Abstract
CTnDOT is a conjugative transposon (CTn) that is found in many Bacteroides strains. Transfer of CTnDOT is stimulated 100- to 1,000-fold if the cells are first exposed to tetracycline (TET). Both excision and transfer of CTnDOT are stimulated by TET. An operon that contains a TET resistance gene, tetQ, and two regulatory genes, rteA and rteB, is essential for control of excision and transfer functions. At first, it appeared that RteA and RteB, which are members of a two-component regulatory system, might be directly responsible for the TET effect. We show here, however, that neither RteA nor RteB affected expression of the operon. TetQ, a ribosome protection type of TET resistance protein, actually reduced operon expression, possibly by interacting with ribosomes that are translating the tetQ message. Fusions of tetQ with a reporter gene, uidA, were only expressed at a high level when the fusion was cloned in frame with the first six codons of tetQ. However, out of frame fusions or fusions ending at the other five codons of tetQ showed much lower expression of the uidA gene. Moreover, reverse transcription-PCR amplification of tetQ mRNA revealed that despite the fact that the uidA gene product, β-glucuronidase (GUS), was produced only when the cells were exposed to TET, tetQ mRNA was produced in both the presence and absence of TET. Computer analysis of the region upstream of the tetQ start codon predicted that the mRNA in this region could form a complex RNA hairpin structure that would prevent access of ribosomes to the ribosome binding site. Mutations that abolished base pairing in the stem that formed the base of this putative hairpin structure made GUS production as high in the absence of TET as in TET-stimulated cells. Compensatory mutations that restored the hairpin structure led to a return of regulated production of GUS. Thus, the tetQ-rteA-rteB operon appears to be regulated by a translational attenuation mechanism.
Many Bacteroides strains carry conjugative transposons (CTns) that are closely related to a CTn called CTnDOT (25, 29, 39). An interesting and unusual feature of many members of the CTnDOT family is that the antibiotic tetracycline (TET) stimulates both excision of the CTn from the chromosome and conjugal transfer of the excised element (24, 31, 34). In fact, without TET stimulation of donor cells, virtually no transfer occurs (33, 38). Previous studies identified a central regulatory region on CTnDOT that is required for TET induction of transfer functions. This region contains a three-gene operon that consists of the TET resistance gene tetQ and two regulatory genes, rteA and rteB (Fig. 1) (29, 34). RteA and RteB are most closely related to the sensor and response regulator components, respectively, of two-component regulatory systems. RteA presumably activates RteB, and activated RteB stimulates the expression of a nearby gene, rteC, which is also required for expression of excision and transfer genes (32-34, 38) (Fig. 1).
FIG. 1.
(A) Organization of CTnDOT. The names of the genes are given above the arrows. The arrows indicate the orientations and positions of genes on CTnDOT. The size of each functional region is given under the diagram. Note, the size is not in ratio. (B) Diagram of the central regulatory region that includes the tetQ rteA rteB operon and the rteC gene and description of their functions. Pq and Pc are the tetQ promoter and the rteC promoter, respectively. Numbers 1, 2, and 3 indicate three levels of regulation. Tc, TET.
The regulatory cascade mediated by RteA, RteB, and RteC is not, however, the first step in TET stimulation of CTn transfer. Expression of the tetQ-rteA-rteB operon itself is controlled by TET. In an earlier study, it was reported that a fusion of tetQ to the reporter gene uidA produced a fusion (pAGF22) whose expression was stimulated by TET (33). The mechanism of this stimulation was not investigated. In a separate study, Bayley et al. (1) compared the sequences of the upstream regions of several Bacteroides genes, including tetQ, and tentatively identified consensus promoter sequences. They also mapped the transcript start sites of these genes and showed that the consensus promoter sequences were located at −7 and −33 relative to the transcript start site, but they did not investigate further the regulatory regions of any of the genes they compared (1). In this report, we provide the first detailed analysis of the tetQ promoter region and provide evidence that TET regulation of the operon is mediated by a translational attenuation mechanism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α MCR or DH10B was grown in Luria broth or on Luria agar. Bacteroides strains were grown in chopped meat (Remel) for subculture or in Trypticase-yeast extract-glucose media (15, 26). The Bacteroides thetaiotaomicron 5482 strains BT4001 and BT4100 are spontaneous rifampin and thymidine auxotrophic mutants, respectively. Strain BT4004 has CTnERL in the chromosome of BT4001. Strain BT4107 has CTnDOT in the chromosome of BT4100. The antibiotic concentrations used were as follows: ampicillin, 100 μg/ml; cefoxitin (FOX), 20 μg/ml; chloramphenicol (CHL), 10 μg/ml; erythromycin (ERY), 10 μg/ml; gentamicin (GEN), 200 μg/ml; rifampin, 10 μg/ml; trimethoprim, 100 μg/ml. TET was used to select for transconjugants at 3 μg/ml and for maximum induction of CTn activity at 1 μg/ml. Thymidine was added to a final concentration of 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant phenotypea | Description (reference) |
|---|---|---|
| Bacteroides strain BT4001ΩQABC | Rifr Tcr | BT4001 with tetQrteABC integrated into chromosome (38) |
| Plasmids | ||
| pMJF2 | Emr (Apr) | pUC19-based vector containing a promoterless uidA (GUS) gene (8) |
| pTUG1 | Emr (Apr) | Blunted BalI-EcoRI fragment spanning from 301 bp upstream of tetQ to 24 bp N-terminal of tetQ DNA cloned into SmaI site of pMJF2 (21) |
| pTUG3 | Emr (Apr) | Blunted EcoNI-EcoRI fragment spanning from 423 bp upstream of tetQ to 23 bp N-terminal of tetQ DNA cloned into SmaI site of pMJF2 (21) |
| pTUG10 | Emr (Apr) | Blunted SstI-EcoRI fragment containing 74-bp γδ sequence 513 bp upstream of and 24 bp N-terminal of tetQ DNA cloned into SmaI site of pMJF2 (21) |
| pTUG11 | Emr (Apr) | SphI-EcoRI fragment spanning from 700 bp upstream of to 20 bp N-terminal of tetQ DNA cloned into SphI-SmaI site of pMJF2 (this study) |
| pTUG12 | Emr (Apr) | SphI-SspI fragment spanning from 700 bp upstream of to 6 bp N-terminal of tetQ DNA cloned into SphI-SmaI site of pMJF2 (this study) |
| pYP44 | Cmr (Apr) | HindIII-HincII-digested P6-Rin1 fragment cloned into HindIII-NruI-digested pC-COW (this study) |
| pNJR24 | Cmr (Knr) | Shuttle vector containing pB8-51 oriV, which can replicate in Bacteroides (16) |
| pAMS9 | Tcr Cmr (Knr) | 7.6-kb tetQ rteA rteB rteC region (including Pq region) cloned into pNJR24 (32) |
| pTC-COW | Tcr Cmr (Ap Tc Cmr) | E. coli-Bacteroides shuttle vector that is mobilizable in E. coli and Bacteroides due to the pB8-51 oriV-mob region (10) |
| pC-COW | Cmr (Apr) | Derivative of pTC-COW shuttle vector containing oriV of pB8-51 |
| pLYL05 | Cefr (Apr) | E. coli-Bacteroides shuttle vector that is mobilizable in E. coli and Bacteroides due to pBI143 oriV-mob region and contains the cfxA gene (4) |
| pYP67 | Cexr (Apr) | SphI-SstI Pq-uidA fragment digested from pTUG15:3-Rin1 cloned into SphI-SstI-digested pLYL05 (this study) |
| pYP54 | Emr (Apr) | SphI-HincII-digested P6-Rin1:4 PCR product cloned into SphI-SmaI-digested pMJF2 (this study) |
| pYP81 | Emr (Apr) | SphI-HincII-digested P6(SM1:4)-Rin1:4 fragment cloned into SphI-SmaI-digested pMJF2 (this study) |
| pYP86 | Emr (Apr) | SphI-HincII-digested P7-Rin1:12 PCR product cloned into SphI-SmaI-digested pMJF2 (this study) |
| pYP116 | Emr (Apr) | SphI-HincII-digested P7(SM1:12)-Rin1:12 fragment cloned into SphI-SmaI-digested pMJF2 (this study) |
| pYP161 | Emr (Apr) | SphI-HincII-digested P7-Rin1:13 PCR product cloned into SphI-SmaI-digested pMJF2 (this study) |
| pYP84TA | (Apr) | P7-Rin1 PCR fragment cloned into pGEMT (this study) |
| pYP220 (Del Hp1) | Emr (Apr) | pMJF2::Pq clone which has a deletion of 11 nucleotides (−117 to −107) in Hp1, Del Hp1 forward and reverse primers and pYP84TA were used as templates for site-directed mutagenesis (this study) |
Phenotypes in parentheses are expressed only in E. coli, and phenotypes outside parentheses are expressed in Bacteroides strains. Abbreviations: Ap, Ampicillin; Cm, CHL; Em, ERY; Kn, kanamycin; Rif, rifampin; Tc, TET; Thy-, thymidine auxotroph; Tp, trimethoprim.
Construction of fusions.
To localize the region needed for regulated expression from the Pq promoter, a nested set of the Pq promoter region segments, generated by PCR, were fused to the uidA reporter gene. The PCR products were each purified with the Promega Wizard DNA clean-up kit. Primers used in this study are listed in Table 2. The forward primers contained an SphI site. The downstream primers contained either an HincII or an SspI site located at the sixth amino acid of tetQ. PCR products were digested by SphI-HincII or SphI-SspI and cloned into the SphI-SmaI site of pMJF2, upstream of the uidA reporter gene (8). This placed the tetQ 5′ end in frame with the ATG start codon of the uidA gene. The resulting plasmids were first transformed into the E. coli DH10B strain and then transferred by conjugation to a Bacteroides recipient. In other experiments, transcriptional fusions were made by the same strategy except that 1 bp was missing from the restriction site, putting the 5′ end of tetQ out of frame with uidA.
TABLE 2.
Primers used in this study
| Primer | Annealing position relative to ATG start codon of tetQ | Sequence of primer (5′-3′) or descriptiona |
|---|---|---|
| P6 | −343 to −324 | CAAATGCATGCCTAAAGAAG |
| P7 | −279 to −260 | TTCAGCATGCTAAAACAGTG |
| P7:3 | −225 to −204 | GTTGAACCTACGCATGCCTAAT |
| P9 | −124 to −104 | GCGGCATGCATAATATACATA |
| Rin1 | 32 to 13 | GTGAGCAAGAGTTAACAAAT |
| Rin1:4 | 32 to −24 | One point mutation at −20 by changing T to A |
| Rin1:12 | 31 to −24 | Three point mutations at −16, −17, and −18 by changing TAT to ATA |
| Rin1:13 | 31 to −24 | Five point mutations at −18 to −12 by changing TATTATT to ATATAAA |
| SM 1:4 | −120 to −104 | One point mutation at −108 by changing A to T |
| SM 1:12 | −130 to −94 | Three point mutations at −112 to −110 by changing ATA to TAT |
| Del Hp1 for | −135 to −82 | GTAATCGTTATGCGGCAGATATTAATACGAGTTAGGAATCCTG |
| Del Hp1 rev | −135 to −82 | CAGGATTCCTAACTCGTATTAATATCTGCCGCATAACGATTAC |
| RTtetQ-R1 | 450 to 425 | CTTGAGACAGATTTGCTTTTATATCC |
The nucleotides in bold indicate the region changed.
DNA sequencing was performed by the University of Illinois Biotechnology Genetic Engineering Facility with an Applied Biosystems model 373A, version 2.0.1A, automated dye terminator. Primers were synthesized by the University of Illinois Biotechnology Genetic Engineering Facility or by Operon Technologies, Inc. (Alameda, Calif.).
Triparental matings.
pMJF2::Pq clones were moved into Bacteroides strains via triparental matings. In each case, the two donors were (i) E. coli DH10B, which contained the pMJF2::Pq derivative to be tested, and (ii) E. coli HB101, which contained the IncPα plasmid RP1. RP1 cannot replicate in Bacteroides spp., but it does mobilize E. coli-Bacteroides vectors from E. coli donors to Bacteroides recipients. Matings were done on nitrocellulose filters as described previously (30). For the pMJF2-based clones, the transconjugants were selected on GEN (200 μg/ml)-ERY (10 μg/ml) plates. For the pTC-COW-based clones, the transconjugants were selected on GEN (200 μg/ml)-CHL (10 μg/ml) plates. For the pLYL05-based clones, the transconjugants were selected on GEN (200 μg/ml)-FOX (20 μg/ml) plates.
GUS assay.
The uidA reporter gene on pMJF2 encodes an E. coli β-glucuronidase (GUS). GUS assays were done by the procedure of Feldhaus et al. (8). One unit was defined as 0.01 A415 U per min at 37°C. Protein concentrations were determined by the method of Lowry et al. (18). All the GUS activities in this study are the average activities of the results from at least three different transconjugants.
Site-directed mutagenesis.
Site-directed mutagenesis was done to test the importance of a possible hairpin structure identified in the DNA sequence. Site-directed mutagenesis was done by the Stratagene QuikChange method. PCR products were treated with DpnI for 3 h, and then 5 μl of the reaction mixture was used to transform E. coli DH10B. Each Pq region mutation was confirmed by sequencing. The Pq mutation was isolated on an SphI-HincII fragment from the vector and cloned into the SphI-SmaI site of pMJF2 to create a translational fusion.
RNA secondary structure prediction.
Predictions of RNA secondary structure and calculations of free energies were determined through the MFOLD, version 3.1, program of M. Zuker (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi).
RT-PCR.
BT4107, which contains a copy of wild-type CTnDOT in the chromosome, was grown in VPI broth without TET treatment or with TET treatment (1 μg/ml). Total RNA was isolated with the Trizol reagent (Invitrogen) and was dissolved in 30 μl of RNA resuspension solution (Ambion). All samples were treated with DNase I (Ambion) to eliminate DNA contamination before using the RNA samples for reverse transcription (RT)-PCR. DNase I-treated RNA samples (1 μl of a 1-ng/μl preparation) served as a template in each reaction (25-μl final volume per reaction). The RT-PCR kit (Promega) was used as described in the manufacturer's directions. The amplified products were electrophoresed on a 1% agarose gel. The forward primer P9 was located −124 bp upstream of the tetQ start codon, and the reverse primer RTtetQ-R1 was located at +450 bp within the tetQ gene (Table 2). The amplified product is 574 bp long.
RESULTS
Translational fusion versus transcriptional fusion with uidA gene.
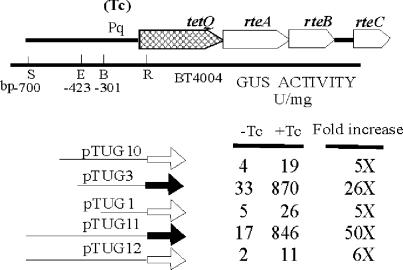
A function map of the conjugative transposon CTnDOT is shown in Fig. 1A, and an enlargement of the Pq-tetQ-rteA-rteB-rteC region is shown in Fig. 1B. In initial experiments to define and characterize the tetQ promoter region, different lengths of the tetQ upstream region were cloned into pMJF2 to construct pTUG1, pTUG3, and pTUG10 (Fig. 2). The cloned Pq regions started from different positions upstream of tetQ, but all were thought to end at the same nucleotide in the blunted EcoRI site that falls at the sixth amino acid of TetQ. These pMJF2::Pq constructs were then moved into BT4004, which contained a copy of CTnERL (CTnDOT minus the ermF region) (37). Surprisingly, the strain containing pTUG3 exhibited a much higher GUS activity and increase than the strains containing pTUG1 and pTUG10 (Fig. 2).
FIG. 2.
GUS activities of different Pq-uidA fusions in strain BT4004 (which contains CTnERL). The increase is the GUS activity detected in TET-induced cells (+Tc) divided by the GUS activity in noninduced cells (−Tc). The thick arrows indicate the reporter gene uidA. The hollow arrows in pTUG10, pTUG1, and pTUG12 indicate an out of frame fusion to the uidA gene. The gray arrows in pTUG3 and pTUG11 indicate an in-frame fusion to the uidA gene. The numbers at the left corner indicate positions relative to the start codon of tetQ. TET (1 μg/ml) was used to induce expression. S, SstI; E, EcoNI; B, BalI; R, EcoRI.
Sequencing of the Pq region in pTUG3, pTUG1, and pTUG10 revealed that one extra nucleotide was missing in pTUG3 at the junction with the reporter gene. Thus, the ATG of tetQ was shifted so that it was in frame with the ATG of the uidA gene, forming a translational fusion. The cloned Pq regions in pTUG1 and pTUG10 were both out of frame with the uidA gene. This indicated that the high GUS activity of the strain containing pTUG3 had resulted from its being a translational rather than a transcriptional fusion, but it was also possible that the extra upstream sequences in pTUG10 or the decreased upstream DNA in pTUG1 were influencing the GUS expression.
To test the hypothesis that the in-frame fusion was responsible for the high activity associated with pTUG3, we made two other constructs, pTUG11 and pTUG12. Both constructs started 700 bp upstream of the start codon of tetQ, but the fusion in pTUG11 generated an in-frame fusion with the uidA gene, whereas the fusion in pTUG12 generated an out of frame fusion with the uidA gene. The results confirmed that only those constructs that had in-frame fusions were associated with high GUS activity and the higher TET induction effect (Fig. 2). Accordingly, unless mentioned specifically, in-frame fusions were used which included the first 6 amino acids (aa) of TetQ in subsequent experiments.
Except for tetQ, no other genes from the CTn have any effect on TET regulation of the tetQ operon.
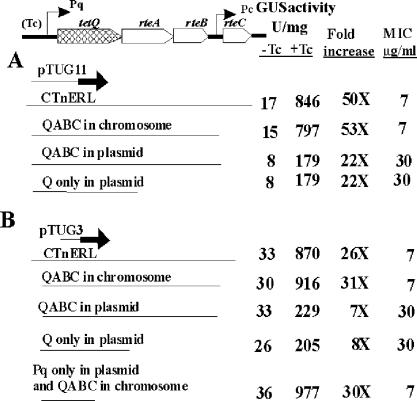
To determine whether any genes from CTnDOT are involved in TET regulation of the tetQ operon expression, an in-frame fusion plasmid (pTUG3 or pTUG11) was moved into different Bacteroides strains, which contained various combinations of CTnDOT genes. The results summarized in Fig. 3 demonstrate that neither genes outside the central regulatory region (QABC) nor the rteA-rteB-rteC genes in the central regulatory region were essential for TET regulation of the tetQ operon expression. We also noted that the expression of the reporter gene was approximately fourfold lower when the QABC region was provided on a plasmid that had a copy number of 5 to 10 than it was if the QABC region was provided in single copy in the chromosome (BT4001ΩQABC). This effect might have been due to the presence of multiple copies of Pq (5 to 10 copies per cell), which led to titration of some host factors, or it could have been due to increased production of TetQ. TetQ is a ribosome protection-type resistance protein that is thought to interact with ribosomes and modify them so that they no longer bind to TET (3, 6). The first possibility was ruled out because when Pq was coresident in the strain that also contained the fusion plasmid, there was no effect on the level of GUS activity. The second possibility was supported by the MIC experiments, which showed that an increase in the copy number of tetQ was associated with a higher MIC, indicating that more TetQ protein was produced in these strains and was affecting the ribosomes (Fig. 3).
FIG. 3.
Determination of which CTnDOT genes affect TET (Tc)-induced expression through the Pq promoter. pTUG11 and pTUG3 were used to determine GUS production in different backgrounds. The strain labeled “QABC in chromosome” (BT4001ΩQABC) had a single copy of QABC integrated into the chromosome of BT4001. The strain labeled “QABC in plasmid” (pAMS9) had 5 to 10 copies of QABC per cell. The strain labeled “Q only in plasmid” (pTC-COW) contained only the tetQ gene on a plasmid (5 to 10 copies per cell). The strain labeled “Pq only in plasmid” (pYP44) had only the Pq region on a plasmid (5 to 10 copies per cell). MIC indicates the minimal inhibition concentration of TET. As in Fig. 2, the increase is the GUS activity detected in TET-induced cells (+Tc) divided by the GUS activity in noninduced cells (−Tc).
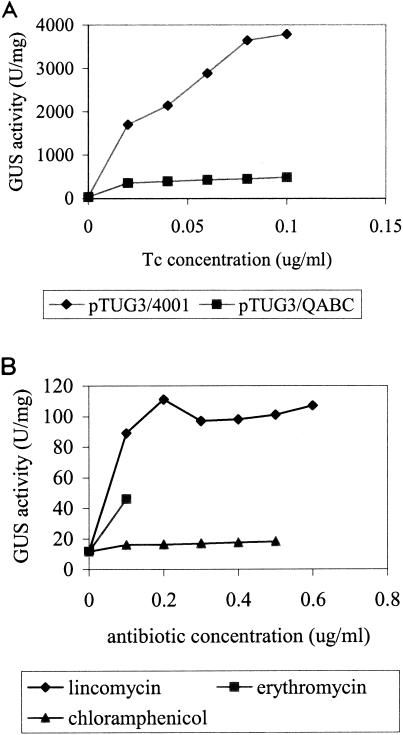
If TetQ causes lower GUS expression, we would expect that GUS activity would rise further in cells that did not contain tetQ. To test this, we compared the GUS activity of a strain that had no copy of tetQ with that of a strain that contained one copy of tetQ in the chromosome (Fig. 4A). The MIC of TET for BT4001(pTUG3), which contained no tetQ gene, was 0.12 μg/ml, so both strains were exposed to concentrations of TET ranging from 0 to 0.10 μg/ml. At 0.10 μg/ml of TET, the GUS production with tetQ in the strain was 480 U/mg compared to 3,800 U/mg in the absence of tetQ (Fig. 4A).
FIG. 4.
GUS production under different conditions. (A) GUS activity of pTUG3/4001 with or without the tetQ gene in the strain. The GUS activity associated with pTUG3 without the tetQ gene was much higher than that associated with tetQ. The MIC for pTUG3/4001 (Genr Emr) is 0.12 μg/ml. Thus, the induction concentration of TET (Tc) ranges from 0 to 0.10 μg/ml. (B) GUS activity of pYP67/4001 (Genr Cefr) induced by LIN (0 to 0.6 μg/ml), ERY (0 to 0.1 μg/ml), and CHL (0 to 0.5 μg/ml). pYP67 contains the Pq-uidA fusion in a vector that carries a FOX resistance gene.
Induction can be mediated by other ribosome-binding antibiotics.
The target of TET is the 30S subunit of the ribosome (5, 11). To test whether other ribosome-targeting antibiotics can also induce GUS expression, ERY, lincomycin (LIN), and CHL were tested as putative inducers. All of these antibiotics are protein synthesis inhibitors that bind to bacterial ribosomes. ERY and LIN block the ribosome exit tunnel, thereby preventing movement and release of the nascent peptide (28), and CHL inhibits peptidyl transferase activity (22, 28).
Previously, ERY was tested to determine whether it can stimulate the transfer of CTnDOT, like TET does. Conjugation experiments were done between BT4107 and BT4001, but the transfer frequency of CTnDOT was similar under different ERY concentrations, indicating that ERY does not function as an inducer (unpublished data). Since CTnDOT contains an ermF gene, which encodes a methylase, ErmF, it was also possible that the methylated ribosomes prevent the ERY induction activity. In this experimental design, no gene which encodes Emr, Lcr, or Cmr was included in the strain. pYP67, which contains the Pq-uidA fusion in a vector that carries a FOX resistance gene, was used in this experiment because FOX does not affect ribosome function (9).
BT4001(pYP67) was induced with low levels of these antibiotics individually. The GUS expression could be induced by LIN and ERY (Fig. 4B). LIN (0.2 μg/ml) caused a 10-fold induction, and ERY (0.1 μg/ml) caused a 3-fold induction. By contrast, CHL did not induce expression at all (Fig. 4B). These results provide further support for the hypothesis that ribosomes are involved in the induction mechanism.
Localization and initial characterization of the tetQ regulatory region.
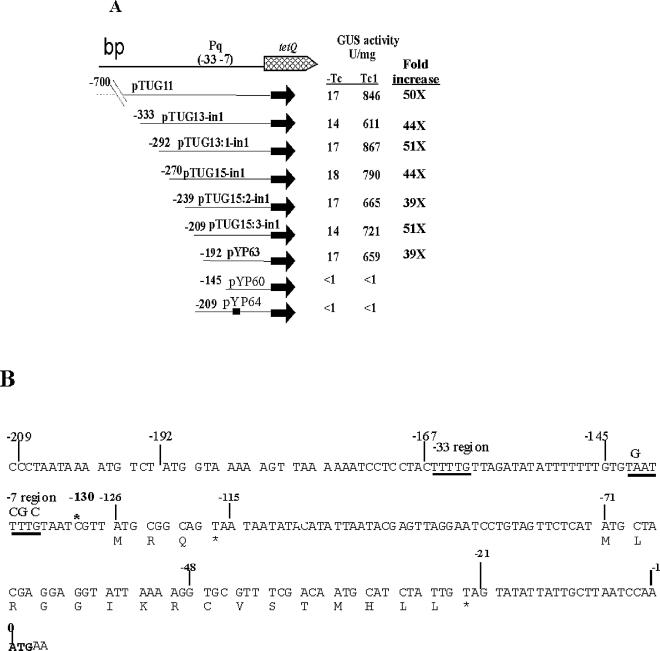
By deleting DNA from the upstream region of tetQ, we were able to narrow down the location of Pq to within 209 bp of the start codon of tetQ (−209) (Fig. 5A). The location of the transcription initiation site (TIS) identified by Bayley et al. (1) is indicated at −130 bp in Fig. 5B. Bayley et al. had identified a possible promoter consensus region (−7 region and −33 region) located immediately upstream of the TIS. Our genetic results agree with this location of the promoter and also indicate that this promoter is the only promoter of tetQ by showing that the activity was abolished in strain pYP64 in which the −7 region was mutated (Fig. 5).
FIG. 5.
(A) Localization of Pq promoter. The number before each fusion indicates the relative position of the fusion joint to the ATG start codon of tetQ. The name of each fusion is labeled behind the number. The consensus tetQ promoter sequence identified by Bayley et al. is indicated by (−33, −7). The thick line in pYP64 indicates that there are four point mutations (Fig. 5B) within the −7 region of the Pq promoter that eliminate the consensus −7 sequence. Tc, TET. (B) Nucleotide sequence of the 209 bp upstream of the ATG start codon of tetQ. The asterisk above the sequence indicates the position of the TIS. The Pq region is indicated by a −33 region and a −7 region. Arrows labeled Hp1 and Hp8 indicate that a hairpin can be formed in the mRNA by Hp1 and Hp8. The numbers above the vertical lines indicate their positions relative to the ATG start codon of tetQ. The deduced amino acid sequences of two leader peptides are given in single-letter code under the nucleotide sequence. The bold ATG is the putative start codon of tetQ. Asterisks indicate stop codons.
There is a 130-bp segment of mRNA between the transcription start site and the first possible start codon of tetQ. Computer analysis of the RNA sequence in this region revealed a complex hairpin structure that included the region immediately upstream of the start codon of TetQ. Such a structure might prevent ribosomes from binding the ribosome binding site (RBS). Although in Bacteroides, no consensus RBS has been identified so far, it is found to be usually located within 20 bp upstream of the following gene based on alignment of known Bacteroides genes (35). If a form of translational attenuation was responsible for controlling operon expression, we would expect that tetQ mRNA would be produced both in the absence and in the presence of TET. RT-PCR, with primers that amplified a portion of tetQ mRNA (P9 and RTtetQ-R1) (Table 2), demonstrated that mRNA was produced even in the absence of TET stimulation, at levels comparable to those seen in TET-stimulated cells (Fig. 6). Since this finding supported the hypothesis that regulation was mediated by a translational attenuation mechanism, we focused our attention on the region between the transcription start site and the start codon of tetQ, the putative leader region.
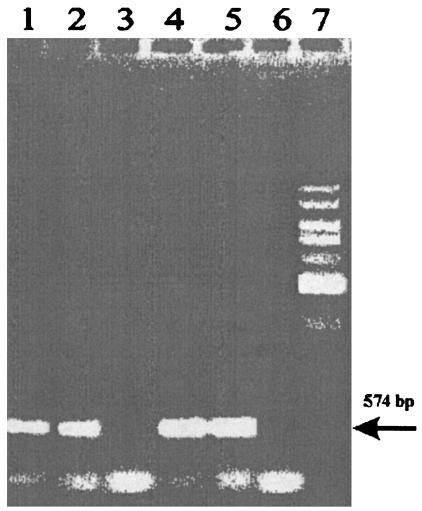
FIG. 6.
RT-PCR to detect the amount of tetQ mRNA in BT4107 under TET-induced and non-TET-induced conditions. Lanes 1 and 2, tetQ product amplified by RT-PCR within the no-TET cells; lane 3, control to which RT was not added; lanes 4 and 5, tetQ product amplified by RT-PCR from the TET-treated BT4107 cells; lane 6, control to which RT was not added; lane 7, 1-kb standard. The arrow indicates the size of the amplified tetQ product (574 bp).
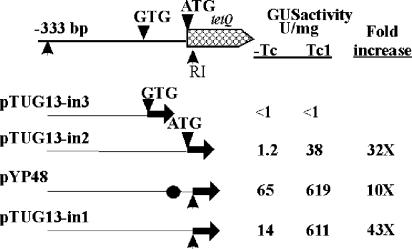
There are two possible start codons for the tetQ gene. One is an ATG at position 0 (Fig. 5B and 7). There is also a GTG 48 bp upstream of the ATG codon. Since the location of the start codon is important if regulation is mediated by an attenuation-type mechanism, a Pq-uidA in-frame fusion was made to each of these codons. There was no detectable GUS activity from the strain in which the Pq region ends at GTG (strain pTUG13-in3), whereas the strain in which Pq ends at the ATG exhibited TET-induced GUS activity (pTUG13-in2) (Fig. 7). Thus, ATG, rather than GTG, is the start codon of tetQ. This was confirmed by introducing two stop codons between GTG and ATG in strain pYP48. TET-regulated GUS activity was still seen in extracts from this strain (Fig. 7).
FIG. 7.
Determination of the start codon of the tetQ gene. The Pq regions in all five of the constructs start from −333 bp upstream of tetQ, but each ends at a different position. The Pq region in pTUG13-in3 ends at the GTG putative start codon of tetQ. In pTUG13-in2, the ATG start codon was fused to the uidA gene. In both pTUG13-in1 and pYP48, the Pq region ends at the sixth amino acid of TetQ and was fused to the uidA gene. The Pq region in pYP48 is the same as that in pTUG13-in1 except that two stop codons, which are indicated by a circle, were put between GTG and ATG, their exact positions are shown in Fig. 8A. Tc, TET.
The GUS activity of the strain containing the fusion to the ATG start codon (pTUG13-in2) was 11- to 16-fold lower than that of the strain containing the first 6 aa of TetQ (pTUG13-in1). This was true for cells grown either with or without TET stimulation. The increases in GUS activity, however, were similar in both cases (30- to 50-fold). To test how the 6 aa made this difference, a series of fusions were made in which the Pq regions start at the same site (−333 bp) but each ends at a different one of the 6 aa. GUS activity of all these strains showed similar TET induction (30- to 50-fold), but the absolute value increased with each additional amino acid. Thus, the difference is probably due to an effect of the extra 6 aa on the stability of the hybrid TetQ-GUS protein. It is also possible that either the bases in that sequence are important for efficient translation initiation or that decoding of those six codons is more efficient.
Involvement of possible RNA hairpin structure in regulation of tetQ operon.
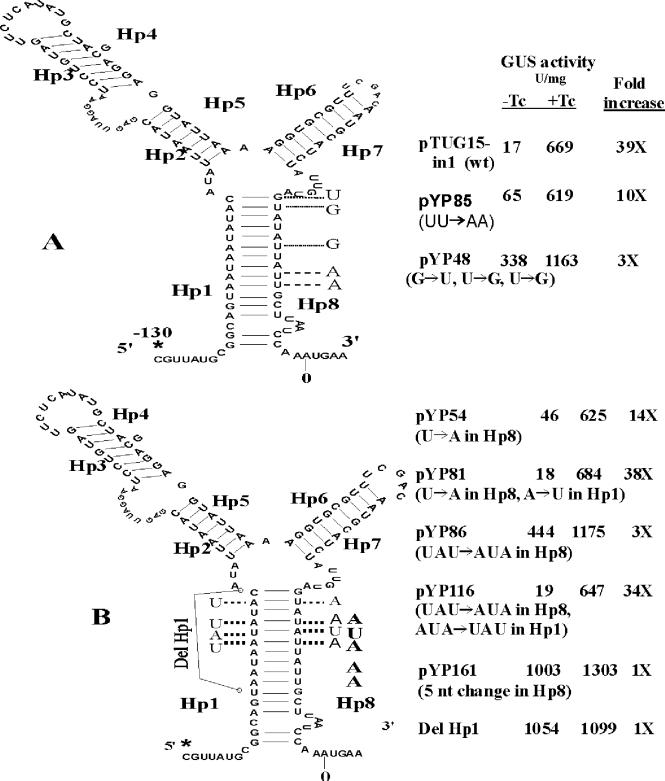
The predicted secondary structure of the mRNA from −130 to +4 is shown in Fig. 8. The program used to predict this structure was developed by M. Zuker et al. (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi). All of the mutations used to test the importance of this predicted structure in TET regulation are shown in Fig. 8. pTUG15-in1 is a strain containing a fusion with the wild-type Pq region. The Pq region in plasmids pYP48 and pYP85 has point mutations on Hp8, the hairpin that would be formed by sequences Hp1 and Hp8 was partially disrupted, and the basal levels of GUS activity of these two strains are higher than that of the wild-type strain (Fig. 8A). In pYP54 and pYP86, the hairpin that would be formed by sequences Hp1 and Hp8 was partially disrupted by changing U to A or UAU to AUA on Hp8 (Fig. 8B). The uninduced levels of GUS activity associated with these mutations were 4- and 25-fold higher, respectively, than the activity in extracts from the strain with the wild-type Pq region. In pYP81 and pYP116, the stem was reestablished by changing the corresponding nucleotides on Hp1 to complementary nucleotides. In strains containing these constructs, the GUS activity returned to that of the strain with the wild-type Pq region. In strain pYP161, when the stem was completely disrupted by changing 5 of 12 nucleotides in Hp8, TET induction was abolished completely and expression was nearly constitutive (Fig. 8B). This result suggests that the hairpin formed by Hp1 and Hp8 is responsible for the full TET induction activity.
FIG. 8.
Evidence for the importance of a secondary structure of the 130-bp tetQ mRNA in TET (Tc) induction. Asterisks indicate the TIS; Hp1 to Hp8 indicate that their mRNA sequences can be part of a hairpin. The ΔG of the secondary structure of the wild-type Pq mRNA (−130 to +4) is −33.24 kcal/mol. The nucleotides following the dotted lines indicate substituted nucleotides to disrupt or reestablish the secondary structure. (A) GUS activity of the wild-type strain and mutants, which have a partially disrupted hairpin in the Pq region. (B) Evidence to show that the Hp1- and Hp8-formed hairpin is responsible for all of the TET induction activity. Strain Del Hp1 is also named pYP220. nt, nucleotide.
In mutant Del Hp1 (pYP220), 11 nucleotides in Hp1 were deleted, and this mutant showed a constitutively high level of GUS expression both without TET and under TET-induced conditions (1,054 and 1,099 U/mg, respectively). These data further support our conclusion that the hairpin formed by Hp1 and Hp8 is responsible for the full TET induction and that disruption of this hairpin caused the release of the RBS in Hp8 and the efficient expression of the downstream gene(s) (Fig. 8B).
DISCUSSION
Our results support a translational attenuation-type regulation of the tetQ-rteA-rteB operon in response to TET. RT-PCR analysis showed that a similar amount of the mRNA from this operon is made regardless of whether TET is present. This result rules out another possible model for attenuational control of gene expression, the binding of TET to a structure in the mRNA itself in such a way as to cause antitermination (12-14). In the case of an antitermination type of control, the concentration of tetQ mRNA should have been much higher under inducing conditions.
Although tetQ mRNA was made both in the presence and absence of TET stimulation, translation of the message, as indicated by GUS activity, was much higher in TET-stimulated cells than in cells not stimulated by TET. This observation, taken together with the fact that a translational fusion with uidA was responsible for much higher activity and a higher increase in TET-stimulated cells than a transcriptional fusion in the same locus (Fig. 2), supports the hypothesis that regulation occurs at the level of translation rather than transcription. There appeared to be slightly more tetQ mRNA produced in TET-treated cells than in untreated cells (Fig. 6). Also, expression of the transcriptional fusion was slightly higher in TET-treated cells. Some antibiotics, including TET, are known to increase the stability of mRNA (2, 7, 27, 36). Thus, it is possible that the apparent increased mRNA level in cells growing in the presence of TET could be due to this type of stabilization rather than to a second stage of regulation at the transcriptional level. Whatever the reason, the increase in TET-treated cells is not nearly high enough to explain the enhancement of GUS activity in TET-treated cells carrying the translational fusion.
Further evidence for translational attenuation came from mutagenesis experiments that were guided by the observation that a hairpin structure could form in the leader region of the mRNA. The predicted hairpin structure was complex. It consisted of a stem composed of Hp1 and Hp8 sequences and intervening sequences Hp2 to Hp7, which could also form hairpin structures. Since Hp8 presumably included an RBS, the Hp1 to Hp8 stem seemed most likely to be involved in regulation. Site-directed mutations that disrupted the Hp1 to Hp8 stem of this hairpin structure made production of the fusion protein as high or nearly as high in the absence of TET as in the presence of TET. Compensating mutations that restored the hairpin structure but changed the sequences of Hp1 and Hp8 restored regulation of protein production. Thus, it appears that the structure of the mRNA is important for TET regulation of protein production from the operon message.
Our finding that ERY and LIN, two other protein synthesis inhibitors, also stimulated higher GUS production supports the contention that the rate at which ribosomes bind the mRNA and move along it might be an important factor in regulation. A finding that argues against this hypothesis, however, is that CHL, another protein synthesis inhibitor, had no stimulatory effect on the production of operon-encoded proteins. Thus, if the rate of ribosome movement along the mRNA is a factor in regulation, CHL must be having some effect on the ribosomes that is different enough from that of TET, ERY, and LIN to explain this difference. It is clear that CHL is capable of participating in translational attenuation because some CHL resistance genes are regulated by a translational attenuation mechanism (17, 23).
If ribosomes are involved in regulation, rather than having a regulatory effect due to binding of TET to a structure in the mRNA, a nontoxic analog of TET ought to be unable to increase GUS production. To test whether TET analogues can cause induced GUS expression, two nontoxic forms of TET, anhydrotetracycline and autoclaved chlorotetracycline, were tested as inducers. Neither of these caused increased production of GUS activity at levels similar to the level of TET used in induction experiments (<10 μg/ml; unpublished data). Autoclaved chlorotetracycline could act as an inducer at very high concentrations (200 μg/ml), but at this concentration, the growth rate of the cells was slowed considerably. This effect could be explained if some intact chlorotetracycline survived autoclaving and was the molecule actually responsible for the induction.
A possible model that explains all of our results is that the effect of TET is to cause ribosomes to move more slowly along the leader sequence and that, in some way, this altered mobility of the ribosomes causes the Hp1 to Hp8 stem of the hairpin to be disrupted, thus exposing the RBS of tetQ so that tetQ mRNA can be translated. Analyzing the leader region with a folding program revealed only the RNA structure shown in Fig. 8. There was no evident alternate stem-loop structure such as is seen in many attenuation systems (17, 19, 20).
In this respect, our results are most similar to those of Rogers et al. (23), who found only a single complex RNA structure in the leader region of the catB gene of Agrobacterium tumefaciens. How such a structure interacts with ribosomes in such a way as to disrupt the structure is unknown. Rogers et al. (23) identified a possible 10-aa leader peptide whose coding sequence extended into the base of the stem-loop structure. Analysis of the tetQ leader region in the vicinity of the hairpin structure revealed only two possible peptide-encoding sequences. One could encode a 3-aa peptide. The sequence of this short peptide overlaps with the base of one side of the hairpin (HP1). The other possible peptide is 16 aa long and is encoded by sequences that lie within the region between Hp1 and Hp8. This sequence does not extend into the Hp1 to Hp8 hairpin on either side and thus is unlikely to play the role of a leader peptide. If GTG is allowed as a start codon, there is a third possible peptide that starts within Hp6 and extends into Hp8. Further work is needed to determine whether any of these putative peptides is involved in attenuation. The role of the sequences between Hp1 and Hp8 also remains to be determined.
Production of RteA and RteB, the other proteins encoded in the tetQ-rteA-rteB operon, should be regulated similarly to production of TetQ. That is, the proteins are produced at detectable levels only when cells are stimulated with TET (unpublished results). Since rteA and rteB have been shown to be in the same operon as tetQ, production of these proteins is presumably due to translational coupling once ribosomes begin to translate the operon mRNA. Our results show clearly that RteA and RteB have no role in TET regulation of operon expression. This was somewhat surprising because RteA and RteB are both essential for CTn excision and transfer functions and resemble components of a two-component regulatory system. Evidently, RteA, the putative sensor protein, is sensing something other than TET.
TetQ was not required for TET stimulation of GUS production, but its presence did have an effect. It actually reduced production of the GUS fusion protein compared to cells that contained no tetQ gene. Moreover, increased levels of TetQ due to increased copy numbers of tetQ caused a greater decrease in protein production. TetQ is a ribosome protection type of TET resistance protein, which is most closely related to elongation factor G. TetQ, presumably like Tet(O) (6), modifies ribosomes so that TET no longer binds as efficiently. The fact that a higher intracellular concentration of TetQ increased the MIC and further decreased translation of operon mRNA suggests that one copy of tetQ is not sufficient to protect all of the ribosomes in the cell. The modulating effect of TetQ on translation may be a means for the cell to limit the production of RteA and RteB and thus the amount of CTn excision and transfer that occurs. Excision of CTnDOT is apparently not a lethal event because the chromosomal site from which the CTn is excised is resealed in the process (4), but too much activity of this sort could have some deleterious effect on fitness that could be important in an environment as competitive as the human colon.
Previously, translational attenuation has been found in the proteobacteria and in the gram-positive bacteria. Finding evidence for this type of regulation in Bacteroides species extends it to a third phylogenetic group, an indication that it is more universal as a regulatory strategy than was previously thought to be the case. It is interesting that so far translational attenuation has been associated almost exclusively with the regulation of antibiotic resistance genes. This focus could be due to the fact that the antibiotics involved target ribosomes, but it could also be due to the fact that this type of regulation has not been considered in other areas, such as the study of slow-growth conditions where ribosomes are also affected. This is the first report of translational attenuation that affects regulatory genes. Preliminary evidence indicates that the excision of CTnDOT, which is controlled by RteA and RteB, might also be regulated in a growth-phase-dependent manner (N. B. Shoemaker, G.-R. Wang, and A. A. Salyers, unpublished data). If so, translational attenuation may affect genes other than antibiotic resistance genes.
Acknowledgments
We thank Ella Rotman for technical assistance and Mikeljon Peter Nikolich for constructing pTUG1, pTUG3, and pTUG10.
This work was supported by NIH grant AI22383.
REFERENCES
- 1.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 2.Bechhofer, D. H., and K. H. Zen. 1989. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol. 171:5803-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burdett, V. 1991. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J. Biol. Chem. 266:2872-2877. [PubMed] [Google Scholar]
- 4.Cheng, Q., Y. Sutanto, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2001. Identification of genes required for the excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625-632. [DOI] [PubMed] [Google Scholar]
- 5.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell, S. R., C. A. Trieber, G. P. Dinos, E. Einfeldt, D. E. Taylor, and K. H. Nierhaus. 2003. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 22:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreher, J., and H. Matzura. 1991. Chloramphenicol-induced stabilization of cat messager RNA in Bacillus subtilis. Mol. Microbiol. 5:3025-3034. [DOI] [PubMed] [Google Scholar]
- 8.Feldhaus, M. J., V. Hwa, Q. Cheng, and A. A. Salyers. 1991. Use of an Escherichia coli beta-glucuronidase gene as a reporter gene for investigation of Bacteroides promoters. J. Bacteriol. 173:4540-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finegold, S. M. 1989. Mechanisms of resistance in anaerobes and new developments in testing. Diagn. Microbiol. Infect. Dis. 12:117S-120S. [DOI] [PubMed] [Google Scholar]
- 10.Gardner, R. G., J. B. Russell, D. B. Wilson, G. R. Wang, and N. B. Shoemaker. 1996. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl. Environ. Microbiol. 62:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geigenmuller, U., and K. H. Nierhaus. 1986. Tetracycline can inhibit tRNA binding to the ribosomal P site as well as to the A site. Eur. J. Biochem. 161:723-726. [DOI] [PubMed] [Google Scholar]
- 12.Grundy, F. J., and T. M. Henkin. 2003. The T box and S box transcription termination control systems. Front. Biosci. 1:20-31. [DOI] [PubMed] [Google Scholar]
- 13.Grundy, F. J., S. C. Lehman, and T. M. Henkin. 2003. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. USA 100:12057-12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henkin, T. M., and C. Yanofsky. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24:700-707. [DOI] [PubMed] [Google Scholar]
- 15.Holdeman, L. V., and W. E. C. Moore. 1975. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg.
- 16.Li, L. Y., N. B. Shoemaker, and A. A. Salyers. 1995. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. J. Bacteriol. 177:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovett, P. S. 1996. Translation attenuation of chloramphenicol resistance in bacteria-a review. Gene 179:157-162. [DOI] [PubMed] [Google Scholar]
- 18.Lowry, O. H., N. J. Rosebrough, A. S. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 143:265-275. [PubMed] [Google Scholar]
- 19.Mayford, M., and B. Weisblum. 1989. Conformational alterations in the ermC transcript in vivo during induction. EMBO J. 8:4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narayanan, C. S., and D. Dubnau. 1987. An in vitro study of the translational attenuation model of ermC regulation. J. Biol. Chem. 262:1756-1765. [PubMed] [Google Scholar]
- 21.Nikolich, M. P. 1994. Tetracycline resistance and its dissemination in the Bacteroides group. University of Illinois at Urbana-Champaign, Urbana.
- 22.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 23.Rogers, E. J., M. S. Rahman, R. T. Hill, and P. S. Lovett. 2002. The chloramphenicol-inducible catB gene in Agrobacterium tumefaciens is regulated by translation attenuation. J. Bacteriol. 184:4296-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salyers, A. A., and N. B. Shoemaker. 1997. Conjugative transposons. Genet. Eng. (New York) 19:89-100. [DOI] [PubMed] [Google Scholar]
- 25.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 27.Sandler, P., and B. Weisblum. 1988. Erythromycin-induced stabilization of ermA messenger RNA in Staphylococcus aureus and Bacillus subtilis. J. Mol. Biol. 203:905-915. [DOI] [PubMed] [Google Scholar]
- 28.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 29.Shoemaker, N. B., R. D. Barber, and A. A. Salyers. 1989. Cloning and characterization of a Bacteroides conjugal tetracycline-erythromycin resistance element by using a shuttle cosmid vector. J. Bacteriol. 171:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoemaker, N. B., and A. A. Salyers. 1987. Facilitated transfer of IncP beta R751 derivatives from the chromosome of Bacteroides uniformis to Escherichia coli recipients by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 169:3160-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemaker, N. B., and A. A. Salyers. 1988. Tetracycline-dependent appearance of plasmidlike forms in Bacteroides uniformis 0061 mediated by conjugal Bacteroides tetracycline resistance elements. J. Bacteriol. 170:1651-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens, A. M., J. M. Sanders, N. B. Shoemaker, and A. A. Salyers. 1992. Genes involved in production of plasmidlike forms by a Bacteroides conjugal chromosomal element share amino acid homology with two-component regulatory systems. J. Bacteriol. 174:2935-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens, A. M., N. B. Shoemaker, L. Y. Li, and A. A. Salyers. 1993. Tetracycline regulation of genes on Bacteroides conjugative transposons. J. Bacteriol. 175:6134-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens, A. M., N. B. Shoemaker, and A. A. Salyers. 1990. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J. Bacteriol. 172:4271-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tribble, G. D., A. C. Parker, and C. J. Smith. 1999. Genetic structure and transcriptional analysis of a mobilizable, antibiotic resistance transposon from Bacteroides. Plasmid 42:1-12. [DOI] [PubMed] [Google Scholar]
- 36.Wei, Y., and D. H. Bechhofer. 2002. Tetracycline induces stabilization of mRNA in Bacillus subtilis. J. Bacteriol. 184:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittle, G., B. D. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Characterization of the 13 kb ermF region of Bacteroides conjugative transposon, CTnDOT. Appl. Environ. Microbiol. 67:3488-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. Identification of two genes involved in the modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 184:3839-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]