b). Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 189 7932–7936 10.1128/JB.00693-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawabe T., Hayashi K., Moriwaki J., Fukui Y., Thompson F. L., Swings J., et al. (2004a). Vibrio neonatus sp. nov and Vibrio ezurae sp. nov., isolated from the gut of Japanese abalones. Syst. Appl. Microbiol. 27 527–534 10.1078/0723202041748154 [DOI] [PubMed] [Google Scholar]
- Sawabe T., Hayashi K., Thompson F. L., Swings J., Potin P., Christen R., et al. (2004b). Vibrio gallicus sp. nov., isolated from the gut of the French abalone Haliotis tuberculata. Int. J. Syst. Evol. Microbiol. 54 843–846 10.1099/ijs.0.02804-0 [DOI] [PubMed] [Google Scholar]
- Sheu S. Y., Jiang S. R., Chen C. A., Wang J. T., Chen W. M. (2011). Vibrio stylophorae sp. nov., isolated from the reef-building coral Stylophora pistillata. Int. J. Syst. Evol. Microbiol. 61 2180–2185 10.1099/ijs.0.026666-0 [DOI] [PubMed] [Google Scholar]
- Shieh W. Y., Chen Y. W., Chaw S. M., Chiu H. H. (2003). Vibrio ruber sp. nov., a red, facultatively anaerobic, marine bacterium isolated from sea water. Int. J. Syst. Evol. Microbiol. 53 479–484 10.1099/ijs.0.02307-0 [DOI] [PubMed] [Google Scholar]
- Stepanauskas R., Sieracki M. E. (2007). Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc. Natl. Acad. Sci. U.S.A. 104 9052–9057 10.1073/pnas.0700496104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall A., Hervio-Heath D., Teillon A., Boisset-Helbert C., Delesmont R., Bodilis J., et al. (2013). Diversity of Vibrio spp. isolated at ambient environmental temperature in the Eastern English Channel as determined by pyrH sequencing. J. Appl. Microbiol. 114 1713–1724 10.1111/jam.12181 [DOI] [PubMed] [Google Scholar]
- Thompson F. L., Gevers D., Thompson C. C., Dawyndt P., Naser S., Hoste B., et al. (2005). Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71 5107–5115 10.1128/AEM.71.9.5107-5115.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson F. L., Gómez-Gill B., Ribeiro Vasconcelos A. T., Sawabe T. (2007). Multilocus sequence analysis reveals that Vibrio harveyi and V. campbelli are distinct species. Appl. Environ. Microbiol. 73 4279–4285 10.1128/AEM.00020-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson F. L., Hoste B., Vandeneulebroecke K., Swings J. (2001). Genomic diversity amongst Vibrio isolates from different source determined by fluorescent amplified fragment length polymorphism. Syst. Appl. Microbiol. 24 520–538 10.1078/0723-2020-00067 [DOI] [PubMed] [Google Scholar]
- Thompson F. L., Li Y., Gomez-Gil B., Thompson C. C., Hoste B., Vandemeulebroecke K., et al. (2003a). Vibrio neptunius sp. nov., Vibrio brasiliensis sp. nov. and Vibrio xuii sp. nov., isolated from the marine aquaculture environment (bivalves, fish, rotifers and shrimps). Int. J. Syst. Evol. Microbiol. 53 245–252 10.1099/ijs.0.02447-0 [DOI] [PubMed] [Google Scholar]
- Thompson F. L., Thompson C. C., Hoste B., Vandemeulebroecke K., Gullian M., Swings J. (2003b). Vibrio fortis sp. nov. and Vibrio hepatarius sp. nov. isolated from aquatic animals and the marine environment. Int. J. Syst. Evol. Microbiol. 53 1495–1501 10.1099/ijs.0.02658-0 [DOI] [PubMed] [Google Scholar]
- Thompson F. L., Thompson C. C., Li Y., Gómez-Gil B., Vanderberghe J., Hoste B., et al. (2003c). Vibrio kanaloae sp. nov., Vibrio pomeroyi sp. nov. and Vibrio chagasii sp. nov., from sea water and marine animals. Int. J. Syst. Evol. Microbiol. 53 753–759 10.1099/ijs.0.02490-0 [DOI] [PubMed] [Google Scholar]
- Thompson F. L., Thompson C. C., Swings J. (2003d). Vibrio tasmaniensis sp. nov., isolated from Atlantic salmon (Salmo salar, L.). Syst. Appl. Microbiol. 26 65–69 10.1078/072320203322337326 [DOI] [PubMed] [Google Scholar]
- Tison D. L., Seidler R. J. (1983). Vibrio aestuarianus: a new species from estuarine waters and shellfish. Int. J. Syst. Bacteriol. 33 699–702 10.1099/00207713-33-4-699 [DOI] [Google Scholar]
- Urbanczyk H., Ast J. C., Higgins M. J., Carson J., Dunlap P. V. (2007). Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int. J. Syst. Evol. Microbiol. 57 2823–2829 10.1099/ijs.0.65081-0 [DOI] [PubMed] [Google Scholar]
- Waechter M., Le Roux F., Nicolas J. L., Marissal E., Berthe F. (2002). Characterization of pathogenic bacteria of the cupped oyster Crassostrea gigas. C. R. Biol. 325 231 10.1016/S1631-0691(02)01428-2 [DOI] [PubMed] [Google Scholar]
- Wang Y. L., Leung P. C., Qian P. Y., Gu J. D. (2006). Antibiotic resistance and plasmid profile of environmental isolates of Vibrio species from Mai Po Nature Reserve, Hong Kong. Ecotoxicology 15 371–378 10.1007/s10646-006-0078-0 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang X.-H., Yu M., Wang H., Austin B. (2010). Vibrio atypicus sp. nov.isolated from the digestive tract of the Chinese prawn (Penaeus chinensis O’sbeck). Int. J. Syst. Evol. Microbiol. 60 2517–2523 10.1099/ijs.0.016915-0 [DOI] [PubMed] [Google Scholar]
- Wietz M., Mansson M., Gotfredsen C. H., Larsen T. O., Gram L. (2010). Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar. Drugs 8 2946–2960 10.3390/md8122946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S., Wada M., Kita-Tsukamoto K., Ikemoto E., Yokota A., Kogure K. (2009). Vibrio azureus sp. nov., a luminous marine bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 59 1645–1649 10.1099/ijs.0.004283-0 [DOI] [PubMed] [Google Scholar]
- Yoshizawa S., Wada M., Yokota A., Kogure K. (2010). Vibrio sagamiensis sp. nov., luminous marine bacteria isolated from sea water. J. Gen. Appl. Microbiol. 56 499–507 10.2323/jgam.56.499 [DOI] [PubMed] [Google Scholar]
- Yoshizawa S., Tsuruya Y., Sawabe T., Yokota A., Kogure K., Higgins M., et al. (2012). Vibrio jasicida sp. nov., a member of the Harveyi clade, isolated from marine animals (packhorse lobster, abalone and Atlantic salmon). Int. J. Syst. Evol. Microbiol. 62 1864–1870 10.1099/ijs.0.025916-0 [DOI] [PubMed] [Google Scholar]