Abstract
Glucarate, an oxidized product of glucose, is a major serum organic acid in humans. Still, its role as a carbon source for a pathogen colonizing hosts has not been studied. We detected high-level expression of a potential glucarate permease encoding gene gudT when Salmonella enterica serovar Typhimurium are exposed to hydrogen gas (H2), a gaseous by-product of gut commensal metabolism. A gudT strain of Salmonella is deficient in glucarate-dependent growth, however, it can still use other monosaccharides, such as glucose or galactose. Complementation of the gudT mutant with a plasmid harbouring gudT restored glucarate-dependent growth to wild-type (WT) levels. The gudT mutant exhibits attenuated virulence: the mean time of death for mice inoculated with WT strain was 2 days earlier than for mice inoculated with the gudT strain. At 4 days postinoculation, liver and spleen homogenates from mice inoculated with a gudT strain contained significantly fewer viable Salmonella than homogenates from animals inoculated with the parent. The parent strain grew well H2-dependently in a minimal medium with amino acids and glucarate provided as the sole carbon sources, whereas the gudT strain achieved approximately 30% of the parent strain's yield. Glucarate-mediated growth of a mutant strain unable to produce H2 was stimulated by H2 addition, presumably owing to the positive transcriptional response to H2. Gut microbiota-produced molecular hydrogen apparently signals Salmonella to catabolize an alternative carbon source available in the host. Our results link a gut microbiome-produced diffusible metabolite to augmenting bacterial pathogenesis.
Keywords: microbial carbon utilization, carbon transport, in vivo pathogen growth, gut microbiome, metabolism and virulence
2. Introduction
d-glucaric acid, an oxidized product of d-glucose, is a natural product found in a variety of fruits and vegetables [1,2], and in mammals [3–6]. It is also used as a dietary supplement in the form of calcium d-glucarate and it has been studied for its potential roles in reducing cholesterol levels and in cancer chemotherapy [7,8]. d-glucaric acid is normally present in the tissues and body fluids of humans [3–5] and it is a major serum organic acid in humans [9]; it approximates the blood levels of the (generally) most abundant organic acid in human serum, pyruvate. Average levels of human d-glucaric acid have been reported to be 9.2 ± 2.0 µg ml−1 in blood [9] and 11 to 12 µg ml−1 in urine [3]. The level of urine d-glucarate is used as an index of hepatic enzyme activity in humans and in experimental animals [10].
The pathogen Salmonella enterica serovar Typhimurium, from here on referred to as S. Typhimurium, are adept at surviving and growing in metabolically diverse environments both within and outside the host animal [11,12]. However, many enzymes that are synthesized and active during host infection exhibit little or no effects in virulence models [13,14]. Researchers have suggested that d-glucarate can serve as a growth substrate for a variety of microorganisms including Escherichia coli and related enteric bacteria [15,16]. In a study identifying some of the genes affecting glucarate utilization in E. coli strain K12, Roberton et al. [17] noted the availability of glucarate in humans at levels near the Km of whole E. coli cells for the organic acid and he subsequently speculated that pathogenic E. coli strains may benefit by using host d-glucarate as an alternative carbon source. While the pioneering studies had indicated that d-glucarate was a potential carbon source for pathogens, its role as a carbon source for pathogenic bacteria was not studied.
From a vast compilation of data that included genome comparisons and Salmonella proteome expression from the pathogen in infected mice, the putative glucarate uptake and catabolism enzymes were among hundreds of enzymes that were assigned into different classes of predicted importance; some of the glucarate catabolic enzymes were expressed in the enteritis model of infection (cecum colonization), and others, but not all, were classified as ‘likely expressed’ in the typhoid fever model [13]. Still, the glucarate uptake gene gudT and others in the catabolic pathway were deemed to be dispensable for causing systemic salmonellosis [13]; this was defined as ‘the gene can be inactivated without significant consequences to virulence’. Also of note is that the glucarate permease and catabolism proteins were not observed among the proteome composition of log phase LB-grown S. Typhimurium [18]. While mutations in many individual permease genes were tested, attenuation of Salmonella mouse typhoid virulence via mutation of a carbohydrate permease was only observed for the mannose PTS transporter [13]; and that permease is thought to transport many different carbohydrates.
S. Typhimurium can colonize host (mice) organs in part by using H2 gas produced by the commensal microbiota; the small diffusible gas is carried in the bloodstream and can achieve levels averaging approximately 50 μM in blood-rich organs, such as liver and spleen of mice, and more than three times that level in the small intestine [19]. A gene-targeted Salmonella mutant strain lacking all H2-oxiding ability was unable to cause mouse mortality when inoculated into mice at a level whereby most of the wild-type (WT)-inoculated animals succumbed to typhoid salmonellosis within 10 days of infection [20]. One role of the Hyb enzyme was assigned via physiological studies; it augments the Salmonella membrane potential [21] and, as in H2-oxidizing Helicobacters, the subsequent energized membrane can facilitate carbon uptake [21,22]. Molecular hydrogen recognition via Hyb and perhaps by one of the other Salmonella hydrogenases stimulates expression of some S. Typhimurium genes associated with carbon acquisition; among the most highly up-expressed by H2 exposure genes was gudT, a gene that encodes a potential glucarate permease GudT [23]. There was 5.7-fold more (via microarray) and 10-fold more (via qRT-PCR) gudT expression in cells exposed to H2, compared with cells in the identical culture and atmospheric condition but without H2 [23]. Additional genes associated with glucarate catabolism, including a putative positive transcriptional regulator of glucarate catabolism genes also increased upon exposure to H2.
The finding that a microbiota-produced dissolved gas within hosts, namely H2, stimulates glucarate catabolism by S. Typhimurium combined with the relatively high levels of glucarate reported in animal serum/tissues led us to hypothesize that the pathogen may use H2 in vivo to express enzymes, and then drive glucarate transport. If so, glucarate catabolism would be expected to augment the bacterium's overall in vivo growth capacity. We thus investigated the involvement of glucarate catabolism in the virulence of S. Typhimurium by studying a non-polar gudT (permease) deletion strain. The gudT strain is deficient in glucarate-dependent growth compared with its parent strain and it exhibits attenuated virulence (i.e. morbidity and organ colonization). An H2 stimulatory effect on glucarate-dependent Salmonella growth was assigned by use of a mutant strain lacking all H2 producing ability. These results link a diffusible metabolic signal from the gut commensals to carbon transport that in turn augments pathogenicity, a link not previously documented.
3. Results
3.1. The genes and enzymes for glucarate catabolism
The genes encoding the enzymes of the d-glucarate catabolic pathway in E. coli have been identified and annotated [24,25]. According to the literature and available gene annotation databases, the enzymes of the d-glucarate catabolic pathways of E. coli and the related bacterium S. Typhimurium share similar functions and 97% homology (NCBI (ncbi.nlm.nih.gov), JCVI CMR (cmr.jcvi.org) BLAST tools, BioCyC (biocyc.org), EcoCyc (ecocyc.org)). d-glucarate is transported into the cells by the enzyme glucarate permease (named GudP in E. coli and GudT in S. Typhimurium) and is dehydrated to 5-dehydro-4-deoxy-d-glucarate by glucarate dehydratase, GudD [26]. d-glucarate aldolase (GarL) next reduces 5-dehydro-4-deoxy-d-glucarate into tartronate semialdehyde. Tartronate semialdehyde is further reduced to d-glycerate by tartronate semialdehyde reductase (GarR). In the final step, d-glycerate is phosphorylated to yield 2-phospho-d-glycerate by glycerate kinase (GarK). The 2-phospho-d-glycerate can be reversibly converted to 3-phospho-d-glycerate by the enzyme phosphoglyceromutase (GpmA).
3.2. Glucarate-dependent growth
Salmonella gene annotation sites list gudT as a gene-encoding putative glucarate transporter based on its high sequence similarity with probable glucarate transporters from other organisms. Its synonyms are gudP and ycgZ. In order to support a role for the putative glucarate permease, GudT, in the uptake of d-glucarate, we constructed a non-polar single gene deletion mutant of S. Typhimurium strain 14028 (JSG210) by using the lambda-Red system. We then compared the growth of the mutant and the parent WT strains in a minimal medium containing 0.4% d-glucarate and 0.2% casamino acids. As expected, the growth of the gudT-deletion strain RLK6 was significantly retarded compared with WT. The WT yielded a CFU count of approximately 1.05 × 109 CFU ml−1 at 8 h, with a growth rate doubling time of 3 h, whereas without glucarate it yielded approximately 4 × 108 CFU ml−1 at about 4 h after incubation and remained at that level. The pattern of RLK6 growth in glucarate-containing medium was nearly identical to that of the WT grown without glucarate (figure 1). Growth of RLK6 in glucose- or galactose-containing medium (0.4% each) was similar to WT (data not shown), suggesting that the gudT mutant is still able to transport and metabolize other monosaccharides. In addition, we complemented the mutant with a plasmid-containing gudT under the control of a (IPTG-inducible) Plac promoter. The growth of the RLK6 (pLac22-gudT) complemented strain in glucarate-containing medium supplemented with 5 μM IPTG was nearly identical to that of WT grown with glucarate (data not shown) confirming that gudT was the only gene affected in strain RLK6. Therefore, conclusions from observations on this mutant strain can be assigned to gudT alone. Taken together, the results indicate that GudT is important for the growth of S. Typhimurium on d-glucarate. In a study by Aghaie et al. [27], mutation of the glucarate permease gene in Acinetobacter baylyi ADP1 resulted in severe growth impairment on d-glucarate, albeit similar results were obtained with (A. baylyi) strains containing mutations in other individual genes involved in the glucarate catabolic pathway.
Figure 1.
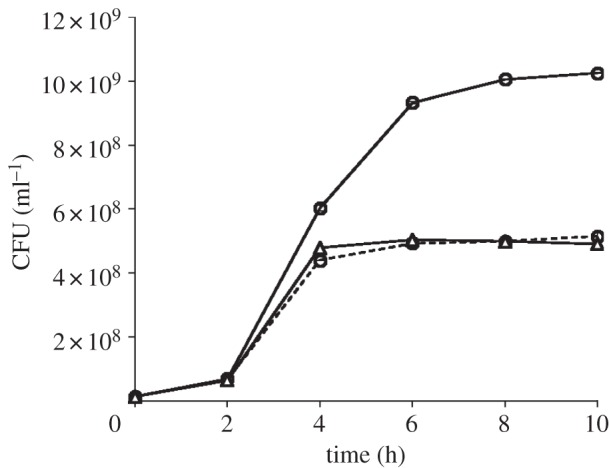
Comparison of aerobic growth of S. Typhimurium WT strain JSG210 (represented as circles) with RLK6/ΔgudT (represented as triangles) in minimal medium with 0.4% d-glucarate. Dashed line indicates WT growth without d-glucarate (control). Data points are the mean from three replicate serum bottles for each strain/condition. The standard deviation was less than 5% of the mean in every case, so that JSG210 with glucarate is greater than the lower two lines for all points at 4 h and greater at p ≤ 0.05 by Student's t-test.
3.3. Effects of added molecular hydrogen on growth
As H2 stimulated the expression of glucarate-use genes more than that of any other carbon acquisition genes [23], we assessed the effects of exogenously added H2 on the growth of S. Typhimurium on d-glucarate. Anaerobic respiratory growth of S. Typhimurium with H2 (reductant) and fumarate (terminal acceptor) was shown previously when amino acids were the only other carbon source provided [21]. WT and RLK6 were grown anaerobically with and without added H2 in a minimal medium containing 0.4% d-glucarate and 0.2% casamino acids. Addition of H2 enhanced the growth of both WT and RLK6, though the total CFU ml−1 of the mutant strain remained much lower than that of the WT in both H2-added and H2-absent conditions. Hydrogen-stimulated growth of the parent with glucarate is likely a consequence of H2 providing both increased glucarate catabolic gene transcription [23] and the ability of H2 oxidation to cause augmented proton motive force in Salmonella [21] that directly facilitates carbon source (e.g. glucarate) uptake. With added H2, RLK6 achieved about 30% of the parent strain's yield (2.0 × 108 CFU ml−1 for RLK6; 6.5 × 108 CFU ml−1 for WT; figure 2) and this was a statistically significant difference.
Figure 2.
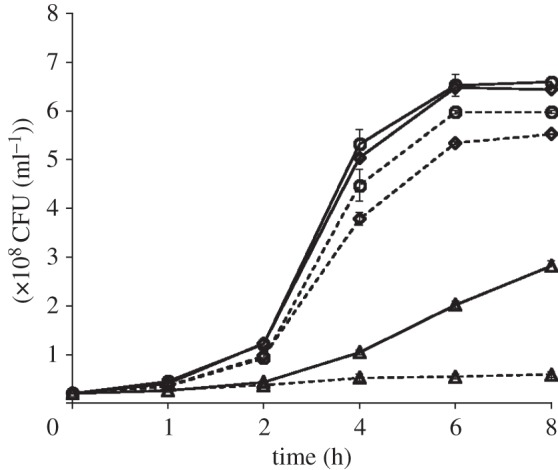
Comparison of H2-facilitated anaerobic growth of S. Typhimurium strains. Anaerobic WT/JSG210 (represented as circles), RLK7/ΔhycC (represented as diamonds) and RLK6/ΔgudT (represented as triangles) in minimal medium with 0.4% d-glucarate. Solid lines indicate growth with added H2 (20% v/v) and dashed lines indicate growth without added H2. The standard deviation was less than 4% of the mean in every case, so that the added H2 condition is significantly greater than without H2 for each individual strain for all points at 4 h and greater at p ≤ 0.05 by Student's t-test. Without H2, there was also a significant difference in JSG210 and the RLK7 mutant at 6 and 8 h points (p ≤ 0.05 by Student's t-test), while with H2, the WT was significantly greater than strain RLK6 only.
S. Typhimurium can use H2 to acquire carbon by transporting amino acids and other carbon sources into the cells from the medium [21,23]. Hence, the H2-augmented growth of RLK6 is likely owing to their acquisition of casamino acids or fumarate present in the medium. Theoretically, an ideal experimental condition to compare the in vitro growth of the strains would be to use a minimal medium containing only d-glucarate and lacking other carbons sources (e.g. casamino acids, fumarate) so that only the effects of d-glucarate could be observed. However, we could not establish such a condition, as WT grew very slowly with glucarate alone and RLK6 failed to grow at all in such a medium.
We also compared the H2-dependent growth of the WT with a hydrogenase mutant (RLK7) containing a deletion in the hycC gene that encodes the HycC subunit of the H2-evolving enzyme Hyc-hydrogenase [28]. This was to enable an assessment of the effects of H2 without interference from internally produced H2 from Salmonella metabolism. Growth experiments were performed with and without added H2 in minimal medium containing 0.4% d-glucarate, 0.2% casamino acids and 0.4% fumarate as described above. Sawers et al. [29] demonstrated that H2 evolution is low (between 0.016 and 0.001 µmol of H2 evolved per minute) when S. Typhimurium cells are grown under anaerobic conditions with fumarate. Added hydrogen stimulated both strains (this was seen previously with the WT, see [21]). Without added H2, RLK7 achieved slightly less yield than the parent, but this difference (see figure 2 legend) was significant. A statistically significant growth stimulation effect by H2 on RLK7 growth was observed at 4, 6 and 8 h points (e.g. for 8 h compared yield of 5.4 ± 0.2 × 108 CFU ml−1 without H2 versus 6.5 ± 0.2 × 108 CFU ml−1 with H2) and this is attributed to a likely positive transcriptional response of H2 on gudT, gudD and/or gar genes, but this was not examined further.
3.4. Mouse colonization and virulence
The ability of RLK6 to survive and cause infection in vivo was assessed by mouse stomach colonization experiments. S. Typhimurium is highly virulent in mice, causing typhoid fever-like disease, with rapid organ colonization and death within days of inoculation. In all mouse experiments (including the repeat experiment), the WT caused infection and death within 9 days of oral inoculation. This is consistent with the observations in our previous S. Typhimurium virulence study with the same WT strain [20]. The mean time of death for mice inoculated with WT was 2 days earlier than for the mice inoculated with RLK6 (figure 3). On day 9 postinoculation, 25% of the RLK6-infected mice still survived. Those remaining infected mice died, one on day 10, another on day 11 and the remaining two at day 12 postinoculation. Similar results (delayed animal mortality by the mutant compared with the parent strain) were obtained from the second experiment repeated at a later time, but using eight mice per strain (data not shown).
Figure 3.
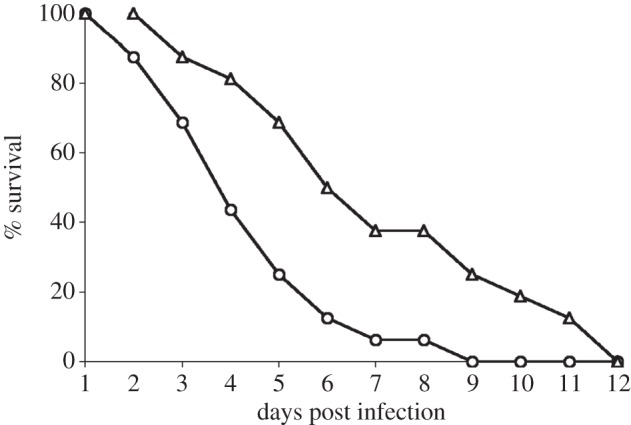
Comparison of virulence of S. Typhimurium strains JSG210/WT (represented as circles) and RLK6/ΔgudT (represented as triangles) in mice. The results shown are for 16 mice infected with each strain. The second experiment with eight mice infected with each strain showed similar results. A Wilcoxon rank-sum statistical analysis of these data was performed, testing that the distribution of dataset A (RLK6) is significantly shifted to the right of dataset B (WT), or H1: A > B using 16 data points for each strain. This test showed significance between the two groups at p ≤ 0.05 for a two-tailed test.
The organ-colonizing abilities of WT and RLK6 were compared from infected mice 4 days (96 h) after oral inoculation. The colonization numbers of bacteria in the livers and spleens were determined from four individual mice for each bacterial strain, as described previously [20]. Two mice injected with sterile PBS were included in each experiment as negative controls, and no Salmonella colonies were recovered from these animals. At 4 days postinoculation, liver and spleen homogenates from mice inoculated with RLK6 contained fewer viable Salmonella than organ homogenates from mice inoculated with WT. The organ burden numbers in mice infected with WT ranged from 1.2 × 106 to 2.7 × 106 (livers) and 1.5 × 106 to 2.6 × 106 (spleens), whereas the colonization numbers of RLK6 ranged from 0.4 × 106 to 1.0 × 106 (livers) and from 0.1 × 106 to 1.5 × 106 (spleens; table 3). The difference in the colony counts of the two strains was statistically significant at p < 0.05. Similar results were observed in the second colonization experiment (see table 3 legend). The results of the organ burden mouse experiments agree with the mortality results and support that the gudT-deleted strain RLK6 is attenuated in virulence compared with WT.
Table 3.
Organ colonization numbers of S. Typhimurium strains JSG210 (WT) and RLK6 (gudT-deleted mutant) in the livers and spleens of infected mice. Numbers indicate ranges of CFU per organ among four mice inoculated with each strain. Statistically significant differences between WT and RLK6 were indicated for both organs by Student's t-test analysis (p < 0.05, n = 4). The second experiment (also with four mice infected with each strain) showed similar results. Wilcoxon rank-sum statistical analysis using each of the latter four data points indicated differences were significant between the two strains for both organs at p ≤ 0.05.
| strain | CFU/liver (×106 cells) | CFU/spleen (×106 cells) |
|---|---|---|
| WT | 1.2–2.7 | 1.5–2.6 |
| RLK6 | 0.4–1.0 | 0.1–1.5 |
4. Discussion
Like many other pathogenic bacteria, Salmonella are catabolically flexible and robust; it is known that they survive diverse environments and can use a variety of carbon sources. From an exhaustive screen of about 600 compounds for ability to be used as possible C or N sources Gutnick et al. [30] found that Salmonella can use more than 80 different metabolites (including glucarate) as carbon sources. It is predicted that 69 of these are available in mouse tissues [14], however, mutational analysis of genes for catabolic enzymes expected to be or known to be expressed in the animal host does not often yield a disease phenotype different from the parent [13,14]. The mutant strain studied herein contained a deletion in the putative glucarate permease and other carbohydrate permeases have been studied recently. Mutation of S. Typhimurium genes for uptake of glucose, glucose 6-phosphate or for mannose uptake yielded little reduced capacity for Salmonella to replicate in epithelial colorectal adenocarcinoma cells [31]. From a broad study [13] measuring proteomic data and mouse typhoid symptoms from hundreds of S. Typhimurium mutant strains it was concluded that the permeases for ribose, fucose, glycerol, galactose and a number of amino acid permeases did not significantly affect virulence; only the mannose permease contributed to virulence but that permease (PTS mannose permease) also transports other carbohydrates. Similarly, from a comprehensive study in which thousands of transposon mutants could be studied via insertion-site sequencing upon animal infection, it was concluded that among transporters, only a proline-specific permease (ProY) and a peptide transporter (SapC) affected systemic typhoid virulence in mice [32]. The putative glucarate permease was not represented among their insertion mutant pool.
Following entry into the intestinal lumen, S. Typhimurium crosses the intestinal epithelium and is engulfed by phagocytic cells, such as dendritic cells, macrophages and granulocytes. Some of them survive this to cause severe disease. Mutational analyses by Bowden et al. [33] showed that the central metabolic pathway of glycolysis is required for the intracellular replication of S. Typhimurium within macrophages. However, mutants of S. Typhimurium defective in glucose uptake were still able to grow and replicate intracellularly, though at a rate lower than that of the WT [34]. While glucose may be the preferred carbon source of S. Typhimurium during some host–pathogen interactions [33–35], S. Typhimurium has enzymes to use alternative carbon sources at times when glucose is unavailable or limiting [35,36]. Considering the large number of carbon sources presumably available to Salmonella within the host, our result indicates that glucarate must rank among the important ones. From studying mutant strain phenotypes, proteomic, transcriptomic and other data, it was concluded [35] that Salmonella must sometimes rely on alternative carbon sources for systemic infections, and these may include both C6 and C3 sources. In our study, the gudT-deletion strain could still cause systemic disease and mortality based on the typhoid model but attenuation was evident; in light of the number of carbon sources thought to be used by Salmonella while residing in the host, obtaining such a phenotype is notable.
Both d-glucarate and H2 can be detected within animals and both substrates likely vary widely in concentration within host tissues. The glucarate catabolic genes are among a number of genes involved in carbon sequestering that are up-expressed by the pathogen in response to molecular hydrogen. This H2-mediated increased gene expression is most likely related to hydrogenases generating a lower potential cellular redox state, such that gene-expression-related effectors are either directly or indirectly activated. Further, some Salmonella appear to have more than one glucarate transporter (JCVI CMR, cmr.jcvi.org); this may mean that Salmonella has flexibility in acquiring the substrate at different concentrations or in different host environments. The glucarate catabolic pathway is stimulated by H2 [35], but we did not know whether this was relevant to colonization of the host. Molecular hydrogen is a substrate which undoubtedly is in dynamic flux within animal tissues, and it likely varies in levels, depending on the microbiome composition, host diet and proximity of the relevant tissue/organ to the source community. The gas may be a signal to Salmonella that it is encountering a nutritionally challenging (carbon-poor?) environment, so the pathogen can begin to catabolize an alternative carbon substrate. Still, levels of the dissolved gas measured several mm within organs (liver, spleen and heart) of live mice were well above the Km for the reductant by membrane-bound bacterial hydrogenases [19]. Glucarate utilization must augment the survival and growth of pathogenic Salmonella at one or more stages of infection within the host and may also be important for colonic survival, and thus transmission. The latter environment is where the host microbial community has exhausted most of the readily usable sugars, and also where H2 levels are the greatest [19]; studying the role of glucarate in Salmonella survival there will require additional animal sampling. Nevertheless, our results link H2-stimulated glucarate catabolism to assisting the virulence of S. Typhimurium and uncover a new link between the commensal gut community and a pathogen.
5. Material and methods
5.1. Strains, growth conditions and reagents
The strains and plasmids used in this study are listed in table 1. Strains were maintained in Luria–Bertani (LB) broth or on LB Agar (LBA) plates. Appropriate antibiotics (100 µg ampicillin ml−1 and 25 µg kanamycin ml−1) were used when necessary. For growth experiments, M9 minimal medium was prepared according to the protocol provided [39] with the following modifications: NiCl2 (5 µM) and FeSO4 (5 µM) were added; d-glucarate, d-glucose or d-galactose (0.4%) was used where indicated and IPTG (5 μM) was supplemented as described. Cells were grown in serum bottles with a large headspace (15 ml culture in 165 ml bottles) at 37°C and shaking at 200 r.p.m. For aerobic growth, the bottles were maintained aseptically via cotton plugs and (loose-fit) aluminium foil. For anaerobic growth experiments, 0.4% fumarate was added to the medium, and bottles were tightly sealed with serum stoppers and aluminium crimps [28]. Anaerobic conditions with H2 were established by sparging the 15 ml of culture medium with inflow and outflow needles in sealed 165 ml bottles with N2 for 15 min, then with anaerobic mix (10% H2, 5% CO2 and 85% N2) for 20 min; H2 gas from a 100% cylinder was then injected to bring the volume of added H2 to 20% partial pressure [21].
Table 1.
Strains and plasmids used in this study. FRT, flippase recombinase recognition target.
| strain/plasmid | genotype/description | reference |
|---|---|---|
| strain | ||
| E. coli | ||
| TOP10 | cloning strain | Invitrogen |
| S. Typhimurium | ||
| JSG 210 | ATCC 14028s (WT) | [28] |
| RLK6 | JSG210ΔgudT::FRT (ΔgudT) | this study |
| RLK7 | JSG210ΔhycC::FRT (ΔhycC) | this study |
| TT22971 | methylating strain | John Roth |
| plasmids | ||
| pCP20 | Ampr; contains flippase gene for λ Red mutagenesis | [37] |
| pKD46 | Ampr; contains λ Red genes γ, β and exo | [37] |
| pKD4 | Kanr; contains kan cassette | [37] |
| pLac22 | Ampr; complementation vector (E. coli Plac) | [38] |
| pLac22-gudT | pLac22 with gudT (BglII-SalI) under Plac control | this study |
5.2. Mutant strain construction
The deletion strain gudT (STM2962 single deletion) was constructed using WT strain S. Typhimurium ATCC 14028 (strain JSG210 or WT) and the lambda-Red system as previously described [28,37]. The gudT-deleted mutant was named strain RLK6, also denoted as ΔgudT in this paper. The deletions made using this system are non-polar and the strains do not contain antibiotic resistance markers. Additionally, a hydrogenase mutant (RLK7 or ΔhycC) with a single deletion of the gene hycC (STM2851) was constructed in a similar manner and was used to assess the effects of exogenous H2 in comparative growth experiments (for figure 2). The deletions were confirmed by PCR using primers complementary to the regions flanking the deleted genes and by sequencing across the deletions (Georgia Genomics Facility or GGF, University of Georgia). The plasmids and strains used in this study are listed in table 1, and the primers used are listed in table 2.
Table 2.
Primers used in this study.
| primer | primer sequence (5′ → 3′) | application |
|---|---|---|
| gudT del-F | TGAGCGTAGCTAACGCGAAATTTCAGGA- GTGCAACATGTGTAGGCTGGAGCTGCTTC |
gudT deletion |
| gudT del-R | CCTTCATGTCCGTAATAACGGGACTGGAT- TGCGTTGTCACATATGAATATCCTCCTTA |
gudT deletion |
| gudT-check-F | GTTTGCTTGCGTTGCCAGTA | gudT-deletion confirmation |
| gudT-check-R | GTTCACAGACCGGCACATTA | gudT-deletion confirmation |
| gudT comp-F | GTCCTAGATCTTATGAGTTCATTAAGTCAC | gudT complementation |
| gudT comp-R | CTGGTGTCGACTCATGATAATTGTCCTGC | gudT complementation |
| hycC del-F | CTTGTTTCAGCAGGCTCAGAGTGGGGA- TGCATATGTGTAGGCTGGAGCTGCTTC |
hycC deletion |
| hycC del-R | GCGCCTGAATTAACGGATAAAACAC- ACTCATTTCATATGAATATCCTCCTTA |
hycC deletion |
| hycC-check-F | GTGAGCTGACGTTTAATACCGA | hycC deletion confirmation |
| hycC-check-R | CGACCGAGCAGTTTGATAATGT | hycC deletion confirmation |
5.3. Mutant complementation
Genomic DNA from WT strain JSG210 and primers gudT comp-F and gudT comp-R (table 2) were used to amplify a 1380-bp-long PCR product containing the whole gudT ORF as well as to introduce BglII and SalI, respectively. The PCR product was gel purified, digested with both enzymes, ligated into similarly digested pLac22 vector [38] (placing gudT under the control of an IPTG-inducible Plac promoter) and transformed into E. coli TOP10. Ampicillin resistant clones were isolated and analysed and the newly generated pLac22-gudT plasmid was sequenced at the GGF, University of Georgia, to ensure that there was no error in the gudT sequence. This plasmid was first introduced in (methylating) strain TT22791 (John Roth, UC Davis) before being finally transformed into RLK6 (gudT) mutant. RLK6 (pLac22-gudT) complemented mutants were used in growth experiments in M9 supplemented with glucarate with or without IPTG (5 μM).
5.4. Growth curves
To confirm the role of the putative glucarate permease GudT in glucarate-dependent growth of Salmonella Typhimurium, the growth rates of RLK6 (ΔgudT) and WT strains in the minimal medium containing d-glucarate as the only carbon source were compared. Bottles (165 ml capacity) containing 15 ml minimal medium with 0.4% d-glucarate were inoculated with 1.0 × 107 WT or deletion mutant S. Typhimurium cells. WT cells inoculated into minimal medium without d-glucarate were also included as controls. Cells were grown for up to 8 h at 37°C with shaking at 200 r.p.m. A600 (OD at 600 nm) was measured in 1- or 2-h intervals and cell numbers were calculated. Optical density for each time point for each strain was measured from three separate bottles, so the data point plotted is the mean ± standard deviation from three independent replicates; those points were appearing without error bars because the error bar was too small to be noted graphically. An A600 of 1.0 corresponds to 6.74 × 108 viable Salmonella per ml. Standard curves of A600 versus CFU ml−1 (plate counts) confirmed that the A600 was proportional to viable cell numbers within the OD range used herein, including for final yield (i.e. saturation growth) numbers. The efficacy of the standard curve was separately verified for each strain. All growth rate studies were performed three times, each time with three replicate serum bottles per strain/condition with results similar to those shown (figures 1 and 2).
5.5. Mouse experiments
The ability of the gudT-deleted strain RLK6 to cause infection in mice was assessed by using the typhoid fever-mouse model [40]. Female BALB/c mice (obtained from National Cancer Institute, Frederick, MD, USA) were orally inoculated (oral gavage into the stomach) individually with cell suspensions of RLK6 and WT, following methods previously described [20]. Sixteen mice were used per strain. Cells were washed and suspended in sterile PBS and 0.1 ml volumes of the cell suspension containing 106 bacterial cells were introduced orally to each mouse. The mice were observed twice daily, and immobile or nearly immobile mice were euthanized and recorded as mortality. The second experiment was performed using eight mice per strain, with results similar to those of the first experiment.
The organ bacterial burdens were determined by performing colony counts of the bacteria harvested from the livers and spleens of infected mice [20]. Four female BALB/c mice were orally inoculated with the mutant (RLK6) or the parent (WT) strains as described above. The mice were euthanized 4 days (96 h) after inoculation. Immediately after euthanizing, the livers and spleens were removed and homogenized in sterile PBS [20]. Dilutions of the homogenate were plated on Bismuth Sulfite Agar, a selective medium for Salmonella (Difco manual, Becton Dickinson and Co.). Colonies were counted after overnight incubation of the plates at 37°C. Two mice inoculated with sterile PBS were included as negative controls and their organs yielded no colonies. The second experiment yielded results similar to that shown in table 3.
Acknowledgements
We thank Katie Miller and Tim Hoover (University of Georgia) for the gift of strain TT22971 and plasmid pLac22.
All animal experiments were approved by the Institutional Animal Care and Use Committee.
Funding statement
This work was supported by award no. 1R21AI073322 from the National Institutes of Health.
References
- 1.Anet EFJ, Reynolds TM. 1954. Isolation of mucic acid from fruits. Nature 174, 930–932 (doi:10.1038/174930a0)13214046 [Google Scholar]
- 2.Whiting GC, Coggins RA. 1960. Organic acid metabolism in cider and perry fermentations. III. Keto-acids in cider-apple juices and ciders. J. Sci. Food. Agric. 11, 337–341 (doi:10.1002/jsfa.2740110608) [Google Scholar]
- 3.Marsh CA. 1985. An enzymatic determination of d-glucaric acid by conversion to pyruvate. Anal. Biochem. 145, 266–272 (doi:10.1016/0003-2697(85)90360-4) [DOI] [PubMed] [Google Scholar]
- 4.Matsui M, Fukuo A, Watanabe Y, Wanibe T, Okada M. 1977. Studies on the glucaric acid pathway in the metabolism of d-glucuronic acid in mammals. IV. Fluorometric method for the determination of glucaric acid in serum. Chem. Pharm. Bull. 20, 845–848 (doi:10.1248/cpb.20.845) [DOI] [PubMed] [Google Scholar]
- 5.Dutton GJ. 1980. Glucuronidation of drugs and other compounds, pp. 83–89 Boca Raton, FL: CRC Press [Google Scholar]
- 6.Moon TS, Yoon S, Lanza AM, Rot-Mayhew JD, Jones-Prather KL. 2009. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl. Environ. Microbiol. 75, 589–595 (doi:10.1128/AEM.00973-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb TE, Abou-Issa H, Stromberg PC, Curley RC, Jr, Nguyen MH. 1993. Mechanism of growth inhibition of mammary carcinomas by glucarate and the glucarate: retinoid combination. Anticancer Res. 13, 2095–2099 [PubMed] [Google Scholar]
- 8.Lajolo C, Sgambato A, Maiorano E, Lucchese A, Favia E, Giuliani M. 2010. Calcium glucarate inhibits DMBA-induced oral carcinogenesis in the hamster: histomorphometric evaluation. Anticancer Res. 30, 843–849 [PubMed] [Google Scholar]
- 9.Blumenthal HJ, Lucuta VL, Blumenthal DC. 1990. Specific enzymatic assay for d-glucarate in human serum. Anal. Biochem. 185, 286–293 (doi:10.1016/0003-2697(90)90294-J) [DOI] [PubMed] [Google Scholar]
- 10.Hunter J, Maxwell J, Carrella M, Stewart DA, Williams R. 1971. Urinary d-glucaric acid excretion as a measure of hepatic enzyme induction in man. Clinic. Sci. 17, 564–571 (doi:10.1042/cs040010P) [DOI] [PubMed] [Google Scholar]
- 11.Foster JW, Spector MP. 1995. How Salmonella survive against the odds. Annu. Rev. Microbiol. 49, 145–174 (doi:10.1146/annurev.micro.49.1.145) [DOI] [PubMed] [Google Scholar]
- 12.Kwon YM, Ricke SC. 1998. Survival of a Salmonella typhimurium poultry isolate in the presence of propionic acid under aerobic and anaerobic conditions. Anaerobe 4, 251–256 (doi:10.1006/anae.1998.0177) [DOI] [PubMed] [Google Scholar]
- 13.Becker D, Selbach C, Rollenhagen MC, Ballmaier M, Meyer TF, Mann M, Bumann D. 2006. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307 (doi:10.1038/nature04616) [DOI] [PubMed] [Google Scholar]
- 14.Bumann D. 2009. System-level analysis of Salmonella metabolism during infection. Curr. Opin. Microbiol. 12, 559–567 (doi:10.1016/j.mib2009.08.004) [DOI] [PubMed] [Google Scholar]
- 15.Trudgill PW, Widdus R. 1966. d-glucarate catabolism by pseudomonadaceae and enterobacteriaceae. Nature 211, 1097–1099 (doi:10.1038/2111097a0) [DOI] [PubMed] [Google Scholar]
- 16.Sharma BS, Blumenthal HJ. 1973. Catabolism of d-glucaric acid to α-ketoglutarate in Bacillus megaterium. J. Bacteriol. 116, 1346–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberton AM, Sullivan PA, Jones-Mortimer MC, Kornberg HL. 1980. Two genes affecting glucarate utilization in Escherichia coli K12. J. Gen. Microbiol. 117, 311–382 (doi:10.1099/0022187-117-2-377) [DOI] [PubMed] [Google Scholar]
- 18.Ansong C, et al. 2013. Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typimurium in response to infection-like conditions. Proc. Natl Acad. Sci. USA 110, 10 153–10 158 (doi:10.1073/pnas.1221210110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maier RJ. 2005. Use of molecular hydrogen as an energy substrate by human pathogenic bacteria. Biochem. Soc. Trans. 33, 83–85 (doi:10.1042/BST0330083) [DOI] [PubMed] [Google Scholar]
- 20.Maier RJ, Olczak A, Maier S, Soni S, Gunn J. 2004. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect. Immun. 72, 6294–6299 (doi:10.1128/IAI.72.11.6294-6299.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamichhane-Khadka R, Kwiatkowski A, Maier RJ. 2010. The Hyb Hydrogenase permits hydrogen-dependent respiratory growth of Salmonella enterica serovar Typhimurium. mBio 1, e00284 (doi:10.1128/mBio.00284-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta NS, Benoit S, Mysore JV, Sousa RS, Maier RJ. 2005. Helicobacter hepaticus hydrogenase mutants are deficient in hydrogen-supported amino acid uptake and in causing liver lesions in A/J mice. Infect. Immun. 73, 5311–5318 (doi:10.1128/AI.73.9.5311-5318.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamichhane-Khadka R, Frye JG, Porwollik S, McClelland M, Maier RJ. 2011. Hydrogen-stimulated carbon acquisition and conservation in Salmonella enterica serovar Typhimurium . J. Bacteriol. 193, 5824–5832 (doi:10.1128/JB.05456-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hubbard BK, Koch M, Palmer DR, Babbitt PC, Gerit JA. 1998. Evolution of the enzymatic activities in the enolase superfamily: characterization of the (d)-glucarate/galactarate catabolic pathway in Escherichia coli. Biochemistry 37, 14 369–14 375 (doi:10.1021/bi981124F) [DOI] [PubMed] [Google Scholar]
- 25.Schmidt DMZ, Hubbard BK, Gerlt JA. 2001. Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of d-glucarate dehydratase from Escherichia coli . Biochemistry 40, 10 054–10 062 (doi:10.1021/bi011640x) [DOI] [PubMed] [Google Scholar]
- 26.Yew WS, Fedorov AA, Fedorov EV, Almo SC, Gerlt JA. 2007. Evolution of the enzymatic activities in the enolase superfamily: l-talarate/Galactarate dehydratase from Salmonella typhimurium LT2. Biochemistry 46, 9564–9577 (doi.10.1021/bi7008882) [DOI] [PubMed] [Google Scholar]
- 27.Aghaie A, et al. 2008. New Insights into the alternative d-glucarate degradation pathway . J. Biol. Chem. 283, 15 638–15 646 (doi:10.1074/jbc.M800487200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zbell AL, Maier RJ. 2009. Role of the Hya hydrogenase in recycling of anaerobically produced H2 in Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 75, 1456–1459 (doi:10.1128/AEM.02064.08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawers RG, Jamieson DJ, Higgins CF, Boxer DH. 1986. Characterization and physiological roles of membrane-bound hydrogenase isoenzymes from Salmonella typhimurium. J. Bacteriol. 168, 398–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutnick D, Calvo JM, Klopotowski T, Ames BN. 1969. Compounds which serve as the sole source of carbon or nitrogen for Salmonella Typhimurium LT-2. J. Bacteriol. 100, 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gotz A, Goebel W. 2010. Glucose and glucose 6-phosphate as carbon sources in extra- and intracellular growth of enteroinvasive Esherichia coli and Salmonella enterica. Microbiology 156, 1176–1187 (doi:10.1099/mic.0.034744-0) [DOI] [PubMed] [Google Scholar]
- 32.Chaudhuri RR, et al. 2013. Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet. 9, e1003456 (doi:10.1371/journal.pgen.1003456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowden SD, Rowley G, Hinton JCD, Thompson A. 2009. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar Typhimurium. Infect. Immun. 77, 3117–3126 (doi:10.11128/AI.00093-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Götz A, Eylert E, Eisenreich W, Goebel W. 2010. Carbon metabolism of enterobacterial human pathogens growing in epithelial colorectal adenocarcinoma (Caco-2) cells. PLoS ONE 5, e10586 (doi:10.1371/journal.pone.0010586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenreich W, Dandekar T, Heesemann J, Goebel W. 2010. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412 (doi:10.1038/nrmicro2351) [DOI] [PubMed] [Google Scholar]
- 36.AbuOun M, Suthers PF, Jones GI, Carter BR, Saunders MP, Maranas CD, Woodward MJ, Anjum MF. 2009. Genome scale reconstruction of a Salmonella metabolic model: comparison of similarity and differences with a commensal Escherichia coli strain. J. Biol. Chem. 284, 29 480–29 488 (doi:10.1074/jbc.M109.005868) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (doi:10.1073/pnas.120163297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warren JW, Walker JR, Roth JR, Altman E. 2000. Construction and characterization of a highly regulable expression vector, pLAC11, and its multipurpose derivatives, pLAC22 and pLAC33. Plasmid 44, 138–151 (doi:10.1006/plas.2000.1477) [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning, a laboratory manual, 2nd edn, vol. III: A.3. Cold Spring Harbor Laboratory Press: New York [Google Scholar]
- 40.Tamayo R, Ryan SS, McCoy AJ, Gunn JS. 2002. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar typhimurium. Infect. Immun. 70, 6770–6778 (doi:10.1128/IAI.70.12.6770-6778.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]


