Abstract
The P1 plasmid addiction operon is a compact genetic structure consisting of promoter, operator, antitoxin gene (phd), and toxin gene (doc). The 73-amino-acid antitoxin protein, Phd, has two distinct functions: it represses transcription (by binding to its operator) and it prevents host death (by binding and neutralizing the toxin). Here, we show that the N terminus of Phd is required for repressor but not antitoxin activity. Conversely, the C terminus is required for antitoxin but not repressor activity. Only a quarter of the protein, the resolution limit of this analysis, was required for both activities. We suggest that the plasmid addiction operon is a composite of two evolutionarily separable modules, an operator-repressor module and an antitoxin-toxin module. Consideration of similar antitoxin proteins and their surroundings indicates that modular exchange may contribute to antitoxin and operon diversity.
Protein-ligand interactions.
The economy and structure of the cell is dependent upon the specificity of myriad protein-ligand interactions. The antitoxin proteins of toxin-antitoxin or plasmid addiction modules are a diverse group of small proteins, typically 72 to 104 amino acid residues in length, with multiple macromolecular ligands. Thus, they are a potentially rich source of novel protein-ligand motifs whose characterization may contribute to our understanding of protein-ligand interactions.
P1 plasmid addiction operon.
Phd is a bifunctional repressor/antitoxin protein encoded by the plasmid addiction operon of the P1 plasmid (which is the plasmid prophage of bacteriophage P1). Escherichia coli grows well without the P1 plasmid. Upon acquiring the P1 plasmid it continues to grow well, but upon losing the plasmid the cells become sick and cease growth. Thus, the cells have acquired a physiological addiction to the plasmid. This effect is mediated by the P1 plasmid addiction operon. This operon encodes a toxin, Doc, which causes death on curing, and an antitoxin, Phd, which prevents host death (33). While the plasmid is retained, the antitoxin is present in sufficient quantity to neutralize the toxin. However, the antitoxin is unstable, due to the action of the host-encoded ClpXP protease (34). When the plasmid is lost, the continuing proteolysis of the antitoxin, unreplenished by new synthesis, unveils the toxin and arrests the cell. By poisoning plasmid-free segregants, the module increases the apparent stability of the plasmid and eliminates competition from plasmid-free sister cells.
Autoregulation of the P1 plasmid addiction operon.
The P1 addiction operon is negatively autoregulated (37). The antitoxin binds as a dimer (18) to two palindromic sequences in the promoter region of the P1 addiction operon and thus inhibits transcription (37). The antitoxin interacts with toxin both in solution and in the repressive complex (17, 38). In the repressive complex, the toxin mediates cooperative interactions between the two palindromic sites and thereby enhances repression (38).
Toxin-antitoxin modules.
Analogous toxin-antitoxin pairs are common in prokaryotes. Some well-studied plasmid-encoded systems include CcdA/CcdB of F, Phd/Doc of P1, PemI/PemK of R100 (identical to Kis/Kid of R1), ParD/ParE of RK2 (and RP4), and HigA/HigB of Rts1 (reviewed in reference 19). Toxin-antitoxin systems can also be found on bacterial chromosomes. The E. coli chromosome, for example, encodes RelB/RelE, a toxin-antitoxin module that appears to be well conserved and widespread among Bacteria and Archaea (21), and two additional systems (including MazE/MazF) that are homologous to PemI/PemK (39). Some strains of E. coli also carry a chromosomal copy of the Phd/Doc system (57).
Phenomena.
Toxin-antitoxin modules have been implicated in diverse and important phenomena, such as plasmid stability (4, 33, 41), plasmid competition (12, 43), programmed cell death (1), growth control (9), and antibiotic sensitivity (53). In each case, if expression of the module is disrupted (by repression, by starvation, by loss of the module, by antibiotics, or even by the activation of other toxin-antitoxin modules [26]), the continuing proteolysis of the unstable antitoxin may liberate the toxin and arrest the cell.
Operon architecture.
The toxin-antitoxin modules share a number of functional, organizational, and regulatory features (reviewed in references 14, 19, 24, 29, 47, 59, and 61). Typically, the gene for the 72- to 104-amino-acid antitoxin immediately precedes the gene for the 92- to 126-amino-acid toxin. An exception to the rule is offered by the HigA/HigB system, in which the typical gene order is reversed (56). Typically, expression of the operon is negatively autoregulated by antitoxin or by an antitoxin-toxin complex (29). An exception is provided by the tripartite Omega-Epsilon-Zeta system, where Epsilon neutralizes Zeta and a third protein, Omega, represses transcription (61). Although these toxin-antitoxin modules share many general features, at the molecular level they are a diverse group comprising more than five families with little or no significant amino acid homology between families (19). Such diversity may indicate great age, positive selection for diversity, or multiple (polyphyletic) origins for the various toxin-antitoxin families. Here, we show that the repressor activity of Phd maps to its N-terminal regions, while the antitoxin activity of Phd maps to its C-terminal regions. We propose that the plasmid addiction operon is a composite of two functional modules, and we discuss evidence that modular exchange has contributed to antitoxin (and operon) diversity.
MATERIALS AND METHODS
Bacteria.
All E. coli strains were ultimately derived from MC1061 (7) by lysogenization with a lambda phage or by CaCl2 transformation with a plasmid. Strains were grown, manipulated, and stored by standard methods (2, 42, 51).
Media.
Bacterial strains were grown, with aeration, at 30°C in Luria-Bertani (LB) broth or on LB agar plates. The media were supplemented with 100 μg of ampicillin/ml, 30 μg of kanamycin/ml, and/or 80 mg of spectinomycin/ml, as indicated, in order to select for transformants or in order to maintain selection for resident plasmids. The medium was supplemented with 50 mM isopropyl-β-d-thiogalactopyranoside (IPTG), as indicated, to induce expression of the toxin Doc from the IPTG-inducible Ptac promoter (6). The medium was supplemented with 0.2% l-(+)-arabinose, as indicated, to induce expression of Phd or Phd mutants from the arabinose-inducible PBAD promoter (22).
Site-directed mutagenesis.
Site-directed mutagenesis of phd was accomplished using standard PCR (44) and recombinant DNA (51) techniques as previously described (37). Briefly, PCR with Taq polymerase was used to amplify all or portions of the phd gene. These phd segments were flanked by primer-encoded restriction enzyme sites, start codons, and stop codons, as indicated (Table 1). The pG3 plasmid (Table 2), which contains the entire P1 plasmid addiction operon, was used as the PCR template. The PCR products and the pBAD24 vector were digested with EcoRI and HindIII restriction enzymes, electrophoresed on low-melting-point agarose gel in Tris-acetate-EDTA buffer, ligated with T4 DNA ligase, and then introduced into E. coli by CaCl2 transformation. Transformants were selected on LB agar in the presence of 100 μg of ampicillin/ml. Four transformants were colony purified and further characterized by restriction digestion, electrophoresis, or dideoxynucleotide sequencing. A set of sequence-validated constructs (Table 2) was selected for further phenotypic characterization. These constructs were introduced into BR7024 and tested for repressor activity. The constructs were also introduced into BR7061 and tested for antitoxin activity.
TABLE 1.
Primers
| Primer nameand description | Primer sequencea (5′ to 3′) |
|---|---|
| Forward PCR primers | |
| JER074 wildtype | GGGAATTCATG CAATCCATTAACTTCCGT |
| JER075 Δ(2-17) | GGGAATTCATG AACAATGTTGAAGCCGG |
| JER076 Δ(2-35) | GGGAATTCATG GCAGTAATTGTCAGCAA |
| JER077 Δ(2-53) | GGGAATTCATG GCTGAATTTGCATCCCTG |
| Reverse PCR primers | |
| JER070 Δ(19-73) | GGGAAGCTTTTA GTTGAGCACTTCAGAAA |
| JER071 Δ(37-73) | GGGAAGCTTTTA TGCTGGCTCACGGCCTC |
| JER072 Δ(55-73) | GGGAAGCTTTTA AGCATCCAGCGCCGCTTT |
| JER073 wildtype | GGGAAGCTTTTA TCGGTTAACCAGTTCCTT |
| Sequencing primers | |
| JAS088F | CCATAAGATTAGCGGATCCTA |
| JAS090R | AGGTGGGACCACCGCGCTACTGC |
All primers are written 5′ to 3′. Primer encoded HindIII (AAGCTT) and EcoRI (GAATTC) sites are underlined. Primer encoded start codons (ATG) are italicized. Similarly, the reverse complements of the TAA ochre codons (TTA) are italicized.
TABLE 2.
Strains, phages, and plasmids
| Bacterial strains, phages, or plasmids | Relevant characteristics and construction | Source or reference |
|---|---|---|
| E. coli strains | ||
| MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (Strr) hsdR2 (rK− mK+) mcrA mcrB1 | 7 |
| BR7024 | λRDM12 in MC1061; used here as the repressor test strain | 37 |
| BR7021 | λRDM 11 (P phd+-lacZYA) placIQ in MC1061; parent of BR7061 | 38 |
| BR7061 | λRDM 11 placIQ pRDM082 (Ptac-doc+) in MC1061; used here as the antitoxin test strain | This work |
| Lambda phages | ||
| λRS45 | A lambda phage carrying a promoterless lacZYA | 54 |
| λRDM12 | P1 plasmid addiction promoter fused to lacZYA; derived from λRS45 | 37 |
| λRDM 11 | P phd+ fused to lacZYA; provides a low level of Phd; derived from λRS45 | 38 |
| Plasmids | ||
| placIQ | lacIq in pACYC177; p15A origin; Kanr; a source of Lac repressor | R. Kolodner |
| pKK223-2 | Ptac expression vector, pMB1 origin, Ampr | 6 |
| pRDM037 | Ptac-doc+ in pKK223-3 | 38 |
| pGB2 | Modest-copy-number cloning vector; pSC101 origin; Specr | 10 |
| pRDM082 | Ptac-doc+ from pRDM037 cloned between BamH1 and HindIII sites of pGB2 | This work |
| pGB2ts | A derivative of pGB2 whose replication is temperature sensitive | 11 |
| pG3 | P phd doc cloned into pGB2ts; used here as a PCR template | 33 |
| pBAD 24 | PBAD expression vector; pBR origin; Ampr | 22 |
| pJS004 | PBADphd+ (PCR of pG3 with JER074 and JER073 cloned into pBAD24) | This work |
| pJS005 | PBADphdΔ(2-17) (PCR of pG3 with JER075 and JER073 cloned into pBAD24) | This work |
| pJS006 | PBADphdΔ(2-35) (PCR of pG3 with JER076 and JER073 cloned into pBAD24) | This work |
| pJS007 | PBADphdΔ(2-53) (PCR of pG3 with JER077 and JER073 cloned into pBAD24) | This work |
| pJS001 | PBADphdΔ(19-73) (PCR of pG3 with JER074 and JER070 cloned into pBAD24) | This work |
| pJS002 | PBADphdΔ(37-73) (PCR of pG3 with JER074 and JER071 cloned into pBAD24) | This work |
| pJS003 | PBADphdΔ(55-73) (PCR of pG3 with JER074 and JER072 cloned into pBAD24) | This work |
Repressor activity.
Repressor activity was assessed by the ability of the test construct to repress transcription of a lacZ reporter fused to the P1 plasmid addiction promoter, using standard techniques (42) as previously described (37, 38).
Antitoxin activity.
Antitoxin activity was assessed by the ability of cells containing the test construct to grow in the presence of an otherwise lethal level of the toxin Doc. Plasmids carrying the phd constructs were introduced into the antitoxin test strain BR7061 (Table 2). The strains were grown overnight with aeration at 30°C in LB broth or LB agar containing 100 μg of ampicillin/ml, 30 μg of kanamycin/ml, and 80 μg of spectinomycin/ml to maintain selection for the resident plasmids. Cultures were then serially diluted into 0.7% NaCl and plated in the presence of antibiotics or in the presence of antibiotics plus 50 mM IPTG to induce expression of Doc and 0.2% l-(+)-arabinose to induce expression of the Phd constructs. Antitoxin activity was indicated by the efficiency of plating (EOP) in the presence and absence of the inducers.
RESULTS AND DISCUSSION
Two Phd activities.
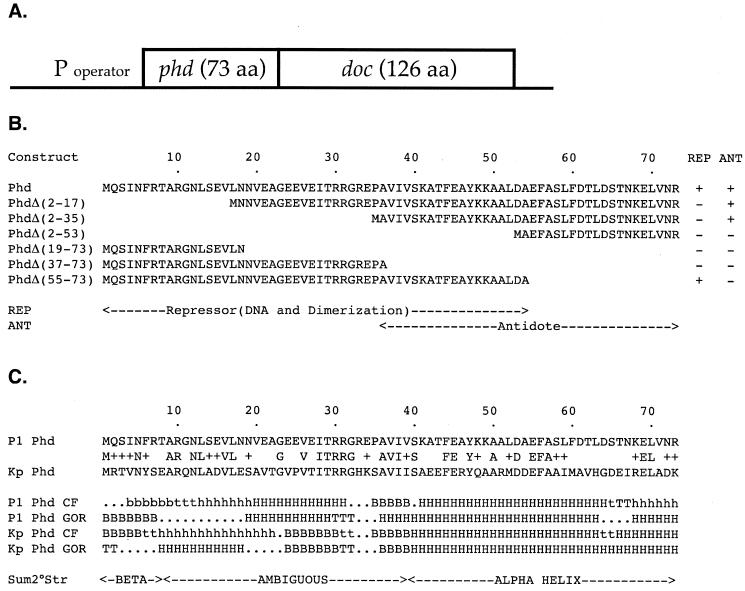
The 73-amino-acid Phd protein regulates transcription (by binding the promoter of the operon) and prevents cell death (by binding and neutralizing the toxin). Thus, it has two distinct and separate functions. Since DNA binding and toxin binding can occur simultaneously, it appears that these interactions involve different surfaces of Phd. In order to further examine the structure-function relationships in Phd, we generated N- and C-terminal deletions of Phd and then tested those deletion constructs for antitoxin activity and repressor activity.
Removal of Phd from its natural context.
Immediately upstream of phd is the operator region which contains a pair of Phd binding sites. Immediately downstream of phd is doc, the gene encoding the toxin which is bound and neutralized by Phd. In order to perform tests on the two activities of Phd, we placed the phd gene under the control of the heterologous arabinose-inducible PBAD promoter. By separating phd from its natural context, we were able (i) to alter the repressor activity of Phd without simultaneously altering the transcription of phd, (ii) to measure the intrinsic repressor activity of Phd in the absence of Doc, and (iii) to construct phd mutations lacking antitoxin activity (without immediately killing the host cell).
Repressor activity.
The phd mutations, encoding N-terminal and C-terminal deletions of Phd, were tested in vivo for repressor activity (Table 3). Repressor activity was indicated by the ability to reduce, relative to a vector control, the β-galactosidase specific activity produced by a transcriptional lacZ reporter fused to the promoter of the addiction operon (37). Deletion of the N-terminal 17 amino acids eliminated the repressor activity but not the antitoxin activity (Table 3), indicating that the N terminus of the protein is specifically involved in repressor functions (Table 3). Deletion of the C-terminal 18 amino acids decreased but did not abolish repression, indicating that this region was not absolutely required for repression. However, the deletion of the C-terminal half of Phd abolished repression, indicating that the third quarter, like the first quarter of Phd, was required for repressor activity (Table 3).
TABLE 3.
Repressor activities of Phd deletions
| Relevant genotypea | β-Galactosidase sp actb | Fold repressionc | Repressor activity |
|---|---|---|---|
| PBAD (no insert) | 5,235 | 1.00 | No |
| PBAD-phd+ | 262 | 19.99 | Yes |
| PBAD-phdΔ(2-17) | 4,672 | 1.12 | No |
| PBAD-phdΔ(2-35) | 4,665 | 1.12 | No |
| PBAD-phdΔ(2-53) | 4,564 | 1.15 | No |
| PBAD-phdΔ(19-73) | 4,625 | 1.13 | No |
| PBAD-phdΔ(37-73) | 4,307 | 1.21 | No |
| PBAD-phdΔ(55-73) | 1,301 | 4.02 | Partial |
The indicated constructs were introduced into BR7024 and tested for their ability to repress a lacZ reporter fused to the P1 plasmid addiction promoter.
β-Galactosidase specific activity is the average from three separate experiments. The standard deviations were less than 10% of the means.
Fold repression is the ratio of the β-galactosidase specific activity of the unrepressed control strain (no insert) divided by the β-galactosidase specific activity of the experimental strain.
Antitoxin activity.
The Phd deletion constructs were tested for antitoxin activity in vivo. Antitoxin activity was indicated by growth (colony formation) in the presence of an otherwise lethal level of toxin. In the test strain for antitoxin activity (Table 2) the toxin gene doc was placed, under the control of the IPTG-inducible Ptac promoter, on a pGB2 plasmid vector. A second plasmid, pLacIQ, provided the Lac repressor, which regulates the Ptac promoter. A chromosomally integrated λ prophage (λRDM11), which carries a single copy of phd under the control of its own promoter, provided sufficient antitoxin to neutralize the basal, uninduced level of Doc. In the absence of IPTG, this strain grows well. In the presence of IPTG, the increased expression of Doc increases, overwhelms the source of Phd, and arrests the cell (Table 4). To test for antitoxin activity, we introduced the Phd deletion constructs into this background and tested the ability of the constructs to neutralize Doc, and thus continue growth, in the presence of IPTG (to induce doc) and arabinose (to induce expression of the phd constructs). Growth was measured qualitatively, by the appearance of a streak (data not shown), and quantitatively, by enumeration of CFU (Table 4) on LB agar with ampicillin, kanamycin, spectinomycin, arabinose, and IPTG. By either measure, the deletion of the first and second quarters of Phd failed to eliminate antitoxin activity (Table 4), indicating that the C-terminal half of the protein was sufficient for antitoxin activity. Further deletion of the third quarter of Phd abolished antitoxin activity (Table 4). Deletion of the last, C-terminal quarter of Phd also abolished antitoxin activity (Table 4). Thus, the third and fourth quarters of Phd are both necessary and sufficient for antitoxin activity.
TABLE 4.
Quantitative antitoxin assay
| Relevant genotypea | CFU/mlb
|
EOPc | Relative EOPd | Antitoxin activity | |
|---|---|---|---|---|---|
| With inducers | Without inducers | ||||
| PBAD (no insert) | 2.33 × 102 | 2.28 × 108 | 1.02 × 10−6 | 1.2 × 10−5 | No |
| PBAD-phd+ | 1.78 × 107 | 2.13 × 108 | 8.35 × 10−2 | 1.0 × 100 | Yes |
| PBAD-phdΔ(2-17) | 3.73 × 107 | 1.27 × 108 | 2.94 × 10−1 | 3.5 × 100 | Yes |
| PBAD-phdΔ(2-35) | 4.95 × 107 | 3.88 × 108 | 1.27 × 10−1 | 1.5 × 100 | Yes |
| PBAD-phdΔ(2-53) | 4.95 × 102 | 1.63 × 108 | 3.03 × 10−6 | 3.6 × 10−5 | No |
| PBAD-phdΔ(19-73) | 4.00 × 102 | 2.33 × 108 | 1.71 × 10−6 | 2.0 × 10−5 | No |
| PBAD-phdΔ(37-73) | 4.03 × 102 | 2.24 × 108 | 2.37 × 10−6 | 2.8 × 10−5 | No |
| PBAD-phdΔ(55-73) | 5.46 × 102 | 2.30 × 108 | 1.79 × 10−6 | 2.1 × 10−5 | No |
Plasmids carrying the phd constructs were introduced into the antitoxin tester strain BR7061, which contains an IPTG-inducible source of the toxin Doc. The strains were grown overnight with aeration at 30°C in LB broth or LB agar containing 100 μg of ampicillin/ml, 30 μg of kanamycin/ml, and 80 μg of spectinomycin/ml to maintain selection for the resident plasmids.
Results are the means of three independent experiments. Standard deviations were less than 40% of the mean in all cases. Results with inducers were determined by serially diluting cultures into 0.7% NaCl and plating in the presence of 50 mM IPTG, to induce expression of Doc, and 0.2% l-(+)-arabinose to induce expression of the Phd constructs, and antibiotics, as described above, to maintain selection for the resident plasmids. Results without inducers were determined by plating and culturing under similar conditions, but without IPTG or arabinose.
The EOP is the ratio of CFU per milliliter in the presence and absence of the inducers.
Relative EOP is the EOP divided by the EOP of the strain with the PBAD-phd+ construct.
Predicted secondary structure.
Since homologs typically have similar structures as well as similar sequences, a comparison of predicted secondary structures for homologous sequences can be used to test or to improve the accuracy of secondary structure prediction. Here, we used two secondary structure prediction methods (those of Chou and Fasman [8] and Garnier, Osguthorpe, and Robson [16]) on two homologous proteins (Phd and a Klebsiella homolog of Phd [Genome Sequencing Center, Washington University, St. Louis, Mo.]). For either method applied to either protein, the secondary structure predictions suggested that the C-terminal half of Phd is predominantly α-helical (Fig. 1). Predictions for the N terminus were less concordant but indicated that the N-terminal half of Phd is likely to contain a β-sheet segment and an α-helical segment. Consistent with these secondary structure predictions, circular dichroism measurements of purified Phd indicate that the protein is ∼45% α-helical (18).
FIG. 1.
(A) Schematic structure of the P1 addiction operon. (B) Deletion analysis of Phd: predicted protein products and summary of their activities. The predicted protein products of phd and various deletion mutants of phd are shown. The repressor (REP) and antitoxin (ANT) activities of each construct are indicated. Repressor activity was indicated by the ability to repress transcription of a lacZ reporter fused to the P1 addiction promoter (Table 3). Antitoxin activity was indicated by the ability of the cells to grow in the presence of an otherwise lethal level of the Doc toxin (Table 4). (C) Secondary structure predictions for Phd and a Klebsiella homolog. The aligned sequences of Phd and a Phd homolog from K. pneumoniae (Kp) are given along with their predicted secondary structures, as determined by the Chou-Fasman algorithm (CF) (8) and by the Garnier-Osguthorpe-Robson algorithm (GOR) (16).
Structure and function of Phd.
It has been previously suggested that Phd may be a β-sheet DNA binding protein and that the N-terminal regions of Phd may form an antiparallel β-sheet that makes specific contacts in the major groove of the DNA (37). Consistent with this hypothesis, secondary structure predictions suggest that the N terminus of Phd may adopt a β-sheet conformation. Furthermore, deletion of the N-terminal quarter of Phd abolishes repressor activity but not antitoxin activity. The second quarter of Phd was also dispensable for antitoxin activity. Although flanked by regions required for repressor activity, we cannot say whether or not the second quarter is itself required for repressor activity, and the secondary structure predictions for the second quarter are ambiguous. The third quarter of Phd was required for both repressor and antitoxin activity and may be thought of as an overlap or transition region. The fourth quarter of Phd was specifically required for antitoxin activity, but not for repressor activity. Secondary structure predictions suggest that the third and fourth quarters of Phd, which are both necessary and sufficient to neutralize the toxin, may consist primarily of a single α-helix.
Modular structure of Phd.
The N-terminal half of Phd was necessary for repression of transcription, but not for neutralization of the toxin. Conversely, the C-terminal quarter of Phd was essential for neutralization of the toxin, but not for repression of transcription. Only a quarter of the protein, the resolution limit of this analysis, was necessary for both activities. Thus, Phd appears to have a modular, rather than a monolithic, structure. Evidence from other systems indicates that such a modular structure may be typical of the antitoxin proteins.
N termini of other antitoxins are also implicated in repression.
Point mutations in the N-terminal region of MazE (36, 60) also reduce or abolish DNA binding. Similarly, point mutations in the N terminus of Kis (52) or short in-frame insertions of five amino acids in the N terminal of ParD (49) disrupted regulation but not antitoxin activity. Finally, and most telling, deletion of the N terminus of CcdA abolished repression but not antitoxin activity (3). Thus, the genetic evidence supports the idea that the N termini of the antitoxins are typically involved in repressor activity.
C termini of other antitoxins are involved in toxin neutralization.
Here, we have shown that the C-terminal 38 amino acids of Phd, preceded by an initial methionine, are sufficient to neutralize Doc. Similar observations have been made regarding the CcdA-Kis-MazE superfamily (31). For example, the C-terminal 41 amino acids of CcdA are sufficient for neutralization of toxin (3). Point mutations in the C terminus of Kis abolish antitoxin activity but not repressor activity (52). Deletion mutations indicate that the C-terminal 45 amino acids of MazE are sufficient to neutralize the MazF toxin (60). X-ray crystallographic studies of the MazE-MazF complex demonstrate that the C-terminal region of MazE makes extensive contacts with the MazF toxin but not with N-terminal moiety of MazE (31). Thus, the evidence supports the idea that the C termini of the antitoxins are typically involved in the neutralization of the toxin and are functionally independent of the N-terminal (repressor) regions.
Toxin-antitoxin families.
It has long been apparent, on the basis of sequence homology, that the Kis (and the identical PemI) antitoxins were homologous to MazE and, more distantly, to CcdA (39, 50). The three-dimensional structural similarities of the corresponding Kid (23), MazF (31), and CcdB (35) toxins have consolidated the evolutionary relationship of these three toxin-antitoxin systems. However, there are still at least four other well-studied systems, including Phd/Doc, RelB/RelE, HigA/HigB, and Omega-Epsilon-Zeta that, on the basis of primary sequence, cannot currently be grouped with the MazE/MazF superfamily or with each other (19). Since the omega repressor (45) and ParD (46) are ribbon-helix-helix (RHH) proteins while MazE has a novel and radically different RRHRRH structure (31, 36), it appears that the antitoxin proteins represent at least two and quite possibly five or more protein folds.
Fission and fusion of protein modules.
In the Omega-Epsilon-Zeta system, the toxin Zeta is neutralized by the antidote, Epsilon (40), while the promoter is repressed by the third protein, Omega (13). Possibly, a bifunctional repressor/antitoxin protein such as Phd could arise from the fusion of two single-function proteins, such as Omega and Epsilon. Conversely, a bifunctional protein could be split (or, more plausibly, duplicated and then differentially reduced) to yield two single-function proteins (58). To the first approximation, the N-terminal and C-terminal functions of Phd appear to be fairly independent and, as demonstrated here, the functions can be easily separated genetically. To the second approximation, however, the functions can and do influence each other. For example, in the physiological context, Doc mediates cooperative interactions between dimers of Phd bound to adjacent DNA sites and thus enhances the affinity, specificity, and cooperativity of the repressive complex (38). Thus, the fusion of repressor and antitoxin domains may enhance repressor function.
Mosaic antitoxins.
We expected Phd homologs and Doc homologs would be found near each other in microbial sequences. Although this expectation was fulfilled in a few instances (for example in Salmonella enterica serovar Typhimurium and Klebsiella pneumoniae), we also found numerous Phd homologs without clear Doc homologs (in, for example, Mycobacterium tuberculosis and Mycobacterium bovis and, more distantly, the pRUM plasmid of Enterococcus faecium) and numerous Doc homologs without clear Phd homologs (in, for example, Brucella suis, Brucella melitensis, and Caulobacter crescentus). Thus, contrary to expectation, toxin-antitoxin systems are susceptible to genetic rearrangements. At least one of these mosaic systems encodes a functional toxin-antitoxin system. Axe-Txe is a recently described antitoxin-toxin system of pRUM, a multidrug-resistant plasmid from a clinical isolate of E. faecium (20). The Axe and Phd antitoxins are clearly, if distantly, related but the corresponding toxins, Txe and Doc, are not, indicating a separate origin (or a separate and even-more-distant evolutionary branching) (20). The N-terminal 46 amino acids of Phd (repressor domain) are similar to Axe (∼25% identity, with two single-amino-acid gaps), but the C-terminal 27 amino acids of Phd (antitoxin domain) show little similarity to Axe (between 0 and 12.5% identity, depending on gaps). Thus, Phd (or Axe, or both) may be a mosaic protein and, as might be expected, the putative recombination junction falls within the repressor-antitoxin transition region of Phd (Fig. 2). A second and much clearer example of a mosaic antitoxin is provided by comparison of the StbD/StbE and the RelB/RelE systems, as noted by Hayes (25). The N-terminal 57 amino acids of StbD are dissimilar to RelB (<10% identity). However, the C-terminal 26 amino acids of StbD and RelB are highly homologous (∼46% identity), as are the corresponding StbE and RelE toxins (∼60% identity) as noted by Hayes (25). The location of this recombinational junction strongly suggests that the antitoxin activities of RelB and StbE are localized to the C termini of these proteins (Fig. 2).
FIG. 2.
(A) Evidence of recombinant antitoxins. Inspection of selected antitoxin alignments provided evidence of recombination events within the antitoxin sequences. The N termini of Phd and Axe are marginally similar, but their C termini, and their corresponding toxins, are dissimilar. The apparent recombination junction revealed by the alignment of Axe and Phd falls within the repressor-antitoxin transition region (indicated by bold type) as defined by deletion mapping of Phd (Table 1). Conversely, the N termini of RelE and StbD are dissimilar, but their C termini, and their corresponding toxins, are very similar, as previously noted by Hayes (25). The complementary RelE/StbD recombination junction noted by Hayes occurs in a comparable position. Protein sequences were globally aligned by using Clustal X (28) with default parameters. Alignments of these proteins have been previously reported (20, 25). (B) Modular model for antitoxin-toxin systems. We propose that toxin-antitoxin systems are a composite of two evolutionarily separable modules: an operator-repressor module and an antitoxin-toxin module. Recombination between modules may contribute to operon and antitoxin diversity. A schematic of two analogous parental genetic structures (the first in plain text, the second in bold text) and a recombinant structure are depicted. Such a recombinant is likely to be functional even if the parental structures are distantly related or heterologous.
Modular evolution of toxin-antitoxin systems.
To explain these observations, we propose that (at least some of) the toxin-antitoxin systems are a composite of two evolutionarily separate modules, an operator-repressor module and an antitoxin-toxin module (Fig. 2B). Recombination in the repressor-antitoxin transition region preserves the operator-repressor association as well as the antitoxin-toxin association and may thus produce a fully functional recombinant. Less precise recombination events might also be functional and undergo later reduction. The production of a functional recombinant from distant or heterologous components is presumably a very rare event (27, 30). However, if similar toxin-antitoxin systems compete for their shared ecological niche within the cell, then the toxin-antitoxin systems may be under positive selection for diversity and the rare functional recombinants may be well favored (12, 38, 43, 48, 55).
A well-ordered operon.
The production of function recombinants from a single crossover requires a well-ordered operon. By well-ordered we mean that the distance between interacting components is minimized. The P1 plasmid addiction operon, for example, appears to be well ordered. Given that an operon is well ordered, the recombinational disruption of coadapted gene complexes is minimized (15) and the probability of producing functional recombinants, with novel and possibly advantageous properties, is maximized (5). Thus, operons that happen to be well ordered might have a slight evolutionary advantage. Alternatively, and more plausibly, multiple rounds of horizontal gene transfer, followed by deletion of intervening nonfunctional or redundant sequences (32), could generate well-ordered operons from poorly ordered ancestral operons.
Acknowledgments
This work was supported by Public Health Service grant 1 R15 GM67668-01 from the National Institute of General and Medical Sciences, National Institutes of Health.
We thank the University of Alabama—Huntsville Summer 2000 Genetics class for their enthusiastic participation in this project. We thank Research Genetics for oligonucleotide primers and sequencing services. We thank the Genome Sequencing Center, Washington University, St. Louis, Mo., for access to their K. pneumoniae sequences.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber, W., L. Enquist, B. Hohn, N. E. Murray, and K. Murray. 1983. Experimental methods for use with lambda, p. 433-466. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 3.Bernard, P., and M. Couturier. 1991. The 41 carboxy-terminal residues of the miniF plasmid CcdA protein are sufficient to antagonize the killer activity of the CcdB protein. Mol. Gen. Genet. 226:297-304. [DOI] [PubMed] [Google Scholar]
- 4.Boe, L., K. Gerdes, and S. Molin. 1987. Effects of genes exerting growth inhibition and plasmid stability on plasmid maintenance. J. Bacteriol. 169:4646-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-490. [DOI] [PubMed] [Google Scholar]
- 6.Brosius, J., and A. Holy. 1984. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. USA 81:6929-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 8.Chou, P. Y., and G. D. Fasman. 1978. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 47:45-148. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 11.Clerget, M. 1991. Site-specific recombination promoted by a short DNA segment of plasmid R1 and by an homologous segment in the terminus region of the Escherichia coli chromosome. New Biol. 3:780-788. [PubMed] [Google Scholar]
- 12.Cooper, T. F., and J. A. Heinemann. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl. Acad. Sci. USA 97:12643-12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Hoz, A. B., S. Ayora, I. Sitkiewicz, S. Fernandez, R. Pankiewicz, J. C. Alonso, and P. Ceglowski. 2000. Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. Proc. Natl. Acad. Sci. USA 97:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, R. A. 1930. The genetical theory of natural selection. Oxford University Press, Oxford, England.
- 16.Garnier, J., D. J. Osguthorpe, and B. Robson. 1978. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 120:97-120. [DOI] [PubMed] [Google Scholar]
- 17.Gazit, E., and R. T. Sauer. 1999. The Doc toxin and Phd antidote proteins of the bacteriophage P1 plasmid addiction system form a heterotrimeric complex. J. Biol. Chem. 274:16813-16818. [DOI] [PubMed] [Google Scholar]
- 18.Gazit, E., and R. T. Sauer. 1999. Stability and DNA binding of the Phd protein of the phage P1 plasmid addiction system. J. Biol. Chem. 274:2652-2657. [DOI] [PubMed] [Google Scholar]
- 19.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47:1419-1432. [DOI] [PubMed] [Google Scholar]
- 21.Gronlund, H., and K. Gerdes. 1999. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J. Mol. Biol. 285:1401-1415. [DOI] [PubMed] [Google Scholar]
- 22.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hargreaves, D., S. Santos-Sierra, R. Giraldo, R. Sabariegos-Jareno, G. de la Cueva-Mendez, R. Boelens, R. Diaz-Orejas, and J. B. Rafferty. 2002. Structural and functional analysis of the kid toxin protein from E. coli plasmid R1. Structure (Cambridge) 10:1425-1433. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, C. S., and R. T. Sauer. 2003. Toxin-antitoxin pairs in bacteria: killers or stress regulators? Cell 112:2-4. [DOI] [PubMed] [Google Scholar]
- 25.Hayes, F. 1998. A family of stability determinants in pathogenic bacteria. J. Bacteriol. 180:6415-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazan, R., B. Sat, M. Reches, and H. Engelberg-Kulka. 2001. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 183:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 28.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 29.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205-210. [DOI] [PubMed] [Google Scholar]
- 30.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 31.Kamada, K., F. Hanaoka, and S. K. Burley. 2003. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell 11:875-884. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehnherr, H., E. Maguin, S. Jafri, and M. B. Yarmolinsky. 1993. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233:414-428. [DOI] [PubMed] [Google Scholar]
- 34.Lehnherr, H., and M. B. Yarmolinsky. 1995. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loris, R., M. H. Dao-Thi, E. M. Bahassi, L. Van Melderen, F. Poortmans, R. Liddington, M. Couturier, and L. Wyns. 1999. Crystal structure of CcdB, a topoisomerase poison from E. coli. J. Mol. Biol. 285:1667-1677. [DOI] [PubMed] [Google Scholar]
- 36.Loris, R., I. Marianovsky, J. Lah, T. Laeremans, H. Engelberg-Kulka, G. Glaser, S. Muyldermans, and L. Wyns. 2003. Crystal structure of the intrinsically flexible addiction antidote MazE. J. Biol. Chem. 12:12. [DOI] [PubMed] [Google Scholar]
- 37.Magnuson, R., H. Lehnherr, G. Mukhopadhyay, and M. B. Yarmolinsky. 1996. Autoregulation of the plasmid addiction operon of bacteriophage P1. J. Biol. Chem. 271:18705-18710. [DOI] [PubMed] [Google Scholar]
- 38.Magnuson, R., and M. B. Yarmolinsky. 1998. Corepression of the P1 addiction operon by Phd and Doc. J. Bacteriol. 180:6342-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meinhart, A., C. Alings, N. Strater, A. G. Camacho, J. C. Alonso, and W. Saenger. 2001. Crystallization and preliminary X-ray diffraction studies of the epsilonzeta addiction system encoded by Streptococcus pyogenes plasmid pSM19035. Acta Crystallogr. D 57:745-747. [DOI] [PubMed] [Google Scholar]
- 41.Miki, T., Z. T. Chang, and T. Horiuchi. 1984. Control of cell division by sex factor F in Escherichia coli. II. Identification of genes for inhibitor protein and trigger protein on the 42.84-43.6 F segment. J. Mol. Biol. 174:627-646. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Mongold, J. A. 1992. Theoretical implications for the evolution of postsegregational killing by bacterial plasmids. Am. Nat. 139:677-689. [Google Scholar]
- 44.Mullis, K. B. 1990. Target amplification for DNA analysis by the polymerase chain reaction. Ann. Biol. Clin. (Paris) 48:579-582. [PubMed] [Google Scholar]
- 45.Murayama, K., P. Orth, A. B. de la Hoz, J. C. Alonso, and W. Saenger. 2001. Crystal structure of omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 Å resolution. J. Mol. Biol. 314:789-796. [DOI] [PubMed] [Google Scholar]
- 46.Oberer, M., K. Zangger, S. Prytulla, and W. Keller. 2002. The anti-toxin ParD of plasmid RK2 consists of two structurally distinct moieties and belongs to the ribbon-helix-helix family of DNA-binding proteins. Biochem. J. 361:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rawlings, D. E. 1999. Proteic toxin-antitoxin, bacterial plasmid addiction systems and their evolution with special reference to the pas system of pTF-FC2. FEMS Microbiol. Lett. 176:269-277. [DOI] [PubMed] [Google Scholar]
- 48.Riley, M. A. 1993. Positive selection for colicin diversity in bacteria. Mol. Biol. Evol. 10:1048-1059. [DOI] [PubMed] [Google Scholar]
- 49.Roberts, R. C., C. Spangler, and D. R. Helinski. 1993. Characteristics and significance of DNA binding activity of plasmid stabilization protein ParD from the broad host-range plasmid RK2. J. Biol. Chem. 268:27109-27117. [PubMed] [Google Scholar]
- 50.Ruiz-Echevarria, M. J., G. de Torrontegui, G. Gimenez-Gallego, and R. Diaz-Orejas. 1991. Structural and functional comparison between the stability systems ParD of plasmid R1 and Ccd of plasmid F. Mol. Gen. Genet. 225:355-362. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Santos-Sierra, S., C. Pardo-Abarrio, R. Giraldo, and R. Diaz-Orejas. 2002. Genetic identification of two functional regions in the antitoxin of the parD killer system of plasmid R1. FEMS Microbiol. Lett. 206:115-119. [DOI] [PubMed] [Google Scholar]
- 53.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simons, R. W., F. Housman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 55.Tan, Y., and M. A. Riley. 1996. Rapid invasion by colicinogenic Escherichia coli with novel immunity functions. Microbiology 142:2175-2180. [DOI] [PubMed] [Google Scholar]
- 56.Tian, Q. B., M. Ohnishi, A. Tabuchi, and Y. Terawaki. 1996. A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 220:280-284. [DOI] [PubMed] [Google Scholar]
- 57.Tyndall, C., H. Lehnherr, U. Sandmeier, E. Kulik, and T. A. Bickle. 1997. The type IC hsd loci of the enterobacteria are flanked by DNA with high homology to the phage P1 genome: implications for the evolution and spread of DNA restriction systems. Mol. Microbiol. 23:729-736. [DOI] [PubMed] [Google Scholar]
- 58.Yanai, I., Y. I. Wolf, and E. V. Koonin. 2002. Evolution of gene fusions: horizontal transfer versus independent events. Genome Biol. 3:research 1-0024.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yarmolinsky, M. B. 1995. Programmed cell death in bacterial populations. Science 267:836-837. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, J., Y. Zhang, and M. Inouye. 2003. Characterization of the interactions within the mazEF addiction module of Escherichia coli. J. Biol. Chem. 278:32300-32306. [DOI] [PubMed] [Google Scholar]
- 61.Zielenkiewicz, U., and P. Ceglowski. 2001. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim. Pol. 48:1003-1023. [PubMed] [Google Scholar]