Abstract
SXT is an integrative and conjugative element (ICE) isolated from Vibrio cholerae. This ∼100-kb ICE encodes resistance to multiple antibiotics and integrates site specifically into the chromosome. SXT excises from the chromosome to form a circular but nonreplicative extrachromosomal molecule that is required for its transfer. Here we found that a significant fraction of freshly isolated SXT exconjugants contained tandem SXT arrays. There was heterogeneity in the size of the SXT arrays detected in single exconjugant colonies. Some arrays consisted of more than five SXTs arranged in tandem. These extended arrays were unstable and did not persist during serial passages. The mechanism accounting for the generation of SXT arrays is unknown; however, array formation was not dependent upon recA and appeared to depend on conjugative transfer. While such arrays did not alter the transfer frequency of wild-type SXT, they partially complemented the transfer deficiency of a Δxis SXT mutant, which is ordinarily unable to generate the extrachromosomal intermediate required for SXT transfer. Exconjugants derived from donor strains that harbored tandem arrays of SXT and R391, an SXT-related element, contained functional hybrid elements that arose from recA-independent recombination between the two ICEs. Thus, arrays of SXT-related elements promote the creation of novel ICEs.
The integrative and conjugative elements (termed ICEs) are a diverse class of mobile elements that share a similar lifestyle in both gram-positive and gram-negative bacteria (5, 26). Like most prophages, ICEs are maintained integrated within the chromosome of their hosts. They excise from the chromosome by recombination between two specific flanking sequences (attL and attR) to form a covalently closed circular molecule that is generally not replicative. Like the conjugative plasmids, ICEs encode conjugation systems that can transfer the excised DNA to a new host, where it integrates once again, often in a site-specific fashion, by recombination between the attP site generated by excision and a target sequence, attB. Different ICEs integrate into a variety of sites and encode diverse recombination, conjugation, and regulation systems (5). They also carry genes encoding a variety of functions, including catabolic pathways (26), antibiotic resistances (21, 29), nitrogen fixation (24), and phage resistance mechanisms (6).
The 99.5-kb SXT element (SXT) is a Vibrio cholerae-derived ICE that was originally identified in a 1993 V. cholerae serogroup O139 clinical isolate, MO10 (28). The MO10-derived SXT, SXTMO10, encodes resistance to sulfamethoxazole, trimethoprim, chloramphenicol, and streptomycin, and its complete nucleotide sequence has been determined (2). SXTMO10-related elements are now present in most (if not all) V. cholerae O139 and V. cholerae O1 clinical isolates from Asia (15). Some of these SXT-related elements contain different antibiotic resistance genes or even lack antibiotic resistance genes. Recently, SXT-related elements were detected in V. cholerae O1 clinical isolates from Africa (10) and in non-O1 and non-O139 clinical isolates from India (25). Furthermore, SXT has also been found in Providencia alcalifaciens clinical isolates from Bangladesh, indicating that SXT elements are not unique to V. cholerae (15). In the laboratory, SXT transfers by conjugation to a variety of gram-negative bacteria, including V. cholerae and Escherichia coli.
SXT encodes a tyrosine recombinase (Int) that catalyzes its integration into the 5′ end of prfC, a gene encoding peptide chain release factor 3 (17). SXT excision from the chromosome also requires Int and is greatly facilitated by a recently described Xis protein (7). Deletion of either int or xis from SXT nearly eliminates SXT transfer, suggesting that the circular extrachromosomal form of SXT is a required intermediate in its transfer (7). Both SXT integration and excision are under the control of SXT-encoded transcriptional activators, SetC and SetD, which also activate the expression of the transfer functions (2, 7). SetC and SetD expression is repressed by a lambda CI-like repressor, SetR. SetR-mediated repression was recently found to be alleviated by the SOS response to DNA damage in a RecA-dependent fashion (3).
R391 is an ICE originally isolated in 1972 in South Africa from Providencia rettgeri that mediates resistance to kanamycin and mercury (9). R391 and several related IncJ elements integrate into prfC, the same chromosomal locus used by SXT. In fact, SXT and R391 are genetically and functionally related (14). Comparison of the nucleotide sequences of these two ICEs showed that they consist of a conserved set of genes that mediate regulation, excision/integration, and conjugative transfer of the respective ICEs (1). Insertions into this shared common backbone confer element-specific properties such as resistance to particular antibiotics. SXT and R391 do not exclude each other, and in cells harboring both elements, SXT and R391 are found integrated in tandem fashion on the chromosome (14). In cells that harbor tandem SXT-R391 arrays, each element can excise and transfer independently of the other.
In the present study, we found that SXT transfer into E. coli and V. cholerae recipients often resulted in the formation of extended tandem SXT arrays. These arrays had variable numbers of constituent SXTs and were sometimes greater than 500 kb in size. Donor strains harboring both SXT and R391 yielded hybrid ICEs in exconjugant cells. Conjugative transfer from donors containing SXT and R391 appeared to be required for formation of hybrid ICEs, but homologous recombination appeared to play no role in their generation. Our findings suggest that tandem ICE arrays may be a source of ICE diversity, creating novel genetic combinations from preexisting elements.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. The ΔprfC mutants E. coli VI31 and VI47 were created as previously described (7). Bacterial strains were routinely grown in Luria-Bertani (LB) broth at 37°C on a roller drum incubator and were maintained at −80°C in LB broth containing 15% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 40 μg/ml; streptomycin, 200 μg/ml; sulfamethoxazole and trimethoprim (SXT), 160 and 32 μg/ml, respectively; tetracycline, 12 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| E. coli | ||
| CAG18439 | MG1655 lacZU118 lacI42::Tn10 | 23 |
| KB1 | MG1655 recA56 gutA52 gutR::Tn10 | K. Bettenbrock, unpublished data |
| HW220 | CAG18439 prfC::SXT | 17 |
| BI533 | MG1655 Nalr | 16 |
| BW25113 | lacIqrrnBt14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBAADLD78 | 11 |
| JO115 | CAG18439 prfC::R391-SXT | 14 |
| JO116 | CAG18439 prfC::SXT-R391 | 14 |
| MW5-39 | MC4100 srl::Tn10 F−araR131 Δlac(U169) rpsL thiA recA1 | 18 |
| VI31 | CAG18439 ΔprfC | This study |
| VI47 | KB1 ΔprfC | This study |
| VI166 | BW25113 prfC::SXT | This study |
| VI200 | MG1655 prfC::SXT Δxis pXis | This study |
| VI227 | KB1 prfC::R391 Δxis-SXT Δxis pSetCD | This study |
| VI228 | KB1 prfC::SXT Δxis-R391 Δxis pSetCD | This study |
| V. cholerae | ||
| MO10 | Toxigenic 1992 serogroup O139 clinical isolate from India, SXTMO10+ | 27 |
| E4 | E1 Tor strain, ΔctxABN4 Kmr | 12 |
| Plasmids | ||
| pVI8A | pCRII-Topo attP | 7 |
| pSetCD | pBAD-Topo setCD | 2 |
| pXis | pBAD-Topo xis | 7 |
Bacterial conjugations.
Conjugation assays were performed by mixing equal volumes of overnight cultures of donor and recipient strains. The cells were harvested by centrifugation and resuspended in a 1/20 volume of LB broth. The cell suspensions were poured onto LB plates, supplemented when necessary with 0.02% arabinose or 0.2% glucose. The conjugation experiments were performed at 37°C for 6 h. Afterwards, the cells were harvested from the plate in 1 ml of LB medium, and serial dilutions of the cell suspensions were plated on the appropriate selective media to determine the numbers of donors, recipients, and exconjugants.
Molecular biology techniques.
Plasmid DNA was prepared with either a Qiaprep Spin miniprep kit or a Qiaprep miniprep kit (Qiagen), and chromosomal DNA was prepared with the G Nome DNA kit (Q-Biogene) as described in the manufacturer's instructions. Southern blotting was performed as described previously (27) with probes conjugated to horseradish peroxidase and detected with a chemiluminescent substrate (Amersham). The attP probe was the 600-bp BstBI fragment of pVI8A (7). The PCR assays to characterize the structure of SXT, R391, or the hybrid elements were performed with the primers described in Table 2 in 20-μl PCR mixtures with a HotStarTaq master mix kit (Qiagen) by using 1 μl of a mixture of a colony resuspended in 8 μl of LB broth as a template. The PCR conditions were as follows: (i) 15 min at 95°C; (ii) 30 cycles of 20 s at 95°C, 30 s at 50°C, and 30 s at 72°C; and (iii) 2 min at 72°C. E. coli chromosomal DNA used for contour-clamped homogenous electric field gel electrophoresis (8) was prepared by following the method of Heath et al. (13). After digestion by restriction enzymes, large E. coli restriction fragments were separated in a 1% agarose gel (Invitrogen) for 30 h by using a CHEF-DR II system (Bio-Rad) with the following settings: initial time, 1 s; final time, 40 s; voltage, 5 V/cm. Electrophoresis buffer was 0.5× Tris-borate-EDTA, which was maintained at 14°C during electrophoresis. Real-time quantitative PCR experiments were carried out as previously described (7) to measure the percentages of cells in a culture that contained unoccupied attB sites (EattBF and EattBR primers) and the SXT or R391 attP sequence (SXTJF and SXTJR primers). The amounts of both attB and attP were normalized to the amount of chromosomal DNA in each sample and expressed as the number of sites per 100 chromosomes.
TABLE 2.
DNA sequences of oligonucleotides used in this study
| Primer | Gene or sequence specified | Nucleotide sequence (5′-3′) |
|---|---|---|
| VISLF | SXT and R391 attL sequence | GAGTACAAATTCCGTTTTAG |
| VISLR3 | SXT attL sequence | GCATTCTCCTGAAAATCAATG |
| VISRF | SXT and R391 setR gene | CTCTCATTAACTGGGTTCAGG |
| VISRR | SXT attR sequence | AATGGTTATCTGATCTGTTACCA |
| VISLR2 | R391 orf1 gene | CGCATCAAACTCCCTAAG |
| VISRR2 | R391 attR sequence | AATGGTTATCTAATCGGCTATCA |
| MER104A | R391 mer gene | GCCTGAACGCTTTAGCT |
| MER103B | R391 mer gene | CTGCTCGGTTCCATCAT |
| 10SF13 | SXT s037 gene | TTGTGGTGGAAAGAGGGTG |
| SXT1-13 | SXT s037 gene | CCAACAAAGAACAGTTTGACTC |
| ORF16 | SXT s052 gene | CATCTACCACTTCATAGGCAGC |
| YND2 | SXT s052 gene | CAGCTTAACTCACCAAGGAC |
| EattBF | attB sequence | GCCGCACTTTTGCCATTATT |
| EattBR | attB sequence | AGCAGCACCTTCTCGGTGAT |
| SXTJF | SXT and R391 attP sequence | GCGAAGGACCTTTGCTATCATC |
| SXTJR | SXT and R391 attP sequence | TGGTTTTAAGCGTTGAAAGGC |
RESULTS
Transfer of SXT promotes transitory formation of SXT tandem arrays in recipients.
We recently developed quantitative PCR assays to measure the amounts of SXT attB and attP sequences in cell cultures (7). We expected that the amount of attB would reflect the number of unoccupied SXT integration sites, that the amount of attP would reflect the number of excised circularized SXT molecules, and that these numbers would be approximately equal. Surprisingly, we found that for exconjugants derived from a single mating, the percentages of attB and attP were not comparable. While the percentages of attB hardly differed between exconjugants, the percentages of attP were highly variable. For example, VI179 and VI180, two exconjugants derived from the same mating, had attP percentages of 8.3 and 114.8, respectively; the attB percentages were 1.7 and 2.9, respectively (Table 3).
TABLE 3.
Quantification of unoccupied attB and attP sites by real-time quantitative PCRa
| Exconjugant | % attB | % attP | Ratiob |
|---|---|---|---|
| HW220 | 1.32 ± 0.06 | 4.98 ± 0.23 | 3.8 ± 0.2 |
| VI179 | 1.69 ± 0.07 | 8.30 ± 0.33 | 4.9 ± 0.3 |
| VI180 | 2.92 ± 0.20 | 114.75 ± 4.44 | 39.3 ± 3.1 |
| VI182 | 3.21 ± 0.16 | 61.61 ± 3.64 | 19.2 ± 1.5 |
| VI184 | 2.15 ± 0.11 | 109.28 ± 5.43 | 50.8 ± 3.6 |
| VI185 | 1.59 ± 0.10 | 7.07 ± 0.31 | 4.4 ± 0.3 |
| VI186 | 1.67 ± 0.12 | 143.87 ± 9.25 | 86.1 ± 8.3 |
The excision frequency was quantified by measuring the percentage of cells with unoccupied attB sites by real-time quantitative PCR. The values presented are the means and standard deviations of the results from triplicate measurements.
The ratio was calculated as percentage of attP/percentage of attB. In the ideal case of a strain harboring a single SXT, the expected value of this ratio is 1, since the excision of a single SXT theoretically generates an attB site and an attP site per chromosome.
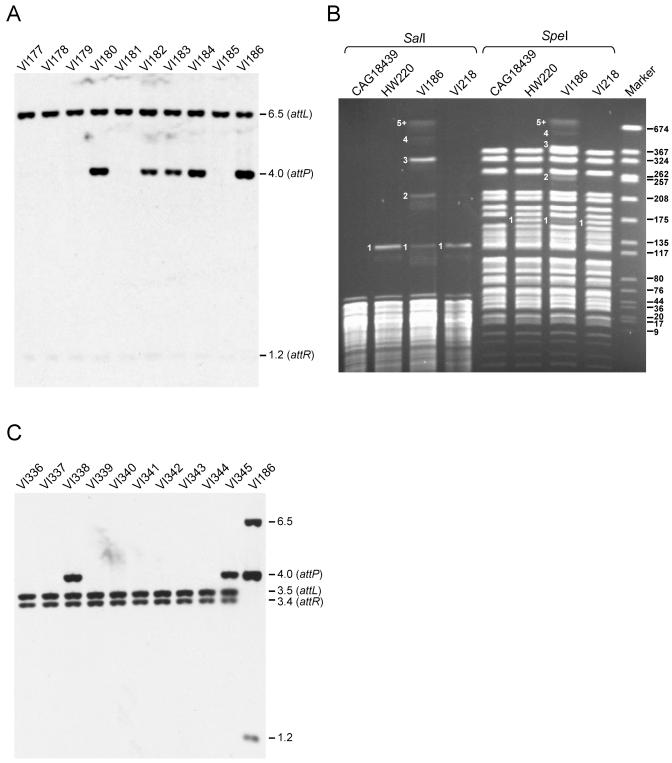
The pronounced variation in the amounts of SXT attP sequences present in freshly derived exconjugants from the same or independent matings was also evident in Southern blots (Fig. 1A). The chromosomal DNAs of 10 exconjugants derived from five independent matings were probed with an SXT attP fragment. Five of ten of the exconjugants studied had a high copy number of a 4-kb SXT attP-specific fragment (Fig. 1A). Furthermore, the intensity of this fragment varied in these five exconjugants and did not correlate with the intensity of the attL- and attR-containing fragments, which remained constant (Fig. 1A).
FIG. 1.
Tandem arrays of SXTs. (A) Southern blot analysis of five pairs of Tetr Sxtr exconjugants resulting from five independent mating experiments between E. coli MG1655 prfC::SXT and E. coli CAG18439. The donor and recipient strains were mated for 6 h at 37°C on LB agar medium without selection. The purified total genomic DNAs of the Tetr Sxtr exconjugants were digested with EcoRI/EcoRV and analyzed by Southern blot hybridization with the 600-bp BstBI fragment of pVI8A containing attP from SXT as a probe. The molecular sizes in kilobases correspond to the fragments containing the attL (6.5 kb, EcoRV), attP (4.0 kb, EcoRI/EcoRV), and attR (1.2 kb, EcoRI/EcoRV) junction fragments. (B) Contour-clamped homogenous PFGE of SalI or SpeI restriction patterns of E. coli CAG18439 (Sxts), HW220 (Sxtr), VI186 (Sxtr), and VI218 (Sxtr). The gel electrophoresis was carried out for 30 h at 5 V/cm. The size marker (Marker) was SmaI-digested chromosomal DNA of Staphylococcus aureus NCTC8325. The molecular sizes indicated to the right of the panel are in kilobases. The numbers indicated to the left of the bands on the HW220, VI186, and VI218 restriction patterns correspond to the copy numbers of SXT in each fragment. The size of the fragments 1 to 5+ observed in the VI186 restriction pattern are, respectively, 125, 224.5, 324, 424.4, and 522.9 kb or more for SalI, and 165.9, 265.4, 364.8, 464.3, and 563.8 kb or more for SpeI. (C) Southern blot analysis of 10 Kanr Sxtr exconjugants (VI336 to VI345) resulting from a mating between V. cholerae MO10 and V. cholerae E4. E. coli VI186 was used as a control. The fragments containing the V. cholerae attL and attR are shown.
The amplified 4-kb attP fragment could be derived from circular extrachromosomal SXT or from tandemly arranged chromosomally integrated SXTs. The former possibility seemed unlikely, as we were unable to extract a circular form of SXT from strains with amplified attP with several alkaline lysis-based techniques (22). Therefore, we used pulsed-field gel electrophoresis to assess whether the amplified attP sequence represented integrated copies of SXT arranged in tandem. For these analyses, DNA from CAG18439, which does not contain SXT, HW220, which contains a single SXT, and VI186, an exconjugant which contains amplified attP (Fig. 1A), were digested with SalI or SpeI, two enzymes which do not cut in SXT. The fragments were separated by contour-clamped homogenous electric field gel electrophoresis (PFGE) (Fig. 1B). Compared to the SalI and SpeI restriction patterns of CAG18439, a single new band was observed in the SalI- and SpeI-digested DNA from HW220, which contains a single SXT. In contrast, five new bands appeared in both the SalI- and SpeI-digested DNA of VI186. The first band from the SalI-digested DNA (Fig. 1B, band 1) had a size identical to the band containing a single SXT integrated into prfC, as seen in HW220 DNA, but was less intense. Bands 2, 3, and 4 had sizes consistent with the presence of two, three, and four SXTs integrated in a tandem fashion into prfC. The band corresponding to the tandem array of three SXTs (band 3) had the strongest intensity in both restriction patterns, suggesting that this was the most prevalent arrangement in the cell population tested. The new band of highest molecular weight (5+) migrated out of the range of resolution of the gel and contained a tandem array of five or more SXTs. Bands 1 to 5+ all hybridized with an attP probe (data not shown), confirming that they contained SXT DNA. A similar distribution of bands was observed with SpeI-digested VI186 DNA; however, these bands differed in their molecular weights, confirming that the arrays were located on the chromosome. Thus, VI186, a population resulting from a single exconjugant, contains different-sized tandem arrays of SXTs. These findings illustrate the remarkable heterogeneity in the population of SXT exconjugants.
We wondered whether tandem SXT arrays formed in SXT's natural host, V. cholerae, as well as in E. coli. Southern blot analysis of exconjugants resulting from a mating between the V. cholerae clinical isolate MO10 (27), used as a donor, and V. cholerae E4 (12), used as a recipient, revealed that 2 of 10 exconjugants harbored tandem SXT arrays (Fig. 1C). This result indicates that tandem SXT arrays form in V. cholerae as well as in E. coli and that E. coli is a suitable SXT host to study this phenomenon.
To investigate the stability of SXT tandem arrays in E. coli, VI186 was cultivated for 5 days with two daily dilutions into fresh LB liquid medium (>200 generations) without selection for the SXT markers. After this time, DNA was isolated from this culture (now called VI218) and the copy number of SXT was analyzed by PFGE as described above. In VI218, the mean copy number of SXT appeared to be one, since only a single band (like that in HW220) was detected (Fig. 1B). This indicates that the high SXT copy number present in VI186 decreased under nonselective conditions. On the other hand, single integrated SXTs were not found to amplify even in the presence of antibiotic selection. Growth of HW220 in LB broth containing various concentrations of sulfamethoxazole (from 160 μg/ml to 2.5 mg/ml) and trimethoprim (from 32 μg/ml to 512 μg/ml) did not promote SXT amplification as measured by attP quantification by real-time quantitative PCR (data not shown).
Formation of SXT arrays does not require recA or prfC.
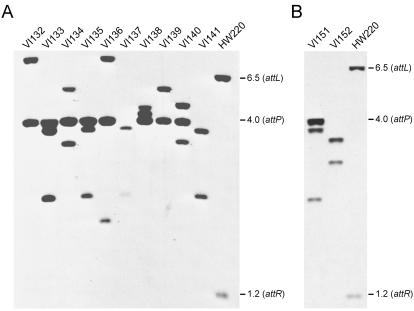
Several mechanisms could account for the formation of tandem SXT arrays. Serial SXT transfer from multiple donor cells (or even a single donor cell) to a single recipient strain could lead to the formation of arrays by recA-dependent recombination between an incoming element and an already integrated SXT. To assess the role of homologous recombination in the formation of tandem SXT arrays, the E. coli recA mutant strain KB1 was used as a recipient in mating experiments with the recA+ donor strain VI166. Quantification of attP percentages in exconjugants revealed that 2 of 10 exconjugants tested carried tandem SXT arrays (data not shown), indicating that recA is not required for the formation of SXT arrays.
We previously found that prfC, the SXT primary integration site, was not absolutely required for SXT integration and that SXT can integrate into alternative sites (7). We excluded the possibility that the SXT integration site within prfC was required for SXT array formation by using VI31, a ΔprfC derivative of E. coli MG1655 as a recipient (Table 1). Eight of ten exconjugants derived from a mating of VI166 and VI31 harbored SXT tandem arrays. This was seen as the amplified attP-containing fragment in Southern blot analysis of DNA from these exconjugants (Fig. 2A). Similarly, when VI47, a recA ΔprfC mutant, was used as a recipient, an amplified attP-containing fragment was detected in 1 of 2 exconjugants tested (Fig. 2B). Together, these data indicate that neither the SXT primary integration site prfC nor homologous recombination is required in the formation of tandem SXT arrays. Instead, we hypothesize that multiple integrations of circular SXTs occur by Int-mediated site-specific recombination into the attL or attR of an already integrated SXT.
FIG. 2.
Tandem arrays in recA and ΔprfC mutant strains of E. coli. Southern blot analyses of Tetr Sxtr exconjugants resulting from mating experiments involving a recA+ ΔprfC strain (VI31) (A) or a recA ΔprfC strain (VI47) (B) as the recipient. E. coli VI166 (BW25113 prfC::SXT) was used as a donor strain in both experiments. The donor and recipient strains were mated for 6 h at 37°C on LB agar medium without selection. The purified total genomic DNAs of the Tetr Sxtr exconjugants were digested with EcoRI/EcoRV and analyzed by Southern blot hybridization with the 600-bp BstBI fragment of pVI8A containing attP from SXT as a probe. The molecular sizes in kilobases correspond to the fragments containing the attL (6.5 kb, EcoRV), attP (4.0 kb, EcoRI/EcoRV), and attR (1.2 kb, EcoRI/EcoRV) junction fragments in E. coli HW220 (CAG18439 prfC::SXT) used as a control strain.
Tandem structures increase the transfer efficiency of a Δxis SXT mutant.
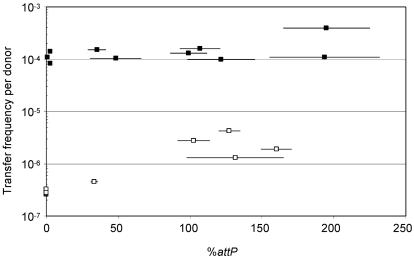
To investigate the effect of tandem SXT arrays on the frequency of exconjugant formation, we isolated 10 randomly selected fresh Sxtr exconjugants derived from a mating where the donor strain harbored a single copy of SXT. These exconjugants were then used as donors in new mating assays, and the percentage of attP was measured for each of these donors. As shown in Fig. 3, there was no significant correlation between the frequency of SXT transfer and the percentage of attP in the exconjugants used as donors.
FIG. 3.
Effect of the tandem arrays on the transfer frequency of SXT Δxis. The frequencies of exconjugant formation per donor were obtained by dividing the number of exconjugants (Nalr Sxtr CFU) by the number of donor cells (Tcr CFU). In all of the mating experiments, the recipient strain was E. coli BI533 (MG1655 Nalr). The donors were E. coli exconjugants obtained from mating VI200 (MG1655 prfC::SXT Δxis pXis) × CAG18439 (□) or from mating VI166 (BW25113 prfC::SXT) × CAG18439 (▪). The expression of xis by pXis in the donor strain VI200 was induced with 0.02% arabinose during the mating experiments on LB agar plates. Real-time quantitative PCR was used to determine the percentage of attP sequences resulting from SXT excision and tandem SXT array formation. Triplicate measurements were performed on each sample, and the means and standard deviations (horizontal bars) are presented for each assay. DNA templates were prepared from overnight LB broth cultures.
Our previous work suggested that, with a Δxis SXT donor, the formation of the extrachromosomal form of SXT was a key step in limiting SXT transfer, since the Δxis SXT cells were 3 orders of magnitude less efficient as donors than cells harboring a single copy of wild-type SXT (7). We wondered whether tandemly arranged Δxis SXTs could at least partially complement the transfer deficiency of a single Δxis SXT donor. An experiment similar to the one described above was carried out to explore this possibility. An initial donor strain harboring a single Δxis SXT in which Xis was provided in trans was used to generate Δxis SXT exconjugants. These exconjugants were then used as donors. The frequency of exconjugant formation from these new donors varied between 2.6 × 10−7 and 4.2 × 10−6 exconjugants/donor (a 16-fold variation). There was a general correlation between the percentage of attP in the donor strain and the frequency of exconjugant formation (Fig. 3). The percentage of attP in 5 of the 10 donors was nearly undetectable (Fig. 3), and these strains were extremely inefficient donors. These strains likely contained a single Δxis SXT, given the very low excision frequency previously observed for the single Δxis SXT (7). The percentage of attP for the five other donors was significantly higher, and these strains were more efficient donors (Fig. 3). The overall trend of the data presented in Fig. 3 shows that tandem Δxis SXT arrays can, at least in part, rescue the transfer deficiency of Δxis SXT mutants. This result could be meaningful in hosts in which SXT is unable to excise.
Tandem SXT and R391 form hybrid elements.
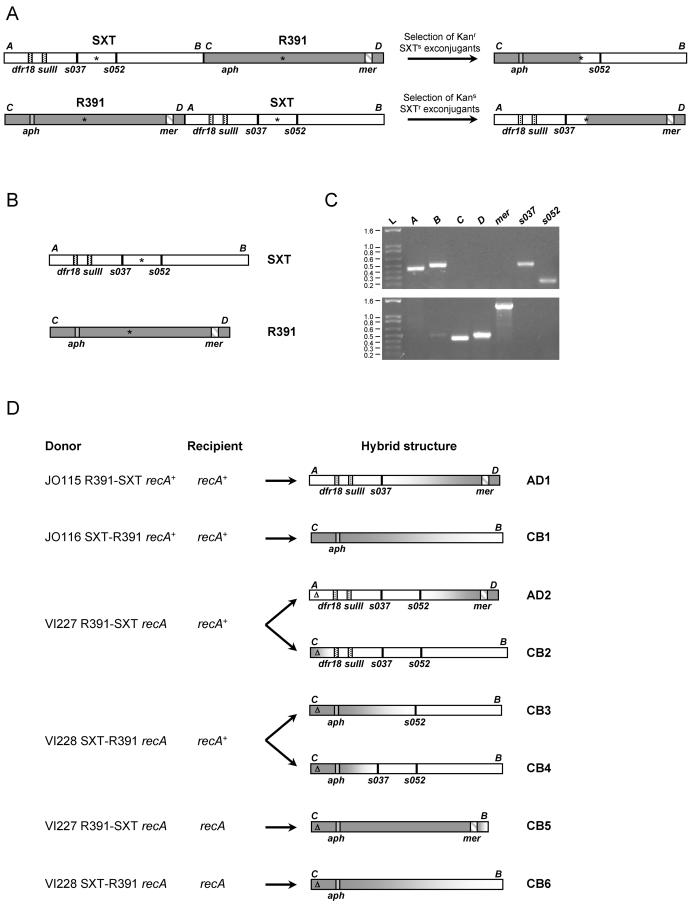
It is not clear how strains harboring Δxis SXT generate extrachromosomal circular SXT DNA or why transfer was higher from strains carrying tandem Δxis SXT arrays. One possibility to explain the latter observation is that the production of Int, required for excision, is higher in cells harboring tandem arrays; however, we previously showed that Int overexpression does not increase the frequency of excision and transfer (7). Another possibility is that tandem arrays allow for different processes for production of the extrachromosomal SXT DNA required for its transfer. For example, extrachromosomal SXT might arise through homologous recombination between adjacent integrated elements. Alternatively, a replicative process, analogous to that used by tandemly arranged CTX prophages (19) might generate extrachromosomal SXT DNA. Both of these processes are expected to yield hybrids containing sequences from adjacent elements. To explore whether either of these models could explain how extrachromosomal SXT DNA is generated by Δxis ICE arrays, we tested whether hybrid ICEs would indeed form by using tandem arrays of SXT and R391 (Kanr Merr). The structure of the hybrids would vary for the two models. If replication initiating at the 5′ oriT and extending to the 3′ oriT in a tandem array generates the extrachromosomal DNA, only one hybrid element would arise from donor cells containing Δxis SXT-Δxis R391 arrays (Fig. 4A). Alternatively, if transfer from xis-deleted arrays depends upon homologous recombination, then many different hybrid elements would arise from donor cells containing Δxis SXT-Δxis R391 arrays, since these ICEs share large segments of identical DNA.
FIG. 4.
Analysis of hybrids formed by recombination between SXT and R391. (A) Schematic representation of the structures of the tandem arrays SXT-R391 and R391-SXT and predicted hybrid ICEs formed by the oriT-to-oriT replication-based mechanism. (B) Schematic representation of the structures of SXT and R391. mer, aph, sulII, and dfr18 encode, respectively, resistances to mercury, kanamycin, sulfamethoxazole, and trimethoprim. The asterisks indicate the positions of the oriT in SXT and the probable oriT in R391. (C) Amplification of sequences specific to SXT and R391. The fragments A (452 bp) and C (449 bp) are specifically amplified from the left part of SXT and R391, respectively, with the primer sets VISLF-VISLR3 and VISLF-VISLR2 (Table 2). The fragments B (509 bp) and D (511 bp) are specifically amplified from the right part of SXT and R391, respectively, with the primer sets VISRF-VIRR and VISRF-VISRR2. mer is a 1,371-bp fragment amplified from R391 withMER104A and MER103B. It covers the middle of the mer operon of R391. The fragments s037 (523 bp) and s052 (242 bp) are specific to SXT and were amplified with the primer sets 10SF13-SXT1-13 and YND2-ORF16, respectively. The sizes of the ladder (L) are given in kilobases. (D) Schematic representation of the structures of the isolated hybrid ICEs. The structure of the hybrids was deduced from the antibiotic resistance phenotype and from the pattern of SXT- and R391-specific fragments detected by PCR, as shown in panel A. The positions of the amplified fragments and the genes encoding the antibiotic resistance are indicated. The precise position of the point of recombination in each hybrid is not known, but it is deduced from the presence or absence of the markers tested. The gradient from white to dark gray indicates the fragment within which the recombination probably took place to generate the hybrids. Δ, Δxis mutation.
For these experiments, the recA+ strains JO115 and JO116 (Table 1), which harbor tandem arrays of wild-type R391-SXT and SXT-R391, respectively (14), were used as donors. We analyzed Sxtr Kans exconjugants when JO115 was used as a donor and Sxts Kanr exconjugants when JO116 was used as a donor because these exconjugants would be more likely to include hybrids that arise from either the replication-based or homologous recombination-based mechanisms outlined above. The structure of exconjugants was analyzed using PCR assays that detect SXT- or R391-specific sequences (Fig. 4B and C). Hybrid elements were readily found. One of the two Sxtr Kans exconjugants harbored a recombinant ICE, AD1, containing the left part of SXT including s037 but not s052 and the right part of R391 (Fig. 4D). One of the six Sxts Kanr exconjugants harbored a recombinant element, CB1, that contained most of R391 and a small region from the right part of SXT. Therefore, donors like JO115 and JO116, where each ICE can excise and transfer independently, can also give rise to hybrid ICEs. While the hybrid AD1 could have arisen from the oriT-to-oriT replication-based mechanism of generation of extrachromosomal DNA, the hybrid ICE CB1 cannot be explained by such a mechanism because it does not include s052, a gene that lies 3′ of the SXT oriT (Fig. 4A and D). Both AD1 and CB1 were functional ICEs and transferred with frequencies similar to that of R391 (Table 4). The process of conjugative transfer appears to be required for generation of recombinant elements from the tandem arrays in JO115 and JO116. We were unable to detect formation of hybrid elements in either of these two strains when they were cultured in the absence of a recipient (data not shown).
TABLE 4.
Mobility properties of hybrid elements
| Elementa | Transfer frequency (10−4)b | Excision frequency (%)c
|
|
|---|---|---|---|
| attB | attP | ||
| Δxis SXT | 0.26 | 2.6 ± 0.2 | 3.9 ± 0.4 |
| Δxis R391 | 2.4 | 3.2 ± 0.6 | 2.0 ± 0.4 |
| AD1 | 2.1 | 7.8 ± 1.3 | 18.6 ± 2.7 |
| CB1 | 5.6 | 1.5 ± 0.2 | 3.5 ± 0.5 |
| AD2 | 0.035 | 5.5 ± 2.6 | 11.4 ± 4.5 |
| CB2 | 2.1 | 3.2 ± 0.6 | 2.9 ± 0.6 |
| CB3 | 4.1 | 5.1 ± 1.2 | 4.5 ± 0.9 |
| CB4 | 3.9 | 6.7 ± 0.6 | 5.2 ± 0.9 |
In the donor strains harboring Δxis SXT, Δxis R391, AD2 CB2, CB3, and CB4, Xis was provided by pXis, expressing xis under control of PBAD to complement the Δxis mutation of the elements.
Transfer frequency was calculated as the frequency of exconjugants per recipient and is the mean value of the results from three independent experiments. The selected markers were aph (Kanr) for R391, CB1, CB3, and CB4 or dfr18 and sulII (Sxtr) for SXT, AD1, AD2, and CB2.
The excision frequency was quantified by measuring the percentage of unoccupied attB sites and the percentage of attP sites by real-time quantitative PCR. The values are the means and standard deviations of the results from three independent measurements.
Next, we evaluated the role of homologous recombination in transfer from tandem arrays of Δxis elements. We constructed a Δxis R391-Δxis SXT array and a Δxis SXT-Δxis R391 array in a recA E. coli strain, yielding VI227 and VI228, respectively (Table 1). We produced SetC and SetD from a plasmid in these strains to bypass RecA's role as a regulator for transfer gene expression. Surprisingly, these two strains were extremely efficient donors. The frequency of transfer of each marker to a recA+ recipient, E. coli BI533, was even higher than that observed with xis+ donors (Table 5). Transfer from these recA donors indicates that homologous recombination is not required for formation of extrachromosomal ICE DNA in Δxis donors containing R391-SXT arrays.
TABLE 5.
Frequency of transfer of the kanamycin and SXT markers
| Donor | Relevant genotypea | Recipient | Relevant genotype | Transfer frequency (10−3)b with selection for:
|
|
|---|---|---|---|---|---|
| Kanamycin | SXT | ||||
| JO115 | R391-SXT recA+ | BI533 | recA+ | 0.43 | 0.18 |
| JO116 | SXT-R391 recA+ | BI533 | recA+ | 0.45 | 0.21 |
| VI227 | R391Δ-SXTΔ recA | BI533 | recA+ | 20.4 | 4.7 |
| VI228 | SXTΔ-R391Δ recA | BI533 | recA+ | 107.6 | 1.8 |
| VI227 | R391Δ-SXTΔ recA | MW5-39 | recA | 10.0 | 5.5 |
| VI228 | SXTΔ-R391Δ recA | MW5-39 | recA | 70.4 | 0.5 |
Δ indicates the deletion of xis. The recA donors carried pSetCD to provide SetC and SetD, which activate the expression of the transfer genes.
The frequency of transfer was calculated by determining the number of exconjugants per recipient, since the overexpression of setC and setD in presence of SXT is detrimental to the growth of the donor cells. The transfer frequencies are the means of the results from three independent experiments. The mating experiments were carried out for 6 h at 37°C, and the medium was supplemented with 0.02% arabinose when the donors carried pSetCD.
Some of the exconjugants derived from both VI227 and VI228 donors contained hybrid elements composed of parts from SXT and R391. For example, two of the Kans Sxtr exconjugants derived from the VI227 donor were found to carry recombinant elements, AD2 and CB2. AD2 contained the left part of SXT and the right part of R391 and CB2 contained the left part of R391 and the right part of SXT (Fig. 4D). Since the hybrids AD2 and CB2 arose from the same donor, the relative position of SXT and R391 in the tandem arrays does not appear to determine the structure of the resulting hybrid element. Two of the Kanr Sxts exconjugants derived from the VI228 donor harbored hybrids, CB3 and CB4, that possessed the left part of R391 and the right part of SXT, although these hybrid ICEs were not identical (Fig. 4D). The identification of hybrids arising from recA donors ruled out the involvement of homologous recombination in the donor. Furthermore, the variety of hybrids detected is additional evidence that a replicative process initiating at the 5′ oriT in a Δxis SXT-Δxis R391 array cannot be the sole explanation for transfer from such arrays nor for the generation of hybrid elements. All of the hybrid ICEs tested were capable of transfer to new hosts (Table 4).
We performed similar experiments with the same donor strains but with a recA E. coli recipient to verify that homologous recombination did not take place in recipient cells. The transfer frequencies of both markers were comparable to those observed in the experiments involving the recA+ recipient (Table 5). We isolated two distinct hybrid elements, CB5 and CB6, from 48 randomly selected Sxts Kanr exconjugants. CB5 was derived from the VI227 donor, and CB6 was derived from the VI228 donor (Fig. 4D). Thus, homologous recombination in recipient cells is not required in the formation of hybrid ICEs.
DISCUSSION
Tandem arrays of SXT were frequently found in freshly derived SXT exconjugants. There was considerable heterogeneity in the size of SXT arrays present in cells from a single colony. Some of these arrays contained more than five copies, indicating that SXT arrays could represent greater than 10% of the E. coli genome. However, these large arrays were unstable and did not persist through serial passage. The mechanism that underlies the formation of SXT arrays is not known, but array formation appears to depend upon conjugative transfer of SXT, since we did not detect arrays following serial passage of a strain harboring a single SXT. Besides illustrating the dynamic nature of chromosome structure in SXT exconjugants, formation of SXT arrays has functional consequences. The presence of SXT arrays can partially rescue the defect in SXT transfer that occurs in the absence of Xis. Furthermore, SXT-R391 arrays facilitate the emergence and evolution of novel ICEs by enabling formation of hybrid elements.
The heterogeneity in SXT copy number in fresh exconjugants was observed in quantitative PCR assays and Southern blots measuring the amount of attP in exconjugants as well as in PFGE analyses. The percentages of attP sites in exconjugants were never integer multiples of 100%, as expected for discrete SXT copy numbers. The fact that we did not detect such values reflects the variation in the sizes of the SXT arrays among cells from individual exconjugant colonies. The attP percentage measured by this PCR assay represents the mean value of the SXT copy number. Thus, even if strains have similar attP percentages, the distribution of SXT copy number in the cell populations may vary. Although an idea about the distribution of SXT copy number cannot be gleaned from the quantitative PCR analyses, the PFGE analysis, as shown in Fig. 1B, provides information regarding this distribution.
We found that the percentage of unoccupied attB sites remains relatively constant, varying from 1.32 to 3.21%, irrespective of the percentage of attP sites (i.e., of the presence or size of SXT arrays) in a strain. The relative stability of the excision frequency may indicate that entire SXT arrays excise as a single circular molecule by recombination between the most distal attL and attR sites flanking the array. Recombination involving internal attP sites between adjacent SXTs in an array could be inhibited due to interactions with Xis, since this protein apparently inhibits recombination between attP and attB, preventing SXT integration (7).
Although formation of tandem SXT arrays appears to occur frequently in exconjugants from both E. coli and V. cholerae, these arrays were transient. Preliminary observations suggest that tandem SXT arrays are stable in the absence of recA (data not shown). Homologous recombination between the elements in a tandem array may lead to reductions in array size. We were unable to find SXT tandem arrays in V. cholerae Sxtr clinical isolates (data not shown), suggesting that arrays are also unstable in natural isolates.
SXT is not the only ICE to form tandem arrays, R391 arrays were also found in freshly derived exconjugants (data not shown). Furthermore, tandem arrays of the 105-kb clc element, an ICE derived from Pseudomonas sp. strain B13, which contains genes for chlorocatechol degradation, have been reported previously (20). Interestingly, Ravatn et al. reported a correlation between the clc copy number in exconjugants and their capability to grow on chlorobenzene as the sole carbon source and suggested that the selective pressure exerted by the substrate influenced the clc copy number (20). In contrast, our work suggests that tandem arrays of SXT or R391 arise as a consequence of the conjugative process itself, since we were unable to detect SXT arrays in a strain that initially contained a single copy of SXT, even on sulfamethoxazole and trimethoprim-supplemented medium.
Several conjugation-dependent mechanisms could explain the formation of SXT arrays. Tandem arrays could form after transfer of several copies of the element from a single donor cell to a single recipient cell. This could occur if a concatemer of several copies of SXT was transferred to a recipient. Alternatively, a single donor may transfer several copies of single SXT to the same recipient. The formation of SXT arrays could also occur following transfer of a single SXT from several donors to a single recipient cell. Given the homology of SXT transfer-related genes to those of conjugative plasmids, we assume that a single-stranded SXT, generated by a rolling-circle-like process, is transferred from donor to recipient.
It was previously shown that SXT and R391 are able to use an attL or attR site for integration (14). Since we found that recA was not required for the formation of SXT arrays, if multiple transfers of SXT account for the generation of the arrays, SXT integration into the recipient chromosome probably occurs by an Int-mediated site-specific recombination event into the attL or attR site of a previously integrated element. We found that prfC, the SXT primary integration site, is not required for the formation of SXT arrays. Therefore, if multiple rounds of SXT transfer occur in a ΔprfC mutant already containing an SXT, the novel attL or attR sites are sufficient for SXT integration. Given the relatively low frequency of transfer of SXT and R391, it seems improbable that multiple donors transfer SXT to the same recipient. However, it is possible that there is some period of time after a recipient has acquired SXT, during which it is transiently highly proficient as a donor. When SXT has just entered a new host cell, the level of the repressor SetR may not be sufficient to repress expression of the tra genes. Epidemic spread of F-like conjugative plasmids from recent exconjugants has been reported (30). Perhaps SXT arrays form after a burst of SXT transfer from recent recipients to new recipients and to former donors.
Tandem SXT-R391 arrays in donor cells can lead to the formation of hybrid ICEs in exconjugants. Each hybrid ICE we identified was able to excise and transfer by conjugation to new recipients. Some of these hybrids, like AD1 and AD2, confer resistance to SXT and likely to mercury as well, two phenotypes specific to each parental element. Therefore, heterogeneous tandem ICE arrays facilitate the generation of ICEs with novel combinations of properties and contribute to the plasticity of this family of elements. Although the mechanism generating these hybrid elements is still unknown, we showed that neither homologous recombination nor oriT-to-oriT replication is required in their formation. Since SXT and R391 both contain orthologues of the Bet recombination protein found in several bacteriophages (54% identity of SXT S065 and R391 Orf68 with Bet protein of phage λ) (2, 4), we hypothesize that these recombinases play a role in the formation of the hybrid ICEs.
Acknowledgments
We thank B. Davis, A. Kane, S. McLeod, and J. Ritchie for critical reading of the manuscript. We are grateful to L. McDermott for help with the PFGE experiments.
This work was supported by funds from NIH grant AI42347, the Howard Hughes Medical Institute, and the NEMC GRASP Center (grant P30DK-34928).
REFERENCES
- 1.Beaber, J. W., V. Burrus, B. Hochhut, and M. K. Waldor. 2002. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell. Mol. Life Sci. 59:2065-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72-74. [DOI] [PubMed] [Google Scholar]
- 4.Boltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 6.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77-97. [DOI] [PubMed] [Google Scholar]
- 7.Burrus, V., and M. K. Waldor. 2003. Control of SXT integration and excision. J. Bacteriol. 185:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu, G., D. Vollrath, and R. W. Davis. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582-1585. [DOI] [PubMed] [Google Scholar]
- 9.Coetzee, J. N., N. Datta, and R. W. Hedges. 1972. R factors from Proteus rettgeri. J. Gen. Microbiol. 72:543-552. [DOI] [PubMed] [Google Scholar]
- 10.Dalsgaard, A., A. Forslund, D. Sandvang, L. Arntzen, and K. Keddy. 2001. Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons. J. Antimicrob. Chemother. 48:827-838. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg, I., and J. J. Mekalanos. 1986. Effect of a recA mutation on cholera toxin gene amplification and deletion events. J. Bacteriol. 165:723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heath, J. D., J. D. Perkins, B. Sharma, and G. M. Weinstock. 1992. NotI genomic cleavage map of Escherichia coli K-12 strain MG1655. J. Bacteriol. 174:558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochhut, B., J. W. Beaber, R. Woodgate, and M. K. Waldor. 2001. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J. Bacteriol. 183:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochhut, B., J. Marrero, and M. K. Waldor. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 182:2043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochhut, B., and M. K. Waldor. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32:99-110. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Moyer, K. E., H. H. Kimsey, and M. K. Waldor. 2001. Evidence for a rolling-circle mechanism of phage DNA synthesis from both replicative and integrated forms of CTXφ. Mol. Microbiol. 41:311-323. [DOI] [PubMed] [Google Scholar]
- 20.Ravatn, R., S. Studer, D. Springael, A. J. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice, L. B. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thungapathra, M., Amita, K. K. Sinha, S. R. Chaudhuri, P. Garg, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2002. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob. Agents Chemother. 46:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Meer, J. R., and V. Sentchilo. 2003. Genomic islands and the evolution of catabolic pathways in bacteria. Curr. Opin. Biotechnol. 14:248-254. [DOI] [PubMed] [Google Scholar]
- 27.Waldor, M. K., and J. J. Mekalanos. 1994. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect. Immun. 62:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldor, M. K., H. Tschape, and J. J. Mekalanos. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178:4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zatyka, M., and C. M. Thomas. 1998. Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiol. Rev. 21:291-319. [DOI] [PubMed] [Google Scholar]