Abstract
The essential genes of microorganisms encode biological functions important for survival and thus tend to be of high scientific interest. Drugs that interfere with essential functions are likely to be interesting candidates for antimicrobials. However, these genes are hard to study genetically because knockout mutations in them are by definition inviable. We recently described a conditional mutation system in Escherichia coli that uses a plasmid to produce an amber suppressor tRNA regulated by the arabinose promoter. This suppressor was used here in the construction of amber mutations in seven essential E. coli genes. Amber stop codons were introduced as “tagalong” mutations in the flanking DNA of a downstream antibiotic resistance marker by lambda red recombination. The drug marker was removed by expression of I-SceI meganuclease, leaving a markerless mutation. We demonstrate the method with the genes frr, gcpE, lpxC, map, murA, ppa, and rpsA. We were unable to isolate an amber mutation in ftsZ. Kinetics of cell death and morphological changes were measured following removal of arabinose. As expected given the wide range of cellular mechanisms represented, different mutants showed widely different death curves. All of the mutations were bactericidal except the mutation in gcpE, which was bacteriostatic. The strain carrying an amber mutation in murA was by far the most sensitive, showing rapid killing in nonpermissive medium. The MurA protein is critical for peptidoglycan synthesis and is the target for the antibiotic fosfomycin. Such experiments may inexpensively provide valuable information for the identification and prioritization of targets for antibiotic development.
The completed genome sequence of Escherichia coli identified open reading frames (ORFs), of which 38% were of unknown function (4). A systematic program to mutagenize every gene of E. coli will be very useful in characterizing the functions and interactions of this large repertoire of genes, as has been demonstrated for Saccharomyces cerevisiae (37). Loss-of-function mutants (knockouts) are the easiest type of mutant to produce at high throughput, and projects to produce collections of transposon knockouts and in-frame deletions are under way in our laboratory and elsewhere (see http://www.genome.wisc.edu and http://ecoli.aist-nara.ac.jp).
Essential genes are of particular interest because they include the core functions upon which basic cellular processes are based, they tend to be widely conserved, and they can serve as a starting point for minimal genome designs (24, 27). The number of essential genes in an organism depends on the conditions used; in this case, we defined them broadly as genes required for growth on rich medium. Different experimental methods have been used to determine the number of essential genes. In E. coli, at least 620 ORFs are essential (11); in Bacillus subtilis, 271 out of ∼4,100 ORFs are essential (21); in Haemophilus influenza, ∼670 out of ∼1,850 ORFs are essential (1); and in Staphylococcus aureus, 658 out of ∼2,630 ORFs are essential (10). Only about 15 essential gene products are targeted by antibiotics. Identification of new targets may help combat the problem of increasingly prevalent antibiotic resistance. Essential gene mutants are important in genomics-based drug discovery for target validation and prioritization and can be used to screen compound libraries and determine the mode of action. Unfortunately, difficulties in mutating essential genes have impeded their genetic study.
Several approaches to conditional mutagenesis can be considered. Temperature-sensitive (TS) mutations offer direct and sometimes reversible control of protein function, but it is extremely difficult to design proteins with a TS phenotype. In some cases, TS alleles may not be possible (33) or activity may be suboptimal even at permissive temperature. Another approach is to place the target gene under the control of an experimentally controllable promoter either on a plasmid (12) or in the genome (17). This allows turning gene expression on and off over a large dynamic range, but the natural expression level and regulation of the gene are overridden by the inducing promoter. Another approach uses antisense RNA in which short pieces of RNA are produced that interfere with translation (10, 15). Antisense mutants are made by using a random whole-genome approach, and it is not clear if a high enough percentage of genes is amenable to this approach to allow disruption of the complete set of essential genes. A fourth approach uses a TS suppressor tRNA to convert amber mutations into a TS phenotype (31) but suffers from a low dynamic range and the relative inefficiency of the TS amber suppressors.
The method we developed uses conditional suppression of amber mutants through inducible expression of the Ala2 amber suppressor tRNA from the arabinose promoter on expression plasmid pBAD/sup2 (see Fig. 1A) (13). The amber codon was chosen because it is the least common of the three stop codons in E. coli, terminating only 326 out of 4,290 annotated ORFs. The Ala2 suppressor was selected because of its high suppression efficiency and its specificity in introducing only the correct amino acid (29). We also have found that expression of this suppressor causes minimal perturbation of global transcription in E. coli (C. D. Herring and F. R. Blattner, unpublished data).
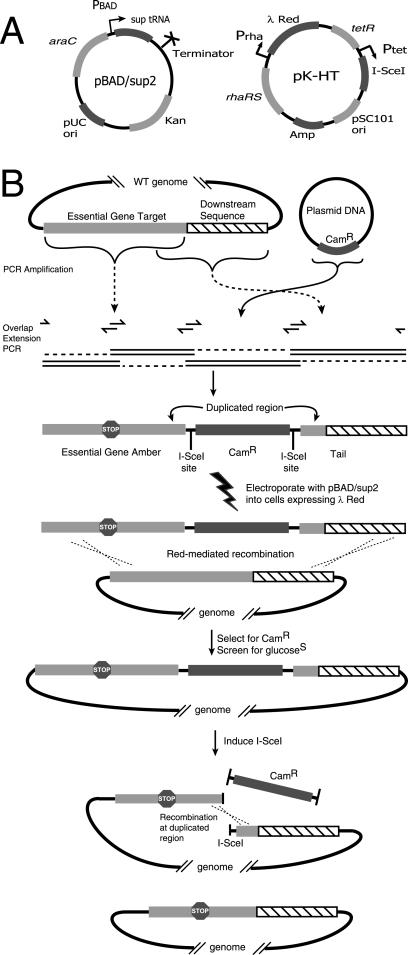
FIG. 1.
(A) Plasmids used in tagalong mutagenesis (not to scale). (B) Mutagenesis strategy. A linear DNA fragment is produced from WT template genomic DNA by overlap extension PCR. The positions of primers are indicated by one-sided arrows. The PCR fusion product is electroporated into cells, and integrations resulting from a double-recombination event are selected. After identification of a clone carrying the tagalong amber stop codon, the I-SceI gene is induced, resulting in removal of the gene encoding Camr. Recombination within a short duplicated region leads to the generation of an amber mutant that is otherwise scarless.
In previous work (13), a method called “gene gorging” was used to introduce amber mutations into the E. coli genome. We demonstrated the ability to turn nonessential genes on and off by changing the growth medium from arabinose to glucose. However, attempts to use gene gorging to obtain conditional mutations in essential genes were nearly fruitless (see below). Here we describe a solution to this problem that differs from the previous method in two main respects. (i) The suppression plasmid is introduced into the cell at the same time as a mutagenic amber PCR fragment by coelectroporation, and (ii) an antibiotic resistance gene is used to select for lambda red recombination of the amber mutant fragment into the genome. The amber fragment is introduced as a “tagalong” mutation, i.e., an unselected mutation in flanking DNA incorporated along with a downstream antibiotic resistance gene.
The genes chosen for this study represent a broad group of essential functions but are restricted to genes at the distal ends of operons. The gene frr encodes ribosome release factor, which is required for the disassembly of ribosomes and mRNA following translational termination (14). FtsZ is a tubulin-like GTPase that polymerizes into a ring that girdles the middle of the cell and leads to cell division. The gene gcpE has been recently reported to encode a terminal step in the non-mevalonate pathway of isoprenoid biosynthesis, which is necessary for the production of diverse metabolites such as quinones and carotenoids (23). The gene lpxC encodes the first committed step in the synthesis of lipid A, which acts as an anchor for lipopolysaccharide in the outer membrane (7). Methionine aminopeptidase is encoded by the gene map and is required for removal of the N-formylmethionine present at the amino terminus of proteins following translation. The gene murA encodes the first committed step in the cytoplasmic synthesis of peptidoglycan, the main component of the cell wall (6). Pi is a central metabolite involved in nearly all areas of cell physiology. The gene ppa encodes inorganic pyrophosphatase, which generates Pi from PPi, a product of nucleic acid polymerization, aminoacylation of tRNAs, and other reactions. The gene rpsA encodes protein S1, the largest ribosomal protein, which is known to have high affinity for mRNA and has been implicated in translation initiation and elongation (34). The new method worked efficiently for all but one of these essential genes of diverse function, arguing that it will be applicable to many of the essential genes of E. coli.
MATERIALS AND METHODS
Abbreviations, strains, media, and oligonucleotides.
E. coli strain MG1655 (4) was the source for all of the mutants generated in this study (frr = FBSC193, gcpE = FBSC195, lpxC = FBSC62, map = FBSC197, murA = FBSC199, ppa = FBSC201, and rpsA = FBSC203). For the sequences of the primer oligonucleotides used, see Table S3 in the supplemental material. Rich defined medium (RDM) was prepared as described by Neidhardt et al. (28) (recipe at http://www.genome.wisc.edu/functional/protocols.htm) or purchased from Teknova (Half Moon Bay, Calif.). Media and plate supplements of 0.2% d-glucose (Glu), 0.2% l-arabinose (Ara), 0.2% l-rhamnose (Rha), 100 ng of anhydrotetracycline (ATC) per ml, 100 μg of ampicillin (AMP) per ml, 50 μg of kanamycin (KAN) per ml, or 25 μg of chloramphenicol (CAM) per ml in various combinations are abbreviated, e.g., RDM/KAN/Ara. For plating, colonies were resuspended and diluted in 0.15 M NaCl.
Construction of pK-HT.
pK-HT (Fig. 1) was derived from pKD46 (8) by inserting the I-SceI gene under tetracycline control, switching the lambda red genes from arabinose to rhamnose control, and replacing the TS replicator with a temperature-stable allele. First we amplified I-SceI, the tetA promoter, and the tetR gene from pST98-AS (32) by using NcoI adapter primers OF428 and OF429 and digested and then ligated the fragment into the NcoI site of pKD46, resulting in pKDTS. Next, the rhaBAD promoter and rhaSR genes were amplified from E. coli genomic DNA with primers OF434 and OF435 and then digested with SacI/BssHII and ligated into the SacI/BssHII site of pKDTS, resulting in pKDTSR. Finally, the single nucleotide mutation responsible for the TS ori phenotype (3) was replaced with the wild-type (WT) allele by megaprimer PCR and ligation as follows. A megaprimer with the desired modification was amplified from pKDTSR with primers OF436 and OF438 and then extended in a second reaction by adding primer OF439 and a small amount of the full-length OF438-OF439 product as the template. It was then reamplified with OF438-OF439. This PCR product was digested with SpeI/PciI and ligated into the SpeI/PciI site of pKDTSR, resulting in pK-HT.
Generation of mutagenic DNAs.
An alanine codon (GCX) was selected near the middle of the coding region of the targeted gene and changed to amber (TAG). If possible, an alanine codon preceded by a C residue was chosen so that each mutant would be marked by CTAG, the relatively rare restriction site for BfaI, so that mutagenesis could be easily confirmed by PCR and restriction digestion. If no codon with an upstream C residue was available, one was selected that could be changed to a C without altering the amino acid sequence.
Four individual pieces were amplified separately with primers designed to overlap one another (Fig. 1B). The ORF was amplified in two halves with primers at the amino and carboxyl termini (called A- and C-term primers in Table S3), paired with overlapping divergent primers in the middle of the gene, both containing the desired amber mutation (mut1 and mut2). The gene for CAM resistance (Camr) was amplified with primers OF105 and OF375, each containing the recognition site for I-SceI. A small part at the end of the ORF and some of the downstream sequence were amplified as a fourth “tail” fragment, with one primer ∼100 nucleotides (nt) before the C terminus (tail start) and another located approximately 500 nt downstream (tail end). The overlapping PCR products were joined in a two-round overlap extension PCR. In the first round, the four primary pieces were joined to a neighboring fragment in three pairwise combinations. In the second round, the pairwise reactions were combined and the complete fusion was amplified. Details of the PCR fusions were as follows. All amplifications were performed with Pfu Turbo DNA polymerase (Stratagene, La Jolla, Calif.). In the first round, the four primary pieces were purified with QIAquick (Qiagen, Valencia, Calif.) and then 4 μl of each neighboring pair was combined with Pfu buffer, deoxynucleoside triphosphates (2.5 mM each), and Pfu Turbo in a 25-μl reaction mixture and subjected to 20 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min. In the second round of fusion, 5 μl of each unpurified product from the three pairwise combinations was combined with 3.5 μl of Pfu buffer, 5 μl of deoxynucleoside triphosphates, 1 μl of Pfu Turbo, and 20.5 μl of H2O and subjected to five cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 6 min. A 2.5-μl volume of each of the A-term and tail end primers (5 μM) was then added, and the reactions were cycled 20 to 25 more times by the same regimen. Smears and multiple bands were often visible on agarose gels, but a strong primary product was the correct size for all of the genes attempted except one, which was not subsequently used. Mutagenic fragments were made for the genes murA and rpsA by this method. For the genes frr, gcpE, map, ppa, and ycfB, amber alleles had previously been generated by megaprimer PCR and cloned (13). These were amplified with the A-term primers and OF169 and then fused to the gene for Camr and the tail by a similar two-round overlap extension PCR. The mutagenic fragments for ftsZ and lpxC were generated by a much less rapid method as follows. The amber stop codons were introduced by megaprimer PCR and cloned adjacent to Camr, and then the amber allele and the gene for Camr were amplified as one unit with the A-term and p14 primers. This PCR product and the tail piece were digested with BamHI, ligated, and then PCR amplified with the A-term and tail end primers. The ftsZ product was further gel purified, cloned, and then amplified again with OF205 and OF364.
Electroporation and integration.
Electrocompetent recombinogenic cells were made by growing E. coli carrying pK-HT in shake flasks in Luria-Bertani medium containing AMP at 37°C. The plasmid-encoded lambda red functions were induced by adding Rha 2 h before harvesting. When the optical density at 600 nm (OD600) reached 0.5, cells were centrifuged, washed twice in chilled water and once in chilled 10% glycerol, resuspended in 10% glycerol at approximately 1/100 of the original volume, and then frozen. Approximately 100 to 200 ng of overlap extension PCR product, purified with QIAquick and G50 spin columns, was electroporated together with ∼70 to 90 ng of pBAD/sup2 (13) plasmid DNA. After electroporation, cells were incubated for 1 h at 37°C in SOC/Ara (SOC made with arabinose rather than glucose) (32a) and grown on RDM/Ara/KAN/CAM/AMP plates for 16 to 24 h at 37°C. Colonies were patched on RDM/KAN/CAM/AMP/Glu and RDM/KAN/CAM/AMP/Ara plates to identify conditional lethal mutants that could grow on Ara but not on Glu.
Conditional lethal mutants were not recovered in ppa except when the growth medium was RDM/Ara or a 1:1 mixture of SOC/Ara and RDM. The medium may be important for full expression of the arabinose-inducible suppressor. For all genes, the conditional lethal clones tended to be small colonies on the transformation plates. For the gene rpsA, conditional lethal colonies were only identified after 24 h of growth. These very small colonies were too small to be detected after only 16 h. The small size may be due to slow growth from incomplete suppression but may also be due to lag time before the suppressor is fully expressed. Very few conditional lethal mutants were obtained when plates were grown at 30°C. For this reason, the plasmid origin from pKD46 was corrected during construction of pK-HT so that it would replicate at 37°C.
Counterselection.
Conditional lethal mutants were resuspended from the Ara patch and plated on RDM/KAN/AMP/ATC/Ara plates at 30°C to induce I-SceI (without inducing lambda red recombinase). Survivors were patched on RDM/KAN/AMP/Ara plates with and without CAM to confirm loss of the Camr marker. Counterselection with I-SceI was more effective at 30°C than at 37°C. The I-SceI allele used here came from pST98-AS and is missing four amino acids near the N terminus (32). Other alleles of I-SceI under control of the Ara promoter used in this laboratory work efficiently at 37°C, so the effect may be due to either the small deletion or the level of I-SceI expressed from the tetA promoter. One gene (gcpE) was also mutated by an alternate counterselection method with the tetracycline resistance gene rather than I-SceI, as described in the supplemental material.
Plasmid curing and sequence verification.
pK-HT was cured from the conditional mutants by streaking and then plating on RDM/KAN/Ara/Rha. Clones that no longer carried the plasmid were identified by patching on RDM/KAN/Ara plates with and without AMP. The targeted essential genes were amplified from one isolate of each mutant with primers located adjacent to the ORF so that only the chromosomal locus could possibly be amplified. Sequencing revealed intact ORFs identical to the WT except for the introduced amber mutation. The primers used for sequencing were OF210, OF215, OF218, OF219, OF242, OF243, OF304, OF305, OF306, OF307, OF308, OF376, OF377, OF378, OF379, OF380, OF381, OF382, OF383, OF410, OF411, OF412, OF413, OF423, OF425, OF443, OF444, OF447, OF448, and OF450.
Growth curves.
The lpxC amber mutant used to determine growth curves was the one mutant made by gene gorging, while all of the others were those made as tagalong mutations as described here. Frozen glycerol stocks of each strain were streaked on RDM/KAN/Ara plates, and single colonies were inoculated into tubes containing 1 ml of RDM/KAN/Ara and grown overnight at 37°C. The overnight cultures were inoculated at ratios of 1:50 to 1:6,400 into 200 μl of RDM/KAN plus either Glu or Ara in 96-well plates with lids (Falcon no. 3072; Becton Dickinson, Franklin Lakes, N.J.). Each lid was taped open about 1 mm to allow aeration, and unused wells were filled with water. The cultures were then grown at 37°C with shaking in a Spectramax 384 plus (Molecular Devices, Sunnyvale, Calif.) and read every 15 min, subtracting the reading for the uninoculated plate as a blank. The plate was removed three times to allow sampling. For light microscopy, bacterial smears were heat fixed, stained with crystal violet, and observed at a magnification of ×1,000 with a Zeiss Axiovert 35 microscope. Viable cells were counted on RDM/KAN/Ara plates incubated at 37°C. The validity of using a small culture volume (200 μl) in these experiments was verified by using the strains with amber mutations in map and lpxC. Very similar growth curves were observed for these mutants in 150-ml cultures. Additionally, WT strain MG1655 grew essentially the same in 200-μl and 100-ml cultures (29- and 28-min doubling times, respectively, in RDM/Glu medium).
RESULTS
Attempts to use gene gorging for essential genes.
An amber allele of lpxC was generated by megaprimer PCR and then cloned into pDHA30. Gene gorging was performed with this plasmid, pACBSR, and pBAD/sup2 as previously described (13). One conditional lethal mutant (strain FBSC62) was identified out of 20 screened on the first try, but further attempts to mutate lpxC or four other essential genes failed despite extensive efforts and many variations on the method. The variations included (i) the use of different media, (ii) separate control of gene gorging and suppression with rhamnose and arabinose, (iii) titration of arabinose induction by counterinduction with glucose, (iv) preinduction of the suppressor, (v) gene gorging for different amounts of time and at different temperatures, (vi) detection of conditional lethal mutants by different methods, (vii) selection of proper suppression by using an amber mutant Camr gene in the mutagenesis plasmid, and finally (viii) cotransformation of the suppressor with the other plasmids. Throughout, the nonessential gene xylA was used successfully as a positive control, indicating that the failure was specific to essential genes. The arabinose promoter is subject to all-or-nothing feedback control (25), and it may be that the suppressor was not correctly expressed in all cells throughout the process of gene gorging.
Mutagenesis strategy.
Conditional amber mutations were introduced into essential genes as outlined in Fig. 1. PCR primers were designed to amplify a target ORF and a region downstream with WT genomic DNA as the template. Four individual PCR fragments with overlapping sequences at the ends were joined by overlap extension PCR, resulting in a DNA fragment containing an amber stop codon in the target gene, a Camr gene flanked by I-SceI sites, a small duplication of the terminus of the target gene, and some of the downstream sequence.
These linear DNAs were then electroporated along with arabinose-inducible suppressor plasmid pBAD/sup2 into competent cells of E. coli MG1655 expressing lambda red from plasmid pK-HT (Fig. 1A). Double recombination spanning the Camr gene resulted in integration of the linear DNA into the targeted gene in the presence of the induced suppressor tRNA. Clones with the amber mutation incorporated along with the selected marker as a tagalong mutation were identified by their dependence on the suppressor with replicate patching on glucose and arabinose media.
Next, the chloramphenicol resistance gene downstream of the essential gene was precisely removed by counterselection with the I-SceI enzyme (32), induced from plasmid pK-HT with ATC. The recognition sequence for I-SceI does not occur in the WT E. coli genome, so the mutant chromosome was cut on both sides of the gene for Camr but nowhere else. Such a double-strand break is lethal, and the majority of the surviving cells were those that had undergone Rec-mediated recombination between the end of the gene and the 100-nt identical region on the other side of the gene for Camr. The essential-gene ORFs were then sequenced and shown to be completely WT except for the introduced mutation.
Mutagenesis results.
Linear DNA fragments were generated for nine genes (Table 1). Camr integrants were obtained for all of the genes, and the percentage that carried the tagalong amber mutation ranged from 2.8 to 53%. Similar integration results were obtained by expressing lambda red from plasmid pKD46 (8), with conditional lethal mutants ranging from 2.5 to 38%. No conditional lethal mutants were recovered with ftsZ. The large variation in the percent amber may be due to incomplete suppression shortly after integration and differing sensitivity to loss of each gene. The highest percentage of amber mutations was observed with the gcpE mutation, which gives rise to reversible bacteriostasis when the suppressor is withdrawn (see below). Conversely, the lowest occurred with the murA mutation, which results in very rapid lysis upon withdrawal of the suppressor.
TABLE 1.
Mutagenesis results
| Gene | Amber locationa | Integrationb
|
Counterselection
|
Plasmid curing
|
|||
|---|---|---|---|---|---|---|---|
| No. tested | % Amberb | No. tested | % Cam− | No. tested | % Amp− | ||
| frr | 94 (186) | 82 | 22 | 10 | 100 | 41 | 9.8 |
| ftsZ | 185 (384) | 69 | 0 | ||||
| gcpE | 196 (373) | 53 | 52.8 | 17 | 58.8 | 21 | 100 |
| lpxC | 165 (306) | 48 | 12.5 | ||||
| map | 136 (265) | 43 | 32.6 | 10 | 90 | 19 | 26.3 |
| murA | 203 (420) | 108 | 2.8 | 10 | 10 | 26 | 15.4 |
| ppa | 104 (177) | 108 | 12.0c | 2 | 100 | 23 | 34.8 |
| rpsA | 269 (558) | 59 | 23.7d | 10 | 100 | 10 | 100 |
| ycfB | 197 (384) | 82 | 8.5 | 11 | 63.6 | 20 | 15 |
The number of the amino acid that was changed to amber is shown, and the length of the gene in amino acids is in parentheses.
Percent amber was calculated from the number of patches that did not grow or grew poorly on glucose divided by the total number tested.
Glucose-sensitive clones were only obtained when cells were grown in RDM/Ara or 1:1 RDM:SOC/Ara.
Glucose-sensitive clones were only obtained when plates contained 0.4% arabinose and cells were grown for 24 h.
After induction of I-SceI, 100- to 3,000-fold fewer colonies grew on plates relative to uninduced controls, indicating effective counterselection. Cams clones were easily identified by screening ∼10 colonies each, and for most genes the majority of colonies were sensitive. The mutants were cured of plasmid pK-HT by taking advantage of the toxic effects of lambda red (35) by inducing lambda red in the absence of selection for the plasmid. After induction, 10 to 100% of the screened colonies were Amps, indicating loss of pK-HT.
Characterization of mutants.
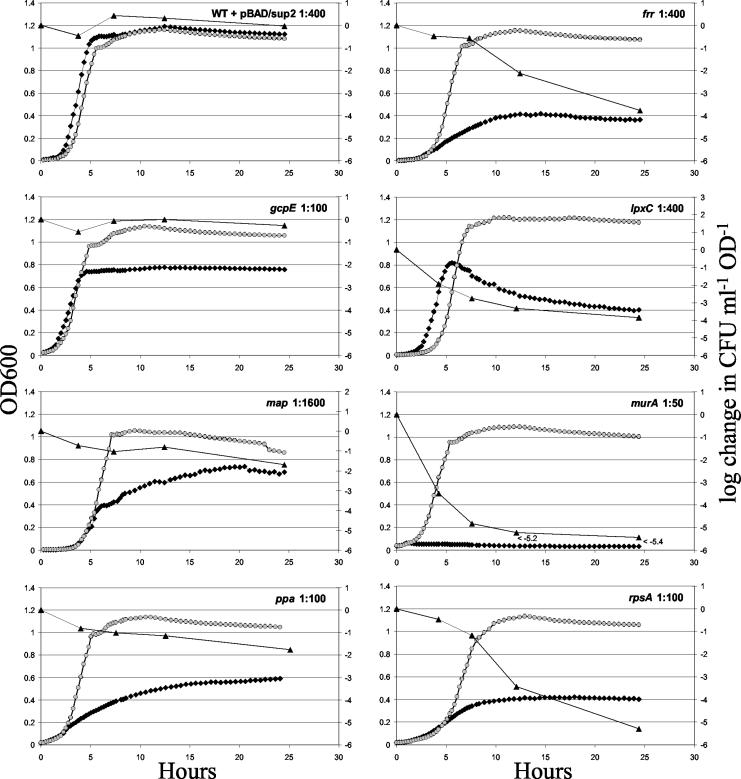
To characterize the behavior of the conditional mutants, growth was monitored by OD under both permissive and nonpermissive conditions (Fig. 2). First, the growth rate was measured in arabinose medium, which permits maximal suppression of the amber mutations. Table 2 shows that the doubling time of most strains was only slightly longer than that of the WT, indicating good suppression. The mutants with amber mutations in rpsA and murA showed longer doubling times, suggesting incomplete suppression. The final culture density of the map mutant in arabinose was reproducibly lower than that of the others.
FIG. 2.
Growth in permissive and nonpermissive media. Overnight cultures were inoculated into glucose (diamonds) or arabinose (circles) medium at the optimal dilution ratios shown in the upper right corner of each panel. OD600 was monitored in a 96-well plate in a shaking Spectramax plate reader at 37°C with periodic removal of samples for microscopy and plating on arabinose plates. The change in viability, given on the right axis (triangles), was calculated by using the concentration of CFU normalized by dividing by the OD600. For each time point, this value was divided by the value at T = 0 and the log of that value is presented here. The numbers of CFU per milliliter per OD unit at T = 0 measured from the saturated inoculum and corrected for dilution were as follows: WT, 4.6 × 109; frr mutant, 3.2 × 109; gcpE mutant, 3.4 × 109; lpxC mutant, 2.2 × 109; map mutant, 2.9 × 109; murA mutant, 8.7 × 108; ppa mutant, 1.5 × 109; rpsA mutant, 1.2 × 109. OD is shown on a linear rather than on a logarithmic scale to best show the dynamics of the faltering cultures, and the scale of the right axis differs for lpxC and map to prevent the trend lines from overlapping. For the last two time points of the murA measurement, no colonies grew.
TABLE 2.
Characterization of mutants
| Gene | Product or function | Doubling timea | Slowb | Stopc | Log killingd | Morphologic defecte | Note(s)f |
|---|---|---|---|---|---|---|---|
| frr | Ribosome release factor | 42.0 ± 4.4 | 3.9 | 6.4 | −3.8 | Moderate | 1, 2, 3 |
| gcpE | Isoprenoid biosynthesis | 38.8 ± 4.7 | 5.2 | 5.4 | −0.3 | Mild | 1, 3 |
| lpxC | Lipid A biosynthesis | 39.5 ± 1.0 | 6.5 | 7.3 | −3.9 | Severe | 2, 4, 5 |
| map | Methionine aminopeptidase | 39.7 ± 2.4 | 7.4 | 9.4 | −1.7 | Severe | 5 |
| murA | Peptidoglycan synthesis | 46.6 ± 6.6 | 0.8 | 0.8 | <−5.4 | Severe | 1, 2, 3 |
| ppa | Pyrophosphatase | 41.5 ± 1.5 | 3.0 | 5.0 | −1.8 | Mild | 3, 6 |
| rpsA | Ribosomal protein S1 | 61.9 ± 2.4 | 3.3 | 4.5 | −5.3 | Moderate | 1, 2 |
| WT + pBAD/sup2 | 35.8 ± 3.0 | NA | NA | 0 | NA | 3 |
Overnight cultures grown in RDM/KAN plus 0.2% Ara were inoculated 1:100 or 1:400 into 200 μl of the same medium and grown in a 96-well format at 37°C while measuring OD600. Data represent three replicates. Average doubling time is given in minutes ± the standard deviation.
Number of doublings before growth slowed down in glucose, calculated as the log2 of the OD600 at which growth slowed down divided by the starting OD, which was estimated from the corrected maximum OD of the Ara control culture divided by the dilution factor.
Number of doublings before growth stopped in glucose, calculated as described in footnote b but using the maximum OD of the culture rather than the OD at slowdown.
Log of the change in the number of CFU per milliliter per unit of OD600 after 25 h of growth in glucose medium relative to T = 0.
Severity of morphological defect from microscopic observations.
1, Cells large and slightly elongated; 2, debris; 3, a few elongated or filamentous cells; 4, chains of large and rounded cells; 5, most cells elongated or filamentous; 6, some short cells.
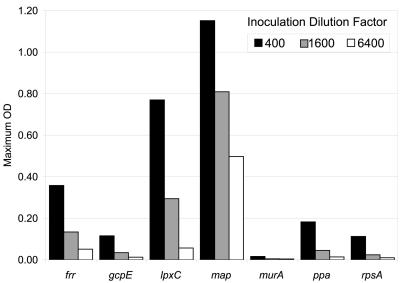
Growth arrest was monitored by diluting cultures from permissive (arabinose) to nonpermissive (glucose) conditions. All mutant strains eventually stopped growing in glucose medium as suppression was lost, but the time that it took for this to happen differed widely among the mutants. The initial dilution factor used for inoculation into nonpermissive medium was varied for each mutant (Fig. 3). In all cases, the maximum OD of the cultures varied with the inoculum size. An optimal amount of inoculum was used for the experiments in Fig. 2 so that growth stopped at an OD high enough for easy observation but prior to saturation. This optimal dilution factor presumably reflects how long it takes after loss of suppression for each essential gene function to drop below the threshold necessary for growth. This factor and the estimated starting density were used to calculate the number of doublings before growth slowed down and stopped (Table 2). The strains carrying amber mutations in lpxC and gcpE stopped growing suddenly, while those with amber mutations in rpsA, ppa, and frr stopped slowly. The shape of the growth curve also varied between strains. The culture turbidity of the mutant carrying an lpxC amber mutation decreased after peaking, suggesting cell lysis, clumping, or a change in shape. The mutant carrying a map amber mutation reproducibly displayed a very interesting S-shaped curve, possibly indicating some adaptation that temporarily allowed growth to resume.
FIG. 3.
Inoculation effects. Overnight cultures of seven amber mutants (listed by gene name) were inoculated into glucose medium at three different dilution ratios as indicated. OD600 was monitored in a 96-well plate in a shaking Spectramax plate reader at 37°C, and the maximum OD that each culture reached is given.
To determine whether the loss of each gene had a bacteriostatic or bactericidal effect, cells were recovered by growth on arabinose plates at various points during growth arrest in glucose (Fig. 2). The number of recovered colonies varied widely, with murA and rpsA amber mutants yielding the fewest viable cells at the last time point. The plating efficiency per OD unit remained high for the gcpE amber strain and the WT control but decreased for all other mutants. This indicates that only the gcpE mutation is bacteriostatic, while the others have some bactericidal effect. In other similar experiments, growth occasionally resumed for the strains with lpxC or frr amber mutations in glucose late in the time course (data not shown). Sequencing of an isolate from one of the lpxC mutant cultures revealed that the alaW tRNA gene had become a chromosomal amber suppressor. The Ala2 suppressor is only 4 nt different from the redundant alaX and alaW tRNA genes, allowing recombination to take place between the suppressor plasmid and the alaW gene. The frequency of cells able to grow without arabinose was measured for each mutant by plating three independent cultures on KAN/Ara and KAN/Glu plates. The frequencies were as follows (average ± standard deviation, listed by gene name): lpxC, 2.6 × 10−6 ± 1.5 × 10−6; frr, 1.4 × 10−7 ± 1.0 × 10−7; gcpE, 6.5 × 10−8 ± 5.1 × 10−8; map, 8.1 × 10−7 ± 4.7 × 10−7; murA, <4.7 × 10−8; ppa, 1.3 × 10−7 ± 1.9 × 10−7; rpsA, 1.6 × 10−7 ± 1.6 × 10−7.
The mutants were observed by light microscopy at various points during growth arrest (Table 2; Fig. S4 in the supplemental material). Abnormally shaped cells were more common in the mutants than in the WT control, but for strains with amber mutations in frr, gcpE, ppa, and rpsA, normal cells were predominant. In contrast, most cells were abnormal in lpxC and murA amber mutants, and the majority were elongated or filamentous in the strain with a map amber mutation. Notably, lpxC and murA encode enzymes required for synthesis of the cell wall. Debris was observed for many strains, probably indicating lysis.
To determine what effect, if any, the gene encoding Camr might have on the growth phenotypes of these mutants, strains were compared before and after the counterselection step. All of them grew the same except for those with mutations in ppa and rpsA, which stopped growing at a lower OD in glucose when the gene for Camr was present (data not shown). Thus, the downstream Camr marker may in some cases have unintended effects and should be removed, as demonstrated here.
DISCUSSION
Here we describe a new approach for making conditional lethal mutations in essential genes that was successful in seven of the eight cases tried and have characterized the behavior of the mutants. An important aspect of the method is that the mutations are directed to specific target genes rather than at random throughout the genome, a desirable feature for researchers investigating a single gene and for systematic-mutagenesis projects. Directed methods are usually more complicated than random methods, and although the method described here has multiple steps, most are simple and do not involve cloning. The method selects for the amber mutation indirectly by association with another marker and takes advantage of the ease of screening for conditional lethal phenotypes. This method also generates a “scarless” mutation; that is, no artifacts are left in the genome after the process is complete, resulting in a clean experimental system. Unlike promoter insertion mutagenesis, it has the advantage of leaving the native promoter intact and thus retains the natural regulation and transcription level of the operon. The ability to completely arrest growth indicates that the suppressor can be tightly shut off by using the arabinose promoter. As an inducer, arabinose control is preferable to TS control because arabinose produces less dramatic changes in cellular physiology. On the other hand, arabinose does have metabolic effects and must be transported, unlike the inducer isopropyl-β-d-thiogalactopyranoside (IPTG). For inducer withdrawal experiments such as those shown here, catabolism of the inducer is an advantage because it reduces residual inducer levels to zero.
Potential disadvantages of the method include the failure to mutate all of the genes targeted, incomplete suppression of some genes, and the presence of a high-copy-number suppressor plasmid, leading to the spontaneous occurrence of constitutive suppressor mutants. Such mutants are infrequent enough to allow growth experiments, as shown here, but the problem might be greatly reduced by deleting one of the redundant Ala2 tRNA genes (alaX or alaW). Also, single-copy suppressors have been used to achieve complete suppression of some genes (30), although such efficiency may not be possible for genes encoding highly abundant proteins. Regardless, this method could be improved by the development of a better inducible suppressor, especially one at single copy. Additionally, it should be noted that an amber mutation in the middle of the gene could result in a partially active product if the truncated protein is still functional. A systematic-mutagenesis project under way in our laboratory has isolated transposon insertions in known essential genes and should identify those where position effects may be a problem (Y. Kang, T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch, K. M. Winterberg, and F. R. Blattner, submitted for publication). Such situations may be resolved by positioning the amber mutation near the beginning of the gene and integrating the selectable marker upstream rather than downstream. Other methods might avoid this problem by complete deletion of the gene and plasmid complementation, but then the native copy number and regulation would be lost, as with a promoter insertion.
A recent study with pathogenic strains of E. coli (26) reported a 10-fold increase in spontaneous rifampin-resistant mutants following overnight expression of lambda red but no increase following a 1-h induction (2 h was used in this study). More study is needed since it is conceivable that rifampin-resistant mutations in RNA polymerase might have been selected by reducing the transcription of toxic lambda red. In any case, the induction time used in future work to make competent cells and for plasmid curing should probably be reduced to minimize the possibility of introducing undesired mutations. The use of I-SceI does not increase the occurrence of spontaneous rifampin mutants (32), and recombinational repair of double-stranded breaks appears to be error free (22).
Polarity effects on the expression of downstream genes can lead to difficulty in interpreting the results of experiments with conditional mutations. Antisense mutations may affect the expression of entire operons, while promoter insertions affect all of the genes downstream of the insertion site. Amber mutations are not expected to have polar effects except in cases of rho-mediated termination. Our difficulty mutating ftsZ may have been due to the presence of lpxC immediately downstream (under the control of its own promoter). Integration of the Camr drug marker and subsequent transcription originating from the Camr promoter might impact genes located downstream. The genes targeted here were at the distal end of transcriptional units or in single-gene operons. On the basis of known or predicted operon structure (5), 468 out of the 684 genes determined to be essential by either Gerdes et al. (11) or the PEC database (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp) occur without other essential genes downstream. We did not test whether this method is limited to such situations. Regardless, it is still widely applicable to most essential genes.
Conflicting results exist for whether ycfB (also named mnmA, asuE, or trmU) is truly essential. Arigoni et al. (2) could not obtain a null mutant, but when the gene was under direct arabinose promoter control, it was insensitive to the absence of arabinose. Kambampati and Lauhon (20) used a different putative translation start site to make an in-frame deletion of this gene, but Gerdes et al. have recently identified it as an essential gene (11). We identified a glucose-sensitive mutation in ycfB on one occasion, but after removal of the gene for Camr and plasmid curing, it was found to grow normally on glucose. Sequencing showed that it was in fact amber, suggesting that ycfB is not essential.
The seven essential gene mutants made here displayed a wide range of behaviors under nonpermissive conditions. For example, growth of the mutant with a murA amber mutation arrested almost immediately, whereas the map amber mutant was relatively unaffected for many generations. When cells are transferred to nonpermissive medium, a number of events must occur before a phenotype is observed: residual arabinose must be metabolized, transcription of the suppressor tRNA must be halted, residual levels of suppressor must degrade or be diluted by cell growth, residual levels of the targeted gene product must turn over, and finally the loss of that product then has a physiological effect that stops growth. Since all but the last two events ought to be the same for all mutants, the wide variety of behaviors that we observed must be due to gene-specific factors, such as protein degradation rates or how closely a protein is expressed relative to the critical threshold necessary for survival.
The protein encoded by murA is the target for the antibiotic fosfomycin (18), and antibacterial agents have been developed against the lpxC gene product (7). The ribosomal protein S1 from rpsA is an essential component of the ribosome that is targeted by many antibiotics, while the products of frr, gcpE, and map are potential targets (16, 19, 36). The behavior of these mutants may give an important indication as to their efficacy as drug targets. If loss of an essential gene product takes many generations to impede cellular functions, then even the most potent drug may be unusable for the treatment of infection. In assessing this, it is important to remember that while a drug may directly inactivate a protein, withdrawal of the suppressor blocks new synthesis.
Strains can be engineered for the titration of arabinose-controlled expression to intermediate levels (25). RNA isolated from conditional lethal mutants at suboptimal suppression levels can be analyzed for expression patterns diagnostic of loss of that essential function. By comparing these patterns with those from cells subjected to an uncharacterized antibacterial compound, the mode of action of that compound may be investigated. Attenuated essential gene mutants are often more sensitive to compounds that target the mutated gene product and can be used to screen for new drugs and to establish the targets of existing drugs (9, 10). The wide variety of behaviors observed here in nonpermissive medium indicates that conditional amber mutants will also be useful in basic research to characterize essential genes of unknown or poorly understood function, such as those identified by Gerdes et al. (11).
Supplementary Material
Acknowledgments
We thank Tim Durfee, Jeremy Glasner, and David Frisch for valuable guidance. Plasmid pST98-AS was generously provided by Gyorgy Posfai, pKD46 was provided by Barry Wanner, and I-SceI was provided by the laboratory of Bernard Dujon.
This work was supported by NIH grant GM35682.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851-856. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, K. A., R. Acosta, E. Ledner, Y. Machida, M. Pancotto, M. McCormick, H. Ohtsubo, and E. Ohtsubo. 1984. A 37 × 103 molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J. Mol. Biol. 175:331-348. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Bockhorst, J., Y. Qiu, J. Glasner, M. Liu, F. Blattner, and M. Craven. 2003. Predicting bacterial transcription units using sequence and expression data. Bioinformatics 19(Suppl. 1):I34-I43. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. D., E. I. Vivas, C. T. Walsh, and R. Kolter. 1995. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 177:4194-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clements, J. M., F. Coignard, I. Johnson, S. Chandler, S. Palan, A. Waller, J. Wijkmans, and M. G. Hunter. 2002. Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob. Agents Chemother. 46:1793-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeVito, J. A., J. A. Mills, V. G. Liu, A. Agarwal, C. F. Sizemore, Z. Yao, D. M. Stoughton, M. G. Cappiello, M. D. Barbosa, L. A. Foster, and D. L. Pompliano. 2002. An array of target-specific screening strains for antibacterial discovery. Nat. Biotechnol. 20:478-483. [DOI] [PubMed] [Google Scholar]
- 10.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, K. G. C., P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu Zy, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herring, C. D., J. D. Glasner, and F. R. Blattner. 2003. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 311:153-163. [DOI] [PubMed] [Google Scholar]
- 14.Janosi, L., S. Mottagui-Tabar, L. A. Isaksson, Y. Sekine, E. Ohtsubo, S. Zhang, S. Goon, S. Nelken, M. Shuda, and A. Kaji. 1998. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 17:1141-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji, Y., B. Zhang, S. F. Van, Horn, P. Warren, G. Woodnutt, M. K. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 16.Jomaa, H., J. Wiesner, S. Sanderbrand, B. Altincicek, C. Weidemeyer, M. Hintz, I. Turbachova, M. Eberl, J. Zeidler, H. K. Lichtenthaler, D. Soldati, and E. Beck. 1999. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285:1573-1576. [DOI] [PubMed] [Google Scholar]
- 17.Judson, N., and J. J. Mekalanos. 2000. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 18:740-745. [DOI] [PubMed] [Google Scholar]
- 18.Kahan, F. M., J. S. Kahan, P. J. Cassidy, and H. Kropp. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 235:364-386. [DOI] [PubMed] [Google Scholar]
- 19.Kaji, A., E. Teyssier, and G. Hirokawa. 1998. Disassembly of the post-termination complex and reduction of translational error by ribosome recycling factor (RRF)—a possible new target for antibacterial agents. Biochem. Biophys. Res. Commun. 250:1-4. [DOI] [PubMed] [Google Scholar]
- 20.Kambampati, R., and C. T. Lauhon. 2003. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42:1109-1117. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolisnychenko, V., G. Plunkett III, C. D. Herring, T. Feher, J. Posfai, F. R. Blattner, and G. Posfai. 2002. Engineering a reduced Escherichia coli genome. Genome Res. 12:640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kollas, A. K., E. C. Duin, M. Eberl, B. Altincicek, M. Hintz, A. Reichenberg, D. Henschker, A. Henne, I. Steinbrecher, D. N. Ostrovsky, R. Hedderich, E. Beck, H. Jomaa, and J. Wiesner. 2002. Functional characterization of GcpE, an essential enzyme of the non-mevalonate pathway of isoprenoid biosynthesis. FEBS Lett. 532:432-436. [DOI] [PubMed] [Google Scholar]
- 24.Koonin, E. V. 2000. How many genes can make a cell: the minimal-gene-set concept. Annu. Rev. Genomics Hum. Genet. 1:99-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan-Kiss, R. M., C. Wadler, and J. E. Cronan, Jr. 2002. Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. Proc. Natl. Acad. Sci. USA 99:7373-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mushegian, A. 1999. The minimal genome concept. Curr. Opin. Genet. Dev. 9:709-714. [DOI] [PubMed] [Google Scholar]
- 28.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Normanly, J., L. G. Kleina, J. M. Masson, J. Abelson, and J. H. Miller. 1990. Construction of Escherichia coli amber suppressor tRNA genes. III. Determination of tRNA specificity. J. Mol. Biol. 213:719-726. [DOI] [PubMed] [Google Scholar]
- 30.Oeschger, M. P., N. S. Oeschger, G. T. Wiprud, and S. L. Woods. 1980. High efficiency temperature-sensitive amber suppressor strains of Escherichia coli K12: isolation of strains with suppressor-enhancing mutations. Mol. Gen. Genet. 177:545-552. [DOI] [PubMed] [Google Scholar]
- 31.Oeschger, M. P., and S. L. Woods. 1976. A temperature-sensitive suppressor enabling the manipulation of the level of individual proteins in intact cells. Cell 7:205-212. [DOI] [PubMed] [Google Scholar]
- 32.Posfai, G., V. Kolisnychenko, Z. Bereczki, and F. R. Blattner. 1999. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 27:4409-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Schmid, M. B., N. Kapur, D. R. Isaacson, P. Lindroos, and C. Sharpe. 1989. Genetic analysis of temperature-sensitive lethal mutants of Salmonella typhimurium. Genetics 123:625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengupta, J., R. K. Agrawal, and J. Frank. 2001. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. USA 98:11991-11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sergueev, K., D. Yu, S. Austin, and D. Court. 2001. Cell toxicity caused by products of the p(L) operon of bacteriophage lambda. Gene 272:227-235. [DOI] [PubMed] [Google Scholar]
- 36.Vaughan, M. D., P. B. Sampson, and J. F. Honek. 2002. Methionine in and out of proteins: targets for drug design. Curr. Med. Chem. 9:385-409. [DOI] [PubMed] [Google Scholar]
- 37.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.