Abstract
HypA and HypB are maturation proteins required for incorporation of nickel into the hydrogenase large subunit. To examine the functions of these proteins in nickel insertion, the hybF gene, which is a homolog of hypA essential for maturation of hydrogenases 1 and 2 from Escherichia coli, was overexpressed, and the product was purified. This protein behaves like a monomer in gel filtration and contains stoichiometric amounts of zinc but insignificant or undetectable amounts of nickel and iron. In filter binding assays radioactively labeled nickel binds to HybF with a KD of 1.87 μM and in a stoichiometric ratio. To identify amino acid residues of HybF involved in nickel and/or zinc binding, variants in which conserved residues were replaced were studied. An H2Q replacement eliminated both in vivo activity and in vitro binding of nickel. The purified protein, however, contained zinc at the level characteristic of the wild-type protein. When E3 was replaced by Q, activity was retained, but an E3L exchange was detrimental. Replacement of each of the four conserved cysteine residues of a zinc finger motif reduced the cellular amount of HybF protein without a loss of in vivo activity, indicating that these residues play a purely structural role. A triple mutant deficient in the synthesis or activity of HypA, HybF, and HypB was constructed, and it exhibited the same responsiveness for phenotypic complementation by high nickel as mutants with a single lesion in one of the genes exhibited. The results are interpreted in terms of a concerted action of HypB and HybF in nickel insertion in which HybF (as well as its homolog, HypA) functions as a metallochaperone and HypB functions as a regulator that controls the interaction of HybF with the target protein.
Escherichia coli possesses four distinct hydrogenases, designated hydrogenases 1, 2, 3, and 4 (1, 5, 6, 47, 48; for a general classification of hydrogenases see reference 53). Isoenzymes 1 and 2 cleave elemental hydrogen into protons and electrons, which are used for energy conservation by feeding into an anaerobic respiratory chain. Hydrogenase 3, on the other hand, is a component of the classical formate-hydrogen lyase complex and reduces protons to elemental hydrogen with the electrons derived from formate (11, 42, 46). Hydrogenase 4 is part of a separate formate-hydrogen lyase system and appears to be a proton-translocating enzyme, thereby contributing to the electrochemical gradient across the cytoplasmic membrane (1); however, its exact physiological role is still unknown (3, 4, 49). The genes coding for structural components and redox partners are located in four operons, hya (hydrogenase 1), hyb (hydrogenase 2), hyc (hydrogenase 3), and hyf (hydrogenase 4) (1, 12, 35, 36). Except for the exceptions discussed below, lesions within these operons lead to loss of activity of the specific isoenzymes, leaving the activities of the other hydrogenases unaffected.
In addition to the products of the genes in the structural gene operons, hydrogenase activity requires the products of genes with functions in enzyme maturation. All four hydrogenases belong to the class of hydrogenases possessing an NiFe metal center coordinated by the thiolates of four cysteine residues from the protein backbone of the large hydrogenase subunit (54). Two of the cysteines function as ligands bridging the Fe and Ni atoms (54, 55; for a review see reference 20). In the classical type of NiFe hydrogenases the Fe carries two cyanide groups and one carbon monoxide group (2, 23). In E. coli, the products of seven genes were determined to have functions in the synthesis of this metal center (25, 28). Four of these products have been demonstrated (HypF and HypE) or supposed (HypC and HypD) to be involved in CN synthesis, in the attachment of the ligands to the iron (9, 41, 43; unpublished data), and in the putative transfer to the apoprotein of the large hydrogenase subunit (9).
The incorporation of nickel into the large subunit is independent of the incorporation of iron and seems to succeed the incorporation of iron. Evidence shows that a precursor of the large subunit synthesized under nickel deprivation conditions can be converted in vitro into the active enzyme by incubation with the metal (30, 34). Mutational studies revealed that insertion of nickel requires the activity of two additional maturation proteins, HypB and HypA. Inactivation of the genes encoding these proteins blocks the formation of active hydrogenases; however, this deficiency can be partially rescued by inclusion of high nickel concentrations in the growth medium (24, 40, 58). It appears, therefore, that nickel incorporation can occur spontaneously, but for functioning at low metal concentrations it is dependent on the activity of the two proteins (25). Protein HypB has been purified from E. coli (31), Rhizobium leguminosarum (45), Bradyrhizobium japonicum (21), and Helicobacter pylori (33). HypB is a guanine nucleotide binding protein, and for functioning in nickel insertion it must possess GTPase activity (32). Intriguingly, there are homologous proteins in other organisms which are involved in nickel incorporation into urease (for a review see reference 37) and CO dehydrogenase (26). HypB from B. japonicum and R. leguminosarum, but not HypB from E. coli and H. pylori, can also act as nickel storage proteins (21, 33, 39, 45). On the other hand, although it has been suggested, it has not been proven unequivocally that HypB is the protein which also donates nickel to the hydrogenase large subunit (32).
The HypA protein has been purified from H. pylori (33). This protein binds nickel stoichiometrically and also appears to interact with the HypB protein, as indicated by detection of heterodimers when stoichiometric mixtures of the two proteins were cross-linked chemically (33). From the results obtained thus far, two models can be proposed for the function of HypA and HypB. Either HypB is the nickel-donating protein and HypA guides HypB to the apoprotein, or HypA is the nickel-binding partner and HypB has a solely regulatory function in facilitating the switch required to release HypA after the nickel is donated. Circumstantial evidence that favors the latter model comes from the fact that purified HypB protein from H. pylori does not bind nickel (33) and the fact that HypA from E. coli functions only in the maturation pathway of hydrogenase 3 and there is a homolog of this protein, HybF, that has a role in the maturation of hydrogenases 1 and 2 instead (24). It is plausible to assume that specific donation of nickel to the metal-free apoprotein requires recognition of the correct target by a protein-protein interaction. The fact that the sequences of the large subunits of isoenzymes 1 and 2 are very similar to each other but very different from that of hydrogenase 3 (53) may have required the evolution of two separate nickel-delivering proteins. HypB from E. coli, on the other hand, is involved in the maturation of all hydrogenase isoenzymes, which reflects a more general role in the maturation process (25). To gain more insight into the function of HybF and thereby also the function of its homolog HypA, we purified and characterized HybF from E. coli.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli strains and plasmids used in this study are listed in Table 1. Strains DPAPB and NIM were obtained by using the gene disruption method of Hamilton et al. (22) with the aid of the pMAK700 derivative pDB705 and pMDHBF. E. coli DH5α was used as the host for cloning procedures.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or phenotype | Reference |
|---|---|---|
| Strains | ||
| DH5α | F− (φ80dlacZΔM15) recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | 59 |
| MC4100 | F−araD139 Δ(argF-lac)U169 ptsF25 deoC1 relA1 flbB5301 rpsL150λ− | 14 |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | 50 |
| SMP101 | MC4100, ATG codon of hypA mutated into TAA | 25 |
| DHP-B | MC4100 ΔhypB | 25 |
| DPABF | SMP101 ΔhybF | 24 |
| DPAPB | SMP101 ΔhypB | This study |
| NIM | DPAPB ΔhybF | This study |
| Plasmids | ||
| pBR322 | Apr Tcr | 13 |
| pACYC184 | Cmr Tcr | 15 |
| pDB705 | pMAK705, ΔhypB (0.8-kb RsaI-AvaII fragment deleted), Cmr | 25 |
| pMDHBF | pMAK700 with 252-bp deletion in hybF, Cmr | 24 |
| pBHBF-Strep | pBR322 hybF-StreptagII, Apr | 24 |
| pBHBF-Strep [H2A] | pBR322 hybF [H2A]-Strep, Apr | This study |
| pBHBF-Strep [H2Q] | pBR322 hybF [H2Q]-Strep, Apr | This study |
| pBHBF-Strep [E3Q] | pBR322 hybF [E3Q]-Strep, Apr | This study |
| pBHBF-Strep [E3L] | pBR322 hybF [E3L]-Strep, Apr | This study |
| pBHBF-Strep [C73A] | pBR322 hybF [C73A]-Strep, Apr | This study |
| pBHBF-Strep [C76A] | pBR322 hybF [C76A]-Strep, Apr | This study |
| pBHBF-Strep [C73/76A] | pBR322 hybF [C73/76A]-Strep, Apr | This study |
| pBHBF-Strep [C89A] | pBR322 hybF [C89A]-Strep, Apr | This study |
| pBHBF-Strep [C89S] | pBR322 hybF [C89S]-Strep, Apr | This study |
| pBHBF-Strep [P90A] | pBR322 hybF [P90A]-Strep, Apr | This study |
| pBHBF-Strep [C92A] | pBR322 hybF [C92A]-Strep, Apr | This study |
| PT7-7 | Apr, T7Φ10 | 51 |
| pThybFStrep | pT7-7 hybF, Apr | This study |
| pThybFStrep [H2Q] | pT7-7 hybF [H2Q], Apr | This study |
| pThybFStrep [C73A] | pT7-7 hybF [C73A], Apr | This study |
| pThybFStrep [C73/76A] | pT7-7 hybF [C73/76A], Apr | This study |
| pThybFStrep [C89A] | pT7-7 hybF [C89A], Apr | This study |
| pThybFStrep [C89S] | pT7-7 hybF [C89S], Apr | This study |
Media and growth conditions.
E. coli cells were grown anaerobically at 37°C in buffered TGYEP medium (7) supplemented either with 15 mM formate plus 0.8% glucose or with 15 mM fumarate plus 0.8% glycerol, as described previously (10); 1 μM sodium molybdate, 1 μM sodium selenite, and 5 μM nickel chloride were used as supplements unless indicated otherwise. The cells were harvested at an optical density at 600 nm of 1.0, and crude extracts were prepared as described previously (17). The antibiotics chloramphenicol and ampicillin were added to the medium at concentrations of 30 and 50 μg per ml, respectively, when they were needed for maintenance of plasmids.
Site-directed mutagenesis and cloning procedures.
To replace amino acid residues of HybF-Strep, site-directed mutagenesis was performed with plasmid pBHBF-Strep by using inverse PCR as described previously (29, 38). To do this, the appropriate primers listed in Table 2 were used. The authenticity of the hybFStrep variant genes was verified by DNA sequencing. Plasmid pThybFStrep and its derivative pThybFStrep [H2Q] were constructed by ligation of the EcoRI- and NdeI-digested vector pT7-7 with the EcoRI- and NdeI-digested PCR fragment carrying the hybFStrep or hybFStrep[H2Q] gene. The hybFStrep and hybFStrep[H2Q] genes were obtained with primers NdelhybFup and tet-inv.+EcoRI and with primers NdeIhybFH2Qup and tet-inv.+EcoRI, respectively; pBHBF-Strep or its [H2Q] derivative was used as the template.
TABLE 2.
Oligonucleotide primers used in this work
| Primer | Sequencea |
|---|---|
| His2Ala-hybF | 5′-GCCGAGCTCTCTCTTTGCCAGAGCGCC-3′ |
| His2Gln-hybF | 5′-CAGGAGCTCTCTCTTTGCCAGAGCGCCG-3′ |
| hybFE3Q-MunI | 5′-GGAGTGGCGAAAATGCATCAATTGTCTCTTTGCCAGAGC-3′ |
| hybFE3L-AflII | 5′-GAGTGGCGAAAATGCATCTCTTAAGCCTTTGCCAGA-3′ |
| hybFH2E3-inv. | 5′-CATTTTCGCCACTCCTGCGACCAAACAGCAGG-3′ |
| Cys89Ala-hybF | 5′-CGGAGCCTGCGCATCGTGCTGATGAATCTCC-3′ |
| Cys89Ser-hybF | 5′-CGGGCTCTGCGCATCGTGCTGATGAATCTCC-3′ |
| Pro90Ala-hybF | 5′-CGCACACTGCGCATCGTGCTGATGAATCTCC-3′ |
| hybFC89P90-inv. | 5′-CTCTGCCATGGCGAGCGGTTGCGTGTCGATACC-3′ |
| HybFC73A | 5′-CCCGCCCAGGCTTGGGCTTGGGATTGCAGCCAGGTG-3′ |
| HybFC76A | 5′-CCCGCCCAGGCTTGGTGCTGGGATGCTAGCCAGGTG-3′ |
| HybFC73/76A | 5′-CCCGCCCAGGCTTGGGCTTGGGATGCTAGCCAGGTGG-3′ |
| HybFC73/76 inv. BstI1071 | 5′-CCAAGCCTGGGCGGGTTTGTATACGATATGTAAATCGC-3′ |
| HybFC92A | 5′-CAACCGCTCGCCGTGAGCGAGCGGACACTGCGCATC-3′ |
| HybFC89/92 inv. SalI | 5′-CACGGCGAGCGGTTGCGTGTCGACACCGGCGATTCG-3′ |
| tet-inv.+EcoRI | 5′-CGGAATTCAAGGCTCTCAAGGGCATCGG-3′ |
| NdeIhybFup | 5′-GGATCCATATGCATGAGTTGTCTCTTTGCC-3′ |
| NdeIhybFH2Qup | 5′-GGATCCATATGCAGGAGTTGTCTCTTTGCC-3′ |
Bases that are different from the bases in the wild-type sequence are underlined; newly generated restriction sites are indicated by boldface type.
Purification of the HybF-Strep protein and its [H2Q] variant.
Strain BL21(DE3) was transformed with plasmid pThybFStrep or pThybFStrep [H2Q] and was grown under anaerobic conditions in TGYEP medium containing 5 μM NiCl2 at a temperature between 16 and 22°C. Expression was induced by adding 0.1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). The cells were harvested after the culture reached an optical density at 600 nm of 1, centrifuged, washed with 50 mM Tris-Cl (pH 7.5), and stored at −20°C. For the purification procedure, the cell pellet was resuspended in 50 mM Tris-Cl (pH 7.5) containing 20 μg of DNase I per ml, 20 μg of phenylmethylsulfonyl fluoride per ml, and 1 mM dithiothreitol (DTT). The cells were broken by two passages through a French press at 118 MPa. The homogenate was clarified by centrifugation at 30,000 × g for 30 min, followed by centrifugation at 100,000 × g for 2 h. The latter centrifugation step was omitted in further purifications of HybF-Strep and HybF-Strep [H2Q] as it had no obvious advantage. The cell extract was loaded onto a Streptactin Sepharose column (Institut für Bioanalytik, Göttingen, Germany), and the column was washed extensively with 50 mM Tris-Cl (pH 8.0)-100 mM NaCl and eluted with 50 mM Tris-Cl (pH 7.5) containing 2.5 mM desthiobiotin. The fractions containing the HybF-Strep protein were concentrated by microcentrifugation (MICROSEP microconcentration system; Pall Gelman Laboratory, Dreieich, Germany) and applied to a HiLoad 16/60 Superdex 75 pg gel filtration column (Amersham Biosciences, Freiburg, Germany). The column was developed with 100 mM Tris-Cl (pH 8.0)-100 mM NaCl with or without 1 mM DTT at a flow rate of 1 ml min−1. Calibration of the column was carried out by using ovalbumin (43 kDa), carbonic anhydrase (29 kDa), chymotrypsinogen (25 kDa), chicken lysozyme (14.4 kDa), and cytochrome c (12.4 kDa) as markers. The fractions containing the HybF-Strep protein were pooled and either analyzed by inductively coupled plasma for the metal content or dialyzed against 25 mM Tris-Cl (pH 8.0)-5 mM NaCl-0.1 mM DTT-50% glycerol and stored at −20°C.
PAGE and immunoblotting.
Proteins in cell lysates or cell extracts were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (27). The gels were subsequently stained with Coomassie blue (Serva Blue R), and specific proteins were visualized by Western blotting; 10% polyacrylamide gels were used in order to detect the large subunits of hydrogenase 2 (HybC) and hydrogenase 3 (HycE), whereas 15% polyacrylamide gels were used for resolution of the HybF-Strep protein. The antisera were used at dilutions of 1:1,500 for anti-HybC and 1:1,000 for anti-HycE and anti-StreptagII (polyclonal serum from Institut für Bioanalytik).
Hydrogenase activity staining in gels.
To localize active hydrogenases 1 and 2 in polyacrylamide gels, cells were grown in TGYEP medium in the presence of 0.8% glycerol and 15 mM fumarate. The proteins in the crude extracts were separated by SDS-PAGE under native conditions, which meant that the loading buffer lacked DTT and 2-mercaptoethanol and that heat treatment was omitted. After separation, the gels were soaked in 100 mM sodium phosphate buffer (pH 7.2) containing 0.5 mM benzyl viologen and 1 mM triphenyltetrazolium chloride and incubated either under an H2 atmosphere at 37°C or in an anaerobic chamber (5% H2, 95% N2) at room temperature. The activity-stained bands were determined to be hydrogenases 1 and 2 by using extracts of E. coli strains HDK103 and HDK203 (25) (data not shown).
In vitro maturation assay.
Crude extracts of the strains were mixed with the appropriate amounts of NiCl2 and incubated for 2 h at 30°C. The newly generated hydrogenase 1 and 2 activities were evaluated by activity staining in polyacrylamide gels and Western blot analysis with antibodies raised against the large subunit of hydrogenase 2 (HybC).
Nickel binding assay.
Filter binding assays were used to investigate the binding of nickel by HybF-Strep or its [H2Q] variant. To do this, 63NiCl2 (specific activity, 697 mCi/mmol; NEN Life Science Products, Inc., Boston, Mass.) was diluted with unlabeled NiCl2 to obtain a 0.5 mM stock solution with a specific activity of 121.6 μCi/mmol. The proteins were incubated with different nickel concentrations for 30 min at room temperature, and this was followed by filtration through nitrocellulose filters (pore size, 0.2 μm; Schleicher & Schuell, Dassel, Germany) and extensive washing. The radioactivity was determined by scintillation counting with a Tri-Carb 2100TR liquid scintillation analyzer (Canberra Packard GmbH, Dreieich, Germany).
Cross-linking experiments.
To investigate a possible interaction between HybF and HypB, cross-linking studies were performed. This was done as described by Mehta et al. (33) by using dimethyl suberimidate, a homobifunctional imidoester cross-linker specific for primary amines (57). Alternatively, the sulfhydryl-specific cross-linking reagent p-phenylenedimaleimide was used (56).
RESULTS
Purification and properties of HybF-Strep.
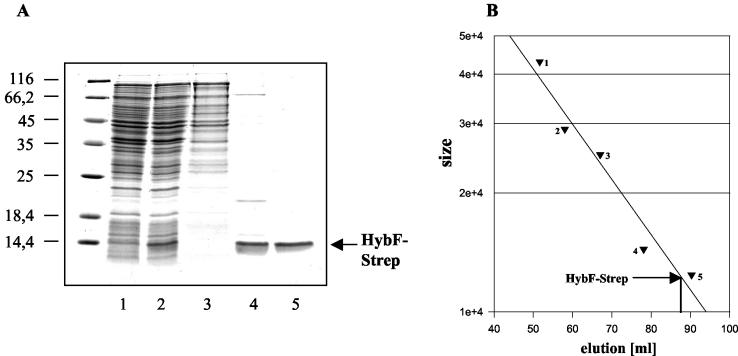
The initial aim of this work was to purify the HypA protein from E. coli. However, despite extensive variation of the expression conditions, our efforts failed since the gene product was present exclusively in an insoluble form. As an alternative, we attempted to express hybF, which codes for the homolog of HypA involved in maturation of hydrogenases 1 and 2 (24). For this we used a variant which contains StreptagII as a C-terminal extension and has been shown previously to be fully active in hydrogenase maturation in vivo (24). As shown in Fig. 1A, most of the HybF-Strep formed was probably insoluble, even when the protein was expressed at a low temperature. The fraction present in the 100,000 × g supernatant, however, was still sufficient to recover a suitable amount of highly purified protein by affinity chromatography on a Streptactin column, followed by gel filtration in a Superdex 75 column and concentration by ultrafiltration. From 28 g (wet weight) of cells 1 mg of HybF-Strep was obtained. The purified protein was amino terminally sequenced and was found to possess an N-terminal methionine residue. In contrast to the HypA protein from H. pylori (33), HybF-Strep from E. coli migrated during gel filtration as an apparent monomer (Fig. 1B). Determination of the metal content of the purified protein (Table 3) showed that it contains an approximately stoichiometric amount of zinc, a very small amount of nickel, and an undetectable amount of iron. Taking into account that the medium in which expression occurred was not supplemented with zinc but was supplemented with 5 μM nickel chloride and that iron, which usually is present as a contaminant together with zinc, was not found, we concluded that HybF is a zinc-containing protein.
FIG. 1.
Purification of HybF-Strep. (A) Proteins from the different purification steps were separated by SDS-PAGE and stained with Coomassie blue. Lane 1, lysate from cells before induction with IPTG; lane 2, lysate from cells after induction with IPTG; lane 3, extract after ultracentrifugation; lane 4, eluate from the Streptactin column; lane 5, concentrated HybF-Strep protein after gel filtration on a Superdex 75 column. (B) Migration behavior of HybF-Strep in the gel filtration experiment. The marker proteins (with sizes in kilodaltons) were ovalbumin (triangle 1), carbonic anhydrase (triangle 2), chymotrypsinogen (triangle 3), lysozyme (triangle 4), and cytochrome c (triangle 5). The position of the HybF-Strep protein is indicated by the arrow.
TABLE 3.
Metal contents of purified HybF-Strep and its [H2Q] variant
| Protein | Metal contenta
|
||
|---|---|---|---|
| Fe | Zn | Ni | |
| HybF-Strep | 0.0 | 1.0 ± 0.1 | 0.0 |
| HybF-Strep [H2Q] | 0.3 ± 0.1 | 0.9 ± 0.2 | 0.0 |
The values are numbers of metal ions per monomer and are means ± standard deviations.
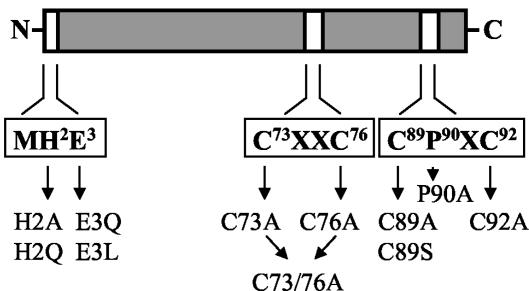
Properties of HybF variants with replacements of amino acids putatively involved in metal coordination.
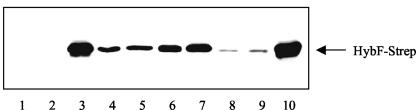
In the search for amino acid residues which are involved in the coordination of nickel, all of the histidine residues of the HypA protein from H. pylori have been replaced by alanine (33). Of the five variant proteins generated and purified, only the H2A form was unable to bind nickel, and the corresponding mutant strain lacked hydrogenase activity (33). Since a single histidine residue is not sufficient to coordinate nickel and in light of the unexpected finding that zinc is a cofactor of the HybF protein, a more extensive search for residues of the protein involved in coordination of metals was conducted. Figure 2 shows the residues of HybF which are conserved in the HypA-HybF protein family and might fulfill such a function. By using inverse PCR the codons for these residues were exchanged in order to insert the amino acids indicated in Fig. 2. The corresponding plasmids were used to transform strain DPABF, which does not express hypA and has a deletion in hybF. The transformants were grown to an optical density at 600 nm of 1, extracts were prepared and separated electrophoretically, and the gels were developed with antiserum directed against StreptagII (Fig. 3). It is evident that compared to the amount in the transformant harboring the wild-type hybF plasmid, the amounts of the majority of the variant proteins were reduced. The most conspicuous effect was due to replacement of Cys89 by either alanine or serine, which caused a drastic reduction in the level of the gene product present (Fig. 3, lanes 8 and 9).
FIG. 2.
Schematic representation of the HybF protein: primary structure of the HybF protein showing the conserved N terminus and the two cysteine motifs. The amino acids that were replaced by site-specific mutagenesis are indicated.
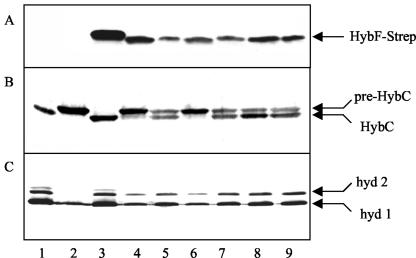
FIG. 3.
Analysis of mutants with amino acid replacements in HybF. An immunoblot analysis of strain DPABF and transformants of this strain was performed with antiserum directed against StreptagII. Each lane was loaded with 30 μg of protein from cell extracts. Lane 1, MC4100; lane 2, DPABF; lane 3, DPABF/pBHBF-Strep; lane 4, DPABF/pBHBF-Strep [H2A]; lane 5, DPABF/pBHBF-Strep [H2Q]; lane 6, DPABF/pBHBF-Strep [E3Q]; lane 7, DPABF/pBHBF-Strep [E3L]; lane 8, DPABF/pBHBF-Strep [C89A]; lane 9, DPABF/pBHBF-Strep [C89S]; lane 10, DPABF/pBHBF-Strep [P90A].
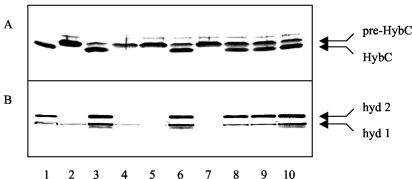
Next, the transformants were analyzed to assess whether the corresponding HybF variants are active in the maturation of hydrogenases 1 and 2. Figure 4A shows the results of the experiment performed to determine the capacities of the mutant forms to allow processing of the large subunit of hydrogenase 2 (HybC), which can only take place when nickel has been inserted. HybC was fully processed in the extract from the wild type (Fig. 4, lane 1), was not processed in the host strain lacking HypA and HypF (lane 2), and was processed in the transformants carrying the wild-type hybF gene and hybF with the replacements E3Q, C89A, C89S, and P90A (lanes 3, 6, 8, 9, and 10). No processing was observed with the variants of HybF having the replacements H2A, H2Q, and E3L (lanes 4, 5, and 7). The results suggest that C89 is not essential since its replacement is not detrimental to activity, although the gene product is present at a very low level. On the other hand, the consequence of replacement of E3 depends on the chemical nature of the newly inserted residue. Replacement by the nonpolar amino acid leucine results in an inactive protein, whereas replacement by glutamine results in retention of activity. Analysis of the generation of active hydrogenases 1 and 2 by activity staining of polyacrylamide gels in which extracts from the transformants had been separated fully corroborated these results (Fig. 4B).
FIG. 4.
Ability of the HybF-Strep protein and its variants to complement the effects of chromosomal deletions of hypA and hybF. (A) Immunoblot analysis with antisera against the large subunit of hydrogenase 2 (HybC) performed to assess the status of processing. (B) Corresponding activity staining in 10% polyacrylamide gels. For this analysis, cells were cultivated in the presence of 50 μM EDTA without addition of nickel to the medium, and 30 μg of protein from crude extract was loaded per lane. The arrows in panel A indicate the positions of the precursor (pre-HybC) or the processed form of the large subunit; the arrows in panel B indicate the migration positions of hydrogenases 1 (hyd 1) and 2 (hyd 2). For lane contents see the legend to Fig. 3.
In order to investigate whether the H2 exchanges had an effect on the binding of zinc, HybF-Strep [H2Q] was purified. The protocol that was used for purification of the wild-type HybF-Strep protein was used, and similar yields of apparently homogeneous proteins were obtained (results not shown). The results of metal analysis are shown in Table 3. Again, approximately one zinc per HybF-Strep [H2Q] monomer was detected. In this preparation, iron was present, although the ratio was substoichiometric; again, nickel was undetectable.
C-terminal cysteine motif is not essential for HybF function.
The replacement of Cys-89 by either alanine or serine showed that this residue has a structural role rather than a functional role, which could involve the coordination of zinc. To obtain further evidence for this hypothesis, the other three cysteine residues of the two motifs were replaced either singly or in pairwise combinations. Analysis of the levels of gene products in the different transformants showed that they all contained reduced amounts of HybF compared with the amount in a transformant carrying the wild-type hybF gene on the plasmid (Fig. 5A). In addition, all variants exhibited HybC processing, albeit to different extents, and formed active hydrogenases 1 and 2 (Fig. 5B and C). The attempt to purify a HybF-Strep variant with altered conserved cysteine motifs in order to confirm the probable role of these motifs in zinc binding was not successful because of the complete insolubility of the products.
FIG. 5.
Stability and function of HybF-Strep protein variants altered in the cysteine motifs at the C terminus. (A) Strain DPABF without and with plasmids encoding HybF-Strep variants was analyzed by immunoblotting with antisera directed against C-terminal StreptagII. Each lane contained 0.15 U of SDS lysate from a culture at an optical density at 600 nm of 1.0. (B) Analysis of the processing of the large subunit of hydrogenase 2 by immunoblotting with HybC-specific antiserum. (C) Activity staining of hydrogenases 1 (hyd 1) and 2 (hyd 2) after separation of extracts in polyacrylamide gels. In panels B and C 30 μg of cell extract was applied per lane. Lane 1, MC4100; lane 2, DPABF; lane 3, DPABF/pBHBF-Strep; lane 4, DPABF/pBHBF-Strep [C73A]; lane 5, DPABF/pBHBF-Strep [C76A]; lane 6, DPABF/pBHBF-Strep [C73/76A]; lane 7, DPABF/pBHBF-Strep [C89A]; lane 8, DPABF/pBHBF-Strep [C89S]; lane 9, DPABF/pBHBF-Strep [C92A].
Maturation defect of a triple mutant devoid of proteins HypA, HypB, and HybF can be phenotypically reversed by a high nickel concentration in the medium.
Waugh and Boxer (58) were the first workers to show that lesions in hypB can be phenotypically suppressed by high nickel concentrations in the medium. This was also demonstrated later for a mutant with defects in both hypA and hybF (24), suggesting that the products of these genes are also involved in nickel insertion. These findings, however, did not rule out the possibility that the products of hypB and of hypA and hybF could mutually compensate for each other in this process. To test this possibility, a mutant was constructed in which the expression of intact products of all three genes was blocked, and this mutant was tested for phenotypic suppression of the defect in hydrogenase maturation by nickel in vivo. It was found that the defect in the triple mutant was rescued by nickel in vivo. An analysis of the approximate concentration of the metal that was sufficient for compensation showed that this concentration was not different from the concentration to which the strain with the hypB deletion responded (data not shown).
Is there an interaction between the HybF and HypB proteins?
The postulated cooperative action of HybF and HypB in nickel insertion into the hydrogenases from E. coli might involve a physical interaction. Formation of such a complex has been demonstrated for the homologous proteins from H. pylori by chemical cross-linking (33). To test whether this is also true for the E. coli proteins, we employed two different cross-linking reagents. This experiment was done by using either the sulfhydryl-specific cross-linking reagent p-phenylenedimaleimide or dimethyl suberimidate, a homobifunctional imidoester cross-linker specific for primary amines. The products of the reaction were separated by SDS-PAGE, and the gels were developed by immunoblotting. However, no evidence for specific cross-linking was obtained (results not shown).
In a second approach to detect an interaction between the two proteins, we analyzed whether purified HybF can modulate the intrinsic GTP hydrolysis catalyzed by the HypB protein. Again, no effect was observed, either in the presence or in the absence of nickel (data not shown), which is in accordance with the results of Mehta et al. (33).
In vitro maturation of hydrogenases.
In vitro processing and maturation systems for hydrogenases have been reported for Azotobacter vinelandii (34) and E. coli (30). These systems require the presence of nickel ions at unphysiologically high concentrations, in the range of several hundred micromolar. Since this could be the consequence of the lack of functional HybF and/or HypA proteins in the extracts, we tested whether addition of the purified proteins made the system functional at low nickel concentrations. However, addition of the proteins, even at high concentrations, in the presence of GTP did not change the maturation pattern (results not shown).
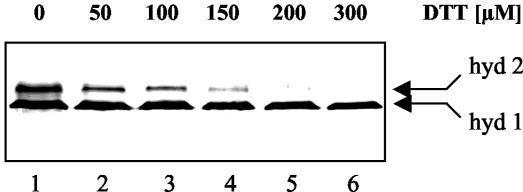
However, a dramatic effect was observed when the amount of thiol compounds in the assay mixture was reduced (Fig. 6). The hydrogenase activity that developed indicated that the in vitro incorporation of nickel is diminished or even eliminated in the presence of equimolar (Fig. 6, lane 3) or excessive (lanes 4 to 6) amounts of DTT.
FIG. 6.
Complexation of free nickel by thiols inhibits in vitro maturation of hydrogenases 1 and 2. The triple mutant NIM lacking HypA, HypB, and HybF was cultivated with glycerol, fumarate, and 50 μM EDTA under anaerobic conditions. Cell extract was used for the in vitro maturation analysis. In the different trials 100 μM nickel chloride was added along with concentrations of DTT ranging from 0 to 300 μM, as indicated at the top. The signals for hydrogenases 1 (hyd 1) and 2 (hyd 2) detected by activity staining are indicated by arrows.
Nickel binding of the HybF-Strep protein.
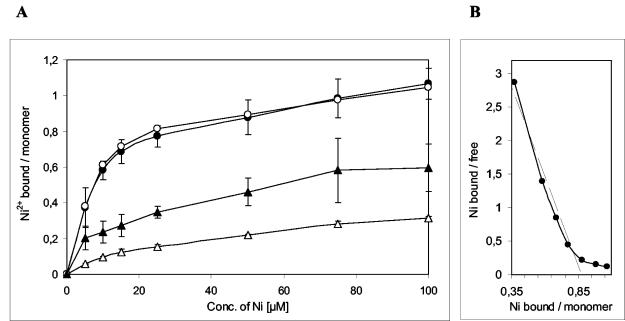
To test the nickel binding capability of HybF, a nickel binding assay was performed by using 63Ni. In this filter binding assay, specific interaction of nickel with the wild-type HybF-Strep protein was observed (Fig. 7A). In contrast, the HybF-Strep [H2Q] variant displayed reduced binding activities which varied greatly in independent experiments. The corresponding Scatchard plot of the binding characteristics of the wild-type protein is shown in Fig. 7B. A KD of 1.87 μM for nickel and a maximal binding of 0.87 nickel atom per HybF-Strep protein were determined. The Scatchard plot of the binding data for HybF [H2Q] (data not shown) indicates that binding to this protein variant is not saturatable, suggesting that there is a nonspecific interaction.
FIG. 7.
Nickel binding abilities of the HybF-Strep protein and its [H2Q] variant. (A) Ni2+ binding ability was determined by filter binding assays with 10 μM HybF-Strep (• and ○) or 10 μM HybF-Strep [H2Q] (▴ and ▵) and different concentrations of 63Ni as indicated on the x axis. ○ and ▵, 25 μM ZnCl2 was also present in the reaction mixture. (B) Scatchard plot for nickel binding by the wild-type HybF-Strep protein. The linear region indicates a KD value of 1.87 μM for nickel and a maximal binding of 0.87 nickel atom per HybF-Strep monomer.
To differentiate between specific binding and nonspecific binding of nickel, competition experiments with zinc as a competitor were performed. Addition of 25 μM ZnCl2 to the incubation mixture had no effect on the nickel binding ability of the HybF-Strep protein (Fig. 7A), whereas it drastically reduced the amount of nickel bound by the HybF-Strep [H2Q] variant (Fig. 7A).
DISCUSSION
The results of genetic and biochemical studies have indicated that nickel insertion into the metal center of NiFe hydrogenases depends on the activity of two proteins, HypA and HypB. HypB from E. coli and other organisms has been thoroughly studied; it is a guanine nucleotide binding protein, and GTP hydrolysis activity is essential for nickel incorporation in vivo (31, 32). Whereas the amino acid residues involved in GTP binding and hydrolysis are strictly conserved in all members of the HypB family, the proteins from several selected organisms also possess a polyhistidine motif close to the N terminus, which appears to have a function in nickel storage. This domain is absent from the HypB protein of E. coli. Whether HypB (apart from nickel storage in some species) also has a function in delivering the metal to the hydrogenase apoprotein has not been determined unequivocally yet. Removal of the polyhistidine stretch from the protein of B. japonicum eliminated the storage function but not the activity in the maturation process (21, 39). These variants still possessed a capacity for stoichiometric binding of nickel. On the other hand, recent studies on the nickel insertion systems involved in urease and hydrogenase formation of H. pylori redirected this function to another member of the maturation system, namely, HypA (33, 40).
HybF binds nickel.
The results for HybF of E. coli, a member of the HypA protein family, presented in this paper support the contention that HypA is a nickel chaperone involved in hydrogenase maturation. As determined by equilibrium dialysis experiments (data not shown) and by the filter binding assays described above, binding of nickel to wild-type HybF is stoichiometric, which correlates with the finding that HypA of H. pylori binds two nickel ions per homodimer. Moreover, the binding is not competed out by zinc, which support the hypothesis that there are separate binding sites. Finally, the H2Q variant of HybF contains the same amount of zinc as the wild-type protein contains, but it has a strongly reduced capacity to bind nickel. The residual activity could be chased out in the presence of zinc, indicating that there is nonspecific binding. Similarly, HypA [H2A] from H. pylori does not bind nickel in equilibrium dialysis experiments (33).
The conclusion based on the results reported previously for HypA from H. pylori (33) and the results presented above for HybF from E. coli is that the H2 residue of these proteins seems to be involved in the coordination of nickel. Replacement of other conserved residues of HybF did not yield any clear evidence that other amino acid side chains serve as ligands for nickel. An exception may be Glu-3 since replacement by Gln, which can provide a coordination site, did not affect activity, whereas replacement by Leu eliminated activity completely. The possibility that two monomers of HybF can interact to provide the binding site(s) cannot be eliminated, although our results show that HybF, in contrast to HypA from H. pylori (33), is a monomeric protein and that the presence of nickel does not stimulate dimerization (results not shown).
One finding that initially contradicted such a nickel binding function was the finding that the purified HybF protein contained zinc stoichiometrically but only insignificant levels of nickel when the cells were cultivated anaerobically, conditions which lead to expression of the high-affinity nickel transport system (19, 60) in a medium supplemented with nickel but not with zinc. The same results were obtained when HybF was overproduced under aerobic conditions (results not shown). An argument could be made that zinc occupies the nickel site in the purified protein, a situation reported for the UreE protein from Bacillus pasteurii (44). UreE from this organism contains one zinc atom per dimer, and this zinc could be replaced by nickel in crystals soaked with nickel chloride. The results presented in this paper, however, although not disproving this possibility, strongly argue in favor of the existence of separate binding sites for nickel and zinc.
HybF contains a zinc finger domain.
Genes coding for HypA-like proteins are present in all microorganisms that synthesize NiFe hydrogenases. Mutational analysis showed that HupA, the homolog in Azotobacter chroococcum, is also essential for maturation of hydrogenases. A Tn5 insertion in hupA could be partially compensated for by high nickel concentrations in the medium, leading to the first suggestion that the gene product is involved in nickel sequestering (18). Polar effects on the expression of downstream genes by the insertion could not be ruled out by Du and Tibelius, however.
The members of the HypA family are rich in cysteine, containing 3.5 to 8.6% cysteine, and the HypA of E. coli is on top of the list. The minimum number of residues is four, and these residues are strictly conserved at their positions in all HypA sequences. They are present in the motif CXXC-X12/13-CXXC, which is characteristic for the formation of a zinc finger domain (8, 16). The mutational screening analysis performed in this study supports the contention that HybF is a zinc finger protein. Single replacements of each of the Cys residues grossly affected the stability and solubility of the variants. The products were, however, still active in the maturation process. It is therefore plausible to assume that these sites are the coordination sites for zinc. This agrees with the conclusion of Vallee and Auld that zinc atoms fully coordinated by four ligands are structurally important, whereas catalytic site zinc is coordinated by three protein ligands (52). A structural, previously purely speculative role of the zinc finger domain could involve the interaction with other components of the maturation system or with the large hydrogenase subunit. In view of the higher propensity of zinc (compared to the propensity of nickel) to interact with thiolate groups, the results obtained in the competition experiments also suggest that the nickel is bound by nonthiolate ligands.
Possible function of HybF.
Nickel can be inserted into hydrogenases chemically both in vivo by supplying hypA, hybF, or hypB mutants with an excess of nickel in the medium (24, 40, 58) and in in vitro maturation assay mixtures challenged with nickel (30, 34). Attempts to demonstrate any activity of each of these gene products under in vitro conditions have failed so far. This may have been due to inactivation during purification, to the lack of a required metabolite, or to the lack of other components of the insertion machinery with which these proteins must interact. It is therefore premature to propose a detailed model for the function of HybF, HypA, and HypB in the maturation process; rather, a working model is presented here. It is plausible to assume that the nickel sequestered by HybF (or HypA) is donated to the apoprotein of the large hydrogenase subunit either directly or via some still unknown macromolecular component. In the former instance it is further plausible to assume that specific recognition of the cognate large subunit must take place, which necessitates a direct interaction between the nickel donor and the target protein. This interaction (docking or release) may be controlled by HypB, which acts as a molecular switch in accordance with the general function of GTPases. Arguments supporting this notion are that the hydrolysis of GTP is required for nickel insertion (31, 32) and that for the H. pylori proteins an interaction between HypA and HypB has been demonstrated by chemical cross-linking (33). Our own attempts to cross-link HybF and HypB from E. coli failed, which may have been due to the fact that E. coli HybF has only 4 Lys residues, compared to the 10 Lys residues in the H. pylori protein. Other reasons could involve the possibility that the interaction requires binding to the precursor of the large subunit.
Since nickel can be inserted chemically into the target proteins, why has such a complex system developed? The answer, of course, requires in vitro demonstration of the activities of these proteins, but it may lie in shielding of the metal from the complexing activity of thiols in the cytoplasmic space. High thiol concentrations can completely bind the trace element and make it unavailable for the insertion process (Fig. 6). Other explanations are that the system guarantees that the correct metal is inserted and that the kinetics of insertion are improved compared to those of the pure chemical process.
Acknowledgments
We thank F. Lottspeich and J. Kellermann for determination of the amino-terminal sequence of HybF-Strep and H. Hartl and P. Klüfers for the ICP measurements.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Andrews, S. C., B. C. Berks, J. McClay, A. Ambler, M. A. Quail, P. Golby, and J. R. Guest. 1997. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogen lyase system. Microbiology 143:3633-3647. [DOI] [PubMed] [Google Scholar]
- 2.Bagley, K. A., E. C. Duin, W. Roseboom, S. P. J. Albracht, and W. H. Woodruff. 1995. Infrared-detectable groups sense changes in charge density on the nickel center in hydrogenase from Chromatium vinosum. Biochemistry 34:5527-5535. [DOI] [PubMed] [Google Scholar]
- 3.Bagramyan, K., N. Mnatsakanyan, A. Poladian, A. Vassilian, and A. Trchounian. 2002. The roles of hydrogenases 3 and 4, and the F0F1-ATPase, in H2 production by Escherichia coli at alkaline and acidic pH. FEBS Lett. 516:172-178. [DOI] [PubMed] [Google Scholar]
- 4.Bagramyan, K., A. Vassilian, N. Mnatsakanyan, and A. Trchounian. 2001. Participation of hyf-encoded hydrogenase 4 in molecular hydrogen release coupled with proton-potassium exchange in Escherichia coli. Membr. Cell Biol. 14:749-763. [PubMed] [Google Scholar]
- 5.Ballantine, S. P., and D. H. Boxer. 1986. Isolation and characterisation of a soluble active fragment of hydrogenase isoenzyme 2 from the membranes of anaerobically grown Escherichia coli. Eur. J. Biochem. 156:277-284. [DOI] [PubMed] [Google Scholar]
- 6.Ballantine, S. P., and D. H. Boxer. 1985. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J. Bacteriol. 163:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Begg, Y. A., J. N. Whyte, and B. A. Haddock. 1977. The identification of mutants of Escherichia coli deficient in formate dehydrogenase and nitrate reductase activities using dye indicator plates. FEMS Microbiol. Lett. 2:47-50. [Google Scholar]
- 8.Berg, J. M. 1990. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J. Biol. Chem. 265:6513-6516. [PubMed] [Google Scholar]
- 9.Blokesch, M., and A. Böck. 2002. Maturation of [NiFe]-hydrogenases in Escherichia coli: the HypC cycle. J. Mol. Biol. 324:287-296. [DOI] [PubMed] [Google Scholar]
- 10.Blokesch, M., A. Magalon, and A. Böck. 2001. Interplay between the specific chaperone-like proteins HybG and HypC in maturation of hydrogenases 1, 2, and 3 from Escherichia coli. J. Bacteriol. 183:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Böck, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C.
- 12.Böhm, R., M. Sauter, and A. Böck. 1990. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol. Microbiol. 4:231-243. [DOI] [PubMed] [Google Scholar]
- 13.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 14.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colbeau, A., P. Richaud, B. Toussaint, F. J. Caballero, C. Elster, C. Delphin, R. L. Smith, J. Chabert, and P. M. Vignais. 1993. Organization of the genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis and identification of two hyp regulatory mutants. Mol. Microbiol. 8:15-29. [DOI] [PubMed] [Google Scholar]
- 17.Drapal, N., and A. Böck. 1998. Interaction of the hydrogenase accessory protein HypC with HycE, the large subunit of Escherichia coli hydrogenase 3 during enzyme maturation. Biochemistry 37:2941-2948. [DOI] [PubMed] [Google Scholar]
- 18.Du, L., and K. H. Tibelius. 1994. The hupB gene of Azotobacter chroococcum hydrogenase gene cluster is involved in nickel metabolism. Curr. Microbiol. 28:21-24. [Google Scholar]
- 19.Eitinger, T., and M. A. Mandrand-Berthelot. 2000. Nickel transport systems in microorganisms. Arch. Microbiol. 173:1-9. [DOI] [PubMed] [Google Scholar]
- 20.Frey, M., J. C. Fontecilla-Camps, and A. Volbeda. 2001. Nickel-iron hydrogenases, p. 880-896. In A. Messerschmidt, R. Huber, T. Poulos, and K. Wieghardt (ed.), Handbook of metalloproteins, vol. 2. John Wiley & Sons, Ltd., Chichester, United Kingdom. [Google Scholar]
- 21.Fu, C., J. W. Olson, and R. J. Maier. 1995. HypB protein of Bradyrhizobium japonicum is a metal-binding GTPase capable of binding 18 divalent nickel ions per dimer. Proc. Natl. Acad. Sci. 92:2333-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Happe, R. P., W. Roseboom, A. J. Pierik, S. P. Albracht, and K. A. Bagley. 1997. Biological activation of hydrogen. Nature 385:126. [DOI] [PubMed] [Google Scholar]
- 24.Hube, M., M. Blokesch, and A. Böck. 2002. Network of hydrogenase maturation in Escherichia coli: role of accessory proteins HypA and HybF. J. Bacteriol. 184:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobi, A., R. Rossmann, and A. Böck. 1992. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch. Microbiol. 158:444-451. [DOI] [PubMed] [Google Scholar]
- 26.Kerby, R. L., P. W. Ludden, and G. P. Roberts. 1997. In vivo nickel insertion into the carbon monoxide dehydrogenase of Rhodospirillum rubrum: molecular and physiological characterization of cooCTJ. J. Bacteriol. 179:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lutz, S., A. Jacobi, V. Schlensog, R. Böhm, G. Sawers, and A. Böck. 1991. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol. Microbiol. 5:123-135. [DOI] [PubMed] [Google Scholar]
- 29.Magalon, A., and A. Böck. 2000. Analysis of the HypC-HycE complex, a key intermediate in the assembly of the metal center of the Escherichia coli hydrogenase 3. J. Biol. Chem. 275:21114-21220. [DOI] [PubMed] [Google Scholar]
- 30.Maier, T., and A. Böck. 1996. Generation of active [NiFe] hydrogenase in vitro from a nickel-free precursor form. Biochemistry 35:10089-10093. [DOI] [PubMed] [Google Scholar]
- 31.Maier, T., A. Jacobi, M. Sauter, and A. Böck. 1993. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J. Bacteriol. 175:630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maier, T., F. Lottspeich, and A. Bock. 1995. GTP hydrolysis by HypB is essential for nickel insertion into hydrogenases of Escherichia coli. Eur. J. Biochem. 230:133-138. [PubMed] [Google Scholar]
- 33.Mehta, N., J. W. Olson, and R. J. Maier. 2003. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J. Bacteriol. 185:726-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menon, A. L., and R. L. Robson. 1994. In vivo and in vitro nickel-dependent processing of the [NiFe] hydrogenase in Azotobacter vinelandii. J. Bacteriol. 176:291-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menon, N. K., C. Y. Chatelus, M. Dervartanian, J. C. Wendt, K. T. Shanmugam, H. D. Peck, Jr., and A. E. Przybyla. 1994. Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J. Bacteriol. 176:4416-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menon, N. K., J. Robbins, H. D. Peck, Jr., C. Y. Chatelus, E. S. Choi, and A. E. Przybyla. 1990. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J. Bacteriol. 172:1969-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olson, J. W., and R. J. Maier. 2000. Dual roles of Bradyrhizobium japonicum nickelin protein in nickel storage and GTP-dependent Ni mobilization. J. Bacteriol. 182:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olson, J. W., N. S. Mehta, and R. J. Maier. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176-182. [DOI] [PubMed] [Google Scholar]
- 41.Paschos, A., A. Bauer, A. Zimmermann, E. Zehelein, and A. Bock. 2002. HypF, a carbamoyl phosphate-converting enzyme involved in [NiFe] hydrogenase maturation. J. Biol. Chem. 277:49945-49951. [DOI] [PubMed] [Google Scholar]
- 42.Peck, H. D., and H. Gest. 1957. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J. Bacteriol. 73:706-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reissmann, S., E. Hochleitner, H. Wang, A. Paschos, F. Lottspeich, R. S. Glass, and A. Böck. 2003. Taming of a poison: biosynthesis of the NiFe-hydrogenase cyanide ligands. Science 299:1067-1070. [DOI] [PubMed] [Google Scholar]
- 44.Remaut, H., N. Safarov, S. Ciurli, and J. Van Beeumen. 2001. Structural basis for Ni2+ transport and assembly of the urease active site by the metallochaperone UreE from Bacillus pasteurii. J. Biol. Chem. 276:49365-49370. [DOI] [PubMed] [Google Scholar]
- 45.Rey, L., J. Imperial, J. M. Palacios, and T. Ruiz-Argueso. 1994. Purification of Rhizobium leguminosarum HypB, a nickel-binding protein required for hydrogenase synthesis. J. Bacteriol. 176:6066-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawers, G. 1994. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Leeuwenhoek 66:57-88. [DOI] [PubMed] [Google Scholar]
- 47.Sawers, R. G., S. P. Ballantine, and D. H. Boxer. 1985. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: evidence for a third isoenzyme. J. Bacteriol. 164:1324-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawers, R. G., and D. H. Boxer. 1986. Purification and properties of membrane-bound hydrogenase isoenzyme 1 from anaerobically grown Escherichia coli K-12. Eur. J. Biochem. 156:265-275. [DOI] [PubMed] [Google Scholar]
- 49.Skibinski, D. A., P. Golby, Y. S. Chang, F. Sargent, R. Hoffman, R. Harper, J. R. Guest, M. M. Attwood, B. C. Berks, and S. C. Andrews. 2002. Regulation of the hydrogenase-4 operon of Escherichia coli by the sigma54-dependent transcriptional activators FhlA and HyfR. J. Bacteriol. 184:6642-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 51.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallee, B. L., and D. S. Auld. 1992. Active zinc binding sites of zinc metalloenzymes. Matrix Suppl. 1:5-19. [PubMed] [Google Scholar]
- 53.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 54.Volbeda, A., M. H. Charon, C. Piras, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
- 55.Volbeda, A., E. Garcin, C. Piras, A. L. de Lacey, V. M. Fernandez, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1996. Structure of the [NiFe] hydrogenase active site: evidence for biologically uncommon Fe ligands. J. Am. Chem. Soc. 118:12989-12996. [Google Scholar]
- 56.Walleczek, J., T. Martin, B. Redl, M. Stoffler-Meilicke, and G. Stoffler. 1989. Comparative cross-linking study on the 50S ribosomal subunit from Escherichia coli. Biochemistry 28:4099-4105. [DOI] [PubMed] [Google Scholar]
- 57.Wang, D., and S. Moore. 1977. Polyspermine-ribonuclease prepared by cross-linkage with dimethyl suberimidate. Biochemistry 16:2937-2942. [DOI] [PubMed] [Google Scholar]
- 58.Waugh, R., and D. H. Boxer. 1986. Pleiotropic hydrogenase mutants of Escherichia coli K12: growth in the presence of nickel can restore hydrogenase activity. Biochimie 68:157-166. [DOI] [PubMed] [Google Scholar]
- 59.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, L. F., and M. A. Mandrand-Berthelot. 1986. Genetic and physiological characterization of new Escherichia coli mutants impaired in hydrogenase activity. Biochimie 68:167-179. [DOI] [PubMed] [Google Scholar]