Abstract
Type a flagellins from two strains of Pseudomonas aeruginosa, strains PAK and JJ692, were found to be glycosylated with unique glycan structures. In both cases, two sites of O-linked glycosylation were identified on each monomer, and these sites were localized to the central, surface-exposed domain of the monomer in the assembled filament. The PAK flagellin was modified with a heterogeneous glycan comprising up to 11 monosaccharide units that were O linked through a rhamnose residue to the protein backbone. The flagellin of JJ692 was less complex and had a single rhamnose substitution at each site. The role of the glycosylation island gene cluster in the production of each of these glycosyl moieties was investigated. These studies revealed that the orfA and orfN genes were required for attachment of the heterologous glycan and the proximal rhamnose residue, respectively.
Protein glycosylation in prokaryotic organisms is now a well-established process, particularly in cell surface-associated or secreted molecules (5). Very recently, a number of examples of glycosylated surface proteins of bacterial pathogens have been described, including surface proteins of Streptococcus sanguis and Mycobacterium tuberculosis (11, 14), the TIB adhesin of Escherichia coli (TIB) (4, 26), flagellum and periplasmic proteins of Campylobacter (12, 40, 43, 44), flagellum proteins of Helicobacter pylori (35), an outer membrane protein of Chlamydia (24), and pilus proteins of Neisseria species (37) and Pseudomonas aeruginosa (8). However, information about the structures of the linked glycans found on prokaryotic glycoproteins, the mechanistic basis of the process, and the biological role of the process in bacterial pathogenesis is still limited.
P. aeruginosa is an opportunistic pathogen which causes life-threatening infections in immunocompromised individuals and burn wound victims and chronic infections in patients with cystic fibrosis (6). This organism produces a number of virulence factors, including toxins, secreted proteins, surface carbohydrates (mucoid exopolysaccharide and lipopolysaccharide [LPS]), and pili (18, 27). A more recent addition to the list of putative virulence factors is the single, polar, nonsheathed flagellum of this organism, which traditionally is considered a motility organ but whose chief constituent, flagellin, is now known to be a potent stimulator of the inflammatory response via Toll-like receptor 5 (19). The flagellin protein can be classified in one of two major types, type a or type b, based on molecular weight and reactivity with specific antisera (1, 25). The type a flagellin appears to have two major subtypes of proteins, designated subtypes A1 and A2 (3). Type a flagellins have been shown to be heterologous by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and to have molecular masses ranging from 45 to 52 kDa, while type b flagellins appear to have conserved sequences and a molecular mass of 53 kDa. The central domain of type a flagellin is described as hypervariable, and this is believed to be responsible for the serological variation among isolates. The flagella of P. aeruginosa strains have been shown to be an important virulence factor, and nonmotile mutants are severely attenuated in the ability to cause disease in animal models (15, 29).
Preliminary evidence for posttranslational modification of Pseudomonas flagellin has been provided previously. Variation in the observed molecular weight as determined by SDS-PAGE of the FliC protein from the predicted sequence was the first evidence for posttranslational modification (41). Subsequently, Brimer and Montie verified that type a flagellins were indeed glycosylated (7). Very recently, a unique genomic island which contained a cluster of 14 genes was identified in strains producing type a flagellin (2). Coinheritance of type a flagellin with this cluster of genes led to protein glycosylation. Based on limited homologies and the finding that the genes in the glycosylation island encoded enzymes involved in the synthesis, activation, or polymerization of sugars necessary for flagellin glycosylation, it was proposed that coinheritance of type a flagellin with this cluster of genes leads to protein glycosylation. Very recently, a comprehensive study of genetic variability within glycosylation islands and flagellar genes in type a strains has been completed (3). The studies described above indicated that antigenic diversity of flagellin protein is probably influenced by glycosylation.
We undertook this study to determine the structural nature of flagellar glycosylation in type a P. aeruginosa strains and to investigate the genetic basis for this process. Here we provide evidence that P. aeruginosa strain PAK produces a heterogeneous complex glycan which is synthesized by the genes of the flagellar glycosylation island. We also show that genetic variation in glycosylation island content leads to changes in the glycosylation profile in type a flagellum-producing P. aeruginosa strains.
MATERIALS AND METHODS
Bacterial strains.
P. aeruginosa strain PAK is a wild-type laboratory isolate and was obtained from D. Bradley. JJ692 is a urinary tract isolate obtained from S. Lory.
Purification of flagella.
Flagella were purified from Pseudomonas strains grown overnight in L broth. Flagella were sheared from the surface of the bacteria and collected by ultracentrifugation as previously described (41).
Mass spectrometry (MS).
Purified flagellins were dialyzed in aqueous 0.2% (vol/vol) formic acid by using a Centricon YM-30 membrane filter (Millipore). The solution was infused into the mass spectrometer at a flow rate of 0.5 μl/min. For identification of the type and location of glycosylation sites, flagellins from strains PAK and JJ692 were digested with trypsin (Promega) overnight. All digests were analyzed with a Waters CapLC apparatus coupled to nanoelectrospray on a Q-TOF Ultima instrument (Waters, Milford, Mass.). Peptides were separated by using a Waters Symmetry C18 precolumn and a homemade Jupiter C18 analytical column (length, 10 cm; inside diameter, 150 μm; particle size, 5 μm; Phenomenex). A linear gradient of 10 to 40% acetonitrile (0.2% formic acid) in 60 min was used.
All MS-MS spectra were obtained in the data-dependent mode with selection for the three most intense ions per survey scan for the MS-MS experiment.
High-performance liquid chromatography collection of fractions of the tryptic digest was performed with a Vydac C18 column (2.1 mm by 20 cm; particle size, 5 μm). A linear gradient of 0 to 100% acetonitrile (0.2% formic acid) in 30 min was used, and fractions were collected every minute. Fractions were screened for the two triply charged ion series by liquid chromatography (LC)-MS. Fractions containing the two ion series were analyzed by nanospray MS-MS.
The glycosylation site was identified precisely by using β-elimination with ammonium hydroxide to leave a modified Ser or Thr residue that could be located by using tandem MS (31). The tryptic digest solution was subjected to alkaline hydrolysis with 800 μl of NH4OH for 18 h at 50°C, evaporated to dryness, and redissolved in H2O (0.2% formic acid) prior to analysis by LC-nanoelectrospray MS-MS. Tryptic peptides showing the characteristic mass shift after β-elimination were subjected to MS-MS analyses to locate the position of the modified residue.
Monosaccharide analysis of flagellar glycan.
Flagellin (5 mg) was hydrolyzed with 3 M CF3COOH for 2 h at 120°C, and the sample was dried under a stream of nitrogen, dissolved in 0.3 ml of water, and treated with 5 mg of NaBH4 for 1 h at 25°C. The NaBH4 reagent was destroyed with 0.5 ml of acetic acid, the sample was dried under a stream of nitrogen, 1 ml of methanol was added, and the sample was dried again under a stream of nitrogen. This was repeated twice, and the sample was then acetylated with 0.5 ml of acetic anhydride for 1 h at 85°C and dried under a stream of nitrogen; 1 ml of water and 0.5 ml of chloroform were added, the sample was shaken, and the chloroform layer was passed through cotton wool. The derivatized carbohydrate extract was then injected into a gas chromatography (GC)-MS instrument (Varian Saturn 2000) equipped with a capillary column (DB-1; 0.25 mm by 50 m) with an ion trap detector. The same procedure was used for preparation of the standards perosamine (4-amino-4,6-dideoxymannose) and viosamine (4-amino-4,6-dideoxyglucose) from the O-specific polysaccharide from E. coli O157 (30) and an LPS core sample from Proteus mirabilis O6, respectively (42).
Generation of isogenic mutants.
orfA and orfN (rfbC) mutants were generated as described previously (2). Briefly, an orfA chromosomal deletion mutant was obtained by allelic exchange in PAK following deletion of 981 bp (327 amino acids) and counterselection with a sacB marker. The chromosomal mutation in orfN was generated by using a plasmid construct of the orfN gene that was missing the portion encoding the N-terminal 526 amino acids and into which a gentamicin cassette was inserted at a unique EcoRV site,. This construct was then used to generate the chromosomal mutation in PAK by marker exchange. Complementation of each mutant to rule out any polar effects was performed as described by Arora et al. (2).
Expression of JJ692 flagellin in PAK.
Chromosomal complementation of the JJ692 flagellin in a PAK background was performed as follows. The complete JJ692 flagellin gene along with its promoter was PCR amplified as a 1.6-kb fragment and cloned into the vector miniCTX1 at the HindIII/SstI sites (20). This construct was mated into the PAKΔfliC strain to obtain tetracycline-resistant single crossovers. These single crossovers were then resolved into double recombinants by introduction of plasmid pflp2, which was then cured. Flagellin was then purified from PAK, JJ692, and the PAKΔfliC strain expressing the JJ692 fliC gene. Purified flagellins were then examined by SDS-PAGE (7.5% acrylamide gel) and were stained with Coomassie blue.
RESULTS
Intact mass analysis of P. aeruginosa PAK flagellin.
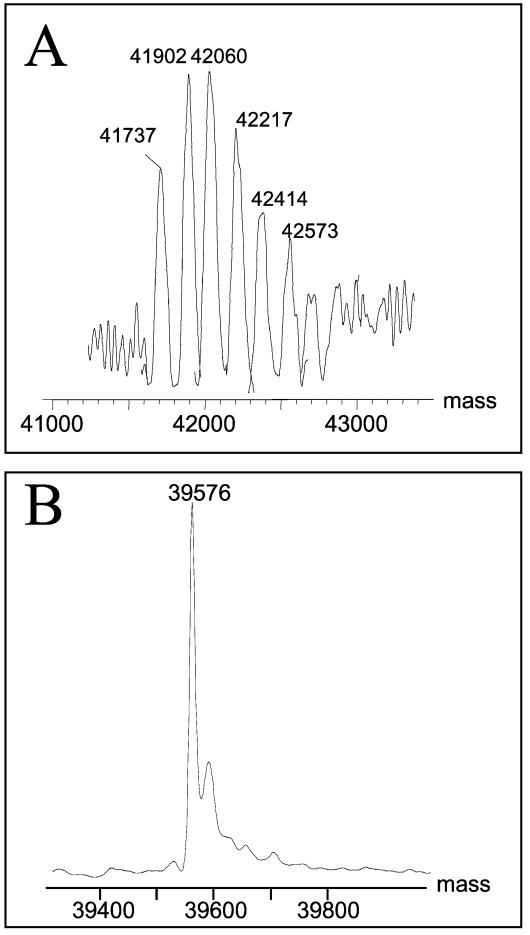
The nanoelectrospray MS analysis of purified flagellin from P. aeruginosa PAK revealed a broad, heterogeneous envelope of multiply charged ions extending from m/z 1,000 to 1,800 (data not shown). The reconstructed molecular mass profile of the corresponding spectrum (Fig. 1A) indicated that the observed flagellin molecular mass was 4 to 7% greater than that predicted from the fliC sequence, 39,905 Da. The molecular mass shift and the heterogeneous pattern of modifications observed here suggest that P. aeruginosa PAK flagellin is expressed with a broad distribution of glycoforms.
FIG. 1.
Intact mass analysis of Pseudomonas flagellins: reconstructed molecular mass profiles of the flagellins from strains PAK and JJ692. (A) PAK flagellin (theoretical molecular mass, 39,905 Da). The broad molecular mass distribution for the PAK flagellin indicates extensive heterogeneity in glycoform distribution. (B) JJ692 flagellin (theoretical molecular mass, 39,288 Da). The flagellin from JJ692 produces a peak at 39,576 Da, corresponding to the attachment of two deoxyhexose molecules to the protein.
In contrast, the reconstructed molecular mass profile of flagellin from a second strain producing type a flagellin, JJ692, showed a sharp, well-delineated peak at 39,576 Da consistent with addition of two deoxyhexose residues on the protein backbone. The second minor peak at a higher molecular mass represents the sodium adduct of the modified flagellin protein. The identification of rhamnose as the O-linked residue was confirmed by tandem MS analyses of the tryptic digest of JJ692 flagellin and by a GC-MS monosaccharide analysis (see below).
LC-electrospray MS-MS analysis of the tryptic digest from the P. aeruginosa PAK flagellin.
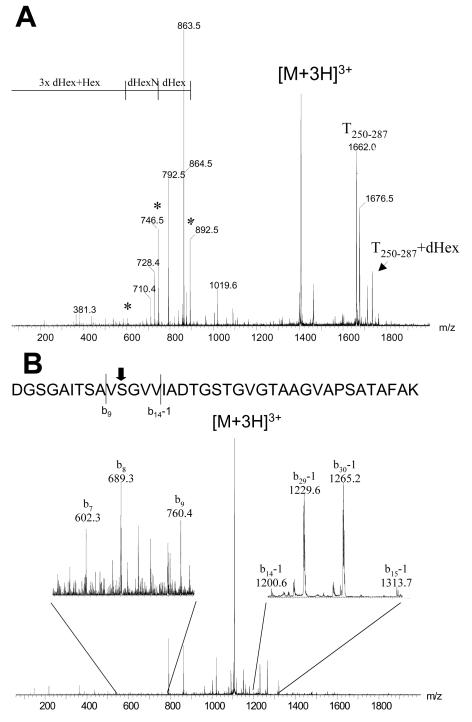
To precisely determine the type and location of the posttranslational modification, PAK flagellin protein was digested with trypsin and then analyzed by capillary LC-nanoelectrospray quadrupole time of flight MS. The MS was performed in a data-dependent mode, and the three most intense multiple ions from each survey scan were selected for MS-MS experiments. An initial Mascot search of the acquired MS-MS spectra was performed, and the results provided sequence coverage of at least 66%; only a few small peptides (<800 Da) and two larger peptides (T175-205 [2,700.4 Da] and T250-287 [3,320.7 Da]) in the central region of the flagellin protein were missing (Fig. 2).
FIG. 2.
Assignment map of PAK flagellin. The sites of O-linked glycosylation are enclosed in boxes. Peptides identified by MS-MS analysis are indicated by boldface type, whereas missing peptides are underlined.
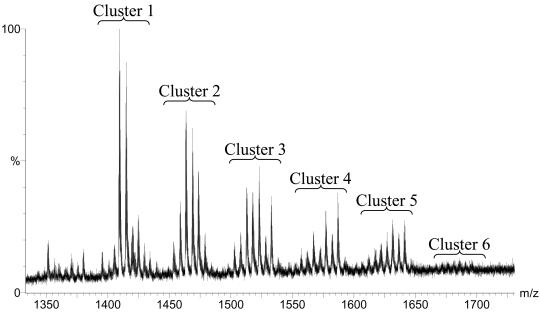
Detailed analyses of the MS data indicated that the two tryptic peptides that were unaccounted for were modified by a heterogeneous oligosaccharide chain, as reflected by a broad series of triply protonated peptide ions, as shown for T250-287 in Fig. 3. Extensive glycosylation of both T175-205 and T250-287 resulted in a broad series of more than 40 different oligosaccharides appended to each peptide. After high-performance liquid chromatography fractionation, glycopeptide ions were subjected to nanospray MS-MS analyses to identify the nature of glycosylation.
FIG. 3.
Ion spectrum of glycosylated tryptic peptide T250-287. Each signal corresponds to a different oligosaccharide attached to the peptide. For information on the oligosaccharide see Fig. 4.
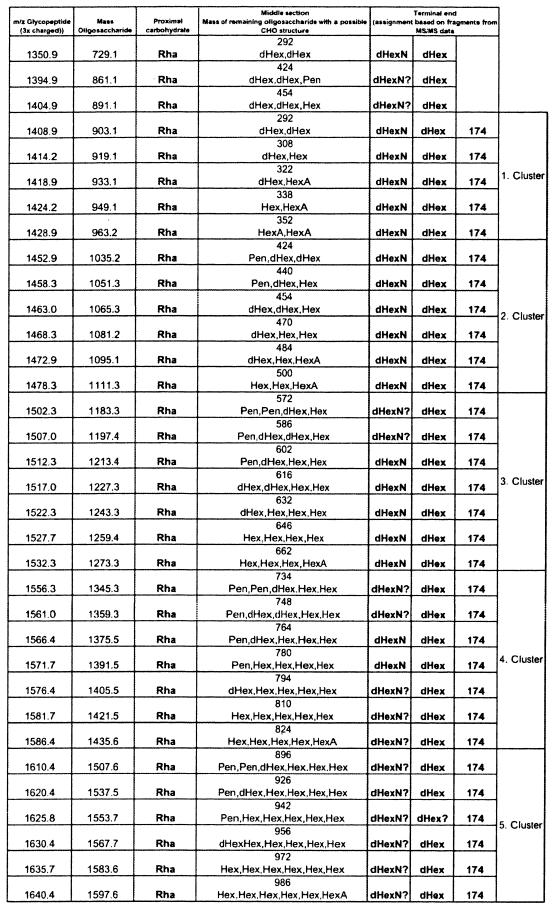
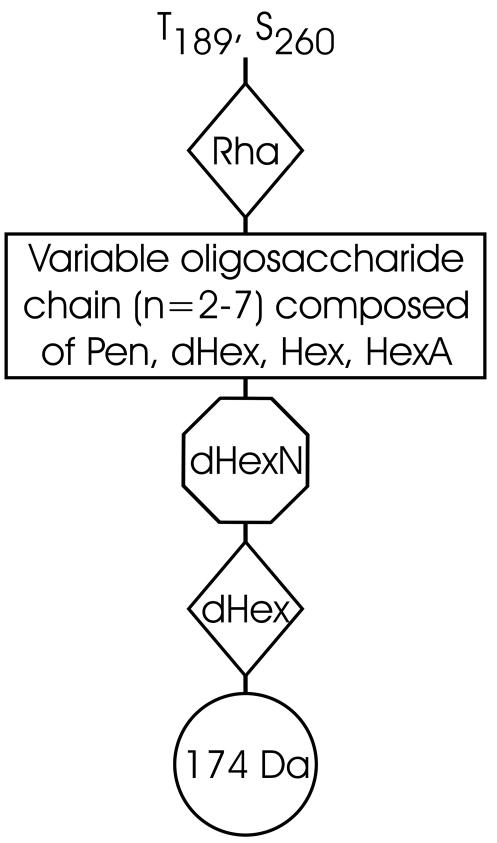
These analyses confirmed that both tryptic peptides were modified with oligosaccharides comprising up to 11 monosaccharide residues with complex microheterogeneity. A molecular mass difference of 14 Da was typically accounted for by substitution of a pentose (132 Da) for a deoxyhexose (146 Da) or by substitution of a hexose (162 Da) for a hexuronic acid (176 Da), whereas a difference of 16 Da was accounted for by substitution of a deoxyhexose (146 Da) for a hexose (162 Da). Rationalization of the glycan structures observed here is shown in Fig. 4. Several structural features characterized the oligosaccharide chains observed in P. aeruginosa PAK flagellin. For example, in all cases the proximal carbohydrate residue through which each glycan moiety was O linked to the peptide backbone had a molecular mass of 146 Da, which corresponded to the molecular mass of a deoxyhexose sugar and which was shown to be rhamnose (see below). Additionally, in the majority of cases the glycan moiety was capped by a trisaccharide consisting of an amino-deoxyhexose, deoxyhexose, and an unknown modification with a molecular mass of 174 Da. The middle region of the glycan exhibited considerable variability in its structural composition and included monosaccharides whose molecular masses corresponded to those of pentose, hexose, deoxyhexose, and hexuronic acid. No fragments from the middle section of the oligosaccharides were observed. The assignments in Fig. 4 are based on the best fits of the unaccounted for mass differences to common sugar combinations and on the GC-MS monosaccharide analysis of the glycan (see below). A structural model accounting for the glycan chains shown in Fig. 4 is shown in Fig. 5.
FIG. 4.
Oligosaccharide chains found in P. aeruginosa PAK flagellin. dHex, deoxyhexose; Pen, pentose; Hex, hexose; HexA, hexuronic acid; dHexN, deoxyhexosamine.
FIG. 5.
Structural assignment of oligosaccharide chains found on PAK flagellin. Rha, rhamnose; Pen, pentose; Hex, hexose; dHex, deoxyhexose; HexA, hexuronic acid; dhexN, deoxyhexosamine.
Monosaccharide analysis of the glycans from PAK and JJ692 flagellins.
The monosaccharide (alditol acetate) GC-MS analysis of PAK flagellin protein resulted in identification of rhamnose, mannose, glucose, and 4-amino-4,6-dideoxyglucose (viosamine) as major components. The presence of viosamine was confirmed by using perosamine and viosamine standards as described in Materials and Methods. The amount of viosamine is unknown as this compound decomposes to a large extent during derivatization. Trace levels of ribose and arabinose were also present. Uronic acids were not determined. Monosaccharide analysis of flagellin from JJ692 confirmed that the deoxyhexose sugar identified by nanospray MS-MS (see above) in this flagellin was indeed rhamnose (data not shown).
Determination of glycan attachment site.
To determine the precise site of attachment of the glycan moiety to the modified peptides, purified glycopeptide fractions were subjected to base-catalyzed hydrolysis in the presence of NH4OH, in which the β-elimination product incorporated a newly formed amino group with a distinct molecular mass (−1 Da) which could be detected by MS. Accordingly, the amino acids Thr189 and Ser260 were modified with an O-linked glycan (Fig. 2). The tandem mass spectra of the tryptic peptide precursor ion T250-287 before and after β-elimination are shown in Fig. 6.
FIG. 6.
Determination of the carbohydrate attachment site on peptide T250-287. (A) Tandem mass spectrum of [M + 3H]3+ at m/z 1,404.9. The spectrum is the spectrum of the tryptic glycopeptide T250-287 modified with an O-linked glycan identified by the fragment ion at m/z 892.5. The naked peptide fragment ion is at m/z 1,661.4 together with a fragment ion at m/z 1,734.5 corresponding to addition of a deoxyhexose (dHex) residue. The type y fragment ions at m/z 792.5, 863.5, 962.5, and 1,019.6 are consistent with the sequence assignment for T250-287, whereas oxonium ions extending from m/z 601.4, 746.5, and 892.5 (identified by asterisks) correspond to backbone cleavage products of the hexasaccharide dHex4-dHexN-Hex. (B) Tandem mass spectrum of [M + 3H]3+ at m/z 1,107.6 corresponding to the β-elimination product of the glycopeptide shown in panel A. The sequence assignment of T250-287 is validated by the type b fragment ion series shown in the expanded region of the mass spectrum (inset). The site of the O-linked attachment to Ser260 was confirmed by identification of type b ions shifted by −1 resulting from substitution of the hydroxyl Ser side chain for an amine group.
Functional characterization of genes involved in flagellar glycosylation.
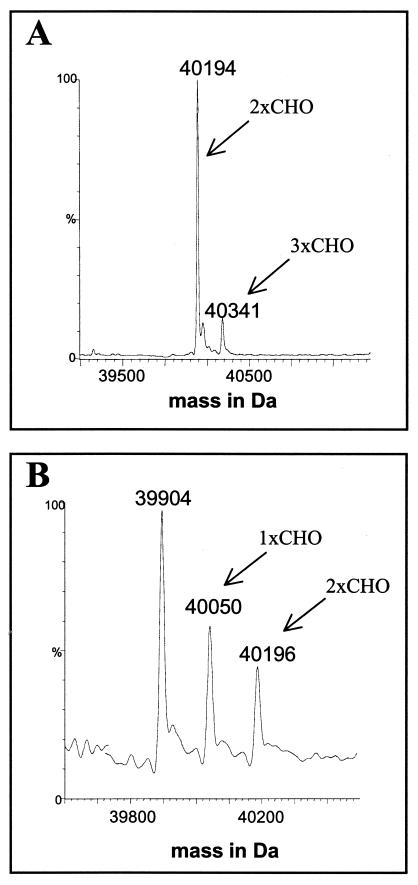
The MS analysis of intact flagellins from PAK and JJ692 revealed significant differences in the extent of glycosylation found in the two flagellins (Fig. 1), yet it has been shown that the two primary amino acid sequences differ by only a few amino acid substitutions and two small deletions of codons for three and four amino acids, respectively, at positions 254 and 279 (GI accession no. AAP33174 and P21184), none of which involves the sites of glycan attachment, as we show here. However, genomic comparisons of the glycosylation islands have revealed a substantial deletion in strain JJ692 (3). Most notably, the JJ692 genomic island is approximately 5.4 kbp shorter, five open reading frames that are present in the PAK island (orfI, orfJ, orfK, orfL, and orfM) are absent in JJ692, and a frameshift mutation in orfE results in premature termination, which results in a 30-amino-acid polypeptide, compared to the predicted 211-amino-acid orfE protein of PAK. Arora et al. (2) suggested that the proteins encoded in the glycosylation island exhibited significant homology to enzymes involved in the synthesis, activation, and polymerization of sugars. In particular, the orfA gene exhibits 58% identity with vioA of E. coli, which encodes a nucleotide sugar aminotransferase involved in the biosynthesis of viosamine. The protein encoded by orfN, which exhibits homology to a number of glycosyltransferases (2), is likely involved in the attachment of a glycan moiety to the flagellin protein. To determine the role of these two genes in flagellin glycosylation, we constructed chromosomal mutations in orfA and orfN in the PAK glycosylation island. Flagellins purified from the PAK isogenic mutants were then examined by MS. Intact mass analysis revealed that glycosylation of the PAK flagellin was considerably reduced and that the extensive heterogeneity observed in wild-type flagellin was not apparent in either mutant (Fig. 7). Instead, in the case of flagellin from the orfA isogenic mutant, the major species of flagellin protein had an intact molecular mass of 40,194 Da, corresponding to the molecular mass of the flagellin protein with two rhamnose molecules. MS-MS analysis of the tryptic digest confirmed that a single rhamnose was attached at the same two glycosylation sites found on PAK flagellin. Additionally, a second minor species (20%) of modified flagellin with a molecular mass of 40,341Da was observed, which corresponded to attachment of a third deoxyhexose. LC-MS analysis of the tryptic digest confirmed that this residue was added to one of the two sites identified by β-elimination (data not shown). It appears, therefore, that insertional inactivation of orfA results in an inability to synthesize the heterogeneous glycan observed in the parent flagellin, but the ability to link rhamnose to serine and threonine is not affected.
FIG. 7.
Intact mass analysis of flagellins from PAK glycosylation island isogenic mutants: reconstructed molecular mass profiles of flagellins from PAK isogenic orfA (A) and orfN (B) mutants. The flagellins from the two isogenic mutants did not have the heterogeneous mass profile of the PAK parent flagellin. The orfA flagellin produced two peaks corresponding to attachment of two or three rhamnose molecules, while the orfN flagellin produced peaks corresponding to unmodified flagellin having the predicted molecular mass (39,905 Da) and smaller amounts of flagellin with either one or two rhamnose molecules.
In the case of the isogenic orfN mutant, which had significant homology to a putative glycosyltransferase gene, rfbC (17), three distinct intact masses were apparent for the flagellin sample (Fig. 7B). The major peak corresponded to flagellin with the predicted molecular mass based on the amino acid sequence with no glycosylation. The two minor peaks corresponded to modification with either a single rhamnose or two rhamnose molecules. In this case, the ability to glycosylate flagellin was severely impaired with only limited addition of the rhamnose monosaccharide.
Previous studies (2) demonstrated that the defect in the ability to glycosylate flagellin in these strains was fully restored, as shown by SDS-PAGE, when a wild-type copy of each gene was provided in trans. This confirmed that the orfA and orfN mutations did not have any polar effects.
Heterologous flagellin glycosylation.
To determine if the heterogeneous glycan found on PAK flagellin was a product of the long glycosylation island, we next examined JJ692 fliC expressed in a PAK background. Flagellin protein produced in this strain (Fig. 8, lane 4) had a higher molecular mass than the parent JJ692 flagellin, as demonstrated by migration on an SDS-PAGE gel (Fig. 8, lane 3). In contrast, when we cloned the missing open reading frames orfIJKLM from the PAK glycosylation island into plasmid pSP329Gm (obtained from S. Lory) and transformed strain JJ692 with this construct, analysis of flagellin revealed no change in the glycosylation pattern (data not shown). This suggests that the inability of JJ692 to extensively glycosylate its flagellin was not only due to the loss of orfIJKLM but also was probably due to other differences in common glycosylation island genes. Most notably, the frameshift mutation in orfE that produces a truncated product containing only 30 amino acids (compare PAK orfE, which encodes 211 amino acids) may be significant.
FIG. 8.
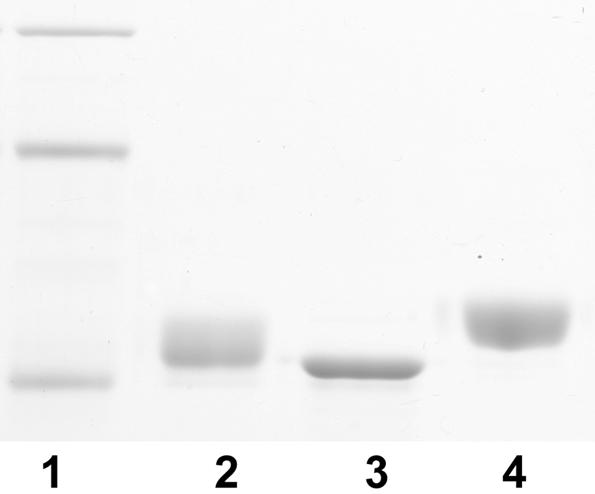
Glycosylation of heterologous JJ692 flagellin: SDS-PAGE of P. aeruginosa flagellin. Lane 1, molecular size markers (97.4, 66.2, and 45 kDa); lane 2, PAK flagellin; lane 3, JJ692 flagellin; lane 4, recombinant JJ692 flagellin expressed in a PAK background.
DISCUSSION
In this study we determined the structural basis of flagellin glycosylation in P. aeruginosa strains producing type a flagellin. Flagellins from two strains, PAK and JJ692, which contain a long glycosylation island and a short glycosylation island, respectively, were examined, and each strain was shown to produce a unique pattern of glycosylation on its flagellin. In both cases the glycosyl moiety is O linked through a rhamnose residue. The long glycosylation island found in PAK contains the additional genetic information responsible for synthesis of a heterogeneous glycan containing up to 11 monosaccharide residues, while the flagellin of strain JJ692 is glycosylated with a single rhamnose residue at each site. Short glycosylation islands has been found in a number of strains, but the open reading frames that are found in JJ692 do not necessarily represent the glycosylation potential of all strains carrying short glycosylation islands due to the mutations found in this strain. Other strains with short glycosylation islands show more extensive glycosylation, as judged by SDS-PAGE of flagellin (3). However, their glycosylation islands have not been sequenced.
The level of glycosylation found in the flagellin of Pseudomonas is notably different from the levels found in flagellins from two other gram-negative, polarly flagellated organisms. Campylobacter flagellin is the most heavily glycosylated prokaryotic protein identified to date, with at least 16 sites/monomer, while Helicobacter flagellin has been shown to have 7 sites of glycosylation on the FlaA protein monomer and 10 sites on the FlaB protein (35, 40). In contrast, the flagellin of P. aeruginosa has only two modification sites, although the modified residues are located in the central, surface-exposed, variable region rather than in the highly conserved N- and C-terminal regions of the protein, as is the case for the Campylobacter and Helicobacter flagellins. The site of attachment again appears not to be based on sequon specificity but may indeed reflect local structural restraints or the surface accessibility of individual serine and threonine residues in the folded protein.
The complex heterogeneity of the glycan of PAK flagellin is indicative of significant differences in the biosynthetic pathway compared to the biosynthetic pathway described for a second major Pseudomonas cell surface glycoprotein, pilin (8, 10). In the pilin glycosylation process no heterogeneity in the glycan structure was observed, and only an intact O-chain repeat unit was present in the pilin monomer. It has been shown that the undecaprenyl-bound O-antigen repeat unit is utilized as the substrate for pilin glycosylation, and so LPS assembly and pilus production are closely connected. In contrast, in the case of Pseudomonas flagellin glycosylation, no serotype O:6 LPS O-antigen repeat unit was found in the flagellar glycan of PAK, and it appears that a novel biosynthetic pathway is utilized for the production of this glycan. Glycosylation of the flagellin protein appears to occur by an alternative mechanism, which at least partially involves sequential attachment of individual sugars. Such a process leads to the complex, heterogeneous glycan composition that was observed at each site in this study. Chemical analysis of the flagellin glycan revealed significant quantities of the deoxyhexose monosaccharide rhamnose, a component also found in the LPS of O:6 serotypes (22). Significant levels of mannose, glucose, and the novel 4-amino-4,6 dideoxyhexose viosamine sugar were also identified by GC-MS analysis. However, both GC-MS analysis of the flagellin glycan and mass measurements of glycan fragment ions confirmed that other O-chain-specific monosaccharides appear not to be components of the heterogeneous flagellin glycan.
Polymorphism in the glycosylation islands of numerous strains of Pseudomonas has been extensively analyzed by microarray analysis, although the precise function of individual genes at this locus is unknown (3). In this study we demonstrated a functional role for glycosylation island genes in flagellar glycan production. Structural analysis of flagellins from strains in which the orfA and orfN genes were mutated in the glycosylation island of PAK clearly demonstrated the role of this locus in the biosynthesis of this glycan moiety. While addition of the rhamnose residue is not affected in an orfA mutant, no heterogeneous glycan is added. orfA exhibits homology to an LPS gene of E. coli which is responsible for the biosynthesis of the 4-amino-4,6-dideoxyhexose viosamine (28) through nucleotide activation, and the presence of this monosaccharide in the flagellar glycan-capping trisaccharide was confirmed in this study. It appears that the orfA-encoded protein of PAK is indeed responsible for activation of this unique sugar.
The protein encoded by orfN appears to be responsible for addition of the deoxyhexose sugar rhamnose to the protein backbone. Identification of rhamnose as the linkage sugar on the flagellin protein expands the known list of sugar-amino acid linkages described by Spiro (36). The flagellin of an orfN mutant was predominantly unglycosylated. We hypothesize that the residual rhamnosylltransferase activity seen is likely due to the activity of a second rhamnosyltransferase elsewhere in the genome, which may indeed utilize the same substrate or a related substrate but has only limited specificity for the protein acceptor. The rhamnsoyltransferases utilized in LPS biosynthesis or, alternatively, the RhlB and -C proteins from the rhamnolipid biosynthetic pathway are potential candidates for this limited activity (32, 33).
In this study we were not able to provide a structural assignment for the unique fragment ion with a molecular mass of 174 Da. Based on the observed mass, a possible assignment could be a diaminohexuronic acid or a trideoxynonulose, although the poor ionization indicates that such an assignment is unlikely. The annotations of the remaining genes in the PAK glycosylation island are based on relatively ill-defined functions, which include fatty acid biosynthesis, the naphthalene catabolic pathway, and nodulation factor biosynthesis, and as such provide few clues to the likely structure of this unique flagellar glycan component. However, P. aeruginosa produces extracellular glycolipids composed of l-rhamnose and 3-hydroxyalkanoic acid known as rhamnolipids, which are recognized as major virulence factors in cystic fibrosis patients (23). The biosynthetic pathway of the glycolipids has recently been investigated, and precursors of the pathway may indeed be utilized in the biosynthesis of the 174-Da modified molecule found in the flagellar glycan linked to rhamnose (9). It is important to note that as the distal residue of the capping structure, this molecule is undoubtedly significant in any type of interaction based on this flagellar glycan.
We have also shown that the composition of the glycosylation island determines the type of glycosylation found in the flagellins. JJ692, which has a truncated version of the glycosylation island, lacks orfI, orfJ, orfK, orfL, and orfM, and has polymorphisms in orfD, orfE, orfH, and orfN, produces flagellin glycosylated with only rhamnose. In contrast, the large glycosylation island found in strain PAK is capable of addition of the heterogeneous glycan to either the subtype A1 flagellin of PAK or the subtype A2 flagellin of JJ692.
Interestingly, in Pseudomonas the absence of glycosylation of the flagellin protein does not lead to a loss of flagellar filament assembly and a loss of motility. In contrast, this is true for two other well-studied polarly flagellated, motile organisms. Both Campylobacter and Helicobacter become nonmotile and are unable to synthesize a flagellar filament when the glycosylation process is prevented (16, 35). In contrast to P. aeruginosa, the flagellar filaments of these two organisms are complex and are comprised of two flagellin monomeric proteins, FlaA and FlaB, and the level of glycosylation per protein monomer is substantially higher. It remains to be determined whether either of these features is the reason for a more dramatic effect on flagellar assembly when glycosylation is inhibited.
The biological significance of each unique flagellar glycan structure can now be explored. As with Campylobacter strains, which also display considerable heterogeneity in glycan structure (38), it is not known if the diversity in glycan structure found in individual Pseudomonas strains confers a unique advantage in particular environments or indeed plays a role in modification of host immune responses to the flagellin protein. Specifically, in terms of the innate immune response, it has been shown that Toll-like receptor 5 recognition sequences are localized within the conserved D1 domain of bacterial flagellins (13, 21, 34). It remains to be established if indeed steric hindrance due to glycan size may prevent this type of receptor interaction in flagellins which are glycosylated. While O-linked glycosylation is now recognized as a common prokaryotic process, the diverse nature of the glycan structures that have been identified suggests that there are unique biological roles. Flagellin glycosylation in the plant pathogen Pseudomonas syringae has recently been shown to be involved in determining plant host specificity (39). In the case of flagellar glycosylation in P. aeruginosa, it is now important to investigate the diversity of flagellar glycan structures in isolates and the role of these structures in clinical outcomes of infection.
Acknowledgments
Financial support for this work was provided to S.M.L. by NRC Genomics and Health Initiative and to R.R. by NIH grant RO1 AI 47852.
REFERENCES
- 1.Allison, J. S., M. Dawson, D. Drake, and T. C. Montie. 1985. Electrophoretic separation and molecular weight characterization of Pseudomonas aeruginosa H-antigen flagellins. Infect. Immun. 49:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora, S. K., M. Bangera, S. Lory, and R. Ramphal. 2001. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. 98:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora, S. K., M. C. Wolfgang, S. Lory, and R. Ramphal. 2004. Sequence polymorphism in the glycosylation island and flagellins of Pseudomonas aeruginosa. J. Bacteriol. 186:2115-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benz, I., and M. A. Schmidt. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-1 adhesin. Mol. Microbiol. 40:1403-1413. [DOI] [PubMed] [Google Scholar]
- 5.Benz, I., and M. A. Schmidt. 2002. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45:267-276. [DOI] [PubMed] [Google Scholar]
- 6.Bodey, G. P., R. Bolivar, V. Fainstein, and L. Jadeja. 1983. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 5:279-313. [DOI] [PubMed] [Google Scholar]
- 7.Brimer, C. D., and T. C. Montie. 1998. Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. J. Bacteriol. 180:3209-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276:26479-26485. [DOI] [PubMed] [Google Scholar]
- 9.Deziel, E., F. Lepine, S. Milot, and R. Villemur. 2003. RhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005-2013. [DOI] [PubMed] [Google Scholar]
- 10.DiGiandomenico, A., M. J. Matewish, A. Bisaillon, J. R. Stehle, J. S. Lam, and P. Castric. 2002. Glycosylation of Pseudomonas aeruginosa 1244 pilin: glycan substrate specificity. Mol. Microbiol. 46:519-530. [DOI] [PubMed] [Google Scholar]
- 11.Dobos, K. M., K. Swiderek, K. H. Khoo, P. J. Brennan, and J. T. Belisle. 1995. Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infect. Immun. 63:2846-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doig, P., N. Kinsella, P. Guerry, and T. J. Trust. 1996. Characterisation of a posttranslational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol. Microbiol. 19:379-387. [DOI] [PubMed] [Google Scholar]
- 13.Donnelly, M. A., and T. S. Steiner. 2002. Two nonadjacent regions in enteroaggregative Escherichia coli flagellin are required for activation of Toll-like receptor 5. J. Biol. Chem. 277:40456-40461. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, P. R., and M. C. Herzberg. 1993. Evidence for the covalent linkage of carbohydrate polymers to a glycoprotein from Streptococcus sanguis. J. Biol. Chem. 268:23780-23783. [PubMed] [Google Scholar]
- 15.Feldman, M., R. Bryan, S. Rajan, L. Scheffler, S. Brunnert, H. Tang, and A. Prince. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 66:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goon, S., J. F. Kelly, S. M. Logan, C. P. Ewing, and P. Guerry. 2003. Pseudaminic acid, the major modification on campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50:659-671. [DOI] [PubMed] [Google Scholar]
- 17.Guo, D., M. G. Bowden, R. Pershad, and H. B. Kaplan. 1996. The Myxococcus xanthus rfbABC operon encodes an ATP-binding cassette transporter homolog required for O antigen biosynthesis and multicellular development. J. Bacteriol. 178:1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn, H. 1997. The type 4 pilus is the major virulence associated adhesin of Pseudomonas aeruginosa—a review. Gene 192:99-108. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 20.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration proficient plasmids for Pseudomonas aeruginosa: site specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 21.Jaccieri, S. G., R. Torquato, and R. R. Brentani,. 2003. Structural study of binding of flagellin by Toll-like receptor 5. J. Bacteriol. 185:4243-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knirel, Y. A., E. V. Vinogradov, A. S. Shashkov, B. A. Dmitriev, N. K. Kochetkov, E. S. Stanislavsky, and G. M. Mashilova. 1985. Somatic antigens of Pseudomonas aeruginosa. The structure of the O-polysaccharide chains of lipopolysaccharides of P. aeruginosa serogroup O4 (Lanyi) and related serotype O6 (Habs) and immunotype 1 (Fisher). Eur. J. Biochem. 150:541-550. [DOI] [PubMed] [Google Scholar]
- 23.Kownatzki, R., B. Tummler, and G. Doring. 1987. Rhamnolipid of Pseudomonas aeruginosa in sputum of cystic fibrosis patients. Lancet i:1026-1027. [DOI] [PubMed] [Google Scholar]
- 24.Kuo, C. C., N. Takahashi, A. F. Swanson, Y. Ozeki, and S. I. Hakomori. 1996. An N-linked high mannose type oligosaccharide expresses at the major outer membrane protein of Chlamydia trachomatis, mediates attachment and infectivity of the microorganism to HeLa cells. J. Clin. Investig. 98:2813-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanyi, B. 1970. Serological properties of Pseudomonas aeruginosa II type specific thermolabile flagellar antigens. Acta Microbiol. Acad. Sci. Hung. 17:35-48. [PubMed] [Google Scholar]
- 26.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:105-160. [DOI] [PubMed] [Google Scholar]
- 28.Marolda, C. L., and M. A. Valvano. 1993. Identification, expression and DNA sequence of the GDP-mannose biosynthesis genes encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O:7K1). J. Bacteriol. 175:148-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montie, T. C., D. Doyle-Huntzinger, R. C. Craven, and I. A. Holder. 1982. Loss of virulence associated with the absence of flagellum in an isogenic mutant of Pseudomonas aeruginosa in the burned mouse model. Infect. Immun. 38:1296-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry, M. B., L. MacLean, and D. W. Griffith. 1985. Structure of the O chain of the phenol phase soluble lipopolysaccharide of Escherichia coli O157:H7. Biochem. Cell Biol. 64:21-28. [DOI] [PubMed] [Google Scholar]
- 31.Rademaker, G. J., S. A. Pergantis, L. Blok-Tip, J. I. Langridge, A. Kleen, and J. Thomas-Oates. 1998. Mass spectrometric determination of the sites of O-glycan attachment with low picomolar sensitivity. Anal. Biochem. 257:149-160. [DOI] [PubMed] [Google Scholar]
- 32.Rahim, R., L. L. Burrows, M. A. Monteiro, M. B. Perry, and J. S. Lam. 2000. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology 146:2803-2814. [DOI] [PubMed] [Google Scholar]
- 33.Rahim, R., U. A. Ochsner, C. Olvera, M. Graninger, P. Messner, J. S. Lam, and G. Soberon-Chavez. 2001. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 40:708-718. [DOI] [PubMed] [Google Scholar]
- 34.Samatay, F. A., K. Imada, S. Nagashima, F. Vonderviszt, T. Kumasaka, M. Yamamoto, and K. Namba. 2001. Structure of the bacterial flagellar proto filament and implications for a switch for supercoiling. Nature 410:321-322. [DOI] [PubMed] [Google Scholar]
- 35.Schirm, M., E. C. Soo, A. J. Aubry, J. Austin, P. Thibault, and S. M. Logan. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579-1592. [DOI] [PubMed] [Google Scholar]
- 36.Spiro, R. G. 2002. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43R-56R. [DOI] [PubMed] [Google Scholar]
- 37.Stimson, E., M. Virji, K. Makepeace, A. Dell, H. R. Morris, G. Payne, J. R. Saunders, M. P. Jennings, S. Barker, M. Panico, I. Blench, and E. R. Moxon,. 1995. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17:1201-1214. [DOI] [PubMed] [Google Scholar]
- 38.Szymanski, C., S. M. Logan, D. Linton, and B. Wren. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi, K., F. Taguchi, Y. Inagaki, K. Toyoda, T. Shiraishi, and Y. Ichinose. 2003. Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J. Bacteriol. 185:6658-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thibault, P., S. M. Logan, J. F. Kelly, J.-R Brisson, C. P. Ewing, T. J. Trust, and P. Guerry,. 2001. Identification and characterization of the carbohydrate moieties and glycosylation motifs in Campylobacter flagellin. J. Biol. Chem. 276:34862-34870. [DOI] [PubMed] [Google Scholar]
- 41.Totten, P. A., and S. Lory. 1990. Characterization of the a-type flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172:7188-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinogradov, E. V., and M. B. Perry. 2000. Analysis of the core region of lipopolysaccharides from Proteus mirabilis serotypes O6, O48 and O57. Eur. J. Biochem. 267:2439-2446. [DOI] [PubMed] [Google Scholar]
- 43.Wacker, M., D. Linton, P. G. Hitchen, M. Nita-Lazar, S. M. Haslam, S. J. North, M. Panico, H. R. Morris, A. Dell, B. W. Wren, and M. Aebi. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790-1793. [DOI] [PubMed] [Google Scholar]
- 44.Young, N. M., J.-R. Brisson, J. Kelly, D. C. Watson, L. Tessier, P. H. Lanthier, H. C. Jarrell, N. Cadotte, F. St. Michael, E. Aberg, and C. M. Szymanski. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the Gram negative bacterium Campylobacter jejuni. J. Biol. Chem. 277:42530-42539. [DOI] [PubMed] [Google Scholar]