Abstract
LuxS-mediated quorum sensing has recently been shown to regulate important physiologic functions and virulence in a variety of bacteria. In this study, the role of luxS of Streptococcus mutans in the regulation of traits crucial to pathogenesis was investigated. Reporter gene fusions showed that inactivation of luxS resulted in a down-regulation of fructanase, a demonstrated virulence determinant, by more than 50%. The LuxS-deficient strain (TW26) showed increased sensitivity to acid killing but could still undergo acid adaptation. Northern hybridization revealed that the expression of RecA, SmnA (AP endonuclease), and Nth (endonuclease) were down-regulated in TW26, especially in early-exponential-phase cells. Other down-regulated genes included ffh (a signal recognition particle subunit) and brpA (biofilm regulatory protein A). Interestingly, the luxS mutant showed an increase in survival rate in the presence of hydrogen peroxide (58.8 mM). The luxS mutant formed less biofilm on hydroxylapatite disks, especially when grown in biofilm medium with sucrose, and the mutant biofilms appeared loose and hive-like, whereas the biofilms of the wild type were smooth and confluent. The mutant phenotypes were complemented by exposure to supernatants from wild-type cultures. Two loci, smu486 and smu487, were identified and predicted to encode a histidine kinase and a response regulator. The phenotypes of the smu486 smu487 mutant were, in almost all cases, similar to those of the luxS mutant, although our results suggest that this is not due to AI-2 signal transduction via Smu486 and Smu487. This study demonstrates that luxS-dependent signaling plays critical roles in modulating key virulence properties of S. mutans.
Streptococcus mutans is recognized as the principal etiological agent of dental caries, one of the most prevalent human infectious diseases. The abilities to adhere to and to form biofilms on the tooth surface, to metabolize carbohydrates, and to survive low pH and other environmental insults are believed to be critical to the cariogenicity of this human pathogen (4). Known for its high degree of acid tolerance (aciduricity) and its high capacity to produce acids (acidogenicity), S. mutans has evolved strategies to survive, grow, and carry out glycolysis at extracellular pH values well below those at which demineralization of tooth enamel begins to take place. As an obligate biofilm-forming bacterium (4), S. mutans lives almost exclusively on the tooth surface at high cell density in a high-diversity ecosystem better known as dental plaque. The structure and composition of the plaque are known to be strongly influenced by such factors as the source and availability of nutrients, the pH in the oral cavity, and the ability of its constituents to adapt to fluctuations in environmental conditions (4, 38).
In a process termed quorum sensing (QS), which was first discovered in the marine bacterium Vibrio fischeri, bacterial populations coordinate gene expression and, thus, the behavior of the community. It is now apparent that both gram-positive and gram-negative bacteria, including pathogens and nonpathogens, possess cell-cell communication mechanisms, and QS systems have evolved as a means to improve access to complex nutrients or environmental niches or for collectively enhancing defense capabilities against other microorganisms or eukaryotic host defense mechanisms (14, 21, 32, 33). Quorum-sensing bacteria convey their presence to one another by releasing and responding to the accumulation of low-molecular-weight chemical signal molecules called autoinducers. In general, gram-negative bacteria use acylated homoserine lactones, which are synthesized by the LuxI type of autoinducer synthases. LuxR-like transcriptional regulators then bind their cognate autoinducers and activate transcription of target genes. In contrast, gram-positive bacteria generally communicate with processed oligopeptides, and the signals are detected via two-component signal transduction systems.
Recent studies of Vibrio harveyi have uncovered a new family of autoinducer synthases, called LuxS (46), which in V. harveyi is responsible for production of the signal molecule autoinducer 2 (AI-2), a furanosyl borate diester (6). Unlike other gram-negative bacteria, V. harveyi produces both AI-2 and acylated homoserine lactones (AI-1), and two QS systems mediated by AI-1 and AI-2 function in parallel to regulate the expression of the luciferase operon (luxCDABE). AI-1 and AI-2 are detected by their respective sensory protein(s), but the signals are then transmitted through a shared phospho-relay cascade reminiscent of QS systems of gram-positive bacteria (40). Proposed as a universal language for interspecies communication (32, 33), AI-2-like activity and the conserved LuxS homologues are found in a wide range of bacterial species (11, 28, 30, 35, 45, 46, 48). Accumulating evidence shows that luxS and AI-2 are involved in global regulation of physiologic functions and virulence, although the actual mechanisms of signal transduction and gene regulation remain to be studied (10-12, 28, 30, 35, 42, 43). In addition to cell density, the production of AI-2 in Escherichia coli was found to be regulated by environmental parameters, such as the availability of preferred carbon sources (9, 46) and acid stresses (20, 44), suggesting that the AI-2-mediated QS is linked to nutritional limitation and stress responses. In fact, luxS of E. coli was found to influence the expression of over 400 genes, including those needed for DNA processing, biofilm formation, carbohydrate metabolism, surface- and outer membrane-associated functions, and virulence (10, 42, 43).
Recent studies of oral bacteria have provided evidence that luxS is involved in interspecies signal responses among oral bacteria and consequently may play important roles in the development of virulence and dental biofilms (3, 8, 11, 12, 30, 48). In Actinobacillus actinomycetemcomitans, AI-2 was shown to be involved in leukotoxin production and iron acquisition, and the addition of cell-free conditioned media of E. coli carrying a plasmid-borne copy of the A. actinomycetemcomitans luxS gene complemented a Porphyromonas gingivalis luxS mutant, as monitored by modulating the expression of the luxS-controlled genes uvrB and hasF (11, 12). The luxS gene of Streptococcus gordonii appeared to be essential for the development of mixed-species biofilms with strains of P. gingivalis lacking luxS (30). Recently, Merritt et al. also reported that luxS-deficient S. mutans strains formed granular biofilms on glass coverslips, while the wild-type biofilms appear smooth and confluent as revealed by phase-contrast microscopy (31).
In this study, we used functional assays, Northern hybridization, reporter gene fusions, scanning electron microscopy (SEM), and proteomic analysis to begin to characterize the role of luxS of S. mutans in the regulation of traits crucial to the cariogenicity of this human pathogen. The data presented here demonstrate that luxS of S. mutans is involved in the regulation of stress tolerance and biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains were grown in Luria-Bertani medium, and when necessary, ampicillin (100 μg/ml), erythromycin (500 μg/ml), kanamycin (20 μg/ml), and/or chloramphenicol (20 μg/ml) were included. S. mutans UA159 and its derivatives were maintained in brain heart infusion (BHI) medium, with or without addition of erythromycin (10 μg/ml) or kanamycin (1 mg/ml). Preparation of competent cells and transformation of S. mutans were done as described previously (5). For enzyme assays and growth studies, S. mutans strains were grown in tryptone-vitamin (TV) base medium with glucose (TVG) or inulin (TVI) (from Dahlia tubers; Sigma) at 0.5% (wt/vol) as the carbohydrate source (5). For AI-2 bioassays, V. harveyi BB170 (luxN::Tn5 AI-1 sensor− AI-2 sensor+) (45) was grown in AB medium (15). For biofilm assays, semidefined biofilm medium (BM) (27) was used with glucose (0.8%, wt/vol) (BMG) or sucrose (0.5%, wt/vol) (BMS) as the supplemental carbohydrate source.
DNA manipulation and construction of reporter gene fusions.
Unless stated otherwise, standard recombinant DNA procedures were used (5, 39). To examine the role of luxS in the regulation of known virulence factors, the promoter regions of the respective genes were amplified by PCRs with gene-specific primers (Table 1) and directly cloned in plasmid pU1 (5) in front of a promoterless chloramphenicol acetyltransferase (cat) gene. After confirmation that the sequences were faithfully amplified, the promoter-cat fusions were released by enzymatic digestions, treated with T4 DNA polymerase to blunt the ends, and then inserted at the SmaI site of integration vector pBGK2, which is a modified version of pBGK (47) generated by deletion of the SspI/ScaI fragment of pBR322 to remove a large portion of the beta-lactamase gene. The resulting constructs were integrated into the chromosome of S. mutans strains UA159 and TW26, an isogenic luxS-deficient mutant (48). Integration of the reporter fusions via double-crossover homologous recombination was confirmed by PCR.
TABLE 1.
Primers used for reporter gene fusions and mutagenesis
| Primer | DNA sequence (5′ → 3′) with incorporated restriction site underlined | Application | Accession no. |
|---|---|---|---|
| spaP5′ | ATGGATCCAGCAAAAATTGACAAATCAAATC | spaP promoter | X17390 |
| spaP3′ | TTCTGCAGCATAAATCCTCCAAATCTGAATAAATC | ||
| gbpB5′ | TAGGATCCTATGAAAGGCGATGTTAAAGC | gbpB promoter | AY046410 |
| gbpB3′ | TTCTGCAGATATAACTCCTTTTTCGATAAGAC | ||
| gtfB5′ | TAGGATCCGACAATTGTGGTGGGTAC | gtfB promoter | M17361 |
| gtfB3′ | TACTGCAGCTTGTCCATTAGGAACCTCC | ||
| gtfC5′ | ATGGATCCGATGCTAACTCTGGAGAACG | gtfC promoter | M17361 |
| gtfC3′ | ATCTGCAGTCCAAAAATAGTTAGAGTTAGTG | ||
| ftf5′ | GGGGATCCTAGCTAGTGGACAGACTCTG | ftf promoter | M18954 |
| ftf3′ | AACTGCAGTTTCCATTAGCAAACCTCC | ||
| fruA5′ | TTGGATCCGAGCATTAATGACATCTGTCATATTAAG | fruA promoter | U78296 |
| fruA3′ | ATCTGCAGTTTTCAAATTTATGAAACTGACAAACTC | ||
| up5′ | CAACATCAAGCGAAGCCTCGAC | 5′ region of smu486 | AE014133 |
| up3′ | TTGAGCTCCATATACGTTCTACCTCCACC | ||
| down5′ | TTAAGCTTCCTGTCATTGAAGCAGGTGC | 3′ region of smu487 | AE014133 |
| down3′ | GAATCAGGCATATAACCCGTATC | ||
| pluxS5′ | AAGGATCCGCATAACCAATAACCAAACC | luxS promoter | AE014133 |
| pluxS3′ | TCCTGCAGAAAGCTTTCAACAGTAACTTC |
Northern blot analysis.
For Northern blot analysis, S. mutans UA159 or its derivatives were grown in BHI medium with the inclusion of antibiotics as needed, and total RNAs were isolated from cultures at early (optical density at 600 nm [OD600] ≅ 0.25) or late (OD600 ≅ 0.5) exponential phase by using the procedure previously described (5). Total RNAs were denatured and transferred to nylon membranes by using standard procedures (39). For probe generation, internal regions of the genes of interest were amplified by PCR with gene-specific primers, and the amplified fragments were confirmed for sequence accuracy by DNA sequencing. Probes were labeled with [α-32P]dATP by using a random primer DNA labeling system (Invitrogen, Inc. Carlsbad, Calif.). Hybridization signals were then quantified by using an IS1000 digital imaging system (Alpha Innotech, San Leandro, Calif.).
Isolation of smu486 smu487 mutants.
The smu486 and smu487 loci were identified by using luxR of V. harveyi to search the genome database of S. mutans (www.genome.ou.edu/smutans.html). A deletion mutant was generated by replacing the entire smu486 gene and a large portion of smu487, from the translation initiation site (TIS) of smu486 to nucleotide 285 of the coding sequence of smu487, with an erythromycin resistance determinant (erm) by using a PCR-ligation-mutation strategy (22). The 5′ fragment of 1,193 bp (from nucleotide 1190 upstream to the TIS of smu486) and the 3′ fragment of 506 bp (from nucleotide 285 to nucleotide 791 downstream of the TIS of smu487) were amplified by separate PCRs with primers listed in Table 1. The amplified fragments were digested with SstI for the 5′ fragment and HindIII for the 3′ fragment and then ligated to erm that was generated with HindIII and SstI enzymatic digestions from pUC:Emr (48). The resulting ligation mixture was transformed directly into S. mutans UA159, and transformants were selected on BHI agar containing 10 μg of erythromycin/ml. Deletion of the smu486/7 loci and replacement with erm as a result of double-crossover recombination were confirmed by PCR and Southern blot analysis.
Acid and hydrogen peroxide killing.
The ability of cells to withstand acid challenge was determined by the method of Belli and Marquis (1). Briefly, S. mutans strains were grown in BHI medium adjusted to pH 7.0 with HCl. Cultures were harvested (at OD600 ≅ 0.3) by centrifugation at 3,800 × g at 4°C for 10 min, washed once with 0.1 M glycine buffer, pH 7.0, and then subjected to killing by incubating the cells in 0.1 M glycine buffer, pH 2.8, for 0, 20, 30, or 45 min. For preparation of cells that had undergone an adaptive acid tolerance response, cultures with an OD600 of ∼0.2 were washed once with 0.1 M glycine buffer, pH 7.0, as described above, and the cell pellets were resuspended in fresh BHI medium that was adjusted to pH 5.0 with HCl. Following two additional hours of incubation, cells were harvested, washed, and subjected to acid killing as described above. The surviving cells were appropriately diluted, plated on BHI plates in triplicate, and incubated in 5% CO2 at 37°C for 24 to 48 h. For hydrogen peroxide challenge, BHI-grown cultures were collected at an OD600 of ∼0.3 and harvested by centrifugation at 4°C at 3,800 × g for 10 min, and the cell pellets were resuspended in 1/5 the original volume in 0.1 M glycine buffer, pH 7.0, containing 0.2% (vol/vol) (58.8 mM final concentration) hydrogen peroxide (H2O2; Fisher Scientific, Fair Lawn, N.J.). Aliquots were collected before and after addition of H2O2 at time points that were experimentally determined to yield appropriate killing. To inactivate hydrogen peroxide, catalase (5 mg/ml; Sigma) was added to samples immediately after collection. As described above, the survival rate was determined by plating in triplicate on BHI plates.
2D gel electrophoresis.
For two-dimensional (2D) gel electrophoretic analysis, cultures of S. mutans UA159 and its otherwise isogenic luxS mutant were grown in BHI medium to an OD600 of ∼0.5, harvested by centrifugation, washed, and homogenized using Bead Beater (Biospec, Bartlesville, Okla.) as previously described (7). Cell extracts were prepared and shipped on dry ice to Kendrick Labs, Inc. (Madison, Wis.), where 200 μg of total protein was subjected to 2D electrophoresis using the method of O'Farrell (34). Briefly, the first-dimension isoelectric focusing was carried out using 2% ampholines, pH 4 to 8 (Gallard-Schlesinger, Garden City, N.Y.), and the proteins were focused for 9,600 V · h (13.75 h at 700 V). The second-dimension electrophoresis was carried out using a 10% sodium dodecyl sulfate-polyacrylamide gel. Gels were stained with Coomassie blue, dried between sheets of cellophane, and digitally scanned, and the protein spots were analyzed by densitometry using software from Alpha Innotech.
Assays.
For assay of chloramphenicol acetyltransferase (CAT) activity, S. mutans strains carrying the reporter gene fusions were grown in TV medium with 0.5% (wt/vol) of either glucose or inulin. Cell extracts were prepared from mid-exponential-phase (OD600 ≅ 0.25) or late-exponential-phase (OD600 ≅ 0.5) cultures, as previously described (7). CAT activity was measured by using the method of Shaw (41). For AI-2 bioassays, culture supernatants of S. mutans UA159 and TW26 grown in TV medium with 0.5% (wt/vol) glucose were prepared by centrifugation and passed through a 0.2-μm syringe filter (Gelman Laboratory) to remove cells. The pH was adjusted to pH 7.0 with KOH. The AI-2 bioassay was carried out using the method of Surette and Bassler (45). Briefly, V. harveyi BB170 was grown at 30°C with aeration in AB medium (15) and diluted 1:5,000 with fresh AB medium, and 1.8 ml of the diluted cultures was added to culture tubes containing 0.2 ml of the cell-free supernatants. Activation of light production was measured with a model TD2020 luminometer. For preconditioned medium assay, cultures grown overnight in BHI medium were diluted by 1:100 into prewarmed TVG medium and allowed to grow until mid-exponential phase (OD600 ≅ 0.4). The cell-free culture supernatants, which were prepared as described above, were corrected with concentrated TV or BM (42, 45). Specifically, 2.5 ml of 35% (wt/vol) tryptone, 0.2 ml of vitamins, and 1.25 ml of 10% (wt/vol) glucose or inulin for preconditioned TVG and TVI media, respectively, were added to 46.05 ml of cell-free culture supernatants (5). For preconditioned BMG and BMS, 5 ml of 10× concentrated BM base medium, 250 μl of 10% MgCl2, 250 μl of 0.3% CaCl2, 0.5 ml of vitamins, 0.5 ml of amino acids, and 1 ml of 1 M glucose or sucrose were added to 42.5 ml of cell-free culture supernatants (27). These preconditioned media were used to grow the luxS-deficient strain TW26, and they were evaluated for their capacity to restore the LuxS-regulated functions or gene expression by using the methods described above.
SEM.
For SEM, biofilms were grown on hydroxylapatite (HA) disks that were deposited in the wells of 24-well culture clusters (Corning Inc., Corning, N.Y.). Briefly, overnight S. mutans cultures grown in BHI medium were diluted 1:100 in BMG or BMS, and 2-ml portions of the diluted cultures were added to each well. Following 24-h incubations at 37°C in a 5% CO2 atmosphere, the HA disks were carefully rinsed with phosphate-buffered saline buffer (50 mM Na2PO4, pH 7.0, 0.85% NaCl) and fixed with Trump fixative solution overnight. Following dehydration through a series of ethanol rinses, the disks were mounted and coated with gold, and the biofilms on the HA disks were then analyzed by SEM in the University of Florida EM core facility laboratory.
RESULTS
AI-2 activity is maximal at the mid-exponential phase of growth.
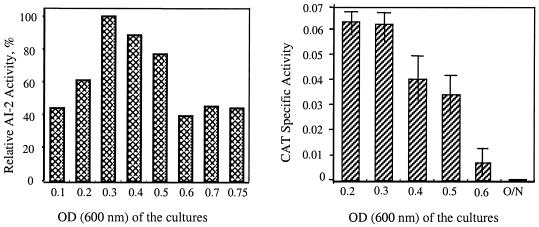
The reporter strain V. harveyi BB170 induces light production exclusively through AI-2 detection. To examine whether S. mutans UA159 and the luxS-deficient mutant produced any AI-2 activity, cell-free supernatants from strains UA159 and TW26 were prepared. The cell-free supernatants of strain UA159 were able to induce light production, and the activity was maximal in the mid-exponential phase of growth (Fig. 1). However, such stimulatory activity was not detected in cell-free supernatants prepared from strain TW26 (data not shown). In support of this bioassay, reporter gene activity under the direction of the luxS promoter (48) was maximal in cells during the early to mid-exponential phases of growth.
FIG. 1.
AI-2 bioassay (left panel) and reporter gene fusion (right panel). For AI-2 bioassays, conditioned cell-free supernatants of S. mutans UA159 grown in TV medium with 0.5% glucose were examined for the capacity to induce light production in V. harveyi BB170 (AI-1 sensor− AI-2 sensor+) by using the method of Surette and Bassler (45). Data are presented as the percentages of AI-2 activity at different growth phases, with the highest activity observed assigned a value of 100%. For reporter gene fusion, the promoter region of luxS was fused with a promoterless chloramphenicol acetyltransferase (cat) gene, and the fusion was integrated into the chromosome by double-crossover recombination using the pBGK2 integration vector. Expression of luxS was then analyzed by CAT assays with cells grown in TV medium (5) plus 0.5% (wt/vol) glucose, and CAT activity was expressed as nanomoles per minute per milligram of protein. O/N, overnight.
Expression of known virulence genes in the luxS-deficient mutant.
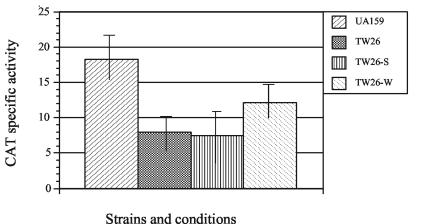
The impact of LuxS on expression of the known virulence factors glycosyltransferase BC (GtfBC), fructosyltransferase (Ftf), glucan-binding protein (GbpB), surface antigen SpaP, and fructanase (FruA) was analyzed by reporter gene fusions. The expression of fruA is induced by its substrates and subject to carbon catabolite repression (5). As expected, the highest CAT activity of cells carrying the PfruA-cat was detected when grown in TV medium with inulin as the sole carbohydrate source. In the luxS-deficient strain, CAT activity was decreased by more than 50% when cells were grown in TVI medium (Fig. 2) and was not detectable when grown in TVG medium (data not shown). The inclusion of cell-free supernatants from wild-type UA159 increased the expression of fruA, albeit not to the level of the wild type because of the need to dilute the spent medium, but cell-free supernatants from the luxS-deficient strain TW26 had no effect (Fig. 2). No significant differences were observed with gftB, gtfC, gbpB, and spaP promoter-reporter gene fusions in the luxS mutant (data not shown). In addition, reporter gene activities were measured in 24-h biofilms that were grown in 24-well polystyrene culture clusters (Corning Inc.) in BM medium with either glucose or sucrose as the supplemental sugar. Again, no major differences were detected with the gftB, gtfC, gbpB, and spaP reporter gene fusions between the wild-type strain and the luxS mutant (data not shown).
FIG. 2.
Effect of LuxS on fruA expression. The expression of cat under the direction of the fruA promoter in wild-type UA159 and the luxS-deficient mutant TW26 were analyzed by CAT assay. All strains were grown in TVI medium (5) with or without inclusion of cell-free supernatants prepared from UA159 (W) and TW26 (S) and harvested for CAT assays when the cells reached the mid-exponential phase of growth (OD600 ≅ 0.3). CAT activity is expressed as nanomoles per minute per milligram of protein.
Acid and oxidative stress tolerance of the luxS-deficient mutant.
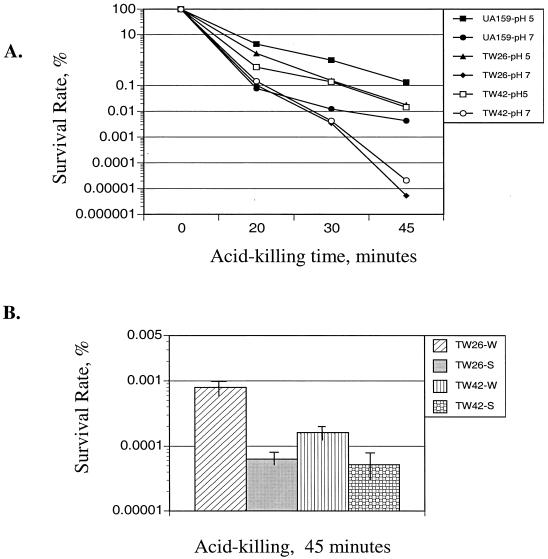
When grown in BHI medium that had been adjusted to pH 5.0 with HCl, the luxS mutant strain displayed a prolonged lag phase and a decreased final OD compared to those of the wild-type strain UA159 (data not shown), indicative of defects in acid tolerance arising from luxS deficiency. After 45 min of incubation at pH 2.8, the survival rate of the strain TW26 was more than 2 logs lower than that of strain UA159 (Fig. 3A). Incubation of the cells in the preconditioned medium from strain UA159 partially restored the resistance of strain TW26 to acid killing, whereas no effect was seen when preconditioned medium from strain TW26 was used (Fig. 3B).
FIG. 3.
Acid tolerance assay. (A) S. mutans strains UA159, TW26, and TW42 were grown in BHI medium adjusted to pH 7.0. Cultures with an OD600 of ∼0.3 were harvested, washed with 0.1 M glycine buffer, pH 7.0, and subjected to acid killing by incubating the cells in 0.1 M glycine buffer, pH 2.8. For preadapted acid killing, cultures with an OD600 of ∼0.2 were harvested and resuspended in fresh BHI medium adjusted to pH 5.0. Following two additional hours of incubation, cells with an OD600 of ∼0.3 were prepared for acid killing as described above. Survival rate was determined by plating in triplicate on BHI agar plates, and results are expressed as percent survival rate versus time at pH 2.8. (B) Cell-free supernatants from UA159 (W) and TW26 (S) were reconstituted with concentrated TV medium to 0.5×, and their impact on acid tolerance was measured by incubating the cells in 0.1 M glycine buffer, pH 2.8, for 45 min as described above.
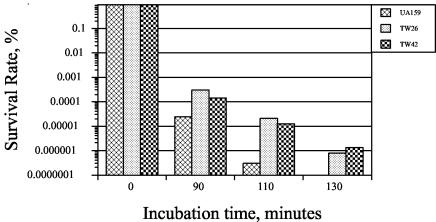
S. mutans is known to adapt to a low pH environment and mount an acid tolerance response (1, 18, 36, 37), as manifested by an increased resistance to acid killing and an elevated capacity to grow and metabolize carbohydrates at lower pH values after initial exposure to mildly acidic conditions. Following 2 h of preincubation in BHI medium that was adjusted to pH 5.0 with HCl to allow adaptation, the survival rate of strain TW26 was elevated in a proportion similar to that of strain UA159, but again the survival rate was lower than that of the acid-adapted UA159 by approximately 10-fold. Surprisingly, compared to UA159, strain TW26 exhibited an enhanced resistance to killing by 0.2% hydrogen peroxide. The survival rate of TW26 was more than 2 logs higher after 110 min of incubation in 0.2% hydrogen peroxide-containing glycine buffer, pH 7.0 (Fig. 4). The adaptation to acid by preincubation for 2 h of either strain in BHI medium that has been adjusted to pH 5.0 did not alter the sensitivity to hydrogen peroxide (data not shown).
FIG. 4.
Sensitivity of S. mutans to hydrogen peroxide. S. mutans strains UA159, TW26, and TW42 were grown to an OD600 of ∼0.3 in BHI medium that was adjusted to pH 7.0. Cells were harvested by centrifugation and resuspended in 0.1 M glycine buffer, pH 7, containing 0.2% (vol/vol) (58.8 mM final concentration) hydrogen peroxide. Surviving cells were plated on BHI plates, and the results were expressed as percent survival versus time.
Inactivation of luxS caused defects in biofilm development.
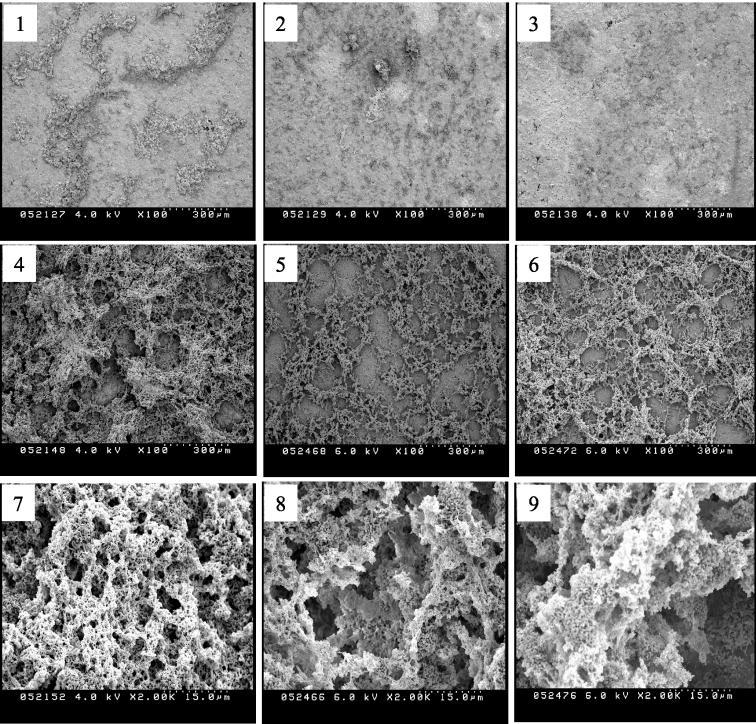
It was previously demonstrated that deficiency of luxS only slightly decreased the formation of biofilms on a polystyrene surface, probably due to defects in cell-cell interaction rather than initial adherence (48). In this study, we used HA disks, a common in vitro model for tooth enamel, as a support matrix to grow biofilms. Compared to UA159, TW26 was found by SEM to have a decreased capacity to form biofilms on HA disks. When grown in BM medium with glucose as the supplemental sugar after 24 h, TW26 formed sporadic microcolonies on HA disks but had less accumulation of biofilms than did UA159 (Fig. 5). The diminished capacity of the mutant to form biofilms was even more evident when sucrose was provided as a supplemental sugar. Preconditioned medium from strain UA159 enhanced the capacity of the mutant strain to form biofilms (Fig. 6). In addition, the architecture of biofilms formed by the luxS mutant appeared to be altered compared to that of the wild type. When grown in sucrose-containing medium, the biofilms formed by the luxS mutant were loose, hive-like, and had large gaps between the biofilm matrix, whereas the wild-type biofilms appeared confluent and more evenly distributed.
FIG. 5.
SEM of S. mutans biofilms. Twenty-four-hour biofilms of S. mutans strains UA159 (panels 1, 4, and 7), TW26 (panels 2, 5, and 8), and TW42 (panels 3, 6, and 9) were grown on HA disks that were deposited in 24-well cell culture clusters in semidefined BM (27) with 0.8% (wt/vol) glucose (panels 1 to 3) or 0.5% (wt/vol) sucrose (panels 4 to 9).
FIG. 6.
Impact of cell-free supernatants on biofilm formation. All strains were grown in BM with sucrose (27) with or without inclusion of cell-free supernatants prepared from UA159 (W) or TW26 (S). Biofilms were grown in the wells of a 96-well (flat-bottom) cell culture cluster and stained with 0.1% crystal violet (48).
Initial characterization of the LuxS regulon: relationship to stress tolerance.
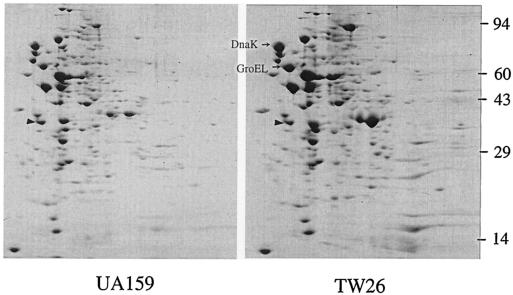
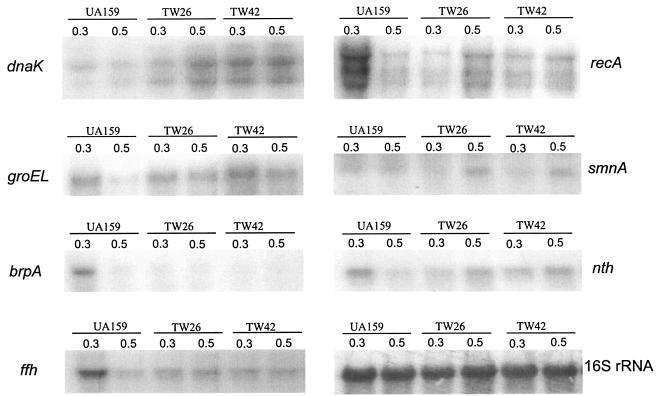
To begin to define the scope of the LuxS regulon, 2D electrophoresis protein patterns in Coomassie blue-stained gels for the wild-type and LuxS-deficient strain were compared. More than 43 proteins were up-regulated and at least 17 were down-regulated as a result of luxS inactivation (Fig. 7). Among the up-regulated proteins were the stress proteins and major molecular chaperones GroEL and DnaK, which we have previously identified by microsequencing spots from 2D gels. In an effort to identify the mechanisms by which luxS regulates stress tolerance and biofilm formation, Northern hybridization was used to analyze the expression of several genes, including those that are involved in stress responses (clpP, dnaK, and groEL) (19, 23, 24), DNA repair (recA, nth, and smnA) (17), biofilm formation (brpA) (48), and integrity of membrane proteins (ffh) (16). As shown in Fig. 8 and Table 2, inactivation of luxS increased expression of dnaK by an average of 250% and groEL by more than 90%. No major differences were observed in the expression of clpP for UA159 and TW26 (data not shown). As a result of luxS inactivation, brpA expression was decreased by as much as 70% in early-exponential-phase cells, although no significant differences were detected in late-exponential-phase cells, when comparisons of the parent and the mutant were made. The level of ffh mRNA in early-exponential-phase cells of the luxS mutant was about 50% of the parental strain, but no significant differences were observed for the ffh expression of UA159 and TW26 in late-exponential-phase cells. Relative to the wild-type strain, the recA transcripts in the luxS mutant were 50% lower than those in UA159 in early exponential phase but increased by up to 25% in late exponential phase. Similar trends were also observed in the transcription of smnA and nth.
FIG. 7.
2D gel analysis. S. mutans UA159 and TW26 were grown in BHI medium to an OD600 of ∼0.5. Cell extracts of 200-μg total proteins were subjected to 2D electrophoresis using 2% ampholines at pH 4 to 8. Gels were stained with Coomassie blue. The triangles indicate the internal standard, tropomyosin, with pI 5.2 and a molecular weight of 32,700.
FIG. 8.
Northern blot analysis. Total RNAs were extracted from S. mutans strains UA159, TW26, and TW42 grown in BHI medium at early exponential phase (OD600 ≅ 0.3) or late exponential phase (OD600 ≅ 0.5). Twelve micrograms of total RNA was loaded to each lane, separated by gel electrophoresis, blotted to a nylon membrane, and then hybridized with radioactively labeled gene-specific probes as indicated.
TABLE 2.
Up- or down-transcription of genes relative to wild-type UA159 at early exponential phasea
| Geneb | Relative level (%) for strain:
|
|||||
|---|---|---|---|---|---|---|
| UA159-0.3 | UA159-0.5 | TW26-0.3 | TW26-0.5 | TW42-0.3 | TW42-0.5 | |
| dnaK | 100 | +20.36 | +103.47 | +174.75 | +227.73 | +201.82 |
| groEL | 100 | −50.00 | +19.57 | −3.16 | +30.95 | −16.84 |
| brpA | 100 | −60.76 | −67.74 | −71.88 | −69.11 | −60.38 |
| ffh | 100 | −63.57 | −45.64 | −43.83 | −48.35 | −41.01 |
| recA | 100 | −31.92 | −32.52 | −9.96 | −42.89 | −27.32 |
| smnA | 100 | +19.12 | −1.33 | +46.81 | −14.56 | +33.51 |
| nth | 100 | −55.64 | −44.11 | −8.37 | −20.11 | +10.37 |
OD600 ≅ 0.3.
For dnaK and recA, the average for each major transcript was used.
Smu486 and Smu487 overlap with the LuxS regulatory circuit.
The smu487 gene is located 13,225 bp upstream of luxS in the S. mutans chromosome. The predicted protein, Smu487, is 215 amino acid residues and most similar to DegU (31% identity), a response regulator of Bacillus subtilis, and to LuxR of Vibrio cholerae (23% identity). The N terminus of Smu487 contains a signal receiver domain, and the C-terminal domain contains a helix-turn-helix motif typical of the LuxR family of transcriptional regulators. Locus smu486 is a 1,002-bp open reading frame located just upstream of, and apparently cotranscribed with, smu487. Smu486 is homologous to a sensor histidine kinase and, in concert with Smu487, constitutes an apparent bacterial two-component signal transduction system. A mutant (strain TW42) that had the entire Smu486 and 95 N-terminal residues of Smu487 deleted was constructed and subjected to functional assays and Northern analysis using the methods described in Materials and Methods. Similar to the luxS-deficient mutant, strain TW42 was more sensitive to killing by acid (Fig. 3A) and more resistant to hydrogen peroxide than S. mutans UA159 (Fig. 4). Also, similar to strain TW26, TW42 formed less biofilm on HA disks, and the biofilms appeared to be sporadic and hive-like compared to those of the parent strain UA159 (Fig. 5). Northern analysis also revealed that expression of both dnaK and groEL were increased in TW42, while brpA and ffh were down-regulated in response to the deficiency of smu486 and smu487 (Fig. 8). Moreover, TW42 also demonstrated decreased expression of smnA in cells of early exponential phase and a slight increase during the late exponential phase. Similar trends were also observed, but to a lesser degree, with the expression of recA and nth in the mutant lacking smu486 and smu487 (Fig. 8). Of note, the inclusion of cell-free supernatants from UA159 enhanced the tolerance of strain TW42 to acid killing (Fig. 3B) and its capacity to form biofilms (Fig. 6).
DISCUSSION
Possession of luxS or AI-2 activity by oral bacteria, including S. mutans, has recently been reported (3, 8, 11, 13, 31, 48). This study has provided further evidence that luxS of S. mutans UA159 is functional and its role in physiologic and virulence regulation is global. The product, AI-2, is capable of stimulating light production in V. harveyi, and such stimulatory activity was maximal during the early to mid-exponential phase of growth, which is similar to what was recently reported by Merritt et al. (31). It is noteworthy that, although S. mutans AI-2 is functional in V. harveyi, the magnitude of light stimulation is relatively low compared to that of AI-2 of the vibrio species (data not shown). This may suggest differences in the structure of AI-2, the absolute amount of AI-2 produced by S. mutans under the conditions tested, or both.
Of the virulence factors analyzed by reporter gene fusions, only fruA is sensitive to carbon catabolite repression. The observation that luxS inactivation decreased fruA expression but had no impact on either gtfBC, gbpB, spaP, or ftf may indicate that LuxS regulates fruA expression through perturbation of the expression of gene products involved in carbon flow or carbon catabolite repression. Although the actual identities remain to be determined, among the altered proteins that were identified by comparing our 2D results (Fig. 7) to the proteome profile of S. mutans H7 (G. Svensater, personal communication) were key enzymes of carbohydrate metabolism. Alteration of these enzymes could certainly have an impact on the process of carbohydrate metabolism, which in turn could influence FruA expression and utilization of extracellular fructose polymers. In support of this concept, regulation of genes or enzymes of carbohydrate metabolism by LuxS has also been reported in E. coli (10, 43) and S. gordonii (30).
The molecular chaperones DnaK and GroEL, which are known for their roles in refolding of nascent and unfolded proteins and in presenting stress-damaged proteins for degradation, are inducible by stresses such as heat and acid shock and serve as the “thermometer” of stress responses in a variety of bacteria, including S. mutans (19, 24). As shown by proteomics and Northern analysis, both DnaK and GroEL are up-regulated in the luxS-deficient strain (Fig. 6 and 7), indicating that cells lacking the AI-2 signal are perceiving stress. These results are consistent with the aberrant stress tolerance characteristics of the LuxS-deficient strains as well as with the alteration in expression of other gene products that appear to contribute to stress resistance. As shown by Northern analysis (Fig. 8), luxS inactivation decreased expression of several genes that encode membrane-associated proteins, including BrpA and Ffh, which have been shown to play roles in envelope integrity and acid tolerance (16, 48). Similarly, selected DNA repair enzymes that are acid inducible and important for acid tolerance in S. mutans (recA, nth, and smnA) (17) (Householder et al., Abstr. 6th Am. Soc. Microbiol. Conf. Strept. Genet., abstr. 109, p. 100, 2002) are down-regulated in the luxS strain. Proton extrusion via the F-ATPase is believed to be a primary mechanism by which mutans streptococci maintain intracellular pH homeostasis (2). However, there was no major difference in the F-ATPase activity of strains UA159 and TW26 (data not shown), indicating that a diminished capacity to move protons from the cell is not the primary reason for altered acid resistance. The maintenance of an adaptive acid tolerance response in the luxS strains indicates that this genetic circuit remains intact in spite of an overall reduction in acid tolerance.
It is known that the capacity of oxidative stress tolerance is important to the survival of oral bacteria, but there is little information available concerning oxidative stress responses induced primarily by deleterious reactive products of oxygen metabolism in S. mutans (29). In contrast to some previous findings (36), no significant protection against hydrogen peroxide was observed in either strain after the acid adaptation of the organisms. Our results reinforce the idea that the acid and oxidative stress tolerance responses likely have overlapping, e.g., DnaK and GroEL, and distinct components that are controlled by multiple regulons, some of which appear to fall under the control of the LuxS circuit.
The capacity of bacteria to form biofilms varies in response to environmental factors. As biofilms mature, modulation of gene expression by environmental sensing and detection of accumulating signal molecules appears essential for the progression from adherent microcolonies to three-dimensional, mature biofilms. The Com system, which controls genetic competence development in response to the concentration of the competence-stimulating peptide, is also involved in biofilm formation and biofilm architecture of S. mutans (25). It is noteworthy that similar phenomena in biofilm formation and acid tolerance by this two-component signal transduction system have also recently been reported by Li et al. (26). The similarities in the behavior of the luxS strain and the strain lacking the two-component signal transduction system are intriguing. However, examination of a strain carrying the smu486 and smu487 mutations and a luxS promoter-cat gene fusion revealed that luxS expression is not affected by the loss of the two-component system (data not shown) and inclusion of cell-free supernatants prepared from strain UA159 could enhance, but only slightly, the capacity of biofilm formation and acid tolerance of strain TW42. This enhancement alone is of interest, and we believe that it is due to the facts that AI-2 production is low in S. mutans and that addition of supernatants from cultures that have accumulated AI-2 can further enhance the biofilm and acid resistance phenotype. Collectively, these data suggest that Smu486 and Smu487 are not directly involved in the LuxS-mediated regulation. Instead, there may be considerable overlap and redundancies between an environmental or small-molecule-sensing two-component system, in addition to the Com system, and the LuxS-mediated signaling system, and all three of these pathways are integral in controlling biofilm maturation and stress tolerance. Comparison of the gene expression profiles of the luxS and smu486 smu487 strains in future studies will be valuable for identifying key factors in biofilm formation and stress tolerance.
In summary, we have shown that LuxS and AI-2 signaling has profound effects on stress tolerance and biofilm formation. The molecular basis for the phenotypes has begun to be revealed by demonstrating defects in the regulation of proteins required for biofilm formation, perturbations in the general stress response pathways, and defects in coping with environmental insults and membrane protein biogenesis. Continued assessment of the impact of the loss of LuxS, Smu486, and Smu487 is ongoing in order to better define critical pathways for establishment and persistence of an important human pathogen and related organisms.
Acknowledgments
We express our gratitude to Bonnie L. Bassler of Princeton University for kindly providing the V. harveyi reporter strains, D. Liao of the University of Florida Department of Anatomy and Cell Biology for access to his luminometer, F. L. Bennett of the EM Core Lab of the University of Florida for his technical assistance with the SEM analysis, and J. Lemos and J. Abranches for their critical evaluation of the manuscript.
This work was supported by NIDCR grants DE13239 and 12236 to R.A.B. and DE15501 to Z.T.W.
REFERENCES
- 1.Belli, W. A., and R. E. Marquis. 1991. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 57:1134-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowden, G. H., and I. R. Hamilton. 1998. Survival of oral bacteria. Crit. Rev. Oral Biol. Med. 9:54-85. [DOI] [PubMed] [Google Scholar]
- 3.Burgess, N. A., D. F. Kirke, P. Williams, K. Winzer, K. R. Hardie, N. L. Meyers, J. Aduse-Opoku, M. A. Curtis, and M. Camara. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763-772. [DOI] [PubMed] [Google Scholar]
- 4.Burne, R. A. 1998. Oral streptococci…products of their environment. J. Dent. Res. 77:445-452. [DOI] [PubMed] [Google Scholar]
- 5.Burne, R. A., Z. T. Wen, Y. M. Chen, and J. E. C. Penders. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y. M., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57.1 urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLisa, M. P., J. J. Valdes, and W. E. Bentley. 2001. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J. Bacteriol. 183:2918-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fong, K. P., L. Gao, and D. R. Demuth. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frias, J., E. Olle, and M. Alsina. 2001. Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 69:3431-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg, E. P., J. W. Hastings, and S. Ulitzer. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 120:87-91. [Google Scholar]
- 16.Gutierrez, J. A., P. J. Crowley, D. G. Cvitkovitch, L. J. Brady, I. R. Hamilton, J. D. Hillman, and A. S. Bleiweis. 1999. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology 145:357-366. [DOI] [PubMed] [Google Scholar]
- 17.Hahn, K., R. C. Faustoferri, and R. G. Quivey, Jr. 1999. Induction of an AP endonuclease activity in Streptococcus mutans during growth at low pH. Mol. Microbiol. 31:1489-1498. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton, I. R., and N. D. Buckley. 1991. Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol. Immunol. 6:65-71. [DOI] [PubMed] [Google Scholar]
- 19.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 20.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleerebezem, M., and L. E. Quadri. 2001. Peptide pheromone-dependent regulation of antimicrobial peptide production in gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579-1596. [DOI] [PubMed] [Google Scholar]
- 22.Lau, P. C. Y., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-201. [DOI] [PubMed] [Google Scholar]
- 23.Lemos, J. A., Y. Y. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y.-H., N. Tang, M. B. Asprias, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y. H., P. C. Lau, N. Tang, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 29.Marquis, R. E. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Ind. Microbiol. 15:198-207. [DOI] [PubMed] [Google Scholar]
- 30.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 33.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 34.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 36.Quivey, R. G., Jr., R. C. Faustoferri, K. A. Clancy, and R. E. Marquis. 1995. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol. Lett. 126:257-261. [DOI] [PubMed] [Google Scholar]
- 37.Quivey, R. G., Jr., R. C. Faustoferri, and S. D. Reyes. 1995. UV-resistance of acid-adapted Streptococcus mutans. Dev. Biol. Stand. 85:393-398. [PubMed] [Google Scholar]
- 38.Quivey, R. G., Jr., W. L. Kuhner, and K. Hahn. 2000. Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 42:239-274. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Schauder, S., and B. L. Bassler. 2001. The language of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 41.Shaw, W. V. 1979. Chloramphenicol acetyltransferase activity from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 42.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen, Z. T., and R. A. Burne. 2001. Construction of a new integration vector for use in Streptococcus mutans. Plasmid 45:31-36. [DOI] [PubMed] [Google Scholar]
- 48.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]