Abstract
Sphingomonas sp. strain A1 has three endotype alginate lyases (A1-I, A1-II [family PL-7], and A1-III [family PL-5]), each of which is encoded by a single gene. In addition to those of these lyases, a gene (the A1-II′ gene) showing significant identity with the A1-II gene was present in the bacterial genome and coded for an alginate lyase with broad substrate specificity. Since no expression of A1-II′ was observed even in bacterial cells grown on alginate, the A1-II′ gene was thought to be a silent gene derived from the A1-II gene, presumably through duplication, modification, and translocation.
Alginate is a linear polysaccharide consisting of α-l-guluronate (G) and its C5 epimer, β-d-mannuronate (M), arranged in three different ways, i.e., in poly-α-l-guluronate [poly(G)], poly-β-d-mannuronate [poly(M)], and heteropolymeric [poly(MG)] regions (7). The polymer produced by brown seaweed is widely used in the food and pharmaceutical industries due to its ability to chelate metal ions and to form a highly viscous solution (23), while some pathogenic bacteria, such as Pseudomonas aeruginosa, produce alginate as a capsule-like biofilm that is responsible for both chronic pulmonary infections and respiratory difficulty in the lungs of patients with cystic fibrosis (3, 4).
Alginate lyase depolymerizes alginate through the β-elimination reaction. A large number of alginate lyases, from sources ranging from bacteria to marine animals, have been characterized (32), although little information on the diversity and evolution of alginate lyases has been accumulated. Based on their primary structures, polysaccharide lyases are classified into 13 families (PL-1 to -13) (B. Henrissat, P. Coutinho, and E. Deleury, http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html). Most alginate lyases are classified into two families, PL-5 and -7. Generally, alginate lyases in families PL-5 and -7 preferably depolymerize poly(M) and poly(G), respectively, though many enzymes can cleave M-G or G-M bonds.
In order to clarify the structure and function of alginate lyases, we have studied the metabolism of alginate in cells of Sphingomonas sp. strain A1 (21). Recently, the complete genome sequence of this bacterium was determined (W. Hashimoto et al., unpublished results). The bacterium incorporates the macromolecule (alginate) through a “superchannel,” consisting of a pit formed on the cell surface and a pit-dependent ABC transporter (13, 22), and depolymerizes the polymer into its constituent monosaccharides through concerted reactions catalyzed by three intracellular endotype alginate lyases (A1-I, A1-II, and A1-III) and an exotype alginate lyase (A1-IV) (8, 36). The three endotype alginate lyases are encoded by a single gene (21), and precursor protein A1-I is autocatalytically processed into A1-II and A1-III (14). A1-II and A1-III are categorized as family PL-7 and -5 lyases, specific for poly(G) and poly(M), respectively. Therefore, A1-I is a fused enzyme with the characteristics of family PL-5 and -7 enzymes. Some pseudomonads, such as P. aeruginosa strain PAO1 (29) and Pseudomonas syringae pv. Tomato strain DC3000 (6) have both family PL-5 and -7 alginate lyases, although their genes are separately located in the bacterial genomes. Therefore, we propose that A1-II and A1-III encoded by the A1-I gene are the original alginate lyases of families PL-7 and -5, respectively, and that the A1-II and A1-III genes that were derived from the A1-I gene independently evolved into various genes belonging to families PL-7 and -5 through duplication, modification, and translocation. As the first step to confirm this hypothesis, we report here the molecular diversity and evolution of alginate lyases in Sphingomonas sp. strain A1.
Occurrence of a gene homologous to the alginate lyase A1-I gene in Sphingomonas sp. strain A1 and its sequence analysis.
Homology analysis of A1-I against the genome database of Sphingomonas sp. strain A1 (Hashimoto et al., unpublished results) was performed with the PSI-BLAST program assisted by the DDBJ server (http://www.ddbj.nig.ac.jp/). As a result, a hypothetical protein, designated A1-II′, showing significant homology with A1-II (55.1% identity) was found, while no open reading frame similar to that of A1-III was observed. A1-II′, which consists of 308 amino acids with a molecular weight of 31,991, is encoded by a gene (the A1-II′ gene) exhibiting high identity (62.3%) with the A1-II gene. In addition to A1-II, A1-II′ is similar to alginate lyases such as PA1176 of P. aeruginosa (34.4% identity in a 227-amino-acid overlap; accession number AE004547) (33), ALYPG of Corynebacterium sp. strain ALY-1 (29.8% identity in a 248-amino-acid overlap; accession number AB030481) (20), and AlyA of Klebsiella pneumoniae subsp. aerogenes (27.7% identity in a 300-amino-acid overlap; accession number L19657-2) (2). However, compared with alginate lyases analyzed so far, A1-II′ has an additional N-terminal extension composed of 80 amino acid residues with serine repeat sequences. Open reading frames up- and downstream of the A1-II′ gene are significantly homologous to those of Neisseria meningitides arginyl-tRNA synthetase (62.2% identity in a 571-amino-acid overlap; accession number AL162756) (30) and Escherichia coli 30S ribosomal protein (64.7% identity in a 553-amino-acid overlap; accession number AB011415-3) (27), respectively.
Overexpression of A1-II′ in E. coli cells.
Overexpression systems for the native A1-II′ [A1-II′(L)] and for a truncated A1-II′ [A1-II′(S)] lacking the N-terminal 80 amino acid residues were constructed in E. coli cells as follows. DNA sequencing and manipulation were carried out as described previously (1, 25). Genomic DNA was isolated from cells of Sphingomonas sp. strain A1 grown in alginate medium (8). To introduce the A1-II′(L) and A1-II′(S) genes into an expression vector, pET21b (Novagen, Madison, Wis.), PCR was performed with KOD polymerase (Toyobo Co., Tokyo, Japan), the bacterial genome DNA as a template, and two synthetic oligonucleotides as primers. The oligonucleotides for A1-II′(L) were 5′-GGCATATGGAAAAGCAGTGCGGATGGTA-3′ and 5′-GGCTCGAGGTTGCTGTGCGACACCGACAGG-3′, withNdeI and XhoI sites, respectively, added to their 5′ regions, and those for A1-II′(S) were 5′-GGCATATGCCGGCTGCCGCACCCGGCAAGA-3′ and 5′-GGCTCGAGGTTGCTGTGCGACACCGACAGG-3′, with NdeI and XhoI sites, respectively, added to their 5′ regions. The pET21b vector is designed to express the proteins with a histidine (His)-tagged sequence at the C terminus. The fragments amplified through the PCR were digested with NdeI and XhoI and then ligated with NdeI- and XhoI-digested pET21b. The resultant plasmids containing the A1-II′(L) and A1-II′(S) genes were designated pET21b-A1-II′(L) and pET21b-A1-II′(S), respectively. The accuracy of the nucleotide sequences of the A1-II′(L) and A1-II′(S) genes was confirmed by DNA sequencing (data not shown). Transformants with the plasmids [pET21b-A1-II′(L) and pET21b-A1-II′(S)] of E. coli BL21(DE3) (Novagen, Madison, Wis.) with no alginate lyase activity were grown at 16°C in Luria-Bertani broth (24) in the presence of isopropyl-β-d-thiogalactopyranoside at 0.1 mM for induction of gene expression. By means of reactions (Western blotting) with anti-His-tagged-sequence antibodies (Bethyl Laboratories Inc., Montgomery, Tex.) as described previously (11), expression of the proteins in cell extracts of the E. coli transformants with pET21b-A1-II′(L) and pET21b-A1-II′(S) was confirmed (Fig. 1A). The assay for alginate lyase was performed with sodium alginate (Nacalai Tesque Co., Ltd., Kyoto, Japan) as a substrate under the conditions described by Yoon et al. (36), with slight modifications (substrate concentration, 0.05%; reaction temperature, 30°C). Both cell extracts exhibited alginate lyase activity (Table 1), suggesting that A1-II′ is an alginate lyase.
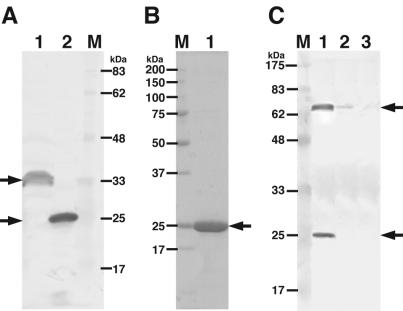
FIG. 1.
Electrophoretic profiles of Sphingomonas alginate lyases. (A) SDS-PAGE, followed by Western blotting with anti-His-tagged sequence antibodies. Lane 1, cell extract of E. coli transformed with pET21b-A1-II′(L); lane 2, cell extract of E. coli transformed with pET21b-A1-II′(S); lane M, molecular mass standards. (B) SDS-PAGE, followed by protein staining with Coomassie brilliant blue. Lane M, molecular mass standards (synthetic polypeptides); lane 1, purified A1-II′(S). (C) SDS-PAGE, followed by Western blotting with anti-A1-II′(S) antibodies. Lane M, molecular mass standards; lane 1, Sphingomonas sp. strain A1 cells grown on alginate; lane 2, cells grown on pectin; lane 3, cells grown on glucose. Arrows indicate alginate lyases.
TABLE 1.
Purification of alginate lyases and expression in E. coli cells
| Stepa | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purifi- cation (fold) |
|---|---|---|---|---|---|
| Cell extract | |||||
| A1-II′(L) | 818 | 2,065 | 2.52 | ||
| A1-II′(S) | 627 | 1,812 | 2.89 | 100 | 1.00 |
| Chelating Sepharose | 8.24 | 442 | 53.6 | 24.3 | 18.5 |
| DEAE-Toyopearl 650M | 5.07 | 437 | 86.2 | 24.1 | 29.8 |
The purification procedures are described in Materials and Methods.
Purification and characterization of A1-II′(S) from E. coli cells.
In order to compare the enzymatic properties of A1-II′ and A1-II, purification of A1-II′(S) lacking the 80 amino acid residues at the N terminus was conducted. The assay for protein was performed as described previously (5). Unless otherwise specified, all operations were carried out at 0 to 4°C. E. coli cells harboring pET21b-A1-II′(S) were grown in 1.5 liters of Luria-Bertani medium (1.5 liters/flask), collected by centrifugation at 6,000 × g and 4°C for 5 min, washed with 20 mM Tris-HCl buffer (pH 7.5), and then resuspended in the same buffer. The cells were ultrasonically disrupted (Insonator model 201 M; Kubota, Tokyo, Japan) at 0°C and 9 kHz for 20 min, and the clear solution obtained by centrifugation at 15,000 × g and 4°C for 20 min was used as the cell extract. The cell extract, after supplementation with 1 mM phenylmethylsulfonyl fluoride and 0.1 μM pepstatin A, was subjected to nickel ion-bound chelating Sepharose Fast Flow column (1.5 by 5 cm) chromatography (Amersham Biosciences, Uppsala, Sweden) after equilibration with 20 mM Tris-HCl buffer (pH 7.5) containing 10 mM imidazole and 0.5 M NaCl. A1-II′(S) was eluted with a linear gradient of imidazole (0.01 to 0.5 M) in 20 mM Tris-HCl buffer (pH 7.5) containing 0.5 M NaCl (30 ml), with a 1-ml fraction being collected every 1 min. The active fractions, which were eluted at around 0.4 M imidazole, were combined and dialyzed against 20 mM Tris-HCl buffer (pH 7.5). The dialysate was added to a DEAE-Toyopearl 650 M gel (Tosoh Co., Tokyo, Japan) equilibrated with 20 mM Tris-HCl buffer (pH 7.5). The gel suspension was centrifuged at 5,000 × g and 4°C for 10 min, and the clear solution containing A1-II′(S) was concentrated to about 2 ml by ultrafiltration with a Centriprep instrument (10-kDa molecular mass cutoff; Millipore Corp., Bedford, Mass.). The concentrate was used as the purified A1-II′(S). Finally, A1-II′(S) was purified 29.8-fold from the cell extract of the E. coli transformant, with an activity yield of 24.1% (Table 1). The purified enzyme was confirmed to be homogeneous by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli (18) (Fig. 1B). The properties of the purified A1-II′(S) were as follows.
(i) N-terminal amino acid sequence.
By using a Procise 492 protein sequencing system (Applied Biosystems, Division of Perkin-Elmer, Foster City, Calif.), the N-terminal amino acid sequence of A1-II′(S) expressed in E. coli cells was determined to be NH2-PAAAP, which corresponds to 81PAAAP85 of the A1-II′ amino acid sequence predicted from the nucleotide sequence. Since A1-II′(S) expressed in E. coli cells is absorbed by nickel ion-bound chelating Sepharose, the protein is thought to contain a C-terminal extension of 8 amino acid residues (LEHHHHHH) derived from the expression vector (pET21b), thus indicating that A1-II′(S) consists of 236 amino acid residues.
(ii) Molecular mass.
The molecular mass of A1-II′(S) was determined to be 25 kDa by SDS-PAGE (Fig. 1B). This value was comparable to the theoretical one (25,763 Da) deduced from the predicted amino acid sequence of the enzyme. On permeation chromatography on Sephacryl S-200HR (Amersham Biosciences), the enzyme was eluted as a protein with a molecular mass of about 25 kDa (data not shown), indicating that the enzyme is in a monomeric form.
(iii) pH and temperature.
Experiments were carried out with 0.05% sodium alginate as a substrate and purified A1-II′(S) (0.298 μg/ml).
(a) Optimal pH.
Reactions were performed at 30°C for 5 min in the following 50 mM buffers; sodium acetate, sodium HEPES, Tris-HCl, and glycine-sodium hydroxide.
(b) Optimal temperature.
Reactions were performed for 5 min at various temperatures in 50 mM Tris-HCl buffer (pH 7.5).
(c) Thermal stability.
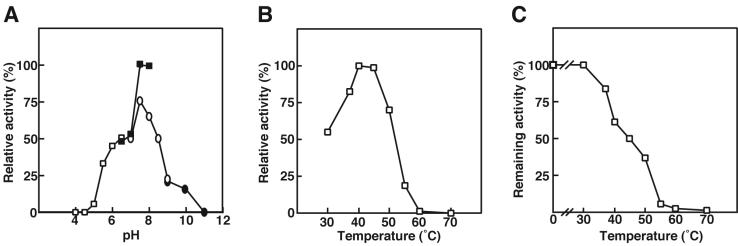
After preincubation of the enzyme (2.98 μg/ml) at various temperatures for 5 min, residual activity was measured at 30°C for 5 min in 50 mM Tris-HCl (pH 7.5). A1-II′(S) was most active at pH 7.5 in 50 mM sodium HEPES buffer (Fig. 2A) and at 40°C (Fig. 2B). Fifty percent of the enzyme activity was lost on preincubation at 45°C for 5 min in 50 mM Tris-HCl (pH 7.5) (Fig. 2C).
FIG. 2.
Effects of pH and temperature on the activity and stability of A1-II′(S). (A) Effect of pH, determined with sodium acetate (open squares), sodium HEPES (closed squares), Tris-HCl (open circles), and glycine-sodium hydroxide (closed circle). The activity at pH 7.5 in sodium HEPES was taken as 100%. (B) Optimal temperature. The activity at 40°C was taken as 100%. (C) Thermal stability. The activity of the enzyme preincubated at 0°C was taken as 100%. The data are averages from two independent experiments and vary within 10%.
(iv) Metal ions and other compounds.
The reaction was carried out in the presence or absence of various compounds, and then the residual activity was measured (data not shown). A bivalent metal ion (Hg2+) partially (30%) inhibited the reaction of A1-II′(S) at 1 mM. Other bivalent metal ions such as Ca2+, Co2+, Mg2+, and Mn2+ had no effects on the enzyme activity at 1 mM. Thiol reagents (dithiothreitol, glutathione [reduced form], 2-mercaptoethanol, and iodoacetic acid), and a chelator (EDTA) showed no significant effect on the reaction of A1-II′(S) at 1 mM.
(v) Substrate specificity.
Since alginate has three block structures [poly(M), poly(G), and poly(MG)], the substrate specificity of A1-II′(S) was investigated with poly(M) (M, 95.1%; G, 4.9%), poly(G) (M, 6.1%; G, 93.9%), and poly(MG) (M, 53.2%; G, 46.8%) (Table 2). A1-II and A1-III purified from recombinant E. coli cells are specific for poly(G) and poly(M), respectively, as described previously (36), whereas A1-II′(S) acted on poly(M), poly(G), and poly(MG) equally. Unlike the alginate-related polysaccharides, pectin, xanthan, and gellan were inert as substrates for A1-II′(S).
TABLE 2.
Substrate specificities of A1-II′(S), A1-II, and A1-III
| Substrate | Sp act (U/mg)
|
||
|---|---|---|---|
| A1-II′(S) | A1-II | A1-III | |
| Poly(M) | 73.2 | 34.8 | 45.2 |
| Poly(G) | 73.2 | 202 | 2.25 |
| Poly(MG) | 78.2 | 80.0 | 15.1 |
(vi) Action mode.
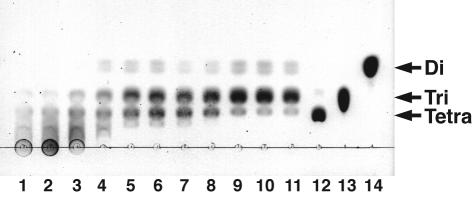
Alginate di-, tri-, and tetrasaccharides were prepared as described previously (10). The enzyme reaction products separated on thin-layer chromatography (TLC) plates (Silica gel 60/Kieselguhr F254; E. Merck, Darmstadt, Germany) were visualized by heating the plates at 130°C for 5 min after spraying with 10% (vol/vol) sulfuric acid in ethanol as described previously (8). Unsaturated saccharides on the TLC plates were stained with thiobarbituric acid as described previously (31). Judging from the results of the TLC analysis, the products in the initial stage of the A1-II′(S) reaction were alginate oligosaccharides with various degrees of polymerization, indicating that the enzyme acts on alginate in an endolytic form (Fig. 3). Alginate was finally depolymerized into di- and trisaccharides through the A1-II′(S) reaction. The alginate oligosaccharides produced from alginate by A1-II′(S) reacted with thiobarbituric acid (data not shown), thus indicating that A1-II′(S) catalyzes the β-elimination reaction.
FIG. 3.
Mode of action of A1-II′(S). Alginate (1.25 mg) was incubated with A1-II′(S) (7.5 μg) at 30°C, and the products were analyzed by TLC with staining with sulfuric acid. The reaction times (hours) were 0 (lane 1), 0.16 (lane 2), 0.33 (lane 3), 0.5 (lane 4), 1 (lane 5), 2 (lane 6), 3 (lane 7), 6 (lane 8), 24 (lane 9), 48 (lane 10), and 72 (lane 11). Lane 12, alginate-tetrasaccharide; lane 13, alginate-trisaccharide; lane 14, alginate-disaccharide. Tera, Tri, and Di, unsaturated tetra-, tri-, and disaccharides derived from alginate, respectively.
Expression of alginate lyases in Sphingomonas sp. strain A1.
In order to determine whether A1-II′ is expressed or not in cells of Sphingomonas sp. strain A1, bacterial cells grown on alginate, pectin, or glucose were subjected to SDS-PAGE, followed by Western blotting with anti-A1-II′(S) antibodies raised in a rabbit. Proteins with molecular masses of 65 and 25 kDa corresponding to A1-I and A1-II, respectively, were expressed in the bacterial cells grown on alginate, while no protein (32 kDa) corresponding to A1-II′ was detected in the bacterial cells cultured under the various conditions (Fig. 1C).
In this work, an alginate lyase gene, the A1-II′ gene, distinct from the A1-I, A1-II, and A1-III genes, was found to be present in the genome of Sphigomonas sp. strain A1. A1-II′ shows the highest identity with A1-II and significant homology with bacterial alginate lyases in family PL-7, thus indicating that A1-II′ is a member of family PL-7. The two sequences YXRSELRE and YFKAGXYXQ are highly conserved in the N- and C-terminal regions of family PL-7 lyases, respectively, and are suggested to be responsible for the catalytic reaction of the enzymes (32). A1-II′ also has two conserved regions, which almost completely match those of A1-II. Furthermore, a tyrosine residue is reported to play a crucial role in the catalytic reaction of polysaccharide lyases such as the family PL-5 alginate lyase A1-III (35) and the family PL-8 hyaluronate (17), chondroitin (15), and xanthan (9) lyases. Therefore, one of the tyrosine residues in the two consensus sequences in family PL-7 alginate lyases is thought to be responsible for the catalytic reaction of the enzymes. A1-II′ (this work) and A1-II (36) are similar in optimal pH, thermal stability, and behavior toward various compounds tested but not in specific activity [A1-II′(S), 86.2 U/mg; A1-II, 109 U/mg], optimal temperature [A1-II′(S), 40°C; A1-II, 75°C], substrate specificity [A1-II′(S), broad; A1-II, poly(G)], and final products [A1-II′(S), di- and trisaccharides; A1-II, tri- and tetrasaccharides]. The enzyme properties of A1-II′(S) are comparable to those of A1-I rather than A1-II. Since it is thought that these differences between A1-II′(S) and A1-II are caused by some substitutions in the amino acid residues, X-ray crystal analysis of A1-II and A1-II′(S) is now in progress in order to identify the amino acid residues responsible for the determination of enzyme characteristics (34). Most endotype alginate lyases analyzed so far show substrate specificity for poly(M) or poly(G) (32), unlike the enzymes of Alteromonas sp. strain H-4 (26) and Pseudoalteromonas sp. strain 272 (16), indicating that A1-II′(S) is an unusual type of enzyme with a broad substrate specificity.
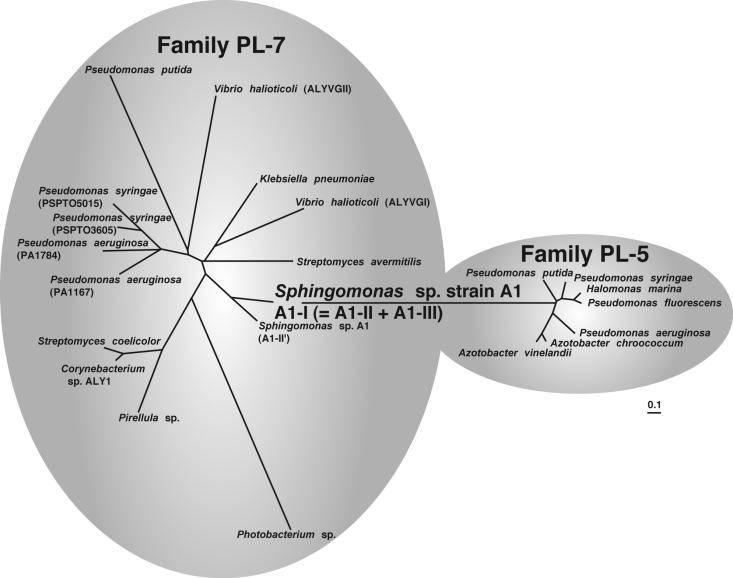
Thus, as described above, Sphingomonas sp. strain A1 was found to have diverse genes for alginate lyases. However, we think that the A1-II′ gene does not play a crucial role in the metabolism of alginate in the bacterium and that the A1-II′ gene arose from the A1-II gene through separation from the A1-I gene, modification, and translocation. The following five facts may support this notion. (i) Little A1-II′ is expressed in bacterial cells. (ii) A1-II′ exhibits the highest identity with A1-II, although A1-II′ has an N-terminal extension (80 amino acid residues) containing serine repeat sequences. (iii) A1-II′ expressed in E. coli cells shows alginate lyase activity. (iv) The locus of the A1-II′ gene is 367 kb away from the gene cluster responsible for the transport and depolymerization of alginate, and no alginate-related genes are found up- or downstream of the A1-II′ gene. (v) No apparent bacterial promoter (12) or ribosome-binding site (28) is present upstream of the A1-II′ gene. As to the molecular evolution of alginate lyases, we propose that A1-II and A1-III encoded by the A1-I gene of Sphingomonas sp. strain A1 are single-module ancestors for family PL-7 and -5 enzymes, respectively, and that the A1-II gene and the A1-III gene derived from the A1-I gene independently evolved into various genes categorized into families PL-7 and -5 through duplication, modification, and translocation. Thus, phylogenetic analysis of alginate lyases categorized into families PL-7 and -5 was conducted (Fig. 4). Alginate lyases belonging to family PL-5, except for A1-III, are similar to each other, while family PL-7 enzymes are widely spread throughout the phylogenetic tree. Almost all of the family PL-5 enzymes exhibit specificity for poly(M) (32). On the other hand, a few family PL-7 enzymes, such as PA1167 of P. aeruginosa (33) and AlxM of Photobacterium sp. (19), show substrate specificities different from those of the general family PL-7 enzymes. Therefore, due to their broad substrate specificities, the family PL-7 enzymes may be spread throughout the tree. As shown in Fig. 4, A1-I of Sphingomonas sp. strain A1 is situated at a central position in the trees, being intermediate between families PL-5 and -7. Since there is no evidence that the A1-I gene evolved through the fusion of the A1-II gene and the A1-III gene independently existing, the alginate lyases categorized into families PL-5 and 7 may be evolved from the ancestral proteins, A1-II and A1-III, forming A1-I.
FIG. 4.
Phylogenetic tree of family PL-5 and -7 alginate lyases. The bacterial producers of the enzymes are indicated. The scale bar indicates approximately 10% sequence difference. Parentheses indicate the isozymes of alginate lyases.
Nucleotide sequence accession number.
The nucleotide sequence of the alginate lyase (A1-II′) gene reported in this paper has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB120939.
Acknowledgments
This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) of Japan.
Poly(M), poly(G), and poly(MG) were obtained from T. Sawabe, Hokkaido University, Hakodate, Japan.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates, New York, N.Y.
- 2.Baron, A. J., T. Y. Wong, S. J. Hicks, P. Gacesa, D. Willcock, and M. J. McPherson. 1994. Alginate lyase from Klebsiella pneumoniae, subsp. aerogenes: gene cloning, sequence analysis and high-level production in Escherichia coli. Gene 27:61-66. [DOI] [PubMed] [Google Scholar]
- 3.Batten, J. C., and D. J. Matthew. 1983. The respiratory system, p. 105-131. In M. E. Hodson, A. P. Norman, and J. C. Batten, (ed.), Cystic fibrosis. Bailliere Tindall, London, United Kingdom.
- 4.Boat, T. F., A. L. Beadet, and M. J. Welsh. 1989. The metabolic basis of inheritated disease, p. 2649-2680. In C. R. Seriver, (ed.), Cystic fibrosis. McGraw-Hill, New York, N.Y.
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gacesa, P. 1988. Alginates. Carbohydr. Polym. 8:161-182. [Google Scholar]
- 8.Hashimoto, W., O. Miyake, K. Momma, S. Kawai, and K. Murata. 2000. Molecular identification of oligoalginate lyase of Sphingomonas sp. strain A1 as one of the enzymes required for complete depolymerization of alginate. J. Bacteriol. 182:4572-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto, W., H. Nankai, B. Mikami, and K. Murata. 2003. Crystal structure of Bacillus sp. GL1 xanthan lyase, which acts on the side chains of xanthan. J. Biol. Chem. 278:7663-7673. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto, W., M. Okamoto, T. Hisano, K. Momma, and K. Murata. 1998. Sphingomonas sp. A1 lyase active on both poly-β-d-mannuronate and heteropolymeric regions in alginate. J. Ferment. Bioeng. 86:236-238. [Google Scholar]
- 11.Hashimoto, W., H. Suzuki, K. Yamamoto, and H. Kumagai. 1995. Effect of site-directed mutations on processing and activity of γ-glutamyltranspeptidase of Escherichia coli K-12. J. Biochem. 118:75-80. [DOI] [PubMed] [Google Scholar]
- 12.Hawley, D. K., and W. R. McClure. 1983. Comparison and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hisano, T., N. Kimura, W. Hashimoto, and K. Murata. 1996. Pit structure on bacterial cell surface. Biochem. Biophys. Res. Commun. 220:979-982. [DOI] [PubMed] [Google Scholar]
- 14.Hisano, T., M. Nishimura, T. Yamashita, K. Sakaguchi, and K. Murata. 1994. On the self-processing of bacterial alginate lyase. J. Ferment. Bioeng. 78:109-110. [Google Scholar]
- 15.Huang, W., L. Boju, L. Tkalec, H. Su, H. O. Yang, N. S. Gunay, R. J. Linhardt, Y. S. Kim, A. Matte, and M. Cygler. 2001. Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis. Biochemistry 40:2359-2372. [DOI] [PubMed] [Google Scholar]
- 16.Iwamoto, Y., K. Iriyama, K. Osatomi, T. Oda., and T. Muramatsu. 2002. Primary structure and chemical modification of some amino acid residues of bifunctional alginate lyase from marine bacterium Pseudoalteromonas sp. strain no. 272. J. Protein Chem. 21:455-463. [DOI] [PubMed] [Google Scholar]
- 17.Jedrzejas, M. J., L. V. Mello, B. L. de Groot, and S. Li. 2002. Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate. J. Biol. Chem. 277:28287-28297. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Malissard, M., C. Duez, M. Guinand, M. J. Vacheron, G. Michel, N. Marty, B. Joris, I. Thamm, and J. M. Ghuysen. 1993. Sequence of a gene encoding a (poly ManA) alginate lyase active on Pseudomonas aeruginosa alginate. FEMS Microbiol. Lett. 110:101-106. [DOI] [PubMed] [Google Scholar]
- 20.Matsubara, Y., R. Kawada, K. Iwasaki, Y. Kimura, T. Oda, and T. Muramatsu. 2000. Cloning and sequence analysis of a gene (aly PG) encoding poly(α-l-guluronate)lyase from Corynebacterium sp. strain ALY-1. J. Biosci. Bioeng. 89:199-202. [DOI] [PubMed] [Google Scholar]
- 21.Mishima, Y., K. Momma, O. Miyake, W. Hashimoto, B. Mikami, and K. Murata. 2002. Super-channel in bacteria: macromolecule uptake and depolymerization systems of Sphingomonas sp. A1 with special cell surface structure. Biotechnol. Genet. Eng. Rev. 19:1015-1019. [DOI] [PubMed] [Google Scholar]
- 22.Momma, K., M. Okamoto, Y. Mishima, S. Mori, W. Hashimoto, and K. Murata. 2000. A novel bacterial ATP-binding cassette (ABC) transporter system that allows uptake of macromolecules. J. Bacteriol. 182:3998-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onsøyen, E. 1996. Commercial applications of alginates. Carbohydr. Eur. 14:26-31. [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawabe, T., M. Otsuka, and Y. Ezura. 1997. Novel alginate lyases from marine bacterium Alteromonas sp. strain H-4. Carbohydr. Res. 304:69-76. [DOI] [PubMed] [Google Scholar]
- 27.Schnier, J., M. Kimura, K. Foulaki, A. R. Subramanian, K. KIsono, and B. Wittmann-Liebold. 1982. Primary structure of Escherichia coli ribosomal protein S1 and of its gene rpsA. Proc. Natl. Acad. Sci. USA 79:1008-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shine, J., and L. Dalgarno. 1974. The 3′ terminal sequence of E. coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosomal binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 30.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 31.Warren, L. 1960. Thiobarbituric acid spray reagent for deoxy sugars and sialic acids. Nature 186:237. [DOI] [PubMed] [Google Scholar]
- 32.Wong, T. Y., L. A. Preston, and N. L. Schiller. 2000. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54:289-340. [DOI] [PubMed] [Google Scholar]
- 33.Yamasaki, M., S. Moriwaki, W. Hashimoto, B. Mikami, and K. Murata. 2003. Crystallization and preliminary X-ray analysis of alginate lyase, a member of polysaccharide lyase family PL-7, from Pseudomonas aeruginosa. Acta Crystallogr. Sect. D 59:1499-1501. [DOI] [PubMed] [Google Scholar]
- 34.Yoon, H.-J., W. Hashimoto, Y. Katsuya, Y. Mezaki, K. Murata, and B. Mikami. 2000. Crystallization and preliminary X-ray crystallographic analysis of alginate lyase A1-II from Sphingomonas species A1. Biochim. Biophys. Acta 1476:382-385. [DOI] [PubMed] [Google Scholar]
- 35.Yoon, H.-J., W. Hashimoto, O. Miyake, K. Murata, and B. Mikami. 2001. Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0Å resolution. J. Mol. Biol. 307:9-16. [DOI] [PubMed] [Google Scholar]
- 36.Yoon, H.-J., W. Hashimoto, O. Miyake, M. Okamoto, B. Mikami, and K. Murata. 2000. Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp. A1 alginate lyases. Protein Expr. Purif. 19:84-90. [DOI] [PubMed] [Google Scholar]