Abstract
Trastuzumab is a successful rationally designed ERBB2-targeted therapy. However, about half of individuals with ERBB2-overexpressing breast cancer do not respond to trastuzumab-based therapies, owing to various resistance mechanisms. Clinically applicable regimens for overcoming trastuzumab resistance of different mechanisms are not yet available. We show that the nonreceptor tyrosine kinase c-SRC (SRC) is a key modulator of trastuzumab response and a common node downstream of multiple trastuzumab resistance pathways. We find that SRC is activated in both acquired and de novo trastuzumab-resistant cells and uncover a novel mechanism of SRC regulation involving dephosphorylation by PTEN. Increased SRC activation conferred considerable trastuzumab resistance in breast cancer cells and correlated with trastuzumab resistance in patients. Targeting SRC in combination with trastuzumab sensitized multiple lines of trastuzumab-resistant cells to trastuzumab and eliminated trastuzumab-resistant tumors in vivo, suggesting the potential clinical application of this strategy to overcome trastuzumab resistance.
Rationally designed targeted therapies could represent the future of personalized medicine for cancer patients. Trastuzumab, one of the most successful examples of these therapies, is a humanized antibody targeting human epidermal growth factor receptor-2 (HER2 or ERBB2). It shows considerable clinical efficacy and extends the overall survival of certain patients with ERBB2-overexpressing breast cancer1,2. However, the overall response rate to trastuzumab-containing therapies remains modest: ~26% when used as a single therapy and 40–60% when used in combination with systemic chemotherapy1,3,4. Many patients do not respond to initial trastuzumab treatment (de novo resistance), and many trastuzumab-responsive patients develop resistance after continuous treatment (acquired resistance)5,6. Although multiple trastuzumab resistance mechanisms have been identified in preclinical studies, no effective regimen has been developed to overcome trastuzumab resistance in patients. More important, inherent tumor heterogeneity and alternative survival pathways developed during drug treatment pose additional challenges to the clinical management of patients with trastuzumab-resistant tumors of different genesis.
Two major categories of trastuzumab resistance mechanisms have been proposed: de novo resistance due to genetic alterations of receptor tyrosine kinases (RTKs) and their downstream signaling targets; and acquired resistance primarily due to the acquisition of alternative RTK signaling activation that compensate for ERBB2 inhibition after trastuzumab treatment. The most prevalent de novo resistance mechanisms include constitutive activation of the phosphoinositide 3-kinase (PI3K) pathway owing to phosphatase and tensin homolog (PTEN) deficiency7 or PIK3CA gene mutations8, and the accumulation of truncated ERBB2 receptors (p95HER2), which lack an extracellular trastuzumab-binding domain9. The overexpression of other RTKs, such as epidermal growth factor receptor (EGFR) family receptors10–12, insulin-like growth factor-1 receptor (IGF-1R)13,14 and hepatocyte growth factor15, also contribute to both de novo and acquired trastuzumab resistance. Heterodimerization between RTKs may redundantly trigger cell proliferation signals and confer resistance when ERBB2 is inhibited by trastuzumab16,17.
One could design targeted therapy corresponding to a specific resistance mechanism. However, it is possible that multiple resistance mechanisms may coexist in patients with late-stage, heterogeneous, metastatic breast cancer. It would be more effective and clinically practical to identify and target the key nodes common to several (if not all) of the aforementioned resistance mechanisms.
Here we report a rationally designed therapy targeting the key node in multiple signaling pathways driving trastuzumab resistance, which might broadly benefit trastuzumab-resistant patients. We identify the nonreceptor tyrosine kinase SRC as a common node, which is hyperactivated in various trastuzumab resistance models. We also show a direct dephosphorylation of SRC by PTEN’s protein phosphatase activity, which is involved in de novo trastzumab resistance, conferred by PTEN deficiency. SRC activation confers trastuzumab resistance in breast cancer cells and is correlated with both lower response and poorer survival in patients who received trastuzumab-based therapy. Notably, targeting this key node with a SRC inhibitor universally sensitized cells bearing distinct resistant mechanisms to trastuzumab treatment in vitro and in vivo. Our data indicate that combinatorial treatment with SRC inhibitor plus trastuzumab could be a clinically useful strategy for overcoming trastuzumab resistance conferred by multiple mechanisms.
RESULTS
SRC activation in acquired trastuzumab-resistant cells
First, we modeled the development of acquired resistance in patients, by treating ERBB2-overexpressing breast cancer cells (BT474, AU565, and SKBR3) with increasing doses of trastuzumab for >6 months to select trastuzumab-resistant sublines (TtzmR). Compared with parental lines, TtzmR cells were significantly (P = 0.003, 0.001 and 0.001) more resistant to trastuzumab treatment in vitro (Fig. 1a and Supplementary Fig. 1). Orthotopic xenograft tumors of BT474. TtzmR cells were significantly (P = 0.001) less responsive to trastuzumab treatment (Fig. 1b). Prolonged treatment with trastuzumab leads to alterations (‘reprogramming’) of various RTKs17. Indeed, we found a substantial downregulation of ERBB2 and a prominent upregulation of EGFR (in all three cell lines), HER3 (in AU565) and IGF-1R (in AU565) in the TtzmR sublines compared with parental cell lines (Fig. 1c,d), suggesting that increased EGFR, HER3 and IGF-1R signaling may be acquired resistance mechanisms. Consistently, EGFR phosphorylation (at Tyr1068) was increased in TtzmR lines, whereas ERBB2 receptor was downregulated (Fig. 1e). Notably, TtzmR lines showed increased SRC phosphorylation at Tyr416 suggesting SRC activation, which was further confirmed by SRC kinase assay (Supplementary Fig. 2). The unchanged PTEN protein and AKT phosphorylation in TtzmR lines (Fig. 1e) indicate that the acquired trastuzumab resistance was not due to selection of a pre-existing sub-population with PI3K pathway alterations.
Figure 1.
SRC hyperactivation is a key signaling alteration in acquired trastuzumab-resistant cells. (a) MTS assay comparing cell proliferation of indicated parental breast cancer cell lines and their corresponding acquired TtzmR sublines upon treatment with freshly added trastuzumab (Ttzm, 2 µg ml−1) for 4 d. (b) Tumor volume of mammary fat pad xenografts derived from either BT474 parental (P) or TtzmR subline upon treatment of IgG control or Ttzm (10 mg per kg body weight, intraperitoneally) weekly. Tumor volume at various times of treatment is presented as percentage of original tumor size at day 0 of treatment. (c) Representative histograms from flow cytometric analysis of EGFR and ERBB2 abundance in BT474 parental and TtzmR cells. (d) Relative amounts of EGFR, ERBB2, HER3 and IGF-1R in the indicated parental and TtzmR cells analyzed by flow cytometry. (e) Immunoblots comparing major cell signaling changes between the indicated parental and TtzmR sublines. P indicates phosphorylation; for example, P-EGFR-Y1068 is EFGR phosphorylated at Tyr1068 (f) Immunoblots assessing the impact of overexpression of EGFR and IGF-1R in BT474 parental (BT474.P) cells on signaling. (g) MTS assay evaluating trastuzumab resistance of BT474 cells overexpressing EGFR or IGF-1R, treated as in a. (h) Immunoblots of EGFR and phosphorylated SRC after shRNA-mediated EGFR knockdown in BT474.TtzmR cells. (i) MTS assay evaluating sensitivity of TtzmR to trastuzumab after EGFR knockdown. Cells were treated as in a. (j) Left, immunoblots of knock-down of SRC in BT474.TtzmR cells by SRC shRNA. Right, MTS assay assessing trastuzumab sensitivity of TtzmR cells after SRC knockdown. All error bars, s.e.m. All quantitative data were generated from a minimum of three replicates.
Next, we investigated the role of alternative RTK pathways in SRC activation by ectopic overexpression of EGFR or IGF-1R in the parental trastuzumab-sensitive BT474 cells. Overexpression of either RTK led to increased phosphorylation of SRC Tyr416 (Fig. 1f) and a trastuzumab-resistant phenotype (Fig. 1g). Knockdown of EGFR in the BT474.TtzmR cells inhibited phosphorylation of SRC Tyr416 (Fig. 1h). shRNA-mediated knockdown of either EGFR (Fig. 1i) or SRC (Fig. 1j) considerably resensitized the TtzmR cells to trastuzumab treatment. Collectively, these data indicate that SRC activation is a critical event downstream of alternative RTK signaling pathways contributing to acquired trastuzumab resistance.
SRC is activated in de novo trastuzumab-resistant cells
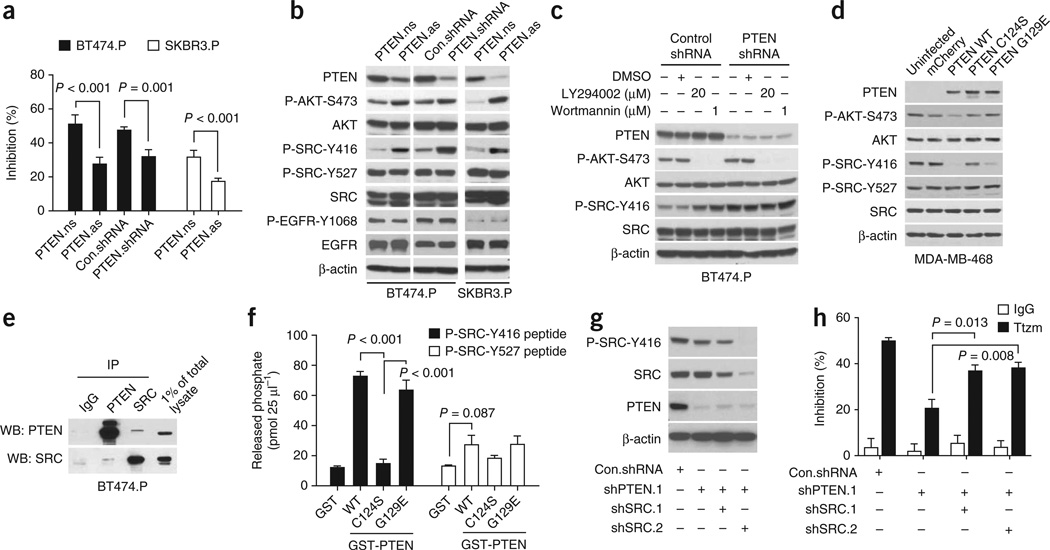
PTEN deficiency confers de novo trastuzumab resistance7. Knocking down PTEN with either antisense oligonucleotides (PTEN.as) or shRNA (PTEN-shRNA) conferred considerable resistance to trastuzumab compared with control cells (Fig. 2a and Supplementary Fig. 3). Notably, although PTEN knockdown had no discernible effect on EGFR, it led to an increase in phosphorylation of SRC Tyr416, indicating that PTEN loss also led to SRC activation (Fig. 2b and Supplementary Fig. 4). Moreover, PI3K inhibitors (LY294002 or wortmannin) did not inhibit SRC activation in PTEN-knockdown cells (Fig. 2c and Supplementary Fig. 5), indicating that SRC activation is not due to PTEN loss–mediated AKT activation.
Figure 2.
SRC is activated in PTEN-deficient de novo trastuzumab-resistant cells. (a) MTS assay comparing cell proliferation of the indicated parental cell lines and their corresponding PTEN knockdown sublines upon trastuzumab treatment (Ttzm, 2 µg ml−1, 4 d). PTEN knockdown by either PTEN antisense (PTEN.as) oligonucleotides or PTEN shRNA is described in Online Methods. PTEN nonsense oligonucleotides, PTEN.ns. (b) Immunoblots assessing the effects of PTEN knockdown on phosphorylation of SRC Tyr416 (P-SRC-Y416) and other signals. (c) Immunoblots examining effects of inhibition of PI3K–AKT pathway on P-SRC-Y416 in PTEN knockdown cells. (d) Immunoblots assessing P-SRC-Y416 after reconstitution of wild-type (WT) PTEN, PTEN C124S or PTEN G129E mutants in PTEN-deficient MDA-MB-468 cells. (e) Coimmunoprecipitation assay to test for interactions between PTEN and SRC in BT474.P cells. (f) In vitro PTEN phosphatase assay detecting capability of purified GST-PTEN proteins (WT, C124S or G129E) to directly dephosphorylate the P-SRC-Y416 and P-SRC-Y527 phosphopeptides. (g) Immunoblots evaluating the efficiency of SRC knockdown by SRC shRNA in BT474 PTEN knockdown cells. (h) MTS assay assessing trastuzumab sensitivity of PTEN knockdown cells with or without SRC knockdown. Cells were treated as in a. All error bars, s.e.m. All quantitative data were generated from a minimum of three replicates.
As PTEN is a dual lipid and protein phosphatase18, we next explored whether SRC Tyr416 is a direct substrate for PTEN protein phosphatase activity and if increased SRC phosphorylation at Tyr416 is a direct result of PTEN loss. In PTEN-deficient MDA-MB-468 cells (Fig. 2d) or PTEN-knockdown BT474 cells (Supplementary Fig. 6), we reconstituted either wild-type PTEN or two PTEN mutants identified from patients’ tumors (a C124S mutant, deficient in lipid and protein phosphatase activities; a G129E mutant, deficient in lipid phosphatase activity only19). The wild-type PTEN and the G129E mutant (but not the C124S mutant) substantially reduced phosphorylation of SRC Tyr416 without affecting Tyr527. Moreover, coimmuno-precipitation (Fig. 2e) and GST pull-down assays (Supplementary Fig. 7) demonstrated a direct interaction between PTEN and SRC proteins. Furthermore, in a cell-free PTEN phosphatase assay, GST-PTEN G129E did not dephosphorylate Phosphatidylinositol (3,4,5)-trisphosphate (PIP3) as wild-type PTEN did (Supplementary Fig. 8). However, the G129E mutant readily released phosphates from the SRC peptide phosphorylated at Tyr416 without strong effect on the SRC peptide phosphorylated at Tyr527 (Fig. 2f). The data indicate that PTEN directly and specifically dephosphorylates SRC at Tyr416 by its protein phosphatase activity. Notably, knockdown of SRC in PTEN-deficient BT474 parental cells significantly (P = 0.013 and P = 0.008 compared with shPTEN) restored the sensitivity of these cells to trastuzumab treatment (Fig. 2g,h). Therefore, loss of PTEN protein phosphatase activity led to SRC activation that contributed to the de novo trastuzumab resistance in PTEN-deficient cells.
SRC activation drives trastuzumab resistance
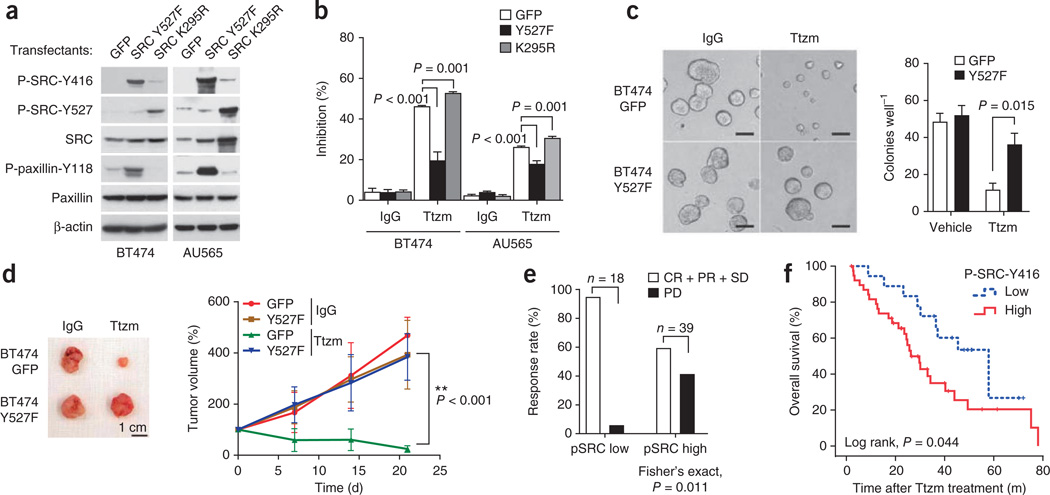
As SRC is commonly activated in EGFR-overexpressing, IGFR-overexpressing and PTEN-deficient cells, SRC activation seems to be a common event downstream of various signaling pathways leading to trastuzumab resistance. To determine whether SRC activation is sufficient to confer trastuzumab resistance, we stably expressed a constitutively active SRC Y527F mutant and a dominant-negative SRC K295R mutant20, in both BT474 and AU565 cells. Indeed, cells expressing SRC Y527F showed increased SRC activation and hyperphosphorylation of SRC Tyr416 and the SRC downstream target paxillin at Tyr118 (Fig. 3a and Supplementary Fig. 9). A notably increased phosphorylation of SRC Tyr527 (an inhibitory site for SRC activity) and reduced phosphorylation of paxillin Tyr118 indicated reduced SRC activity in SRC K295R–transfected cells (Fig. 3a). Compared with controls, SRC Y527F cells were highly resistant to trastuzumab-mediated growth inhibition whereas SRC K295R cells were more sensitive (Fig. 3b). Similarly, overexpression of wild-type SRC also conferred SRC activation and trastuzumab resistance (Supplementary Figs. 10 and 11).
Figure 3.
SRC is a key modulator of trastuzumab response. (a) Immunoblots comparing SRC phosphorylation status in the indicated cells expressing a constitutively active SRC mutant (Y527F) or a kinase-dead SRC mutant (K295R). (b) MTS assay assessing trastuzumab sensitivity of cells transfected with SRC Y527F or SRC K295R mutant. Cells were treated as in Figure 1a. Error bars, s.e.m. (c) 3D tumor spheroid assay comparing response to trastuzumab treatment of BT474.GFP and BT474.SRC Y527F. 3D tumor spheroid assay was carried out as described in Online Methods. Scale bar, 100 µm. (d) Top, representative BT474 orthotopic xenograft tumors. Scale bar, 1 cm. Bottom, volume of mammary fat pad xenograft tumors derived from either GFP-labeled BT474 parental (GFP) or SRC Y527F–expressing cells upon treatment with IgG or Ttzm. Tumor volume at various times of treatment is percentage of original tumor size at day zero of treatment. Error bars, s.e.m. Ttzm-treated GFP group versus SRC Y527F group (ANOVA, P < 0.001, two-sided). (e) Correlation between clinical response rate and amount of tumor phospho-SRC-Y416 (pSRC) in patients who received first-line trastuzumab-based therapy. Complete response (CR), partial response (PR) and stable disease (SD) were grouped together and compared with SD. Patient response was compared by Fisher’s exact test (P = 0.011, two-sided). (f) Low versus high tumor phospho-SRC-Y416 abundance and overall survival of patients who received first-line trastuzumab-based therapy. Difference of overall survival was analyzed by Kaplan-Meier survival model with log-rank test (P = 0.044, two-sided).
Additionally, SRC Y527F cells responded poorly to trastuzumab in a three-dimensional (3D) tumor spheroid assay (Fig. 3c). Furthermore, trastuzumab treatment did not inhibit the growth of BT474.SRC Y527F mammary fat pad tumors (Fig. 3d). Collectively, these data indicate that SRC activation is sufficient to confer trastuzumab resistance.
SRC activity correlates with patient trastuzumab response
To examine whether SRC activation is involved in clinical trastuzumab resistance, we retrospectively evaluated the relationship between SRC activation and patient response to trastuzumab-based therapies. We assessed SRC activation in primary breast tumors from 57 patients, who were subsequently treated with trastuzumab-based therapies (Supplementary Table 1), by immunohistochemistry staining using a validated antibody for SRC phosphorylated at Tyr416 (pSRC-Y416; Supplementary Fig. 12).
In general, the phosphorylation of SRC Tyr416 is positively correlated (Pearson r = 0.297, P = 0.025) with total SRC protein abundance (data not shown). Notably, patients with high amounts of phosphorylated SRC (pSRC) in tumors showed a lower clinical response rate and a higher progressive disease rate after trastuzumab treatment than patients having low pSRC tumors (Fisher’s exact test, P = 0.011, Fig. 3e). The overall survival of patients with high pSRC tumors (median 34.2 months) was significantly (log-rank test, P = 0.044, Fig. 3f) lower than that of patients with low pSRC tumors (median 57.9 months) after trastuzumab-based therapy (Fig. 3f). These data suggest that SRC activation is correlated with clinical trastuzumab resistance.
SRC activates RTKs that induce trastuzumab resistance
To study the mechanism of SRC activation–mediated trastuzumab resistance in TtzmR cells, we modulated SRC activity using either the small-molecule inhibitor saracatinib or SRC shRNA in TtzmR cells. In TtzmR cells, compared with parental cells, the amount of total EGFR and receptor dimerization increased under EGF stimulation (Supplementary Fig. 13). SRC knockdown inhibited EGF-induced EGFR dimerization (Fig. 4a) and suppressed EGFR phosphorylation at Tyr845, a known SRC-dependent EGFR phosphorylation site21, and at Tyr1068, an autophosphorylation site, under both basal and EGF-stimulated conditions (Fig. 4b). Saracatinib treatment also inhibited EGFR phosphorylation in TtzmR cells (Fig. 4c). These data indicate an essential role of SRC in mediating EGFR signaling and the existence of a positive feedback loop between EGFR and SRC22. Notably, trastuzumab treatment transiently induced ERBB2 receptor dimerization and phosphorylation, which led to a further increase of SRC activation in TtzmR cells (Supplementary Figs. 14 and 15). Inhibition of SRC activity by either saracatinib or knocking down SRC (shRNA.SRC2) diminished trastuzumab-induced transient ERBB2 phosphorylation (Fig. 4d,e). Additionally, SRC inhibition potentiated trastuzumab-mediated inhibition of phosphorylation of HER3, a critical dimerization partner of ERBB2 (refs. 17,23,24; Fig. 4d,e), although SRC inhibition had no discernable effect on ERBB2-HER3 heterodimerization (Supplementary Fig. 16). These data indicate that SRC activation induces trastuzumab resistance by facilitating EGFR, ERBB2 and HER3 activation in TtzmR cells.
Figure 4.
SRC inhibition induced signaling alterations in multiple trastuzumab-resistant models. (a) Immunoblots detecting EGF-indu3ced EGFR dimerization in BT474. TtzmR cells stably infected with control shRNA or SRC shRNA. Cross-linking of cell membrane EGFR is described in Online Methods. MW, molecular weight. (b) Immunoblots comparing EGFR phosphorylation upon EGF treatment in BT474.TtzmR cells with or without SRC knockdown. (c) Immunoblots assessing EGFR signaling in BT474.TtzmR cells upon treatment with the SRC inhibitor saracatinib with or without EGF treatment. Cells were pretreated with saracatinib or vehicle 1 h before EGF stimulation. (d) Immunoblots assessing abundance of HER3 phosphorylation and other signaling events upon treatment with saracatinib alone, trastuzumab alone or combination treatment. (e) Immunoblots evaluating HER3 activation in response to trastuzumab treatment after stable knockdown of SRC in TtzmR cells by shRNA. (f–h) Immunoblots showing the impact of trastuzumab plus saracatinib on AKT signaling in multiple trastuzumab-resistant models: BT474.TtzmR (f), PTEN knockdown (g), or constitutively active SRC mutant (SRC Y527F) expressing (h).
Furthermore, we examined the effects of saracatinib and trastuzumab treatment on the signaling events downstream of RTKs. Compared with a complete inhibition of AKT phosphorylation by trastuzumab in parental cells (Supplementary Fig. 17), TtzmR cells were largely resistant to trastuzumab-mediated AKT inhibition (Fig. 4f), perhaps owing to alternative RTK activation. Concurrent treatment with saracatinib inhibited AKT activation in TtzmR cells (Fig. 4f). Moreover, this combination also inhibited AKT phosphorylation, along with SRC phosphorylation, in PTEN-deficient de novo trastuzumab-resistant cells (Fig. 4g), SRC.Y527F cells (Fig. 4h) and EGFR-overexpressing cells (Supplementary Fig. 18). Together, these data indicate that RTK activation and their downstream signaling events (AKT activation) involved in trastuzumab resistance can be effectively blocked by inhibiting SRC.
SRC inhibition overcomes trastuzumab resistance in vitro
As our data indicate that SRC sits on the signaling hub mediating multiple types of trastuzumab resistance of different mechanisms (both acquired and de novo), we hypothesized that targeting this key node could be a more effective strategy to overcome trastuzumab resistance conferred by multiple mechanisms. To target SRC, we used saracatinib, an orally available small-molecule SRC inhibitor tolerated well in phase 1 and 2 clinical trials25. Saracatinib treatment universally sensitized trastuzumab-resistant cells to trastuzumab treatment in all models tested, including acquired resistance, PTEN deficiency, EGFR overexpression, IGF-1R overexpression (Fig. 5a) and SRC constitutive activation (Fig. 5b). Combination treatment also suppressed 3D tumor spheroid formation in both BT474 acquired resistant cells (Fig. 5c) and PTEN-deficient cells (Supplementary Fig. 19). We also observed similar inhibitory effects in clonogenic assays (Supplementary Fig. 20).
Figure 5.
Trastuzumab treatment plus SRC inhibition overcomes multiple resistance mechanisms in vitro. (a) MTS assay examining the effect of SRC inhibition in combination with trastuzumab treatment in the indicated four trastuzumab-resistant models. BT474.TtzmR cells, cells overexpressing IGF-1R and EGFR, and PTEN.shRNA cells were treated as described in Online Methods. (b) MTS assay evaluating the effects of trastuzumab, saracatinib or combination treatment in control BT474-GFP and trastuzumab-resistant cells overexpressing SRC Y527F. (c) 3D tumor spheroid assay comparing the cell proliferation of BT474. TtzmR cells upon treatment with trastuzumab alone, saracatinib alone or combination treatment. Tumor spheroid assay was carried out as described in Online Methods. Scale bar, 100 µm. (d) MTS assay comparing SRC inhibition by saracatinib and AKT inhibition by triciribine on overcoming trastuzumab resistance. (e) TUNEL assay examining induction of apoptosis by saracatinib and trastuzumab combined treatment in control (Con.shRNA) and PTEN knockdown (PTEN.shRNA) cells. TUNEL-positive cells were detected by flow cytometry. (f) The induction of DNA fragmentation (indicated by sub-G1 population detected by flow cytometry) by combined treatment with saracatinib and trastuzumab in BT474.PTEN knockdown cells (PTEN.as) and BT474.SRC constitutively active (Y527F) cells. All error bars, s.e.m. All quantitative data were generated from a minimum of three replicates.
Next, we examined whether the AKT inhibitor triciribine could have a similar sensitization effect to saracatinib. In all models we tested, saracatinib was more effective in sensitizing resistant cells to trastuzumab compared with triciribine (Fig. 5d and Supplementary Fig. 21). As treatment with a combination of trastuzumab and saracatinib effectively suppressed AKT signaling (Fig. 4f,g) and induced cell blebbing in 3D drug response assays (Supplementary Fig. 19), we examined whether SRC inhibition could sensitize trastuzumab-resistant cells to treatment partially through induction of apoptosis. Indeed, combinatorial treatment considerably increased the number of TUNEL-positive cells in PTEN knockdown cells (Fig. 5e) and increased sub-G1 populations in PTEN-deficient and SRC Y527F cells (Fig. 5f). These consistent data from multiple trastuzumab resistance models demonstrate that inhibition of SRC enhances trastuzumab-mediated growth inhibition by promoting apoptosis.
Targeting SRC overcomes trastuzumab resistance in vivo
To test whether targeting SRC could be an effective strategy to overcome trastuzumab resistance in vivo, we first knocked down SRC in trastuzumab-resistant, PTEN-deficient mammary fat pad tumors by intratumoral injection of viral particles carrying SRC shRNA. Trastuzumab treatment led to a significant (P = 0.0003) shrinkage of tumors injected with SRC shRNA but not control shRNA (Fig. 6a). In the acquired resistance model, trastuzumab plus saracatinib greatly inhibited BT474.TtzmR tumors, whereas saracatinib alone at 25 mg per kg body weight daily (a dosage tolerated well by humans25,26) was not effective. This tumor inhibition by trastuzumab plus saracatinib was more substantial than trastuzumab plus triciribine (Fig. 6b). Notably, in the PTEN-deficiency model, combinatorial treatment eliminated tumors and most tumors were barely detectable 21 d after treatment (Fig. 6c). Phosphorylation of AKT in PTEN.shRNA xenograft tumors was also decreased by combinatorial treatment but not by single treatment alone (Fig. 6d). Notably, although combinatorial treatment showed a statistically insignificant trend of decreased Ki67 staining (data not shown) compared with controls, there was a significant (P = 0.039) increase of TUNEL-positive staining in these tumors, suggesting an induction of apoptosis by the combinatorial treatment (Fig. 6e). Collectively, these data indicate that SRC inhibition by saracatinib sensitizes trastuzumab-resistant tumors to trastuzumab in vivo, at least partially through induction of apoptosis.
Figure 6.
Trastuzumab plus saracatinib combinatorial treatment overcomes trastuzumab resistance in vivo. (a) Top, representative immunofluorescence images of SRC knockdown in PTEN.shRNA xenografts using intratumoral injection of SRC.shRNA-containing virus. Scale bar, 100 µm. Bottom, volume of trastuzumab-resistant PTEN-deficient tumors with or without SRC knockdown upon treatment with IgG or Ttzm. Tumor volume at various times of treatment is presented as percentage of original tumor size at day zero of treatment. (b) Top, representative immunohistochemistry (IHC) images of in vivo inhibition of SRC-Y416 phosphorylation by saracatinib or AKT-S473 phosphorylation by triciribine in BT474.TtzmR xenograft tumors. Scale bar, 100 µm. Bottom, TtzmR xenograft tumor volume in response to different treatments. (c) Left, representative in vivo luciferase images of mice at day 0 and 21 days after indicated treatment. Left side of animal, BT474 control.shRNA tumors; right side, BT474 PTEN.shRNA tumors. Right, tumor volume in response to different treatments. (d) Left, representative IHC staining of AKT-S473 phosphorylation after different treatments (vehicle or trastuzumab plus saracatinib) in BT474.PTEN.shRNA xenograft tumors. Scale bar, 50 µm. Right, overall AKT-S473 phosphorylation IHC staining intensity between trastuzumab-alone group and combination-treatment group. Phospho-AKT (pAKT) staining was compared between each group by Fisher’s exact test (P = 0.049, two-sided). (e) Top, representative tumor sections with TUNEL staining. Scale bar, 50 µm. Bottom, in situ TUNEL staining of apoptotic cells in tumors treated as indicated. All error bars, s.e.m. All in vivo data were generated from a minimum of five replicates. (f) Model of SRC as a common node downstream of multiple resistance pathways and conquering trastuzumab resistance by targeting SRC.
DISCUSSION
Recent studies have highlighted a scenario in which, upon prolonged treatment by a highly specific targeted therapy (such as trastuzumab), tumor cells reprogram themselves to develop alternative compensatory pathways to sustain cell proliferation, ultimately leading to drug resistance17,27. Additionally, pre-existing genetic alterations of the RTK-PI3K–AKT pathway (such as PTEN loss and IGF-1R overexpression) make tumor cells inherently more resistant to trastuzumab7,8,13. Owing to the heterogeneous nature of tumors, different resistance mechanisms may coexist in the same patient. Thus, targeting one molecule or one resistance mechanism is usually ineffective27. Targeting multiple altered signaling pathways that confer resistance is warranted. However, targeting multiple pathways by simply combining several US Food and Drug Administration–approved drugs against each pathway may not always be feasible and may cause severe side effects in some patients28. To develop more effective regimens to overcome drug resistance, we need to further dissect the signaling events in resistant tumors. Identification of the ‘common signaling nodes’ involved in multiple resistance mechanisms may help researchers to rationally design new combinatorial therapies.
SRC interacts with multiple RTKs through its SH2 domain and facilitates RTK-mediated signaling20,29–34. Small-molecule inhibitors targeting multiple kinases including SRC (such as PP2 and Dasatinib) potentiate the initial efficacy of anti-RTK therapies in naive tumor cells35–37. A recent study also suggested that SRC activation may contribute to transient trastuzumab resistance induced by TGF-β treatment36. Here we demonstrate that, in both acquired and de novo trastuzumab-resistant tumors, SRC has a universal role in mediating multiple resistance pathways including alternative activation of RTK pathways and PTEN deficiency. This places SRC at a common signaling node for trastuzumab resistance (Fig. 6f). Thus, we propose that instead of targeting each of the RTK or PI3K-AKT pathway alterations individually, researchers may effectively overcome trastuzumab resistance resulting from multiple mechanisms by inhibiting this common key node. Our data support this new concept. Targeting SRC universally sensitized trastuzumab-resistant cells to trastuzumab treatment and significantly suppressed tumor growth in vivo in multiple preclinical resistance models (Fig. 6f). This new combination strategy warrants further clinical investigation in patients with trastuzumab-resistant tumors having different de novo or acquired resistance mechanisms. Furthermore, similar strategies may also be applied to overcoming resistance to other RTK-targeting drugs.
Previously, we tested whether we could overcome de novo trastuzumab resistance conferred by PTEN deficiency by combining trastuzumab with PI3K-AKT pathway inhibitors38. In our side-by-side comparison of trastuzumab plus SRC inhibitor versus trastuzumab plus AKT inhibitor in vitro and in vivo, we observed that combination with the SRC inhibitor saracatinib was more effective in overcoming trastuzumab resistance in both TtzmR and PTEN deficiency models38. Notably, from a clinical perspective, toxicity is a critical consideration in prescribing further treatment for trastuzumab-resistant patients who have already been heavily treated. The extensively documented side effects of the clinically applied AKT inhibitor triciribine39 may preclude the combination of this drug with trastuzumab in trastuzumab-resistant patients, although this combination showed some efficacy in our TtzmR xenograft model. In contrast, the SRC inhibitor saracatinib is tolerated well in phase 1 and 2 clinical trials25. Thus, combination with SRC-targeting agents is a clinically more applicable treatment for trastuzumab-resistant patients.
In addition to the AKT pathway40, SRC also modulates many downstream targets, including focal adhesion kinase41, signal transducer and activator of transcription-3 (ref. 42), mitogen-activated protein kinases43,44, the protein kinase C family45 and c-MYC46. These signaling pathways have diverse roles in regulating tumor cell survival47–49 and metastasis50,51. Combinatorial therapy with trastuzumab and saracatinib could have a broad impact on tumor progression. Particularly, this combination treatment inhibited migration of trastuzumab-resistant cells (Supplementary Fig. 22), suggesting a potential benefit of this regimen in preventing further metastatic recurrence for patients with metastatic breast cancer.
In conclusion, our studies identified SRC activation as a key convergence point of multiple trastuzumab resistance mechanisms. The combinatorial regimen of SRC inhibitor plus trastuzumab is effective in overcoming various de novo and acquired resistance mechanisms. Our findings may directly impact the clinical management of patients with ERBB2-overexpressing breast cancer.
ONLINE METHODS
Cell culture, vectors and virus
We obtained AU565, SKBR3 and MDA-MB-468 cells from American Type Culture Collection. We obtained the tumorigenic BT474.m1 subline (BT474) as previously described7. We purchased pLKO.1-based shRNAs from Sigma-Aldrich and obtained the SRC mutants (Y527F and K295R) and GST-PTEN constructs from Addgene. We established acquired trastuzumab-resistant cells as described52. We carried out 3D tumor spheroid culture as described53. We subcloned human EGFR cDNA into pcDNA6 vector.
In vitro cell proliferation assay, cell cycle analysis and apoptosis assay
We carried out cell proliferation assay using the CellTiter 96 AQueous cell proliferation assay kit (Promega). We calculated the percentage of inhibition of cell proliferation as [1 – (treated cells / untreated cells)] × 100. We carried out cell cycle analysis and APO-BrdU TUNEL assay (Invitrogen) on the basis of the manufacturer’s protocols.
Protein cross-linking and coimmunoprecipitation
To detect receptor dimerization, we cross-linked membrane RTKs by resuspending the cells in 5 mM Sulfo-EGS prepared in PBS on ice for 2 h followed by adding 10 mM Tris (final concentration). For coimmunoprecipitation experiments, we incubated the total cell lysate with first antibody overnight at 4 °C and immunoprecipitated the immunocomplex with protein G–agarose. For detection of ERBB2 and HER3 interaction, we covalently immobilized antibody to ERBB2 on UltraLink Biosupport resin (Thermo Fisher Scientific) before carrying out immunoprecipitation.
PTEN phosphatase assay
We reconstituted the synthetic SRC peptides phosphorylated at Tyr416 and Tyr527 (SRC-Y416 (RRLIEDNE-pY-TARG) and SRC-Y527 (TSTEPQ-pY-QPGENL); AnaSpec) in phosphate-free water. We carried out the phosphatase assay by incubating 250 ng of purified GST-PTEN protein with 100 µM phosphorylated SRC peptide in 25 µl phosphatase assay buffer (25 mM Tris-HCl, pH 7.4, 140 mM NaCl, 2.7 mM KCl, 10 mM DTT) for 30 min at 37 °C. We measured the released free phosphate from substrate using Malachite Green reagent (Echelon Biosciences).
Patient samples
We collected primary breast cancer specimens from women at MD Anderson Cancer Center from 1996 to 2005 under a protocol approved by the MD Anderson Cancer Center Institutional Review Board. Informed consent was obtained from all subjects. We included patients who received first-line trastuzumab-based therapies (without prior chemotherapy) in our retrospective analysis. We evaluated the clinical response to trastuzumab-based therapy every 8–12 weeks using response evaluation criteria in solid tumors with modifications. We defined clinical benefit as complete response, partial response or stable disease longer than 6 months. We defined overall survival time after trastuzumab treatment as the time from initiation of trastuzumab treatment to death.
Immunohistochemistry evaluation
We used the following primary antibodies for immunohistochemistry staining: PTEN (138G6, Cell Signaling), phospho-SRC-Y416 (2101, Cell Signaling), phospho-AKT-S473 (736E11, 1:150, Cell Signaling) and Ki67 (MIB-1, 1:75, Dako). Retrieval butters were: 0.01 M citrate retrieval buffer, pH 6.0, for Ki67 staining; 1 mM EDTA, pH 8.0, for PTEN, phospho-SRC-Y416 and phospho-AKT-S473 staining. We scored staining by H-score according to the staining intensity and percentage of positive cells within the whole tissue section. We considered cases with phospho-SRC-Y416 H-score > 20 (>10%, ++; 20%, +) to have high phospho-SRC-Y416 staining. We classified phospho-AKT-S473 staining intensity into four groups (0, 1, 2 and 3+) by H-score. Phospho-AKT staining with H-score ≥ 250 is defined as 3+. We quantified PTEN expression using the immunoreactive score as described7.
Tumor xenograft studies
All mouse experiments were carried out in accordance with approved protocols from the Institutional Animal Care and Use Committee of MD Anderson Cancer Center. We implanted 6-week-old female severe combined immunodeficiency mice with 0.72 mg 60-d release 17β-estradiol pellets (Innovative Research). Then we established mammary fat pad tumors by injection of 8 × 106 BT474.m1 cells in 100 µl of PBS-Matrigel mixture (1:1 ratio) orthotopically into the mammary fat pads. When xenografts reached about 100–150 mm3, we started to treat the mice with trastuzumab (10 mg per kg−1) or vehicle control (IgG) intraperitoneally once per week. We treated mice with saracatinib daily at a dose of 25 mg per kg body weight in vehicle (0.5% wt/vol hydroxypropyl-methylcellulose with 0.1% vol/vol Tween 20) via oral gavage. We treated mice with triciribine by intraperitoneal daily at dose of 0.5 mg per kg body weight. For intratumoral lentivirus injection, we concentrated lentiviral particles containing pLKO.con.shRNA or pLKO.SRC shRNA using PEG-it virus precipitation solution (System Biosciences) to reach ~2.6 × 107 transduction units (TU) ml−1. We carried out intratumoral injection of 50 µl of concentrated virus every 3 d. We calculated tumor volume as length × width2/2. For tissue TUNEL staining, the apoptotic index was expressed as a percentage of TUNEL-positive cells per section. We excluded necrotic areas in the tumors from evaluation.
Statistical analyses
We analyzed quantitative results either by one-way analysis of variance (multiple groups) or t test (two groups). For patient samples, we calculated correlation by Fisher’s exact test. We carried out survival analysis using Kaplan-Meier log-rank test. We considered a difference with P< 0.05 (two-sided) statistically significant.
Additional methods
Detailed methodology is described in Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank AstraZeneca for providing us with saracatinib and W. Zhang in the Department of Pathology, MD Anderson Cancer Center for providing us with the IGF-1R expression vector. We also thank C. Neal, J. Lu, S. Rehman and F.J. Lowery for insightful comments on this manuscript. This work was supported by US National Institutes of Health grants P30-CA 16672 (MD Anderson Cancer Center), RO1-CA112567 (D.Y.), PO1-CA099031 project 4 (D.Y.), MD Anderson Cancer Center Breast SPORE P50 CA116199 (G.N.H.) project 4 (D.Y.), US Department of Defense Center of Excellence subproject W81XWH-06-2-0033 (D.Y.), Susan G. Komen Breast Cancer Foundation promise grant KG091020 (D.Y.), Cancer Prevention Research Institute of Texas grant RP100726 (D.Y.), the MD Anderson Cancer Center Breast SPORE Career Development Award (S.Z.) and the Susan G. Komen Breast Cancer Foundation postdoctoral fellowship (S.Z.). D.Y. is the Hubert L. and Olive Stringer Distinguished Chair in Basic Science at MD Anderson Cancer Center.
Footnotes
Note: Supplementary information is available on the Nature Medicine website.
AUTHOR CONTRIBUTIONS
S.Z., W.-C.H. and D.Y. designed experiments and analyzed data; S.Z., W.-C.H., H.G., P.L., S.-B.P., S.W.B., Y.X., L.-M.T. and Z.D. carried out experiments; S.Z., H.G. and S.-H.L. did statistical analysis of clinical data; A.A.S. collected tumor samples and evaluated immunohistochemistry staining with H.G.; G.N.H. and F.J.E. collected clinical patient information and analyzed patient response data; S.Z., S.W.B. and D.Y. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 2.Romond EH, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 3.Vogel CL, et al. Effcacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J. Clin. Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 4.Seidman AD, et al. Randomized phase III trial of weekly compared with every-3-weeks paclitaxel for metastatic breast cancer, with trastuzumab for all HER-2 overexpressors and random assignment to trastuzumab or not in HER-2 nonoverexpressors: final results of Cancer and Leukemia Group B protocol 9840. J. Clin. Oncol. 2008;26:1642–1649. doi: 10.1200/JCO.2007.11.6699. [DOI] [PubMed] [Google Scholar]
- 5.Lan KH, Lu CH, Yu D. Mechanisms of trastuzumab resistance and their clinical implications. Ann. NY Acad. Sci. 2005;1059:70–75. doi: 10.1196/annals.1339.026. [DOI] [PubMed] [Google Scholar]
- 6.Piccart M. Circumventing de novo and acquired resistance to trastuzumab: new hope for the care of ErbB2-positive breast cancer. Clin. Breast Cancer. 2008;8(Suppl 3):S100–S113. doi: 10.3816/cbc.2008.s.006. [DOI] [PubMed] [Google Scholar]
- 7.Nagata Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 8.Berns K, et al. A functional genetic approach identifes the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Scaltriti M, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J. Natl. Cancer Inst. 2007;99:628–638. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 10.Moulder SL, et al. Epidermal growth factor receptor (HER1) tyrosine kinase inhibitor ZD1839 (Iressa) inhibits HER2/neu (erbB2)-overexpressing breast cancer cells in vitro and in vivo. Cancer Res. 2001;61:8887–8895. [PubMed] [Google Scholar]
- 11.Dua R, et al. EGFR overexpression and activation in high HER2, ER negative breast cancer cell line induces trastuzumab resistance. Breast Cancer Res. Treat. 2009;122:685–697. doi: 10.1007/s10549-009-0592-x. [DOI] [PubMed] [Google Scholar]
- 12.Ritter CA, et al. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clin. Cancer Res. 2007;13:4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- 13.Lu Y, Zi X, Zhao Y, Mascarenhas D, Pollak M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin) J. Natl. Cancer Inst. 2001;93:1852–1857. doi: 10.1093/jnci/93.24.1852. [DOI] [PubMed] [Google Scholar]
- 14.Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005;65:11118–11128. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- 15.Shattuck DL, Miller JK, Carraway KLIII, Sweeney C. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res. 2008;68:1471–1477. doi: 10.1158/0008-5472.CAN-07-5962. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, et al. Heterotrimerization of the growth factor receptors erbB2, erbB3, and insulin-like growth factor-I receptor in breast cancer cells resistant to herceptin. Cancer Res. 2010;70:1204–1214. doi: 10.1158/0008-5472.CAN-09-3321. [DOI] [PubMed] [Google Scholar]
- 17.Narayan M, et al. Trastuzumab-induced HER reprogramming in “resistant” breast carcinoma cells. Cancer Res. 2009;69:2191–2194. doi: 10.1158/0008-5472.CAN-08-1056. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Yu D. PI(3)king apart PTEN′s role in cancer. Clin. Cancer Res. 2010;16:4325–4330. doi: 10.1158/1078-0432.CCR-09-2990. [DOI] [PubMed] [Google Scholar]
- 19.Liaw D, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 20.Kmiecik TE, Shalloway D. Activation and suppression of pp60c-src transforming ability by mutation of its primary sites of tyrosine phosphorylation. Cell. 1987;49:65–73. doi: 10.1016/0092-8674(87)90756-2. [DOI] [PubMed] [Google Scholar]
- 21.Tice DA, Biscardi JS, Nickles AL, Parsons SJ. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc. Natl. Acad. Sci. USA. 1999;96:1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishizawar R, Parsons SJ. c-Src and cooperating partners in human cancer. Cancer Cell. 2004;6:209–214. doi: 10.1016/j.ccr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 24.Junttila TT, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 25.Tabernero J, et al. Phase I study of AZD0530, an oral potent inhibitor of Src kinase: First demonstration of inhibition of Src activity in human cancers. ASCO Meeting. 2007;25:3520. [Google Scholar]
- 26.Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 27.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 28.Tol J, et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N. Engl. J. Med. 2009;360:563–572. doi: 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]
- 29.Bromann PA, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004;23:7957–7968. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- 30.Parsons JT, Parsons SJ. Src family protein tyrosine kinases: cooperating with growth factor and adhesion signaling pathways. Curr. Opin. Cell Biol. 1997;9:187–192. doi: 10.1016/s0955-0674(97)80062-2. [DOI] [PubMed] [Google Scholar]
- 31.Biscardi JS, et al. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J. Biol. Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 32.Karni R, Jove R, Levitzki A. Inhibition of pp60c-Src reduces Bcl-XL expression and reverses the transformed phenotype of cells overexpressing EGF and HER-2 receptors. Oncogene. 1999;18:4654–4662. doi: 10.1038/sj.onc.1202835. [DOI] [PubMed] [Google Scholar]
- 33.Belsches-Jablonski AP, et al. Src family kinases and HER2 interactions in human breast cancer cell growth and survival. Oncogene. 2001;20:1465–1475. doi: 10.1038/sj.onc.1204205. [DOI] [PubMed] [Google Scholar]
- 34.Ishizawar RC, Miyake T, Parsons SJ. c-Src modulates ErbB2 and ErbB3 heterocomplex formation and function. Oncogene. 2007;26:3503–3510. doi: 10.1038/sj.onc.1210138. [DOI] [PubMed] [Google Scholar]
- 35.Nautiyal J, et al. ErbB-inhibitory protein: a modified ectodomain of epidermal growth factor receptor synergizes with dasatinib to inhibit growth of breast cancer cells. Mol. Cancer Ther. 2010;9:1503–1514. doi: 10.1158/1535-7163.MCT-10-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang SE, et al. Transforming growth factor beta induces clustering of HER2 and integrins by activating Src-focal adhesion kinase and receptor association to the cytoskeleton. Cancer Res. 2009;69:475–482. doi: 10.1158/0008-5472.CAN-08-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seoane S, Montero JC, Ocana A. Pandiella, A. Effect of multikinase inhibitors on caspase-independent cell death and DNA damage in HER2-overexpressing breast cancer cells. J. Natl. Cancer Inst. 2010;102:1432–1446. doi: 10.1093/jnci/djq315. [DOI] [PubMed] [Google Scholar]
- 38.Lu CH, et al. Preclinical testing of clinically applicable strategies for overcoming trastuzumab resistance caused by PTEN deficiency. Clin. Cancer Res. 2007;13:5883–5888. doi: 10.1158/1078-0432.CCR-06-2837. [DOI] [PubMed] [Google Scholar]
- 39.LoPiccolo J, Blumenthal GM, Bernstein WB, Dennis PA. Targeting the PI3K/AKT/mTOR pathway: effective combinations and clinical considerations. Drug Resist. Updat. 2008;11:32–50. doi: 10.1016/j.drup.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson D, et al. Regulation of both apoptosis and cell survival by the v-Src oncoprotein. Cell Death Differ. 2000;7:685–696. doi: 10.1038/sj.cdd.4400700. [DOI] [PubMed] [Google Scholar]
- 41.Webb DJ, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 42.Ren Z, Schaefer TS. ErbB-2 activates Stat3α in a Src- and JAK2-dependent manner. J. Biol. Chem. 2002;277:38486–38493. doi: 10.1074/jbc.M112438200. [DOI] [PubMed] [Google Scholar]
- 43.Kennedy SG, et al. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 44.Olayioye MA, Badache A, Daly JM, Hynes NE. An essential role for Src kinase in ErbB receptor signaling through the MAPK pathway. Exp. Cell Res. 2001;267:81–87. doi: 10.1006/excr.2001.5242. [DOI] [PubMed] [Google Scholar]
- 45.Tan M, Li P, Sun M, Yin G, Yu D. Upregulation and activation of PKC alpha by ErbB2 through Src promotes breast cancer cell invasion that can be blocked by combined treatment with PKCα and Src inhibitors. Oncogene. 2006;25:3286–3295. doi: 10.1038/sj.onc.1209361. [DOI] [PubMed] [Google Scholar]
- 46.Bowman T, et al. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc. Natl. Acad. Sci. USA. 2001;98:7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franke TF, Hornik CP, Segev L, Shostak GA, Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene. 2003;22:8983–8998. doi: 10.1038/sj.onc.1207115. [DOI] [PubMed] [Google Scholar]
- 48.Chang F, et al. Regulation of cell cycle progression and apoptosis by the Ras/Raf/ MEK/ERK pathway. Int. J. Oncol. (Review) 2003;22:469–480. [PubMed] [Google Scholar]
- 49.Al Zaid Siddiquee K, Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008;18:254–267. doi: 10.1038/cr.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang XH, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyer B, Bourgeois Y, Poupon MF. Src kinase contributes to the metastatic spread of carcinoma cells. Oncogene. 2002;21:2347–2356. doi: 10.1038/sj.onc.1205298. [DOI] [PubMed] [Google Scholar]
- 52.Nahta R, Takahashi T, Ueno NT, Hung MC, Esteva FJ. P27kip1 down-regulation is associated with trastuzumab resistance in breast cancer cells. Cancer Res. 2004;64:3981–3986. doi: 10.1158/0008-5472.CAN-03-3900. [DOI] [PubMed] [Google Scholar]
- 53.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.