Abstract
The purpose of this study was to determine the effect of enrofloxacin in the carrier stage of Haemophilus parasuis in naturally colonized weaned pigs. Twenty-three pigs colonized by H. parasuis received either 7.5 mg/kg body weight (BW) of enrofloxacin or a saline solution intramuscularly at weaning. Nasal and tonsillar swab samples were collected daily throughout the study and at necropsy and tested by quantitative polymerase chain reaction (qPCR). The H. parasuis isolates obtained from samples collected at necropsy were subjected to genotyping by enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) and a multiplex PCR for the detection of the virulence-associated trimeric autotransporter (vtaA) genes. Haemophilus parasuis was detected in the nasal cavity and tonsils of pigs in the control group throughout the study. Antibiotic-treated pigs tested negative for H. parasuis at 1 d post-treatment and the proportion of nasal samples that tested positive was higher for control pigs than for treated pigs at 1, 2, 3, 4, 5, 6, and 7 d post-treatment and at 2, 4, and 5 d post-treatment for tonsil samples (P < 0.003). Genotyping by ERIC-PCR demonstrated that pigs were colonized with a common H. parasuis strain at the end of the study. Isolates were negative for the vtaA gene, which indicates the absence of vtaA virulence factor. In conclusion, enrofloxacin significantly reduced the H. parasuis load in naturally colonized pigs, but was unable to completely eliminate the organism.
Résumé
Cette étude avait comme objectif de déterminer l’effet de l’enrofloxacin dans le portage d’Haemophilus parasuis chez des porcs sevrés colonisés naturellement. Vingt-trois porcs colonisés par H. parasuis ont reçu au moment du sevrage par voie intramusculaire soit de l’enrofloxacin à un dosage de 7,5 mg/kg de poids vif (BW) ou une solution saline. Des écouvillons nasaux ou des amygdales ont été prélevés quotidiennement durant l’étude et à la nécropsie et testés par réaction d’amplification en chaîne par la polymérase quantitative (qPCR). Les isolats d’H. parasuis obtenus des échantillons prélevés lors de la nécropsie ont été soumis à une analyse génotypique par PCR des séquences intergéniques consensus répétitives des entérobactéries (ERIC-PCR) ainsi qu’à une épreuve PCR multiplex pour la détection des gènes auto-transporteurs trimériques associés à la virulence (vtaA). La présence d’H. parasuis fut détectée dans la cavité nasale et les amygdales des porcs du groupe témoin tout au long de l’étude. Les porcs traités aux antibiotiques étaient négatifs pour la présence d’H. parasuis au jour 1 post-traitement et la proportion d’échantillons nasaux qui ont testé positifs était plus élevée pour les porcs témoins que pour les porcs traités aux jours 1, 2, 3, 4, 5, 6 et 7 post-traitement et aux jours 2, 4 et 5 post-traitement pour les échantillons d’amygdales (P < 0,003). Le génotypage par ERIC-PCR a permis de montrer qu’à la fin de l’étude les porcs étaient colonisés par une souche commune d’H. parasuis. Les isolats étaient négatifs pour la présence du gène vtaA, ce qui indique l’absence du facteur de virulence vtaA. En conclusion, l’enrofloxacin a diminué significativement la charge d’H. parasuis chez des porcs colonisés naturellement, mais a été incapable d’éliminer complètement le microorganisme.
(Traduit par Docteur Serge Messier)
Introduction
Haemophilus parasuis is an economically significant Gram-negative organism that colonizes the upper respiratory tract of pigs soon after birth (1,2). The presence of humoral immunity generally prevents pigs from developing systemic disease (3,4), which is commonly characterized by fibrinous polyserositis, arthritis, and meningitis (5). Stress conditions coinciding with decay of maternal immunity, such as weaning and transport (6), and coinfections with immuno-suppressive agents, such as porcine reproductive and respiratory syndrome (PRRS) virus (7), have been suggested as risk factors for systemic invasion of H. parasuis.
Alterations in the carrier stage of H. parasuis at a young age have also been associated with the development of Glasser’s disease during the post-weaning period (8). Most of the studies have focused on the effect of early weaning in the disruption of the colonization patterns under the presence of maternal immunity. In these studies (2,4,9), H. parasuis disease was exacerbated when pigs were colonized late and maternal immunity was waning. There is limited information, however, on what other factors may alter the carrier stage of H. parasuis.
Antibiotics are commonly used to mitigate the effects of bacterial disease in pigs in order to limit bacterial challenge. A recent study has reported that marbofloxacin treatment was able to reduce the nasal carriage of H. parasuis in weaned pigs (10). Another fluoroquinolone, enrofloxacin, is a common antimicrobial used to treat Glasser’s disease on farms in North America. Enrofloxacin is among the products approved by the United States Food and Drug Administration Center of Veterinary Medicine for treating and controlling disease associated with H. parasuis. There is no information on the specific effect that enrofloxacin may have on the carrier state of H. parasuis in naturally colonized pigs and whether the carrier state is affected at all. The purpose of this study was therefore to evaluate the effect of enrofloxacin in reducing H. parasuis colonization in weaned pigs.
Materials and methods
Animals and animal housing
Forty-five, 1-week-old pigs with a history of Glasser’s disease, porcine reproductive and respiratory syndrome (PRRS) virus, porcine cirovirus type 2 (PCV2), and Mycoplasma hyopneumoniae were identified on a conventional North American farm and screened for the presence of H. parasuis in the upper respiratory tract using 16S ribosomal ribonucleic acid (rRNA) gene polymerase chain reaction (PCR) (11). The pigs received PCV2 vaccine at 4 d of age and at weaning and M. hyopneumoniae vaccine at weaning. Of those 45 pigs, twenty-four 3-week-old weaned pigs that tested positive for H. parasuis were selected and moved to the University of Minnesota research isolation facility. Pigs were randomly divided into treatment (n = 12 pigs) and control groups (n = 12 pigs) and housed in 2 separated isolation rooms (1 pig in the control group died shortly after arrival due to an unrelated H. parasuis cause). The pigs were fed ad libitum and had free access to water. Pigs were cared for according to the guidelines of the Institutional Animal Care and Use Committee of the University of Minnesota.
Experimental design
On the day of arrival at the research facility, blood samples and nasal and tonsil swabs were collected from all pigs and tested by 16S rRNA gene PCR (11) and qPCR as described in the next section. Pigs in the treatment group were treated with a single dose of injectable enrofloxacin (7.5 mg/kg BW Baytril; Bayer Animal Health, Shawnee, Kansas, USA) at 24 h post-arrival and those in the control group received saline solution intramuscularly. Tonsillar and nasal swabs were collected daily. At 3, 7, and 14 d post-treatment, 4 pigs from each group were randomly chosen and euthanized. At necropsy, blood samples and swabs from the nasal cavity, interior of the tonsil, trachea, lung, and peritoneal and pleural serosa were collected in duplicate. One swab from each organ was used for bacterial isolation and the other swab was tested by quantitative PCR (qPCR) as outlined in the next section. Haemophilus parasuis isolates were further characterized by ERIC-PCR (12) and virulence-associated trimeric autotransporter polymerase chain reaction (vtaA-PCR) (13).
Quantitative PCR
Deoxyribonucleic acid (DNA) from swabs was extracted using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) and then tested individually by qPCR (14), with modifications. Briefly, primers forward (5′-CGACTTACTTGAAGCCATTCTTCTT-3′) and reverse (5′-CCGCTTGCCATACCCTCTT-3′) were based on the infB gene of H. parasuis, which is also considered to be a genetic marker for phylogenetic studies of species, separating H. parasuis from other closely related species. The FAM-labelled TaqMan probe with a TAMRA quencher (5′-6FAM-ATCGGAAGTATTAGAATTAAGTGC-TAMRA-3′) was supplied by Applied Biosystems (Carlsbad, California, USA). The 25-μL reaction mix consisted of 7 μL of H2O, 10 μL of master mix, 100 nmol/L of CTinfF1, 400 nmol/L of reverse primer, 100 nmol/L of forward primer, and 5 μL of template. A standard curve was established by a 10-fold serial dilution of known quantities of the H. parasuis reference strain Nagasaki extracted DNA. The 7500 Fast System SDS software (Applied Biosystems) was used to calculate the quantity of unknown target sequences from the standard curve for the detector of that target. The reaction was run with the following cycling conditions: first cycle at 95°C for 2 min, followed by 30 cycles of 95°C for 20 s and 58°C for 60 s, followed by 1 cycle at 28°C for 1 min. All reactions, including the standard curve, were run in duplicate. Results are shown in CFU/reaction.
Haemophilus parasuis isolation and characterization
For isolation of H. parasuis, samples were plated onto sheep blood agar (BA) streaked with a nurse Staphylococcus sp. strain, cultured, and incubated at 37°C in a 5% carbon dioxide (CO2) atmosphere for 24 to 48 h. Up to 5 H. parasuis-like colonies per agar plate were selected for further identification (15). Enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) (12) was used to better characterize H. parasuis isolates.
A multiplex PCR based on vtaA genes (13) was used to differentiate the non-virulent from potentially virulent H. parasuis isolates. Haemophilus parasuis possesses virulence-associated trimeric auto-transporters (vtaA): group 1 vtaA has been associated with virulent strains, while group 3 vtaA gene is highly conserved among H. parasuis strains (16).
Enzyme-linked immunosorbent assay (ELISA)
Blood samples were tested for H. parasuis antibodies using an enzyme-linked immunosorbent assay (ELISA) based on the H. parasuis species-specific protein OppA (17), using the Swine SK104 Haemophilus parasuis (OppA) Antibody Test Kit (BioChek, Scarborough, Maine, USA). Samples testing sample to positive ratio (S/P) > 0.5 were considered positive.
Statistical analysis
Differences between the proportions of H. parasuis-positive pigs in treated versus control groups at each sampling time point were calculated using Fisher’s exact probability test. The Bonferroni correction was used to address multiple comparisons (α = 0.003).
Results
Polymerase chain reaction (PCR) targeting the 16S rRNA was used to screen naturally colonized pigs at the farm of origin and to confirm their H. parasuis colonization status when the pigs arrived at the research facility. Tonsil and nasal swabs from all pigs tested positive for H. parasuis by 16 rRNA PCR on the day of arrival at the research facilities. No clinical signs of H. parasuis disease (fever, anorexia, lameness, thumping) were observed during the experiment.
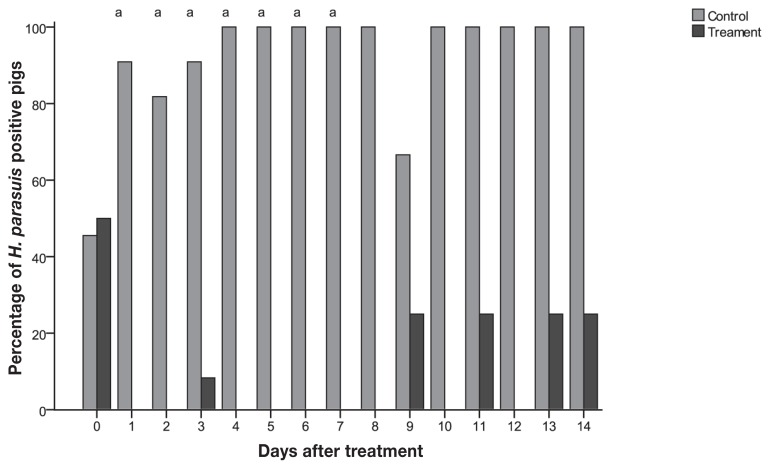
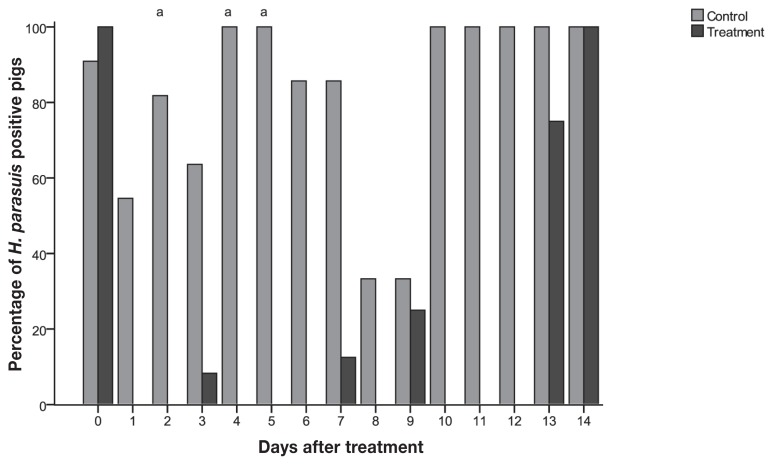
Quantitative PCR was used to detect and quantify H. parasuis in the tonsil and nasal cavity during the study period. Before treatment, 22 out of 23 pigs (95.7%) tested positive by qPCR from tonsil swabs, while only 11 pigs (48%) tested positive from nasal swabs (Figures 1 and 2). Among the pigs that tested positive before treatment, the average H. parasuis load detected in tonsils and nasal cavities was 2.8 × 105 and 2.3 × 104 colony-forming units (CFUs)/reaction, respectively. Pigs in the control group tested positive throughout most of the study, while all treated pigs tested negative for H. parasuis by qPCR at 1 d post-treatment (Figures 1 and 2, Table I). The proportion of nasal samples that tested positive was higher for control pigs than for treated pigs at 1, 2, 3, 4, 5, 6, and 7 d post-infection and at 2, 4, and 5 d post-treatment for tonsil samples (P < 0.003) (Figures 1 and 2).
Figure 1.
Percentage of pigs tested positive by nasal swabs qPCR from control (n = 11) and treatment (n = 12) groups by day.
a Differences between treatment and control (P < 0.003).
Figure 2.
Percentage of pigs tested positive pigs by tonsil swabs qPCR from control (n = 11) and treatment (n = 12) groups by day.
a Differences between treatment and control (P < 0.003).
Table I.
Daily average colony-forming units (CFUs)/reaction and standard deviation (SD) from pigs in treated and control groups
| Days after treatment | Average CFU/reaction ± (SD) | |||
|---|---|---|---|---|
|
| ||||
| Tonsil | Nasal | |||
|
|
|
|||
| Control | Treatment | Control | Treatment | |
| 0 | 1.8 × 105 (1.5 × 105)a | 3.5 × 105 (4.3 × 105) | 1.0 × 104 (1.9 × 104) | 1.1 × 104 (2.7 × 104) |
| 1 | 4.0 × 104 (8.0 × 104) | 0c | 6.4 × 103 (8.2 × 103) | 0 |
| 2 | 2.0 × 105 (3.8 × 105) | 0 | 1.2 × 104 (1.1 × 104) | 0 |
| 3 | 1.5 × 105 (3.3 × 105) | 6.8 × 105b | 6.4 × 104 (1.5 × 105) | 0 |
| 4 | 2.3 × 103 (1.7 × 103) | 0 | 3.8 × 104 (2.2 × 104) | 0 |
| 5 | 4.6 × 103 (2.8 × 103) | 0 | 4.0 × 104 (3.3 × 104) | 0 |
| 6 | 8.0 × 103 (1.3 × 104) | 0 | 7.6 × 103 (1.0 × 104) | 0 |
| 7 | 1.8 × 103 (1.8 × 103) | 3.9 × 104 | 1.1 × 104 (1.6 × 104) | 0 |
| 8 | 1.8 × 103b | 0 | 1.2 × 105 (9.3 × 103) | 0 |
| 9 | 7.0 × 103 | 3.0 × 105 | 5.1 × 104 (4.2 × 104) | 0 |
| 10 | 2.7 × 104 (2.7 × 104) | 0 | 2.0 × 105 (1.2 × 105) | 0 |
| 11 | 4.6 × 104 (3.5 × 104) | 0 | 2.3 × 105 (8.8 × 104) | 1.8 × 104 |
| 12 | 1.9 × 103 (2.5 × 103) | 0 | 2.9 × 105 (2.7 × 105) | 0 |
| 13 | 3.7 × 103 (3.2 × 103) | 2.6 × 103 (3.1 × 103) | 4.0 × 105 (1.5 × 105) | 3.7 × 104 |
| 14 | 2.2 × 103 (1.9 × 103) | 2.1 × 103 (1.0 × 103) | 2.6 × 105 (3.5 × 104) | 9.3 × 104 |
Average ± standard deviation.
Only one pig positive in the group.
No positive pigs in the group.
At necropsy, no gross lesions were observed and H. parasuis was detected by qPCR in 9 out of 11 control pigs in at least 1 of the 5 samples tested (Table II). In contrast, only 4 out of 12 pigs in the treatment group tested positive. Interestingly, all 4 H. parasuis isolates obtained were recovered from samples collected at necropsy at 15 d post-infection, but not in the necropsies before that. Enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) profiles were obtained from the 4 isolates. One single cluster could be distinguished for the 4 isolates based on coverage of 90% agreement (18).
Table II.
Number of pigs detected positive for H. parasuis positive by qPCR and bacterial culture at necropsy
| Control | Treatment | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Groups | 4 | 8 | 14 | 4 | 8 | 14 |
| Day post-treatment | (n = 4) | (n = 4) | (n = 3) | (n = 4) | (n = 4) | (n = 4) |
| Nasal | 2a (0b) | 4 (0) | 3 (2) | 0 (0) | 0 (0) | 1 (1) |
| Tonsil | 2 (0) | 3 (0) | 2 (0) | 0 (0) | 0 (0) | 3 (0) |
| Trachea | 1 (0) | 1 (0) | 3 (0) | 0 (0) | 0 (0) | 1 (0) |
| Lung | 1 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 0 (0) |
| Serosae | 0 (0) | 0 (0) | 0 (1) | 0 (0) | 0 (0) | 0 (0) |
qPCR.
Bacterial culture.
The putative virulence factor, the group 1 vtaA gene, was not detected in any of the 4 isolates obtained, while the group 3 vtaA, highly conserved among H. parasuis strains, was detected in all isolates.
All anti-OppA serum antibody levels, as measured by ELISA S/P on arrival (mean S/P = 0.01) and at necropsy (mean S/P control pigs 0.001 versus mean S/P treated pigs 0.03) were very low and under the cutoff value of 0.5. All values were considered negative.
Discussion
In this study, we demonstrated that enrofloxacin reduced the number of H. parasuis in the tonsil and nasal cavity of pigs and decreased the number of pigs that were positive for H. parasuis by qPCR during the first week after treatment.
The presence of H. parasuis was demonstrated by gel-based PCR in samples from tonsils and nasal cavity in all pigs before treatment, while qPCR detected 95.7% positives on tonsils but only 48% positives in nasal cavity. Differences in results might be due to the lower sensitivity of the qPCR (4 × 103 CFU/mL) compared with the gel-based PCR (1 × 102 CFU/mL) (11). However, qPCR was preferred in order to quantify the effect of the antibiotic on bacterial load. The fact that the pigs tested negative after treatment and that some pigs then became positive again suggests that the treatment decreased the level of H. parasuis below the threshold of qPCR detection, but did not completely eliminate H. parasuis.
These results are in agreement with a study investigating the effect of marbofloxacin in the H. parasuis carrier state in pigs at the farm level (10). Both marbofloxacin as well as enrofloxacin in the present study showed a similar effectiveness after treatment, reducing the amount of H. parasuis in the nasal cavity of pigs, but not completely eliminating it.
The rapid and relatively long effect of enrofloxacin on H. parasuis in the nasal cavity and tonsil of pigs can be attributed to enrofloxacin’s tissue penetration, extended half-life, and rapid bactericidal effect in a concentration-dependent manner and prolonged post-antibiotic effect (19). Possible explanations for the antibiotic’s inability to fully eliminate H. parasuis are that the dose of antibiotic may not have reached an adequate concentration at the colonization sites or that enrofloxacin induced the emergence of resistant strains. Even though the concentration of enrofloxacin in the nasal cavity or tonsil was not assessed in this study, the concentration of enrofloxacin in nasal secretions of pigs is considered nearly equal to plasma concentrations, which suggests that it was not a dose-effect issue (20). Additionally, at necropsy, even when cutting the tonsils and collecting swabs from inside the tonsils, pigs treated with enrofloxacin tested negative for H. parasuis at 4 and 8 d post-infection, which demonstrates that bacteria was not concentrated deep in the tonsils soon after treatment. Furthermore, antibiotic resistance was not likely since 3 of the 4 H. parasuis isolates recovered at necropsy were tested for antimicrobial susceptibility (data not shown) and all 3 isolates were susceptible to enrofloxacin. The fourth isolate would not grow in the test medium.
All pigs remained clinically healthy throughout the study and, as expected, most of the control pigs tested positive in samples from the upper respiratory tract. Furthermore, at necropsy, 5 pigs tested positive in the trachea and 2 pigs tested positive in the lung, which confirms that H. parasuis can be detected in the trachea and lung of healthy pigs, as reported elsewhere (21). In contrast, swabs collected at necropsy from pigs treated with antibiotics only tested positive when collected on day 15 of the study.
Pigs in both groups remained serologically negative. The OppA ELISA kit was used to monitor H. parasuis systemic infection during the study, since it measures the amount of antibodies to the H. parasuis species-specific antigen OppA in pigs systemically infected by all H. parasuis serotypes or vaccinated with H. parasuis. Healthy (colonized) pigs are therefore not expected to have anti-OppA antibodies. While outside the scope of this study, further investigation is required to determine whether H. parasuis strains colonizing the upper respiratory tract are able to trigger an immune response.
Characterization of all H. parasuis isolates recovered at necropsy indicated that both groups were colonized with the same H. parasuis strain. In addition, the isolates were negative by the vtaA group 1 PCR, which indicates that these isolates did not have the vtaA virulence factor. This result was expected since H. parasuis is a commensal organism of the upper respiratory tract and most of the isolates recovered from the nose are considered non-pathogenic (22). One isolate that was vtaA negative was obtained from systemic sites (serosas) in a clinically healthy control pig, however, which requires further investigation.
In conclusion, the results of this study indicate that enrofloxacin can reduce the levels of H. parasuis in naturally colonized pigs. Reduction of H. parasuis in the upper respiratory tract of pigs may help to control the disease during susceptible stages such as the weaning period, possibly delaying the infection to a point when the pigs are able to develop their own active immunity against this organism. Further research is needed, however, to evaluate the lasting effect of enrofloxacin in the colonization patterns and disease dynamics of H. parasuis.
Acknowledgments
This study was funded by Bayer Animal Health. The authors thank Dr. Barry Kelly for his input and discussions.
Footnotes
Conflict of interest statement
MT had served in an advisory role for Bayer Animal Health and AH is a company employee. The results from this study have not been influenced because of that. NM, SO, and AR declared no conflicts of interest.
References
- 1.Smart NL, Miniats P, Maclnnes JI. Analysis of Haemophilus parasuis isolates from southern Ontario swine by restriction endonuclease fingerprinting. Can J Vet Res. 1988;52:319–324. [PMC free article] [PubMed] [Google Scholar]
- 2.Oliveira S, Batista L, Torremorell M, Pijoam C. Experimental colonization of piglets and gilts with systemic strains of Haemophilus parasuis and Streptococcus suis to prevent disease. Can J Vet Res. 2001;65:161–167. [PMC free article] [PubMed] [Google Scholar]
- 3.Solano-Aguilar GI, Pijoan C, Rapp-Gabrielson V, Collins J, Carvalho LF, Winkelman N. Protective role of maternal antibodies against Haemophilus parasuis infection. Am J Vet Res. 1999;60:81–87. [PubMed] [Google Scholar]
- 4.Cerdà-Cuéllar M, Naranjo JF, Verge A, et al. Sow vaccination modulates the colonization of piglets by Haemophilus parasuis. Vet Microbiol. 2010;145:315–320. doi: 10.1016/j.vetmic.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Amano H, Shibata M, Kajio N, Morozumi T. Pathologic observations of pigs intranasally inoculated with serovar 1, 4, and 5 of Haemophilus parasuis using immunoperoxidase method. J Vet Med Sci. 1994;56:639–644. doi: 10.1292/jvms.56.639. [DOI] [PubMed] [Google Scholar]
- 6.Kirkwood RN, Rawluk SA, Cegielski AC, Otto AJ. Effect of pig age and autogenous sow vaccination on nasal mucosal colonization of pigs by Haemophilus parasuis. J Swine Health Prod. 2001;9:77–79. [Google Scholar]
- 7.Solano GI, Bautista E, Molitor TW, Segales J, Pijoan C. Effect of porcine reproductive and respiratory syndrome virus infection on the clearance of Haemophilus parasuis by porcine alveolar macrophages. Can J Vet Res. 1998;62:251–256. [PMC free article] [PubMed] [Google Scholar]
- 8.Pijoan C, Torremorell M, Solano G. Colonization patterns by the bacterial flora of young pigs. Am Assoc Swine Pract. 1997:463–464. [Google Scholar]
- 9.Blanco I, Galina-Pantoja L, Oliveira S, Pijoan C, Sánchez C, Canals A. Comparison between Haemophilus parasuis infection in colostrum-deprived and sow-reared piglets. Vet Microbiol. 2004;103:21–27. doi: 10.1016/j.vetmic.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Vilalta C, Galofre N, Aragon V, Rozas AMP, Fraile L. Effect of marbofloxacin on Haemophilus parasuis nasal carriage. Vet Microbiol. 2012;159:123–129. doi: 10.1016/j.vetmic.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 11.Oliveira S, Galina L, Pijoan C. Development of a PCR test to diagnose Haemophilus parasuis infections. J Vet Diagn Invest. 2001;13:495–501. doi: 10.1177/104063870101300607. [DOI] [PubMed] [Google Scholar]
- 12.Rafiee M, Bara M, Stephens CP, Blackall PJ. Application of ERIC-PCR for the comparison of isolates of Haemophilus parasuis. Aust Vet J. 2000;78:846–849. doi: 10.1111/j.1751-0813.2000.tb10507.x. [DOI] [PubMed] [Google Scholar]
- 13.Olvera A, Pina S, Macedo N, Oliveira S, Aragon V, Bensaid A. Identification of potentially virulent strains of Haemophilus parasuis using a multiplex PCR for virulence-associated auto-transporters (vtaA) Vet J. 2012;191:213–218. doi: 10.1016/j.tvjl.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Turni C, Pyke M, Blackall PJ. Validation of a real-time PCR for Haemophilus parasuis. J Appl Microbiol. 2010;108:1323–1331. doi: 10.1111/j.1365-2672.2009.04526.x. [DOI] [PubMed] [Google Scholar]
- 15.Rapp VJ, Ross RF, Young TF. Characterization of Haemophilus spp. isolated from healthy swine and evaluation of cross-reactivity of complement-fixing antibodies to Haemophilus pleuropneumoniae and Haemophilus taxon “minor group”. J Clin Microbiol. 1985;22:945–950. doi: 10.1128/jcm.22.6.945-950.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pina S, Olvera A, Barceló A, Bensaid A. Trimeric autotransporters of Haemophilus parasuis: Generation of an extensive passenger domain repertoire specific for pathogenic strains. J Bacteriol. 2009;191:576–587. doi: 10.1128/JB.00703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macedo N, Oliveira S, Van Esch E, Rush P. Novel Haemophilus parasuis ELISA test: Tracking systemic exposure and protective immunity. Proc Allen D Leman Swine Conference. 2010;82 [Google Scholar]
- 18.Oliveira S, Blackall PJ, Pijoan C. Characterization of the diversity of Haemophilus parasuis field isolates by use of serotyping and genotyping. Am J Vet Res. 2003;64:435–442. doi: 10.2460/ajvr.2003.64.435. [DOI] [PubMed] [Google Scholar]
- 19.Walker RD, Dowling PM. Fluoroquinolones. In: Giguere S, Prescott JF, Baggot JD, Walker RD, Dowling PM, editors. Antimicrobial Therapy in Veterinary Medicine. Ames: Blackwell Publishing; 2006. pp. 263–284. [Google Scholar]
- 20.Bimazubute M, Cambier C, Baert K, et al. Penetration of enrofloxacin into nasal secretions and relationship between nasal secretions and plasma enrofloxacin concentrations after intramuscular administration in healthy pigs. J Vet Pharmacol Therap. 2009;33:183–188. doi: 10.1111/j.1365-2885.2009.01123.x. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira S. Improving rate of success in isolating Haemophilus parasuis from clinical samples. J Swine Health Prod. 2004;12:308–309. [Google Scholar]
- 22.Oliveira S, Galina L, Blanco I, Canals A, Pijoan C. Naturally-farrowed, artificially-reared pigs as an alternative model for experimental infection by Haemophilus parasuis. Can J Vet Res. 2003;67:146–150. [PMC free article] [PubMed] [Google Scholar]