Abstract
Purine nucleotides are ubiquitous molecules that play vital roles in all kingdoms of life, not only as components of nucleic acids, but also participating in signaling and energy storage. Cellular pools of purines are maintained by the tight control of several complementary and sometimes competing processes including de novo biosynthesis, salvage and catabolism of nucleotides. While great strides have been made over the past sixty years in understanding the biosynthesis of purines, we are experiencing a renaissance in this field. In this feature article we discuss the most recent discoveries relating to purine biosynthesis, with particular emphasis upon the dynamic multi-protein complex called the purinosome. In particular we highlight advances made towards understanding the assembly, control and function of this protein complex and the attempts made to exploit this knowledge for drug discovery.
1. Introduction
Purines are essential molecules that serve a variety of functions and are utilized by all forms of life. Purines are components of a myriad of biomolecules that are vital for many cellular processes such as genetic transfer (DNA), translation and transcription (RNA), energy storage and transfer (ATP and GTP), signaling (cyclic AMP and GMP) and also act as cofactors (NADH, NADPH and coenzyme A) in varied biochemical reactions. While much is known about the metabolism of purines 1–3, particularly from work on prokaryotic pathways, and despite several decades of research, new and surprising findings are regularly being reported 4–11. The recent discoveries have led to renewed interest in the study of purine biosynthesis, its regulation and the relationship this pathway and its intermediates have with other fundamental cellular processes.
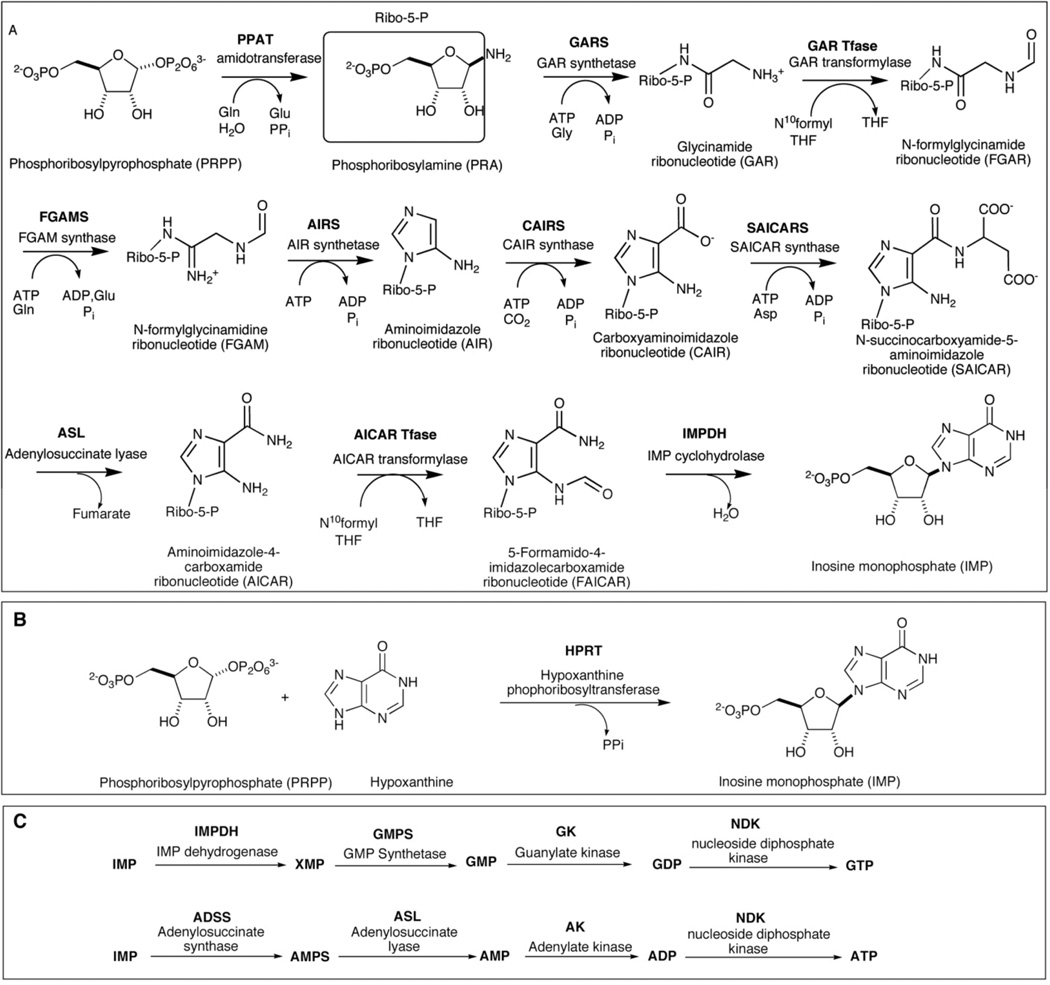
Cells access purine nucleotides through two separate mechanisms, de novo biosynthesis of inosine monophosphate (IMP) from 5-phosphoribosyl-1-pyrophosphate (PRPP, Fig. 1A) or through a salvage pathway that utilizes hypoxanthine, adenine or guanine (Fig. 1B). IMP is the point of convergence for the two pathways, as the purine nucleotides adenine and guanine are both separately synthesized from IMP in two additional steps (Fig. 1C). When the level of hypoxanthine is sufficient for cell growth (30 µM in one case 12), purine nucleotides are synthesized preferentially by the salvage pathway. The regulatory capacity of the de novo pathway, however, is significant larger than that of the salvage pathway and has a much greater effect upon growth rate 12, 13. This increased production of purines via the de novo pathway is intricately linked to cell growth and malignant transformation 14, 15. The level of purine de novo biosynthesis varies widely amongst different human tissues. While liver and skeletal muscle tissue were reported to have relatively high rates of biosynthesis, the activity of the pathway enzymes in brain cells is only 25 – 30% of that in liver. Bone marrow cells, however, are believed to have limited capacity for de novo biosynthesis of purines 16–19. Those tissues that have inherently limited biosynthetic capacity would be expected to rely upon the purine salvage pathway or obtain purines from other tissues.
Figure 1.
Purine biosynthesis pathways. (A) The de novo purine biosynthetic pathway in human. The pathway contains 10 chemical steps and 6 human enzymes are involved. The trifunctional enzyme: TGART including GARS, GAR Tfase and AIRS, catalyzes the 2, 3 and 5 steps. The bifunctional enzyme PAICS contains CAIRS (6th step) and SAICARS (7th step). The last 2 steps are catalyzed by another bifunctional enzyme: ATIC, which is composed of AICAR transformylase and IMP cyclohydrolase. (B) The purine salvage pathway. HPRT transfers the hypoxanthine onto PRPP to form IMP. GMP or AMP can be synthesized by the same pathway from guanine or adenine using GPRT or APRT, respectively. (C) A simplified representation of the biosynthesis of GTP and ATP downstream of the de novo biosynthesis of IMP. GTP and ATP are synthesized from IMP via four steps in two separate pathways
There are numerous disorders of purine and pyrimidine metabolism that affect humans and result, primarily, from genetic deficiencies of metabolic enzymes. There have been at least thirty different defects of purine and pyrimidine metabolism identified and seventeen of these are known to cause human disease 20. One of the most common of these, stemming from a deficiency of the salvage enzyme hypoxanthine phosphoribosyl transferase (HPRT), leads to hyperuricemia, a condition where the level of uric acid in the blood is higher than the normal range (360 µmol/L for women, 400 µmol/L for men), and a disorder called Lesch-Nyhan disease 21, 22. A similar hyperuricemic metabolic disorder results from mutation of phosphoribosylpyrophosphate synthetase (PRPPS). In this case, the overactivity of this enzyme that delivers purine precursors to the pathway causes the hyperuricemia 22. Hyperuricemia can also lead to debilitating inflammatory arthritis, called gout 23, 24. The hyperuricemia characteristic of gout is believed to stem from a variety of metabolic defects, including HPRT deficiency and accelerated purine biosynthesis or transport 24, 25. Several disorders of purine metabolism can also lead to immunodeficiency. Multiple mutations on the gene encoding adenosine deaminase have been identified and are believed to be the major cause of severe combined immunodeficiency disease (SCID 26). A deficiency of purine nucleoside phosphorylase can also lead to a clinical syndrome similar to SCID 27. Mutations in the enzyme that catalyzes the eighth step in purine biosynthesis, adenylosuccinate lyase (ASL), can lead to mental retardation and seizures 28. Similarly, a more recently discovered disorder, which results from a buildup of aminoimidazole carboxamideribonucleotide (AICAR), is caused by a defect in the transformylase activity of ATIC and can lead to impaired development or neurologic disease 29.
Despite the serious health implications that can result from defects, the redundancy in purine metabolism resulting from the presence of both a de novo and salvage pathway has precipitated the development of many clinically important drugs that target these pathways. In fact, greater than 20% of all approved oncology drugs are purine or pyrimidine antimetabolites 30. Examples of these include thiopurines, used to treat acute lymphocytic leukemia 31, and purine analogues such as fludarabine, nelarabine, cladribine and others 31. In addition to drugs that target purine metabolic enzymes directly, antifolates have also been employed to disrupt purine biosynthesis by interfering with the biosynthesis of folate cofactors. These molecules, which include methotrexate and pemetrexed, are anticancer agents and are also employed as a treatment for autoimmune disorders including rheumatoid arthritis 32–34.
2. The de novo biosynthesis of purines
The de novo biosynthesis of purines is a highly conserved pathway that forms inosine monophosphate (IMP) from the 5-phosphoribosyl-α-1-pyrophosphate (PRPP) and glutamine with the help of several cofactors and four to six molecules of ATP. Seminal discoveries by Buchanan and coworkers starting in the 1950s † 35, 36 established that ten enzymatic activities are required to carry out this conversion in chicken liver extracts. More recent studies, in particular the availability of complete genomes for a variety of organisms have allowed for the further characterization of this pathway in both prokaryotes and eukaryotes. In bacteria, the ten chemical transformations of the pathway employ as many as twelve enzymatic activities and require additional ATP to complete. Humans, however, make use of several multifunctional proteins to carry out the complete transformation from PRPP to IMP.
The ten, highly conserved, chemical steps of de novo purine biosynthesis in humans are catalyzed by six enzymes (Fig. 1A). These enzymes include phosphoribosylpyrophosphate amidotransferase (PPAT), a trifunctional enzyme (TGART) which is composed of glycinamide ribonucleotide synthetase (GARS), GAR formyltransferase (GART) and aminoimidazole ribonucleotide synthetase (AIRS), formylglycinamidine ribonucleotide synthase (FGAMS), a bifunctional enzyme (PAICS) which is composed of carboxyaminoimidazole ribonucleotide synthase (CAIRS) and succinoaminoimidazolecarboxamide ribonucleotide synthetase (SAICARS), adenylosuccinate lyase (ASL) and a bifunctional enzyme (ATIC) which is composed of aminoimidazolecarboxamide ribonucleotide transformylase (AICART) and inosine monophosphate cyclohydrolase (IMPCH). The first step in this pathway, catalyzed by PPAT, is rate-limiting and is known to be under allosteric control 13, 37. Two distinct nucleotide binding sites, both overlapping the PPAT active site, bind AMP, ADP, GMP and GDP and provide a mechanism of feedback control for the pathway 37, 38.
Most of what we know about the structures and mechanisms of the enzymes of the de novo purine biosynthetic pathway come from studies carried out on the prokaryotic proteins 2. The human enzymes range in size from approximately 57 kDa for PPAT to nearly 150 kDa for FGAMS and assume varied quaternary structures. While the structures of PPAT from both E. coli and B. subtilis 38, 39 reveal a tetrameric structure for this enzyme, work on the human enzyme suggests that there may be an equilibrium between the dimeric and tetrameric states 40. X-ray crystal structures have been solved for each of the three domains of human TGART; however an intact, full-length structure still eludes researchers 41–43. Small angle X-ray scattering analysis has suggested that in solution this trifunctional enzyme assumes a relatively disordered, extended conformation 42. This observation may indicate that an additional protein, such as FGAMS or PPAT, may bind to TGART and interact with two or all three of the domains. The octameric crystal structure of human PAICS 44 and dimeric human ATIC 45, 46 have also been reported. While the structure of ATIC reveals an extensively intertwined obligate dimer, there is little structural evidence that channeling of the intermediate could traverse the approximately 50 Å between the active sites of this bifunctional enzyme. Conversely, the structure of PAICS reveals a potential tunnel system within the enzyme to accommodate channeling of the intermediate, CAIR.
3. The purinosome
For several decades there has been a great deal of speculation that the enzymes of the de novo purine biosynthetic pathway come together to form a functional complex. Several lines of evidence, accumulated over the years, support this hypothesis. One of the first indications of this phenomenon was the co-purification of enzymes from native sources 47. Sequence information on the genes in the pathway provided additional support for complex formation. Not only does the mammalian pathway utilize several multifunctional enzymes, the trifunctional protein TGART catalyzes three non-sequential steps in the pathway (Fig. 1A). The most logical explanation for the retention of such a fusion protein in the genome is an evolutionary driving force that favors an important interaction between this protein and the protein that catalyzes the intermediate step, FGAMS.
Additional evidence that these proteins assemble into a complex comes in the form of kinetic arguments. An investigation of the degradation of the first intermediate in the pathway, PRA (Fig. 1A), suggests that this molecule is unstable and possesses a short half life in solution 48. In-depth kinetic analysis of the reaction catalyzed by the first two enzymes in the bacterial pathway argues in favor of complexation and presents a model that is not consistent with free diffusion of the reaction intermediate 49. More recently, several proteomics initiatives have identified interactions between members of the de novo purine biosynthetic pathway. Havigimana et al. identified nearly 14,000 physical interactions in human cells using an integrative global proteomic profiling approach based upon chromatographic separation 50. One of the interactions identified in this study was between PAICS and TGART. Zhang et al. employed a structure based prediction of protein-protein interactions to identify more than 300,000 interactions in humans with high confidence 51. Among these results were the prediction that FGAMS interacts with both TGART and PAICS.
3.1 Discovery of the purinosome
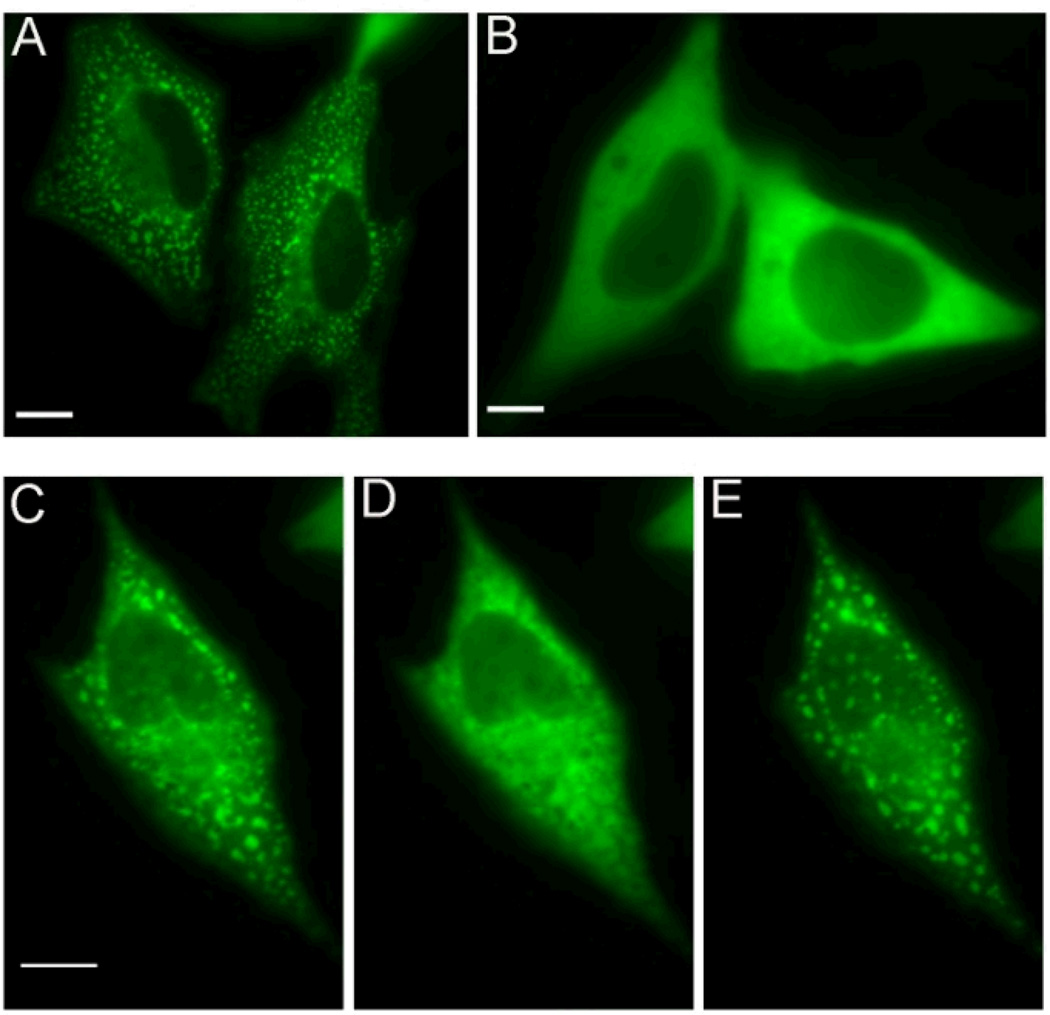
Despite the mounting evidence that the proteins in this pathway assemble in some functional manner, it wasn’t until recently that direct evidence for such a complex was presented. Using fluorescence microscopy, the reversible compartmentalization of all six enzymes of the purine biosynthetic pathway was observed in the cytoplasm of several cancer cell lines 5. The phenomenon was demonstrated to be dependent on the level of purines in the media and was shown to be reversibly dynamic (Fig. 2). Immunofluorescence imaging of this protein assembly also demonstrated that the complex was observed in non-transfected cells with only endogenous levels of proteins present. A control protein that is involved in a related pathway, C1-Tetrahydrofolate Synthase (C1-THF), did not cluster with the purine biosynthetic proteins. These observations provided strong evidence that a multi-enzyme complex, termed the purinosome, forms in cells under conditions of purine depletion.
Figure 2.
Images of the purinosome. The pathway 4th enzyme FGAMS was fused with green fluorescent protein (GFP): FGAMS-GFP. (A) Purinosomes formed in HeLa cells transiently transfected with FGAMS-GFP in purine-depleted medium. (B) Diffuse fluorescence signal of FGAMS-GFP in HeLa cells in purine-rich medium. Reversible formation of clusters by FGAMS-GFP is shown in C, D and E. (C) Purinosome formed when HeLa cells are cultured in purine-depleted medium. (D) Purinosomes disperse within 2 hours upon incubation with purine-rich medium. (E) Purinosomes reformed after returning to purine-depleted medium for 1 hour.
While providing the first direct evidence for protein complexation in the de novo purine biosynthetic pathway, the purinosome is not a unique form of intracellular structure. In E. coli, proteinaceous microcompartments that sequester metabolic pathways involving unstable or toxic metabolites have recently been characterized 52, 53. A screen of yeast strains yielded a total of 180 proteins that formed discrete physical structures upon nutrient starvation 54. Similarly, 4 different filament-forming proteins were identified in screens of S. cerevisiae, D. melanogaster, and E. coli 55, 56. In all of the above studies, the protein complexes are believed to provide temporal and spatial regulation as well as to provide protection from degradation of the protein components and pathway intermediates.
3.2 Purinosome control, interactions and assembly
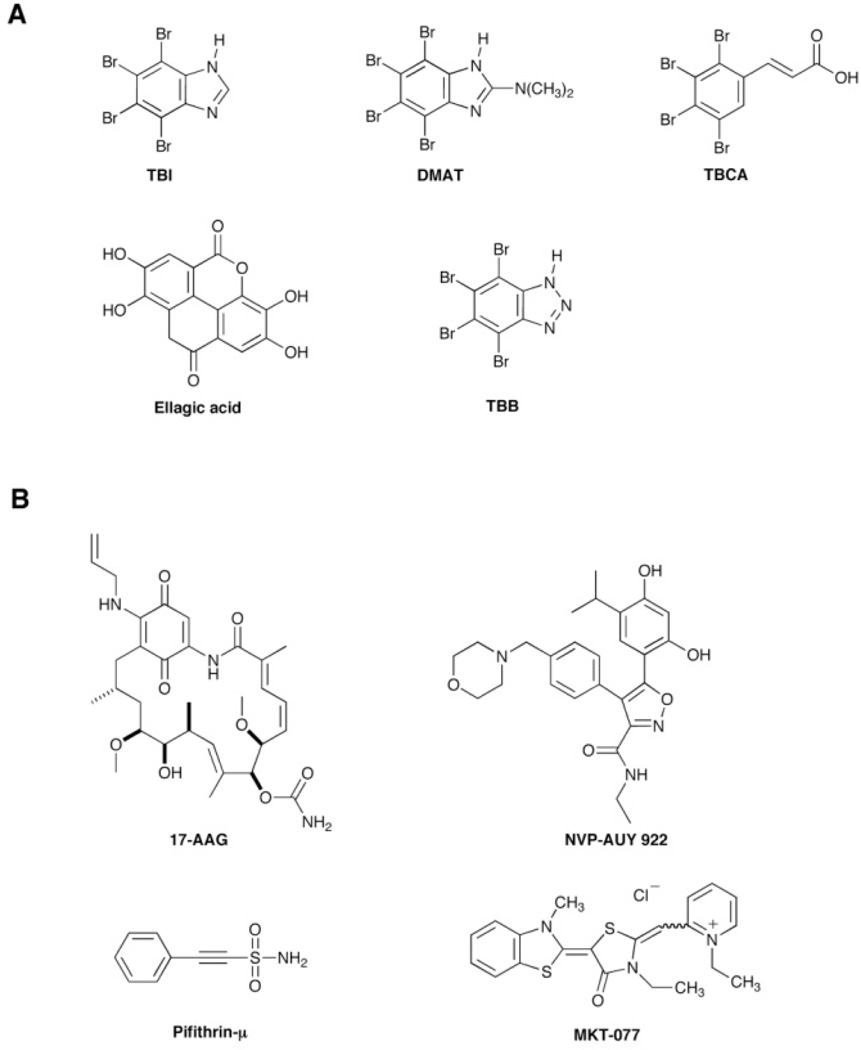
A more detailed investigation of the purinosome led to several interesting findings. Several effectors of human protein kinases were found to trigger the association or dissociation of purinosomes 57. The complex formation was promoted by the addition of one of several small molecule casein kinase II (CK2) specific inhibitors (Fig. 3A). The inhibitors 4,5,6,7-tetrabromo-1H–benzimidazole (TBI), 2-dimethylamino-4,5,6,7-tetrabromo-1H–benzimidazole (DMAT), tetrabromocinammic acid (TBCA), and 4,4’,5,5’,6,6’ - hexahydroxydiphenic acid 2,2’,6,6’ – dilactone (ellagic acid) all stimulated the formation of purinosomes. This result was confirmed by silencing endogenous CK2 using siRNA. Strangely, treatment with the CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) resulted in the converse effect and caused the dissociation of purinosomes. While it is not known whether the pathway proteins themselves are phosphorylated by this kinase, it is clear that at least CK2 or a CK2-mediated signaling cascade participates in the reversible regulation of purinosome assembly.
Figure 3.
Chemical structures of small molecules that affect the purinosome assembly inside cell. (A) Inhibitors of casein kinase II (CK2). (B) Inhibitors of Hsp90 and Hsp70 that have demonstrated to disrupt purinosomes.
A more recent study also implicated the Heat Shock Protein (Hsp) 90 and Hsp-70 chaperone machinery in the formation of the purinosome 58. Mass spectrometry analysis of an immunoprecipitation of FGAMS and associated proteins after treatment with a chemical cross-linking reagent yielded several chaperones and co-chaperones. Investigation of these proteins using fluorescence microscopy indicated that Hsp90 and Hsp70, as well as a set of co-chaperone proteins, co-localized dynamically with purinosomes. The abatement of Hsp70 or Hsp90 function through mutation or the decrease of co-chaperone expression using siRNA both led to decreases in purinosome content in the cell. These results suggested a possible means to disrupt purinosomes via inhibition of chaperone function. As expected, treatment of cells with several different Hsp90 or Hsp70 inhibitors: 17-N-allylamino-17-demethoxygeldamycin (17-AAG), 5-(2,4-dihydroxy-5-isopropyl-phenyl)-N-ethyl-4-[4-(morpholinomethyl) phenyl] isoxazole-3-carboxamide (NVP-AUY922), 2-phenylethynesulfonamide (Pifithrin-µ), 1-ethyl-2-[[3-ethyl-5-(3-methylbenzothiazolin-2-yliden)]-4-oxothiazolidin-2-ylidenemethyl] pyridinium chloride (MKT-077) (Fig. 3B) led to disruption of purinosomes. In fact, co-treatment of cancer cells with a combination of a known inhibitor of purine biosynthesis, methotrexate 32, and an Hsp90 inhibitor gave a synergistic cytotoxic effect. This work validates the purinosome as a viable target for novel cancer chemotherapeutics and opens up the possibility for the development of new combination therapies that block purine biosynthesis by targeting both individual enzymes within the pathway and the formation of the purinosome complex.
The organization of purine biosynthetic proteins into purinosomes in cells suggests some form of intracellular spatial control of these proteins. Indeed, a microtubule-assisted mechanism for functional purinosome formation was proposed based upon results of fluorescent live-cell imaging studies 59. This work determined that cytosolic purinosome clusters associated with microtubule filaments in the cell, but did not colocalize with the actin network. In addition, treatment with nocodazole, a known inhibitor of microtubule formation, not only caused the disruption of purinosomes but also led to a decrease in flux through the de novo purine biosynthetic pathway.
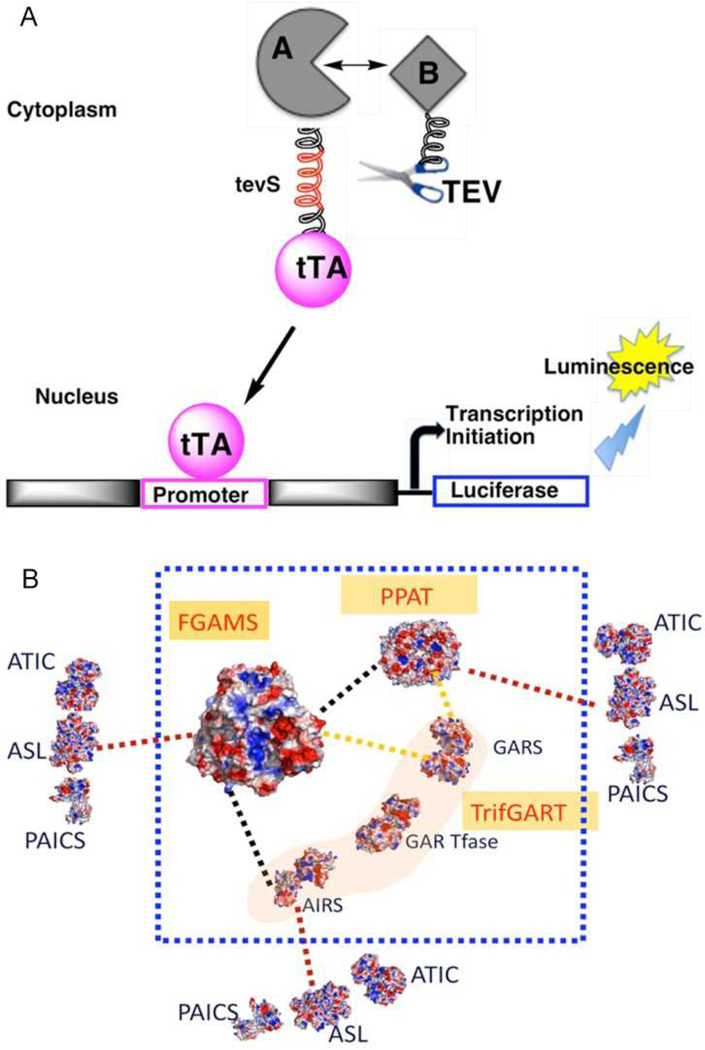
A further investigation into the organization of the purinosome employed a novel variation of the Tango assay 60 in order to examine potential protein-protein interactions. In this assay a transcription factor is tethered to the target protein with a linker that contains the cleavage site for a protease (Fig. 4). The protease, which is linked to a second protein, will cleave off the transcription factor only when the two proteins are in close proximity. Using this assay interactions between all six enzymes of the de novo purine biosynthetic pathway were detected and evidence for a core of the proteins that catalyze the first three steps was uncovered 61. PPAT, TGART and FGAMS exhibited the strongest degree of interaction and presumably nucleate formation of the overall complex. The remaining three enzymes, PAICS, ASL and ATIC interact individually with the core enzymes, but also were determined to interact with one another.
Figure 4.
A modified Tango assay was employed to detect and quantify protein-protein interactions among purinosome proteins. (A) Bait protein A is fused with the transcription factor, tTA linked by a peptide containing the recognition site (tevS) for a modified TEV protease. The prey protein, B, is linked to the modified TEV protease. When a protein-protein interaction occurs between proteins A and B, the TEV protease cleaves at tevS and releases tTA. This transcription factor enters the nucleus and turns on transcription of a luciferase gene. A representation of the interactions measured using this technique is provided in (B). The color of the lines represents the relative strength of the interaction detected between the two protein partners (from strongest to weakest – black, yellow, red). This work demonstrated that FGAMS, PPAT and TGART form a strong core complex that the other enzymes interact with more transiently.
The modified Tango assay employed to investigate the purinosome provided details about the interactions between proteins, but was unable to answer any questions about the signals that drive the assembly or disassembly of this complex.
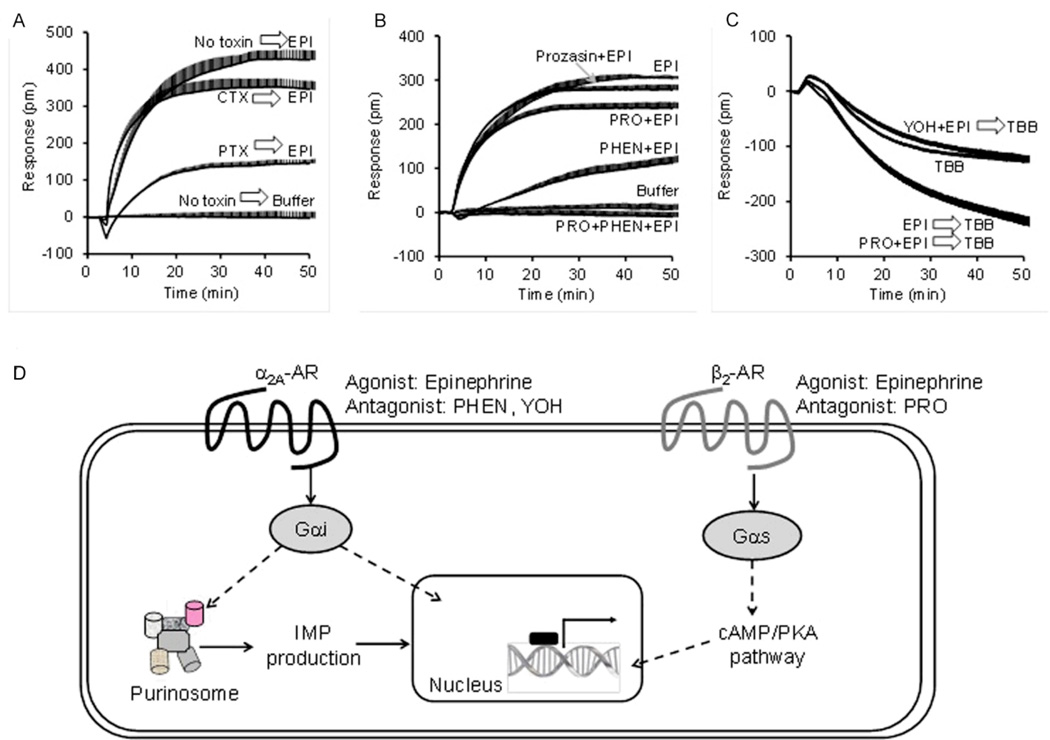
In addition to what has been reported about the involvement of kinases 57, Verrier et al. reported that GPCRs can also regulate the assembly of the purinosome 62. The regulation of purinosome by small molecules targeting intracellular signaling proteins such as casein kinases suggests that the purinosome dynamics is under control of cell signaling 57. To discover the upstream receptor signaling pathways, a methodology combining a label-free biosensor-enabled dynamic mass redistribution (DMR) assay with immunofluorescence imaging and molecular biology techniques was developed 62. The DMR assay is capable of translating an agonist-activated signal transduction process into a real-time whole cell phenotypic response, leading to a characteristic DMR signal 63, 64. Using this label-free technique in conjunction with fluorescent live-cell imaging, Verrier et al. determined that GPCR signaling was involved in the regulation of purinosomes 62. Changes in DMR responses arising from the purinosome modulators DMAT and TBB were observed for several known adrenergic receptor agonists. Investigation with agents specific for α2A-AR or β2-AR confirmed that purinosome formation was specifically associated with α2A-AR activation (Gαi pathway, Fig 5). The ability to promote purinosome formation was found to be a general mechanism for several endogenous Gαi - coupled receptors including purinergic P2Y receptors, LPA receptors, and prostaglandin receptors in HeLa cells. These findings suggest that, in addition to direct regulation of growth promoting genes in the nucleus, GPCR signaling also regulates purinosome assembly, an essential step in cell mitotic proliferation 65.
Figure 5.
GPCR signaling regulates purinosome formation. (A) The epinephrine (EPI) DMR contains contributions from both Gαi and Gαs pathways, as evidenced by the pathway deconvolution data using G protein-specific toxins. (B) Pharmacological profiling using co-stimulation of EPI with known AR antagonists showing that prazosin had little effect, propranolol slightly suppressed, phentolamine or yohimbine markedly suppressed, but the propranolol and phentolamine combination completely inhibited the EPI signal. Prazosin is a potent α1-selective blocker, while propranolol (PRO) is a β-blocker, phentolamine (PHEN) and yohimbine (YOH) are two α2-selective blockers. Together with quantitative real time-PCR, it was demonstrated that HeLa cells endogenously express functional Gαs-coupled β2-AR and Gαi-coupled α2A–AR. (C) 4,5,6,7-tetrabromobenzotriazole (TBB), which is known to dissociate the preassembled purinosome via a non-casein kinase II pathway, triggered a negative DMR. Pretreatment with EPI alone potentiated the TBB DMR, suggesting that EPI increases the number of purinosome complexes for TBB to dissemble. The co-pretreatment of EPI with YOH inhibited the EPI-induced potentiation, but propranolol had no impact, suggesting that EPI induces the formation of purinosome via the α2A–AR. (D) Schematic representation of the expression and signaling of α2A-AR and β2-AR in HeLa cells. Using both DMR and imaging, formation of the purinosome was determined to be promoted by the Gαi-coupled α2A-AR, but not Gαs-coupled β2-AR in HeLa cells.
In addition to being a potential target for cancer chemotherapeutics, the purinosome has also been implicated in AICA-ribosiduria and ASL deficiency 66, 67. Cultured skin fibroblasts from patients with these diseases harbored mutations that destabilized purinosomes. These mutations cause structural instability in the enzymes which are known to be correlated with the disease phenotype. The results demonstrate that formation of purinosomes are dependent upon structurally intact ATIC and ASL, presumably in addition to the other members of the pathway.
3.3 Linking purine biosynthesis and the purinosome to other metabolic pathways
At two separate steps within the de novo purine biosynthetic pathway a formyl group is added to the intermediate using a tetrahydrofolate-derived cofactor (Fig. 1A). The necessity for these cofactors in purine biosynthesis is evident in the widespread use of anti-folates employed as anti-cancer agents to disrupt purine biosynthesis 68. The supply of these one-carbon donors is maintained by a folate-dependent one-carbon metabolic network that facilitates the interconversion of tetrahydrofolates 69, 70. Considering the importance of this pathway to purine biosynthesis, it was not surprising that one of its members, methenyltetrahydrofolate synthetase (MTHFS), was found to colocalize with purinosomes 71. Functional MTHFS was shown to be necessary for flux through the purine biosynthetic pathway, as mice deficient in MTHFS activity showed reduced levels of purine biosynthesis. In addition, the MTHFS protein appeared to require sumoylation in order to be trafficked to the purinosome. As well as linking the one-carbon metabolic network to the purinosome, these results provide additional evidence that signaling events, such as post-translational modifications, may be necessary for purinosome formation.
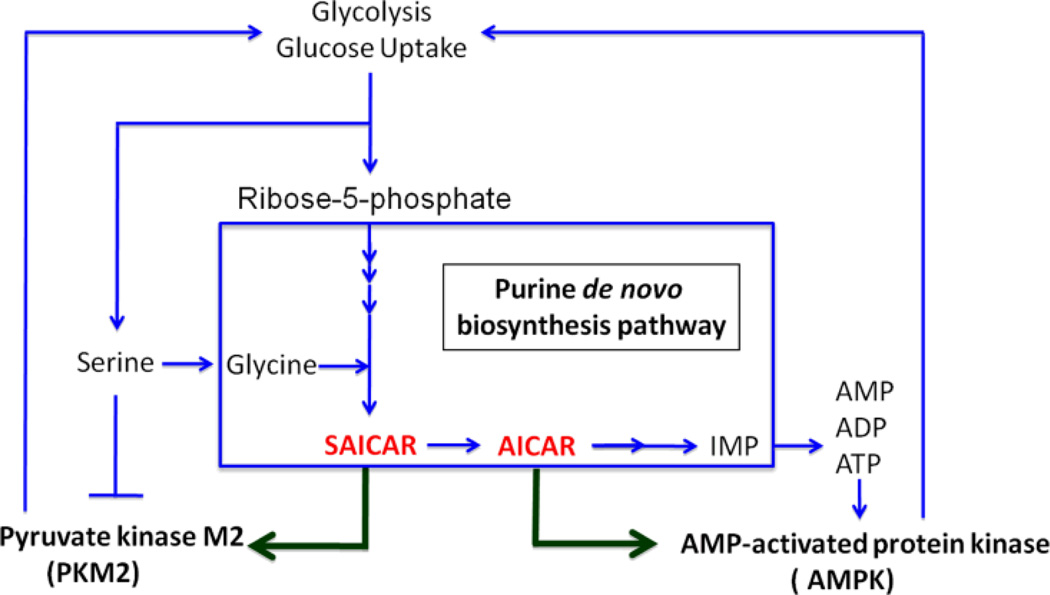
The growth of cancer cells is dependent upon metabolic reprogramming to increase glucose uptake and lactic acid fermentation, a phenomenon called the Warburg effect 72, 73. One of the enzymes involved in glycolysis, pyruvate kinase, may serve as a bridge between this pathway and purine biosynthesis. This kinase, which plays a vital role in the growth and metabolic reprogramming that occurs in cancer cells, was shown to be allosterically controlled by the purine biosynthetic pathway intermediate SAICAR 8. These studies suggest that the accumulation of SAICAR under conditions of glucose depletion is related to an increase in de novo purine biosynthesis. This hypothesis is consistent with the observation that in cells where the PAICS activity was knocked-down, levels of SAICAR were not inducible, whereas cells that were deficient in the downstream enzyme ASL had constitutively high levels of SAICAR. These results provide evidence for a possible link between purine biosynthesis and glycolysis. The starting material for purine biosynthesis, ribose-5-phosphate, is produced by the pentose phosphate pathway. The control of PKM2 by SAICAR-mediated allosteric stimulation may thus allow cells to adjust the generation of energy and metabolic flux in response to nutritional and energetic demands.
The de novo purine biosynthetic intermediate AICAR also plays a role in metabolic control, primarily through its effect on AMP kinase (AMPK). The AMPK system acts as a sensor of energy status in vivo and is activated by changes in AMP:ATP ratio 74. AICAR, which acts as an AMP analog, is a known agonist for AMPK. AMPK acts as a regulatory switch for several intracellular processes including glucose uptake, glycolysis, insulin signaling, lipogenesis, cell cycle control and others 74–76. Activators of AMPK, such as metformin, are widely used to treat diabetes and related metabolic disorders 74. Considering the cellular implications of AMPK activation it is clear that tight control over levels of activators, such as AICAR, be maintained within the cell. Formation of the purinosome represents a potential means to minimize AICAR-mediated AMPK activation during de novo nucleotide biosynthesis. Coordination of the enzymes of purine biosynthesis into such an assembly could limit the accumulation of AICAR in order to prevent undesired metabolic consequences. In this fashion, the purinosome may provide a further level of metabolic control and could represent a novel means to control AMPK signaling via modulation of AICAR levels.
In a systematic characterization of cancer cell metabolite consumption and release, glycine was shown to be highly correlated with rates of cancer cell proliferation 77. The more rapidly dividing cell lines showed a much higher dependence on the availability of glycine. This high degree of dependence of these cancer cells on glycine metabolism was demonstrated to be due, at least in part, to its role in supporting de novo purine biosynthesis. In addition, serine levels may also influence purine metabolism in cancer cells, both via its role as an allosteric activator of PKM2 and as a precursor to glycine 78. The regulation of PKM2 by serine levels provides a feedback mechanism for control of various metabolic pathways within the cell. Figure 6 summarizes some of the important relationships between intermediates in the purine biosynthetic pathway and several of the kinases responsible for metabolic control. While the significance of these metabolites in metabolic reprogramming and cancer progression are only beginning to be understood, it is clear that the regulation of purine levels in rapidly dividing cells is intimately tied to these processes and under very tight control. The fact that purinosomes are sensitive to purine levels as well as modulation by various kinases suggests that these protein assemblies may also play an important role in metabolic reprogramming.
Figure 6.
Summary of some kinase-mediated interactions between the purine biosynthetic pathway, its intermediates and energy metabolism in the cell.
Conclusions
The discovery of the purinosome has heralded a paradigm change in our understanding of nucleotide metabolism. The modulation of the assembly, dynamics and trafficking of this structure represent novel entry points for drug discovery efforts. With the validation of the purinosome as a means to improve the cytotoxicity of currently employed cancer chemotherapeutics, the potential has arisen for an entirely new class of purine antimetabolites that specifically target the purinosome or those factors that govern its assembly. Future challenges involve elucidating the role and extent of post-translational modifications on the de novo purine biosynthetic proteins and how these changes impact purinosome assembly. The interplay and impact of the purinosome with other metabolic pathways is only just beginning to be understood, and its role in metabolic reprogramming is an area that is bound to yield many exciting new insights. Similarly, the relationship between purinosome formation and the control of various important cellular processes such as the cell cycle, DNA replication, or glycolysis are yet to be fully elucidated. In addition, the level of local control over metabolite concentrations that is afforded by the purinosome is a topic that is still not well understood. These investigations will likely require new technologies that enable the measurement of local metabolite concentrations within cells at a very high degree of spatial and temporal resolution. Some imaging technologies that are likely to have a profound effect on how we study the purinosome and similar structures are already beginning to emerge. Super resolution imaging techniques such as Stochastic Optical Reconstruction Microscopy (STORM) and Stimulated Emission Depletion (STED) are examples of tools that will find great utility for the study of such phenomena. Finally, the characterization of the molecular details of the structure of the purinosome and its components is necessary to fully understand its function and to drive drug discovery efforts. Structural elucidation will certainly require a multidisciplinary approach that combines a myriad of biophysical techniques such as x-ray crystallography, NMR, mass spectrometry, super-resolution microscopy, label-free methods and computational approaches. In addition, as we begin to appreciate how important such dynamic multi-protein complexes are to myriad cellular processes, new tools will no doubt be needed to probe the functions, interactions and dynamics of such transient protein complexes in vivo.
Footnotes
Buchanan et al. published a series of 35 reports from 1952 through 1971 in the Journal of Biological Chemistry entitled ‘The Biosynthesis of Purines’. These seminal contributions laid the foundation for our current understanding of the enzymes and chemsitry invovled in purine biosynthesis.
References
- 1.Carter NS, Yates P, Arendt CS, Boitz JM, Ullman B. Adv Exp Med Biol. 2008;625:141–154. doi: 10.1007/978-0-387-77570-8_12. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Morar M, Ealick SE. Cell Mol Life Sci. 2008;65:3699–3724. doi: 10.1007/s00018-008-8295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zrenner R, Stitt M, Sonnewald U, Boldt R. Annu Rev Plant Biol. 2006;57:805–836. doi: 10.1146/annurev.arplant.57.032905.105421. [DOI] [PubMed] [Google Scholar]
- 4.Hicks KA, O'Leary SE, Begley TP, Ealick SE. Biochemistry. 2012;52:477–487. doi: 10.1021/bi301262p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An S, Kumar R, Sheets ED, Benkovic SJ. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 6.Treusch S, Hamamichi S, Goodman JL, Matlack KE, Chung CY, Baru V, Shulman JM, Parrado A, Bevis BJ, Valastyan JS, Han H, Lindhagen-Persson M, Reiman EM, Evans DA, Bennett DA, Olofsson A, DeJager PL, Tanzi RE, Caldwell KA, Caldwell GA, Lindquist S. Science. 2011;334:1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French JB, Ealick SE. Biochemistry. 2010;49:5975–5977. doi: 10.1021/bi1006755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller KE, Tan IS, Lee YS. Science. 2012;338:1069–1072. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji Y, Clark DV. Genetics. 2006;172:1621–1631. doi: 10.1534/genetics.105.045641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long H, Cameron S, Yu L, Rao Y. Genetics. 2006;172:1633–1642. doi: 10.1534/genetics.105.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng A, Uribe RA, Yieh L, Nuckels R, Gross JM. Development. 2009;136:2601–2611. doi: 10.1242/dev.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaoka T, Kondo M, Honda S, Iwahana H, Moritani M, Ii S, Yoshimoto K, Itakura M. J Biol Chem. 1997;272:17719–17725. doi: 10.1074/jbc.272.28.17719. [DOI] [PubMed] [Google Scholar]
- 13.Yamaoka T, Yano M, Kondo M, Sasaki H, Hino S, Katashima R, Moritani M, Itakura M. J Biol Chem. 2001;276:21285–21291. doi: 10.1074/jbc.M011103200. [DOI] [PubMed] [Google Scholar]
- 14.Natsumeda Y, Prajda N, Donohue JP, Glover JL, Weber G. Cancer Res. 1984;44:2475–2479. [PubMed] [Google Scholar]
- 15.Weber G, Lui MS, Natsumeda Y, Faderan MA. Adv Enzyme Regul. 1983;21:53–69. doi: 10.1016/0065-2571(83)90008-0. [DOI] [PubMed] [Google Scholar]
- 16.Brosh S, Boer P, Zoref-Shani E, Sperling O. Biochim Biophys Acta. 1982;714:181–183. doi: 10.1016/0304-4165(82)90143-x. [DOI] [PubMed] [Google Scholar]
- 17.Howard WJ, Kerson LA, Appel SH. J Neurochem. 1970;17:121–123. doi: 10.1111/j.1471-4159.1970.tb00509.x. [DOI] [PubMed] [Google Scholar]
- 18.Murray AW. Annu Rev Biochem. 1971;40:811–826. doi: 10.1146/annurev.bi.40.070171.004115. [DOI] [PubMed] [Google Scholar]
- 19.Watts RW. Adv Enzyme Regul. 1983;21:33–51. doi: 10.1016/0065-2571(83)90007-9. [DOI] [PubMed] [Google Scholar]
- 20.Jurecka A. J Inherit Metab Dis. 2009;32:247–263. doi: 10.1007/s10545-009-1094-z. [DOI] [PubMed] [Google Scholar]
- 21.Lesch M, Nyhan WL. Am J Med. 1964;36:561–570. doi: 10.1016/0002-9343(64)90104-4. [DOI] [PubMed] [Google Scholar]
- 22.Nyhan WL. Mol Genet Metab. 2005;86:25–33. doi: 10.1016/j.ymgme.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 23.Jin M, Yang F, Yang I, Yin Y, Luo JJ, Wang H, Yang XF. Front Biosci. 2012;17:656–669. doi: 10.2741/3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.So A. Arthritis Res Ther. 2008;10:221. doi: 10.1186/ar2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson JF, Rosenbloom FM, Kelley WN, Seegmiller JE. J Clin Invest. 1968;47:1511–1516. doi: 10.1172/JCI105844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giblett ER, Anderson JE, Cohen F, Pollara B, Meuwissen HJ. Lancet. 1972;2:1067–1069. doi: 10.1016/s0140-6736(72)92345-8. [DOI] [PubMed] [Google Scholar]
- 27.Giblett ER. Ann N Y Acad Sci. 1985;451:1–8. doi: 10.1111/j.1749-6632.1985.tb27090.x. [DOI] [PubMed] [Google Scholar]
- 28.Jaeken J, Van den Berghe G. Lancet. 1984;2:1058–1061. [PubMed] [Google Scholar]
- 29.Marie S, Heron B, Bitoun P, Timmerman T, Van Den Berghe G, Vincent MF. Am J Hum Genet. 2004;74:1276–1281. doi: 10.1086/421475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker WB. Chem Rev. 2009;109:2880–2893. doi: 10.1021/cr900028p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allegra CJ, Hoang K, Yeh GC, Drake JC, Baram J. J Biol Chem. 1987;262:13520–13526. [PubMed] [Google Scholar]
- 33.Cronstein BN. Pharmacol Rev. 2005;57:163–172. doi: 10.1124/pr.57.2.3. [DOI] [PubMed] [Google Scholar]
- 34.Rollins KD, Lindley C. Clin Ther. 2005;27:1343–1382. doi: 10.1016/j.clinthera.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Buchanan JM, Hartman SC. Adv Enzymol Relat Areas Mol Biol. 1959;21:199–261. [Google Scholar]
- 36.Hartman SC, Buchanan JM. Annu Rev Biochem. 1959;28:365–410. doi: 10.1146/annurev.bi.28.070159.002053. [DOI] [PubMed] [Google Scholar]
- 37.Smith JL. Curr Opin Struct Biol. 1998;8:686–694. doi: 10.1016/s0959-440x(98)80087-0. [DOI] [PubMed] [Google Scholar]
- 38.Smith JL, Zaluzec EJ, Wery JP, Niu L, Switzer RL, Zalkin H, Satow Y. Science. 1994;264:1427–1433. doi: 10.1126/science.8197456. [DOI] [PubMed] [Google Scholar]
- 39.Chen S, Tomchick DR, Wolle D, Hu P, Smith JL, Switzer RL, Zalkin H. Biochemistry. 1997;36:10718–10726. doi: 10.1021/bi9711893. [DOI] [PubMed] [Google Scholar]
- 40.Holmes EW. Adv Enzyme Regul. 1980;19:215–231. doi: 10.1016/0065-2571(81)90017-0. [DOI] [PubMed] [Google Scholar]
- 41.Dahms TE, Sainz G, Giroux EL, Caperelli CA, Smith JL. Biochemistry. 2005;44:9841–9850. doi: 10.1021/bi050307g. [DOI] [PubMed] [Google Scholar]
- 42.Welin M, Grossmann JG, Flodin S, Nyman T, Stenmark P, Tresaugues L, Kotenyova T, Johansson I, Nordlund P, Lehtio L. Nucleic Acids Res. 2010;38:7308–7319. doi: 10.1093/nar/gkq595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Desharnais J, Greasley SE, Beardsley GP, Boger DL, Wilson IA. Biochemistry. 2002;41:14206–14215. doi: 10.1021/bi020522m. [DOI] [PubMed] [Google Scholar]
- 44.Li SX, Tong YP, Xie XC, Wang QH, Zhou HN, Han Y, Zhang ZY, Gao W, Li SG, Zhang XC, Bi RC. J Mol Biol. 2007;366:1603–1614. doi: 10.1016/j.jmb.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 45.Greasley SE, Horton P, Ramcharan J, Beardsley GP, Benkovic SJ, Wilson IA. Nat Struct Biol. 2001;8:402–406. doi: 10.1038/87555. [DOI] [PubMed] [Google Scholar]
- 46.Wolan DW, Greasley SE, Beardsley GP, Wilson IA. Biochemistry. 2002;41:15505–15513. doi: 10.1021/bi020505x. [DOI] [PubMed] [Google Scholar]
- 47.Smith GK, Mueller WT, Wasserman GF, Taylor WD, Benkovic SJ. Biochemistry. 1980;19:4313–4321. doi: 10.1021/bi00559a026. [DOI] [PubMed] [Google Scholar]
- 48.Schendel FJ, Cheng YS, Otvos JD, Wehrli S, Stubbe J. Biochemistry. 1988;27:2614–2623. doi: 10.1021/bi00407a052. [DOI] [PubMed] [Google Scholar]
- 49.Rudolph J, Stubbe J. Biochemistry. 1995;34:2241–2250. doi: 10.1021/bi00007a019. [DOI] [PubMed] [Google Scholar]
- 50.Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, Babu M, Craig SA, Hu P, Wan C, Vlasblom J, Dar VU, Bezginov A, Clark GW, Wu GC, Wodak SJ, Tillier ER, Paccanaro A, Marcotte EM, Emili A. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, Maniatis T, Califano A, Honig B. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka S, Sawaya MR, Yeates TO. Science. 2010;327:81–84. doi: 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]
- 54.Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O'Connell JD, Mirrielees J, Ellington AD, Marcotte EM. Proc Natl Acad Sci U S A. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noree C, Sato BK, Broyer RM, Wilhelm JE. J Cell Biol. 2010;190:541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. Nat Cell Biol. 2010;12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.An S, Kyoung M, Allen JJ, Shokat KM, Benkovic SJ. J Biol Chem. 2010;285:11093–11099. doi: 10.1074/jbc.M110.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.French JB, Zhao H, An S, Niessen S, Deng Y, Cravatt BF, Benkovic SJ. Proc Natl Acad Sci U S A. 2013;110:2528–2533. doi: 10.1073/pnas.1300173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. Proc Natl Acad Sci U S A. 2010;107:12872–12876. doi: 10.1073/pnas.1008451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, Axel R, Lee KJ. Proc Natl Acad Sci U S A. 2008;105:64–69. doi: 10.1073/pnas.0710487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng Y, Gam J, French JB, Zhao H, Benkovic SJ. J Biol Chem. 2012;287:36201–36207. doi: 10.1074/jbc.M112.407056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verrier F, An S, Ferrie AM, Sun H, Kyoung M, Deng H, Fang Y, Benkovic SJ. Nat Chem Biol. 2012;7:909–915. doi: 10.1038/nchembio.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang Y. Drug Discov Today Technol. 2010;7:e5–e11. doi: 10.1016/j.ddtec.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fang Y, Ferrie AM, Fontaine NH, Mauro J, Balakrishnan J. Biophys J. 2006;91:1925–1940. doi: 10.1529/biophysj.105.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohr K, Kostenis E. Nat Chem Biol. 2011;7:860–861. doi: 10.1038/nchembio.716. [DOI] [PubMed] [Google Scholar]
- 66.Baresova V, Skopova V, Sikora J, Patterson D, Sovova J, Zikanova M, Kmoch S. Hum Mol Genet. 2012;21:1534–1543. doi: 10.1093/hmg/ddr591. [DOI] [PubMed] [Google Scholar]
- 67.Duval N, Luhrs K, II TGW, Beresova V, Skopova B, Kmock S, Vacano GN, Zikanova M, Patterson D. Mol Gen Metab. 2013;108:178–189. doi: 10.1016/j.ymgme.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McGuire JJ. Curr Pharm Des. 2003;9:2593–2613. doi: 10.2174/1381612033453712. [DOI] [PubMed] [Google Scholar]
- 69.Fox JT, Stover PJ. Vitam Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 70.Shane B. Food Nutr Bull. 2008;29:S5–S16. doi: 10.1177/15648265080292S103. discussion S17–19. [DOI] [PubMed] [Google Scholar]
- 71.Field MS, Anderson DD, Stover PJ. Front Genet. 2011;2:36. doi: 10.3389/fgene.2011.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 73.Kim JW, Dang CV. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 74.Towler MC, Hardie DG. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- 75.Hardie DG, Ross FA, Hawley SA. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O'Neill LA, Hardie DG. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 77.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, Frezza C, O'Reilly M, Gottlieb E. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]