Abstract
The definitive phage types (DT) 2 and 99 of Salmonella enterica serotype Typhimurium are epidemiologically correlated with a host range restricted to pigeons, in contrast to phage types with broader host ranges such as epidemic cattle isolates (DT104 and DT204). To determine whether phage types with broad host range possess genetic islands absent from host-restricted phage types, we compared the genomes of four pigeon isolates to serotype Typhimurium strain LT2 using a DNA microarray. Three of the four isolates tested caused fluid accumulation in bovine ligated ileal loops, but they had reduced colonization of liver and spleen in susceptible BALB/c mice and were defective for intestinal persistence in Salmonella-resistant CBA mice. The genomes of the DT99 and DT2 isolates were extremely similar to the LT2 genome, with few notable differences on the level of complete individual genes. Two large groups of genes representing the Fels-1 and Fels-2 prophages were missing from the DT2 and DT99 phage types we analyzed. One of the DT99 isolates examined was lacking a third cluster of five chromosomal genes (STM1555 to -1559). Results of the microarray analysis were extended using Southern analysis to a collection of 75 serotype Typhimurium clinical isolates of 24 different phage types. This analysis revealed no correlation between the presence of Fels-1, Fels-2, or STM1555 to -1559 and the association of phage types with different host reservoirs. We conclude that serotype Typhimurium phage types with broad host range do not possess genetic islands influencing host restriction, which are absent from the host-restricted pigeon isolates.
Salmonella enterica is the cause of an estimated 1.4 million cases of enterocolitis annually in the United States, and it is the most common cause of food-borne illness with lethal outcome, resulting in approximately 500 deaths annually (21). Transmission to humans occurs largely due to the ingestion of meat, meat products, eggs, and egg products that are contaminated by fecal material. S. enterica serotype Typhimurium is the serotype currently most frequently associated with enterocolitis in humans, accounting for 26% of cases reported to the Centers for Disease Control and Prevention (CDC) in 1998 (6), although 70% of over 2,000 serotypes of Salmonella have been isolated from patients with enterocolitis (13). This statistic highlights the remarkable ability of serotype Typhimurium to persist in the human food supply, particularly in cattle, causing significant human and animal disease.
S. enterica serotypes form a group of pathogens that vary in their host ranges within mammals and birds. Some members of this group, of which S. enterica serotype Typhi is the prototype (9), have a strict and narrow host range, while others are able to infect a broader spectrum of mammals and birds. Serotype Typhimurium is typically thought of as the pathogen of this group that can infect and persist in a broad range of host species. Past attempts to define factors responsible for host adaptation between S. enterica serotypes by moving genes from a broad-host-range pathogen into a narrow-host-range pathogen have, as yet, been unsuccessful (28; L. Pascopella, S. Falkow, and P. L. C. Small, Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996, abstr. B-109, p. 173, 1996). This type of analysis is complicated by the large amount of serotype-specific DNA in these bacterial genomes (20, 23).
Serotype Typhimurium consists of a collection of isolates that are more closely related to each other than they are to other S. enterica serotypes (3, 4) and are distinguishable from each other by phage typing and other epidemiologic typing methods (27). Epidemiologic evidence indicates that certain phage types of serotype Typhimurium have a narrow host range and are very frequently isolated from particular host species and rarely from other mammals or birds. The serotype Typhimurium phage types DT2 and DT99 are frequently isolated from pigeons, where they cause paratyphoid (5), but are rarely isolated from other bird or mammalian species (27). On this basis, the existence of a serotype Typhimurium “pigeon type” has been proposed (22, 32, 35, 37). Recent in vivo experiments testing the virulence in pigeons of pigeon (DT99; Copenhagen) and porcine (also Copenhagen) isolates of serotype Typhimurium showed that porcine isolates are less virulent in pigeons, causing lower mortality, a less severe clinical syndrome, and lower bacterial load in internal organs (24). A reasonable hypothesis based on these observations is that DT2 and DT99 pigeon isolates have additional genetic determinants, not shared by other serotype Typhimurium phage type isolates, that allow them to be better pathogens in pigeons. This hypothesis is not testable by genomic approaches until complete genome sequence data become available for any DT2 or DT99 isolates. By analogy, we can postulate that DT2 and DT99 pigeon isolates may lack genetic determinants present in serotype Typhimurium phage types associated with disease in livestock and food-borne infections in humans. With the advent of microarray analysis for genomic content and the availability of the complete genome sequence of broad-host-range isolates, this hypothesis is now testable.
Phage types of serotype Typhimurium associated with disease in livestock generally have a broader host range and are frequently transmitted to humans via food products. These phage types include DT49, DT104, and DT204, which have been associated with epidemics among livestock and humans in Europe and the United States (27). While differences in host range among different serotype Typhimurium phage types are well established through epidemiological surveys, the genetic basis for this phenomenon is currently unknown.
The serotype Typhimurium Lilleengen type 2 (LT2) strain is commonly studied in the laboratory and originated as a human isolate from a food-borne disease outbreak in the United Kingdom in 1948 (18). The sequenced isolate of serotype Typhimurium LT2 belongs to phage type DT4 in the Anderson system (20), and this phage type was frequently isolated from livestock, domestic fowl, and humans in Germany during the 1980s (W. Rabsch, unpublished data). While some LT2 sublines remain virulent for mice (12) and humans (2), others have become avirulent for mice since description of the original isolate in 1948. This loss of mouse virulence of some LT2 sublines is not due to deletion of virulence genes but is due to the acquisition of a point mutation in rpoS during laboratory passage (34, 36). The gene complement of the LT2 strain could thus be seen as representative for serotype Typhimurium strains associated with livestock and enterocolitis in humans. Sequencing of the serotype Typhimurium LT2 genome and development of genomic microarrays for this serotype provide a unique opportunity to directly compare the genomes of serotype Typhimurium isolates of different phage types and host ranges (8, 20, 26, 27).
We compared two serotype Typhimurium DT2 and two serotype Typhimurium DT99 pigeon isolates to serotype Typhimurium LT2 (DT4) using microarray analysis to determine whether the host restriction of serotype Typhimurium pigeon isolates is attributable to the loss of discrete genes present in a clinical isolate from a human food-borne disease outbreak. We extended this analysis using Southern blotting against genomic DNA of a large collection of serotype Typhimurium clinical isolates, in an attempt to develop a correlation between the presence of certain genes in particular phage types and host restriction. In addition, we tested the ability of the serotype Typhimurium pigeon isolates analyzed by microarray in this study to cause fluid accumulation in bovine ligated ileal loops and to compete with virulent serovar Typhimurium strain ATCC 14028 in susceptible and resistant mouse models of infection.
MATERIALS AND METHODS
Bacterial strains.
Serotype Typhimurium LT2 is a standard laboratory strain, first described in 1948 and used in early studies of phage-mediated transduction (18). It was obtained from the American Type Culture Collection (ATCC 700720). IR715 is a virulent, nalidixic acid-resistant derivative of serotype Typhimurium strain ATCC 14028 and has been previously described (33). AJB715 is a phoN mutant derivative of IR715 and has been previously described (15). Serotype Typhimurium strain SR11 has been previously described (31) and was used in bovine ligated ileal loop experiments.
The clinical isolates described in this study were obtained from the collection of the Robert Koch Institute. These isolates were collected at necropsies over a 28-year period, and all isolates have been phage typed. The collection is fully described in Table 1. The four serotype Typhimurium variants analyzed using the microarray were from this collection and are clinical isolates collected post mortem from pigeons in Germany. Two phage type DT2 strains (R14 and R16) and two phage type DT99 strains (HA4 and R33) were used in this study and were chosen on the basis of collection at the widest time intervals available. Of the DT2 strains, R14 was collected in 1988 and R16 was isolated in 1994. Of the DT99 strains, R33 was collected in 1988 and HA4 was collected in 1996.
TABLE 1.
Clinical isolate collectiona
| Strain | Isolate name | Date of isolation | Phage type | Host species | Location |
|---|---|---|---|---|---|
| R1 | STM55 | 1990 | DT104 | Bovine | Jessen |
| R2 | STM174 | 1994 | DT104 | Bovine | Jena |
| R3 | STM3794 | 1996 | DT104 | Bovine | Nuremberg |
| R4 | STM6199 | 1996 | DT104 | Bovine | Neubrandenburg |
| R5 | STM10666 | 1997 | DT104 | Bovine | Rodach |
| R6 | STM2081 | 1997 | DT104 | Bovine | Kronach |
| R7 | STM2120 | 1997 | DT104 | Bovine | Lichtenfels |
| R8 | STM6335 | 1997 | DT104 | Bovine | Tirschenreuth |
| R9 | STM760 | 1998 | DT104 | Bovine | Frauenroth |
| R10 | STM2447 | 1998 | DT104 | Bovine | Waidhaus |
| R11 | STM16603 | 1997 | DT104 | Bovine | Grimannsberg |
| R12 | STM8663 | 1999 | DT104 | Bovine | Lengenfeld |
| R13 | STM515 | 1999 | DT104 | Bovine | Kulmbach |
| R14* | STM138 | 1988 | DT2 | Pigeon | Karl-Marx-Stadt |
| R15 | STM2058 | 1992 | DT2 | Pigeon | Potsdam |
| R16* | STM213 | 1994 | DT2 | Pigeon | Neumünster |
| R17 | STM3406 | 1994 | DT2 | Pigeon | Dresden |
| R18 | STM16 | 1996 | DT2 | Pigeon | Neubrandenburg |
| R19 | STM3043 | 2000 | DT2 | Pigeon | Dresden |
| R20 | STM9652 | 2001 | DT2 | Pigeon | Dresden |
| HA8 | STM9034 | 1999 | DT2 | Pigeon | Ellersleben |
| HA9 | STM4752 | 2000 | DT2 | Pigeon | Neumünster |
| HA10 | STM1025 | 2001 | DT2 | Pigeon | Dresden |
| R21 | STM1190 | 1974 | DT204 | Human | Gera |
| R22 | STM1491 | 1975 | DT204 | Bovine | Dresden |
| R23 | STM1666 | 1979 | DT204 | Bovine | Halle |
| R24 | STM2166 | 1990 | DT204 | Bovine | Borna |
| R25 | STM1749 | 1996 | DT204 | Bovine | Schwerin |
| R26 | STM10011 | 2000 | DT204 | Bovine | Kiel |
| R27 | STM10022 | 2000 | DT204 | Bovine | Kiel |
| R28 | STM210 | 1994 | DT204c | Bovine | Neumünster |
| R29 | STM1582 | 1994 | DT204c | Bovine | Potsdam |
| R30 | STM6203 | 1997 | DT204c | Bovine | Celle |
| R31 | STM6209 | 1997 | DT204c | Bovine | Celle |
| R32 | STM16448 | 1997 | DT204c | Bovine | Giessen |
| R33* | STM222 | 1988 | DT99 | Pigeon | Berlin |
| R34 | STM2799 | 1992 | DT99 | Pigeon | Saalkreis |
| R35 | STM217 | 1994 | DT99 | Pigeon | Neumünster |
| R36 | STM2708 | 1994 | DT99 | Pigeon | Stendal |
| R36 | STM2708 | 1994 | DT99 | Pigeon | Stendal |
| R37 | STM1453 | 1996 | DT99 | Pigeon | Berlin |
| R38 | STM951 | 2000 | DT99 | Pigeon | Potsdam |
| R39 | STM2134 | 2000 | DT99 | Pigeon | Bad Langensalza |
| HA4* | STM7896 | 1996 | DT99 | Pigeon | Unterhaching |
| HA5 | STM951 | 2000 | DT99 | Pigeon | Berlin |
| HA6 | STM2134 | 2000 | DT99 | Pigeon | Hesserode |
| HA7 | STM514 | 2001 | DT99 | Pigeon | Bad Langensalza |
| R40 | STM2870 | 1991 | DT49 | Bovine | Bautzen |
| R41 | STM747 | 1994 | DT49 | Bovine | Bad Langensalza |
| R42 | STM444 | 1996 | DT49 | Bovine | Nuremberg |
| R43 | STM8260 | 1996 | DT49 | Bovine | Nuremberg |
| R44 | STM1052 | 1988 | DT193 | Bovine | Fuerstenwalde |
| R45 | STM4865 | 1996 | DT193 | Bovine | Stendal |
| R46 | STM6343 | 1997 | DT193 | Bovine | Nuremberg |
| R47 | STM376 | 1988 | DT15A | Bovine | Halle |
| R48 | STM575 | 1988 | DT1 | Bovine | Bischofswerda |
| R49 | STM1522 | 1989 | DT1 | Bovine | Zschopau |
| R50 | STM958 | 1988 | DT10 | Bovine | Gotha |
| R51 | STM2691 | 1991 | DT10 | Bovine | Forst |
| R52 | STM1520 | 1988 | DT17 | Bovine | Wanzleben |
| R53 | STM1301 | 1989 | DT17 | Bovine | Freital |
| R54 | STM245 | 1990 | DT17 | Bovine | Kloetze |
| R55 | STM1666 | 1988 | DT12 | Bovine | Meiningen |
| R56 | STM1100 | 1992 | DT12 | Bovine | Eisenbach |
| R57 | STM2942 | 1990 | DT66 | Bovine | Freiberg |
| R58 | STM2848 | 1991 | DT66 | Bovine | Forst |
| R59 | STM3251 | 1994 | DT66 | Bovine | Oberschleissheim |
| R60 | STM8360 | 1996 | DT66 | Bovine | Stendal |
| R61 | STM1215 | 1991 | DT22 | Bovine | Zossen |
| R62 | STM8710 | 1996 | DT7 | Bovine | Dresden |
| R63 | STM1855 | 1991 | DT41 | Bovine | Sangerhausen |
| R64 | STM7080 | 1996 | DT106 | Bovine | Neumünster |
| R65 | STM2547 | 1992 | DT12A | Bovine | Querfurt |
Asterisks indicate strains subjected to microarray analysis.
Strains were routinely cultured in Luria-Bertani (LB) broth and plates, supplemented with 50 mg of nalidixic acid/liter and 30 mg of kanamycin/liter where appropriate. For the detection of PhoA expression, 20 mg of 5 bromo-4-chloro-3-indolyl-β-d-galactophospate (XP)/liter was added to LB agar plates. All strains were grown aerobically at 37°C in LB broth for genomic DNA isolation and for infection of susceptible and resistant mice (BALB/c and CBA). For bovine ligated ileal loop experiments, all strains were grown for 16 to 18 h statically at 37°C in LB broth.
Genomic DNA preparation and probe labeling.
Genomic DNA from each bacterial strain was isolated using one of the following techniques. Bacteria were grown overnight in LB broth at 37°C with aeration and harvested by centrifugation at 3,300 × g (Eppendorf 5804 R; A-4-44 rotor). Pellets were resuspended in 10 ml of phosphate-buffered saline (pH 7.4) containing 1.8% sodium dodecyl sulfate and 0.1 mg of proteinase K/ml. This mixture was incubated at 37°C overnight or until clear. A 7-ml volume of phenol-chloroform was added, and the solution was inverted until well mixed. The aqueous phase was separated by centrifugation for 3 min at 3,300 × g and collected. Five milliliters of chloroform-isoamyl alcohol (24:1) was added, and the sample was mixed by gentle inversion. The aqueous phase was separated by centrifugation for 3 min at 3,300 × g and collected. In order to precipitate the DNA, a 1/10 volume of 3 M sodium acetate (pH 4.8) and 2.5 volumes of absolute ethanol were added, and samples were gently inverted. DNA was hooked out and briefly washed in 70% ethanol. Samples were air dried for several minutes and resuspended in 0.5 ml of distilled H2O. RNase A was added to a final concentration of 0.4 mg/ml. All genomic DNA used for making probes was prepared in this fashion. For Southern blotting, genomic DNA was prepared by the hexadecyltrimethyl ammonium bromide procedure in some instances (1).
Aliquots of 1.5 μg of genomic DNA were used as the templates for generating probes using P. Brown's protocol as previously described (http://cmgm.stanford.edu/pbrown/protocols/4_genomic.html) (8). Aliquots of 12 μg of random hexamers (Sigma Genosys, Houston, Tex.), 10 U of Klenow fragment (NEB, Beverly, Mass.), and 2 nmol of Cy3-dCTP or Cy5-dCTP (Amersham, Piscataway, N.J.) were included in the labeling reaction that was incubated for 16 to 20 h at 37°C. Probes were purified using a QIAquick PCR purification kit (Qiagen), dried, and solubilized in 10 μl of sterile distilled water. Probes were resuspended in hybridization buffer (4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.2% sodium dodecyl sulfate, and 200 μg of salmon sperm DNA [Stratagene, La Jolla, Calif.]/ml) prior to use. Probes were denatured by boiling for 2 min immediately prior to use.
Microarray construction, hybridization, image acquisition, and data analysis.
S. enterica serovar Typhimurium (LT2) open reading frame microarray construction was done as previously described (8, 26). The genomic DNA of each serotype Typhimurium isolate used was labeled with Cy3 or Cy5 and mixed with an equal amount of LT2 genomic DNA labeled with the opposite fluorophore. This mixture was hybridized to the microarray. Two slides containing identical triplicate arrays were prepared for each serotype Typhimurium clinical isolate tested, using reciprocal fluorophore combinations, generating six data points for each open reading frame. Probes were hybridized to the microarray in hybridization chambers (Telechem International) overnight submerged in a 55°C water bath and protected from light. Prior to scanning, slides were washed according to the manufacturer's instructions and dried by spinning at 500 rpm for 5 min at room temperature. Slides were scanned using a ScanArray 5000 laser scanner (Packard BioChip Technologies, Billerica, Mass.) using ScanArray 3.1 software. Signal intensities were quantified using QuantArray 3.0 software (Packard BioChip Technologies). Intensities of individual spots were analyzed by adaptive quantification methods using local background subtraction, the percentage of contribution of each spot to total signal in each channel was calculated, the ratios of these percentages were identified, and the median of the six ratios per gene was recorded as previously described (8, 26). Scattered data graphs with the log10 of median ratio values of test strains against the LT2 strain were produced. Genes displaying the lowest hybridization signals (lowest 5%) with LT2 strain probes were excluded prior to graphical representation. Log values lower than −0.4 were taken as defining the absence of a gene in test strains. This cutoff is greater than 3 standard deviations from the mean of the ratios for a set of genes known to be shared by Salmonella and three other enterobacteria (20). A log value of +0.4 was chosen to identify a gene amplification (8, 26).
Southern analysis for verification of microarray data.
Genomic DNA was prepared as described above, from a large group of clinical isolates obtained from the collection of the Robert Koch Institute. Genomic DNA was digested with EcoRI for 2 to 3 h at 37°C and diluted with DNA loading buffer. Samples were run on 0.7% agarose gels at 18 V for 18 to 20 h at room temperature and were blotted to positively charged nylon membranes by standard techniques (Southern transfer). Blots were probed using probes prepared with a Renaissance random primed labeling kit, and Southern blots were probed and developed according to kit instructions (no. NEL803; Perkin-Elmer, NEN).
Probe generation.
Probes against the Fels-1 and Fels-2 prophages were generated in the following manner. Genes on Fels-1 and Fels-2 prophages were chosen based on a high likelihood that they would be specific for the particular prophage under study. The criteria for this consisted of choosing genes within the prophage that lacked homology to previously described phage genes. For Fels-1, STM879 was chosen as an open reading frame within the phage genome that lacks significant homology to genes from other phages. STM2724 was chosen from the Fels-2 prophage based on its lack of homology to genes of other known phages. STM2724 is located near the 3′ end of Fels-2. All of the genes in region III were used to generate probes for Southern analysis of this region (STM1555 to -1559).
In order to generate probes, the designated open reading frames were generated by PCR and cloned into pCR2.1-TOPO (Invitrogen). Open reading frames of interest were cut from these plasmids using EcoRI and gel purified, and the DNA was purified from the agarose using a Qiaex II agarose gel extraction kit (Qiagen). Purified fragments were used as templates in a random-primed labeling reaction (NEN) overnight at room temperature, protected from light.
Testing fluid accumulation using bovine ligated ileal loops.
Four- to 6-week-old male Holstein calves were obtained, fed, and tested for salmonellosis prior to surgery as described previously (29). Ligated ileal loop surgery was performed as described previously (29). Loops were injected with either 109 virulent serovar Typhimurium SR11 or with equal numbers of DT2 isolates R14 and R16 or DT99 isolates R33 or HA4. Infected ligated loops were returned to the abdominal cavity and incubated for 8 h. At this time, loops were excised, the volume of accumulated fluid was determined, and tissue samples were collected to determine tissue-associated serovar Typhimurium CFU. The amount of fluid per centimeter of ileum was calculated, and the mean fluid accumulation (in milliliters per centimeter) of three independent experiments was determined. Statistical significance was determined using Student's t test and a P value of <0.05.
Organ colonization in Salmonella-susceptible BALB/c mice.
Clinical isolates analyzed on the microarray were tested for virulence in 6- to 8-week-old female BALB/c mice (Jackson Labs) in mixed infections with virulent S. enterica serotype Typhimurium 14028 (AJB715) using the following protocol. S. enterica serotype Typhimurium strains used as inocula were grown overnight at 37°C with aeration, diluted 1:10 in LB, and mixed in a 1:1 ratio of clinical isolate to AJB715. Inocula were serially diluted, and titers were determined for bacterial CFU and to determine the exact ratio of both strains in the competitive infection.
Groups of four mice were inoculated intragastrically by gavage with approximately 2 × 107 bacteria in 200 μl of LB. Infected mice were observed daily for signs of illness and were euthanized after the development of signs, usually in about 4 days (inactivity and reluctance to move, ruffled fur, crouching together). Immediately after euthanasia, livers and spleens of infected mice were excised and homogenized in 5 ml of ice-cold phosphate-buffered saline. Homogenates were serially diluted and plated to determine the ratio of pigeon variant CFU versus virulent AJB715 from the liver and spleen of infected animals. Data were converted logarithmically and displayed graphically, and statistical significance was determined using Student's t test and a P value of <0.05.
Persistence in Salmonella-resistant CBA mice.
Clinical isolates analyzed on the microarray were tested for the ability to persist in Salmonella-resistant CBA mice in competitive infections with virulent 14028 derivative AJB715. S. enterica serotype Typhimurium strains were grown to stationary phase at 37°C with aeration and were mixed 1:1 prior to inoculation. Twelve-week-old CBA mice were infected intragastrically by gavage with an equal mixture of pigeon variant and AJB715, approximately 2 × 108 in 200 μl of LB in groups of five mice (100 μl of each stationary-phase culture). Approximately 100 mg of feces was collected at various time intervals (1, 3, 5, 7, 10, 15, 20, and 30 days), serially diluted, and plated for CFU of pigeon variant versus AJB715 on LB plus XP plates containing nalidixic acid.
RESULTS
Comparison of genomic content by microarray analysis.
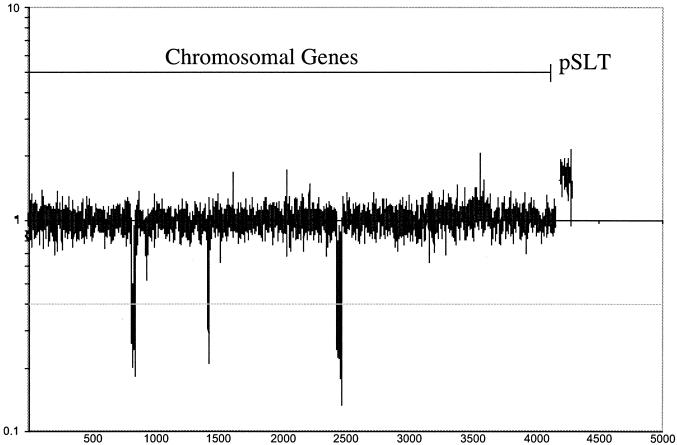
Microarray data were collected and analyzed for four clinical isolates (R14, R16, R33, and HA4) whose phage type had previously been identified as DT2 or DT99 (Table 1). These two phage types are commonly associated with infection of pigeons but are very rarely associated with disease in other warm-blooded species. The data for all four strains were very similar with respect to chromosomally located and virulence plasmid-associated genes of LT2 (Table 2). Presence or absence of genes was evaluated as previously described (Materials and Methods). All of the clinical pigeon isolates examined lacked genes in two chromosomal regions (Fig. 1), and one of the strains examined lacked a third chromosomal region.
TABLE 2.
Regions of genetic differences between S. typhimurium LT2 and phage type DT2 and DT99 isolates
| Region | Genes | Function (no. of genes involved) |
|---|---|---|
| I | STM893 to STM929 | Fels-1 prophage, complete deletion (37 genes) |
| II | STM2694 to STM2739 | Fels-2 prophage, deletion (46 genes) |
| III | STM1555 to 1559 | Various putative functions (4 genes): putative transcriptional regulator STM1555, putative Na+/H+ antiporter STM1556, putative aminotransferase STM1557, and putative glycosyl hydrolases STM1558 and -1559 |
FIG. 1.
Quantitative results of genomic comparison using a serotype Typhimurium LT2 microarray to strain R16, a DT2 pigeon isolate from the German city of Neumünster (1994). Three areas of deleted genes in strain R16 relative to strain LT2 are marked I, II, and III. The y axis denotes the medians of the DT2/LT2 signal ratio plotted on a logarithmic scale. The grey line represents the cutoff value for presence or absence of a gene; peaks that reach below this line represent genes that are absent from the test strain.
Region I included genes STM0893 to STM929 that encompassed the entire genome of the Fels-1 prophage present in LT2 (Fig. 1). Several of the genes in region I did not fall below the cutoff value of −0.4 for one or more of the pigeon isolates tested and thus should be scored as present. However, of these genes, at least six (STM907, STM908, STM911, STM922, STM923, and STM927) are known to cross-hybridize with other open reading frames in serotype Typhimurium LT2 (S. Porwollik, unpublished data). Cross-hybridization of the remaining 11 open reading frames of the Fels-1 prophage likely occurred due to the presence of close prophage homologs in pigeon isolate strains DT2 and DT99. The complete genome sequence of a pigeon isolate DT2 or DT99 phage type will be needed in order to identify additional cross-hybridizing open reading frames.
Region II included genes STM2695 to STM2739, encompassing the Fels-2 prophage genes in LT2. Although, within the prophage, 25 of the genes in region II did not fall below the cutoff value for absent genes and therefore were scored as present. Of these genes, four open reading frames have known cross-hybridizing homologs in LT2 (STM2703, STM2712, STM2713, and STM2714). Presumably, the pigeon isolate strains contain close homologs of the remainder of these prophage genes that cross-hybridize with the LT2 open reading frames on the microarray.
In addition, one of the clinical isolate strains examined (R16) lacked a cluster of five genes (STM1555 to STM1559). However, the other clinical isolates tested on the microarray all possessed these genes. Analysis of the presence of genes in all three regions was extended across our large strain collection using Southern analysis.
Southern analysis.
Regions of genetic differences between LT2 and the clinical pigeon isolates tested on the microarray were examined more closely by Southern analysis over a large strain collection containing over 26 different phage types (Table 1). Of the clinical isolates, the majority fell into two groups of phage types. The first group included the pigeon isolates: 21 samples were DT2 or DT99 and fell into this group. The second group of 29 isolates, DT104, DT204, and DT49, are the epidemic cattle isolates. The remainder of the clinical isolates represent 13 additional phage types. Probes were generated to individual genes in each region (I, II, and III) identified as missing by microarray analysis and were used to confirm the results from the microarray analysis and to study the distribution of genomic areas of interest across numerous serotype Typhimurium isolates of various phage types. In order to avoid cross-hybridization between similar phage genes located at different positions in the genome, the genes used as probes for regions I and II (Fels-1 and -2) were chosen because they had very little homology to genes of other phage.
STM897 was used as the probe for region I (Fels-1 prophage). This gene was predicted to be absent from all four clinical isolates tested based on microarray analysis. The STM897 probe was used to analyze 74 serotype Typhimurium clinical isolates (including the strains tested on the array) and two LT2 isolates, including that used to construct the microarray. Southern analysis confirmed that STM897 was indeed absent from the clinical isolates tested on the microarray (data not shown). Southern analysis indicated that the STM897 probe did not hybridize with any clinical isolates tested, but it did hybridize to both LT2 isolates tested (data not shown). This Southern analysis confirmed the prediction of the microarray analysis and extended this analysis to show that the Fels-1 prophage was likely absent from all of the serotype Typhimurium isolates analyzed.
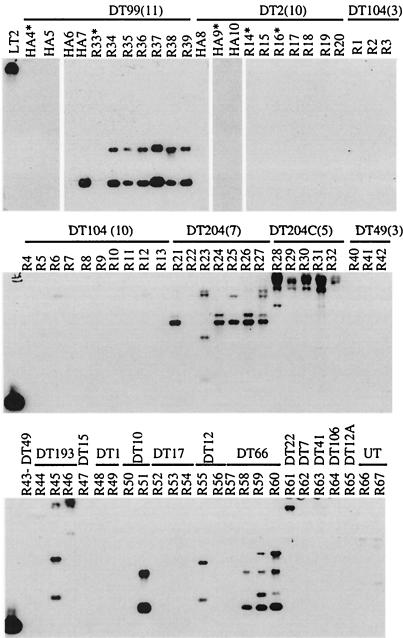
STM2724 was used as a probe for region II (Fels-2 prophage). This gene was predicted to be absent from the DT2 and DT99 pigeon isolates analyzed by microarray. STM2724 is also one of very few genes on the Fels-2 prophage that is not homologous to any genes of the sopE prophage, which is generally highly homologous to the Fels-2 prophage (25). Southern analysis confirmed the absence of STM2724 in strains HA9, R14, R16, and R33 (Fig. 2). Southern analysis indicated that both of these genes, or close homologs, were present in the majority of DT99 pigeon isolates and in the majority of DT204 cattle isolates, but they were not present in DT2 pigeon isolates or in DT104 epidemic cattle isolates (Fig. 2). Thus, the presence or absence of these genes in particular phage types, as defined by Southern analysis, was not correlated with host restriction of serotype Typhimurium pigeon isolates.
FIG. 2.
Southern analysis for the presence of STM2724, a gene located on the LT2 Fels-2 prophage (region II). Using this open reading frame, we probed genomic DNA of a collection of serotype Typhimurium clinical isolates of various phage types and host origins. The asterisks denote strains that were analyzed by hybridization to serotype Typhimurium LT2 microarray prior to this Southern analysis.
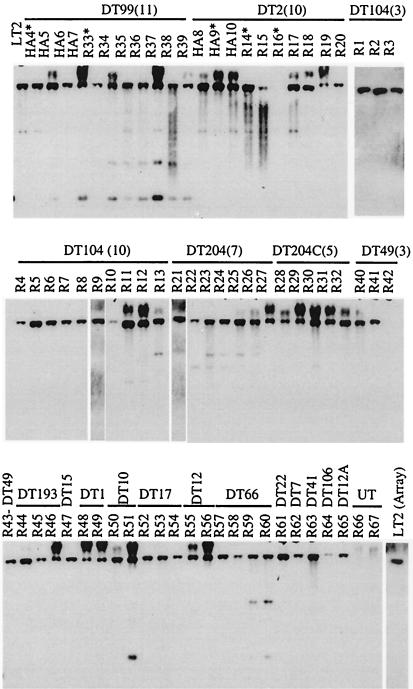
The distribution of genes in region III, which are absent in strain R16 by microarray analysis, was also examined across a large strain collection, using several of the individual genes in this region as probes for Southern analysis (Fig. 3, STM1555). Probes generated to STM1555, -1556, -1557, and -1559 were used to probe genomic DNA of 74 clinical isolates as described previously. As predicted by the microarray analysis, STM1555 was missing from strain R16 and was present in LT2 by Southern analysis (Fig. 3). However, STM1555 was present in all of the other phage types tested by Southern analysis, and identical results were obtained for STM1556, STM1558, and STM1559 (data not shown). These results indicated that the presence or absence of genes in region III has no correlation with phage type or host restriction.
FIG. 3.
Southern analysis for the presence of STM1555, a gene in region III. This gene was predicted to be absent from the pigeon clinical isolate R16, based on comparison with serotype Typhimurium LT2 genomic DNA by microarray analysis. STM1555 was predicted to be present by microarray analysis in other pigeon isolates tested on the LT2 microarray.
Bovine ligated ileal loops and mouse models of infection.
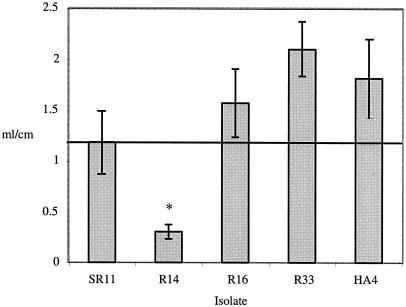
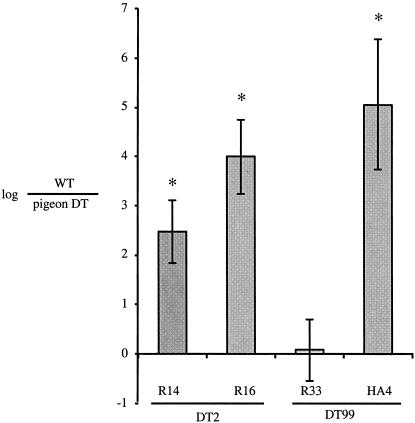
In order to define the pathogenesis of S. enterica serovar Typhimurium DT2 and DT99 isolates (R14, R16, R33, and HA4) in mammalian models, two isolates of each type were tested for fluid accumulation in bovine ligated ileal loops and for virulence and persistence in susceptible and resistant mouse models of infection. Both DT99 isolates tested, R33 and HA4, and one DT2 isolate, R16, were able to stimulate fluid accumulation to similar levels as virulent serotype Typhimurium strain SR11 in three identical experiments (Fig. 4). The DT2 isolate R14, however, consistently failed to elicit as much luminal fluid accumulation as virulent SR11 (Fig. 4). These observations indicated that many pigeon phage type isolates are capable of causing enterocolitis in mammals.
FIG. 4.
Fluid accumulation in bovine ligated ileal loops caused by DT2 and DT99 pigeon isolates. Loops were inoculated with 109 bacteria in 3 ml of L broth, and at 8 h postinoculation loops were excised and fluid accumulation was measured. The asterisk indicates a significant difference from inoculum using Student's t test, based on a P value of <0.05.
These four serotype Typhimurium pigeon isolates were also tested for their ability to compete with virulent serotype Typhimurium strain AJB715 in susceptible BALB/c mice. Only one DT99 isolate, R33, was fully capable of colonizing the spleens of infected BALB/c mice as efficiently as virulent AJB715 (Fig. 5). R14, R16, and HA4 pigeon isolates colonized the spleens of infected BALB/c mice to a significantly lesser extent than virulent strain AJB715 (Fig. 5). The colonization ability in the liver for these four strains was identical to colonization of the spleen in these animals (data not shown). Thus, the majority of the serotype Typhimurium pigeon isolates tested were defective for organ colonization in BALB/c mice.
FIG. 5.
Competitive infection of susceptible mice (BALB/c) with an equal mixture of either DT2 or DT99 pigeon isolates and virulent serotype Typhimurium AJB715. Bacteria from spleens of infected mice were enumerated. Three to four mice were used per pigeon isolate used. Values are expressed as log AJB715 per pigeon isolate, and error bars represent standard error. The asterisk indicates a significant difference from inoculum using Student's t test, with a P value of <0.05.
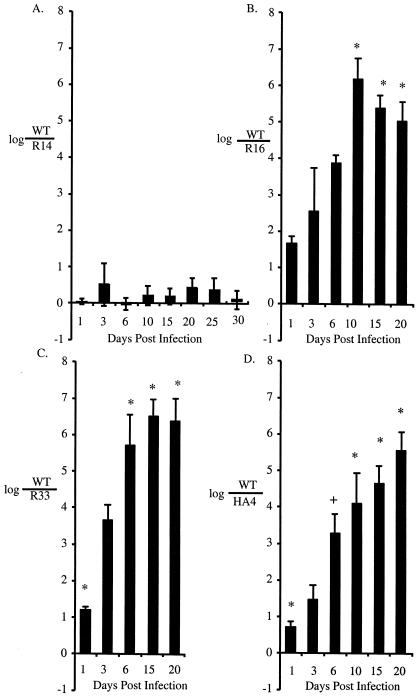
Competitive infections in serotype Typhimurium-resistant CBA mice were used to test the intestinal persistence of the DT2 and DT99 pigeon isolates over 20 to 30 days of infection. Only one isolate tested, R14 (DT2), was able to persist in the intestine of CBA mice to levels similar to virulent serotype Typhimurium strain AJB715 (Fig. 6A). The remaining three isolates tested, R16, R33, and HA4, were unable to persist in CBA mice from very early time points of infection (Fig. 6B, C, and D).
FIG. 6.
Intestinal persistence of DT2 and DT99 pigeon isolates in competitive infection with AJB715 in a resistant mouse model (CBA). Groups of five mice were infected with an equal mixture of a DT2 or DT99 isolate and virulent serovar Typhimurium AJB715. Feces were collected, diluted, and plated for serotype Typhimurium CFU at the indicated time intervals. Values are expressed as log AJB715 per pigeon isolate, and error bars represent standard errors. The asterisk indicates a significant difference from the inoculum using Student's t test and a P value of <0.05. A “+” indicates a significant difference from the inoculum using Student's t test and a P value of <0.1.
DISCUSSION
The genetic basis for host range restriction within S. enterica has been a topic of interest that has not been successfully resolved (28; Pascopella et al., Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996). We have attempted to address this question by comparing the genomes of very closely related clinical isolates of serotype Typhimurium from pigeons, with phage types that have a narrow host range, to serotype Typhimurium LT2, the sole broad-host-range isolate for which the complete genome sequence is available. The genomic differences between DT2 or DT99 and serotype Typhimurium LT2 that we were able to detect by microarray were localized in three clusters of genes. Two of these regions, I and II, represented the Fels-1 and Fels-2 prophages that are present in LT2 and are missing from the pigeon strains we studied using the microarray. The probe we generated for STM897, a gene in Fels-1 with very little homology to genes of other phages, failed to hybridize with any of our clinical isolates but hybridized with the positive control serotype Typhimurium LT2 by Southern analysis. These data indicated that the Fels-1 prophage is likely missing in all the clinical isolates we tested. Testing additional genes in this region by Southern analysis is somewhat difficult, as genes involved in phage functions are often highly homologous between different phages. In any case, the uniform absence of Fels-1 across our entire strain collection indicated that the presence or absence of this prophage is not correlated with a particular phage type or type and is thus unlikely to influence host restriction of different serotype Typhimurium pigeon isolates.
The probe used to study the distribution of the Fels-2 prophage, STM2724, revealed a different pattern of distribution of this prophage between different phage types of serotype Typhimurium. STM2724 is a gene of Fels-2 that does not have homology to any genes on the SopE prophage, a very closely related bacteriophage that has recently been completely sequenced (25). The STM2724 probe hybridized with genomic DNA of the majority of DT99 pigeon isolates, but with none of the DT2 pigeon isolates. This finding indicated that the absence of Fels-2 does not correlate with a host range restricted to pigeons. Furthermore, the STM2724 probe hybridized with all of the DT204 and DT204c cattle isolates, but with none of the DT104 clinical isolates from cattle. In addition, STM2724 did hybridize with several other phage types of serotype Typhimurium clinical isolates and the positive control serotype Typhimurium LT2, which is a DT4 phage type isolate. Collectively, these data indicate that the STM2724 distribution does seem to be clustered within particular phage types, but its distribution does not correlate with either host-restricted or broad-host-range phage types. Sequence analysis of prophages from each isolate may be required to reliably determine the distribution of a particular prophage, since it may be difficult to distinguish between closely related variants. Indeed, the Fels-1 prophage has not been found by array analysis in any strain examined except serotype Typhimurium LT2 (8, 26) (Porwollik, unpublished).
STM1555, the probe used for region III, hybridized with all of the clinical isolates tested with the exception of R16, one DT49 isolate (R42), and two untyped isolates. The results were very similar for the other genes in this cluster, STM1556 to STM1559 (data not shown). Thus, the absence of region III did not correlate with particular phage types or host restriction of pigeon isolates.
Two basic elements are important for a pathogen to circulate in a population: first, the pathogen has to be able to cause disease; second, the pathogen has to be able to persist within a population. Our data show that the majority of serotype Typhimurium pigeon isolates tested here are capable of causing fluid accumulation in bovine ligated ileal loops. Our microarray data confirmed that the DT2 and DT99 isolates possess all of the genes present in LT2 (a natural isolate from a human outbreak of enterocolitis) with the exception of the Fels-1 and Fels-2 prophages. The results of our bovine ligated ileal loop studies suggested that Fels-1 and Fels-2 are unlikely to be involved in the development of enterocolitis, and recent evidence suggests that at least the Fels-1 prophage is LT2 specific (Porwollik, unpublished). Thus, there are no genetic islands present in the broad-host-range isolate LT2, whose loss is predicted to result in host restriction. It seems reasonable to suggest that most serotype Typhimurium DT2 and DT99 pigeon isolates possess the genetic determinants that are necessary and sufficient to cause disease (enterocolitis) in calves, the best model currently available for Salmonella-induced enterocolitis in humans. The fact that DT2 and DT99 rarely cause disease in humans is therefore more likely to be a result of their scarcity in food animal reservoirs (i.e., livestock and domestic fowl) and is probably not due to an inability of these phage types to cause enterocolitis.
The use of mouse models to study the human pathogenesis of serotype Typhimurium is limited by the fact that infected mice do not develop enterocolitis. Nonetheless, mouse models of salmonellosis are useful to study the systemic phase of disease and the ability of the organism to persist in the intestine (15). In addition, mice are important vectors for introducing and spreading Salmonella serotypes among farm animals (10, 11, 14, 16, 30). In our experiments, three of four pigeon isolates tested in competitive infections in susceptible BALB/c mice were severely defective for colonization of the spleen and liver. Three of four pigeon isolates we tested were also unable to persist in the intestine of CBA mice, as measured by fecal excretion of the organism in competitive infection with a mouse virulent serotype Typhimurium isolate. This defect in intestinal persistence of the DT2 and DT99 pigeon isolates occurred as early as 6 days postinfection, and these isolates were undetectable by the conclusion of the experiment at 20 days postinfection. The reduced ability of pigeon isolates to colonize mice may thus contribute to their infrequent isolation from animals through which they may infect humans.
Our organ colonization and persistence data suggest that the DT2 and DT99 pigeon isolates lack factors present in LT2 that are necessary for systemic infection and persistence in mice. However, our microarray and Southern analysis data indicate that the genetic differences responsible for the inability of DT2 and DT99 isolates to colonize spleen or liver and to persist in mice are unlikely to be at the level of absence of discrete genes. It is worth reemphasizing that the only genes that appear to be consistently missing by microarray analysis from the pigeon isolates examined are the genes of the Fels-1 and Fels-2 prophages. By Southern analysis, these genes are also absent from isolates from many phage types, including epidemic cattle isolates DT49, DT104, and DT204, leaving no correlation between phage type, the presence of these prophages, and the ability to persist in mammals. The fact that broad-host-range isolates do not possess unique genetic islands that influence host restriction makes it likely that host restriction is due to point mutations or small deletions resulting in loss of function, although the possibility that pigeon isolates may possess genetic islands that affect the expression or function of genes that are shared with LT2 cannot currently be ruled out. A similar conclusion was recently reached by comparison of the genomes of the host-restricted serotype Typhi strain CT18 and the broad-host-range serotype Typhimurium strain LT2. The serotype Typhi genome contains a considerably larger number of pseudogenes than the genome of serotype Typhimurium, suggesting that host restriction may be in part explained by the accumulation of loss-of-function mutations in the strictly human-adapted serotype Typhi (23). The hypothesis that the reduced ability of DT2 and DT99 isolates to colonize mice may be due to point mutations and/or small insertions or deletions is furthermore consistent with previous observations that the loss of mouse virulence of some serotype Typhimurium LT2 sublines is caused by acquisition of a point mutation in rpoS (34, 36).
Several previous studies have examined genomic differences between isolates belonging to the same species of the family Enterobacteriaceae (7, 8, 17, 19). Comparison of the serotype Typhimurium LT2 genome with that of the pigeon isolate phage types DT2 and DT99 by microarray analysis supports several lines of previously published work. First, the results of the microarray analysis showed that the genetic differences between these isolates are small, and they confirmed previous results that different isolates of the same serotype are very closely related (3, 4). A recent pulsed-field gel electrophoretic analysis of 38 DT99 pigeon isolates and 18 isolates from other species indicated that the DT99 isolates examined were essentially clonal (24).
Second, analysis of polymorphisms found by comparing the genomes of different Escherichia coli O157:H7 isolates suggests that differences between strains are primarily due to insertions and deletions, not single-nucleotide polymorphisms (17, 19). Our analysis indicated that large insertions or deletions distinguishing serotype Typhimurium isolates are mainly caused by the presence or absence of prophages. However, these differences did not correlate with the host range restriction of serotype Typhimurium isolates. Our data suggest that genetic differences that cannot be detected by microarray analysis (e.g., point mutations and small deletions or insertions) may contribute to the epidemiological success of individual clones within different animal reservoirs. Although the insertion of additional prophage or isolated genes in serotype Typhimurium pigeon isolates is undetectable by analysis using the serotype Typhimurium LT2 microarray, it is clear that the reduced ability of DT2 and DT99 isolates to colonize murine organs is not due to the absence of discrete genes.
Finally, the importance of point mutations and small deletions or insertions as a source of strain variation is revealed by comparison of genomic sequences of serotype Typhi strains Ty2 and CT18. This genomic comparison shows that while large insertions or deletions are due to the presence or absence of prophages, point mutations and small deletions or insertions have resulted in silencing of different gene functions in both strains (7). That is, Ty2 contains 11 pseudogenes that are intact in CT18, while the CT18 genome contains 9 pseudogenes that are intact in strain Ty2.
Acknowledgments
We thank Caleb Dorsey for critical reading of the manuscript and Marcel de Zoete and Lindsay Garza for technical assistance.
Work in A.B.'s laboratory is supported by USDA/NRICGP grant 2002-35204-12247 and Public Health Service grants AI40124 and AI44170. H.A. is supported by Public Health Service grant AI52250.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. J. Wiley & Sons, New York, N.Y.
- 2.Baumberg, S., and R. Freeman. 1971. Salmonella typhimurium strain LT-2 is still pathogenic for man. J. Gen. Microbiol. 65:99-100. [DOI] [PubMed] [Google Scholar]
- 3.Beltran, P., J. M. Musser, R. Helmuth, J. J. Farmer III, M. W. Frerichs, K. Wachsmuth, K. Ferris, A. C. McWorter, J. G. Wells, A. Cravioto, and R. K. Selander. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltran, P., S. A. Plock, N. H. Smith, T. S. Whittam, D. C. Old, and R. K. Selander. 1991. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J. Gen. Microbiol. 137:601-606. [DOI] [PubMed] [Google Scholar]
- 5.Brandis, H., J. Posch, G. Oberhoffer, L. Andries, and U. Lehmacher. 1980. Contributions to the epidemiology of Salmonella typhimurium, analyzed according to the results of phage typing in the period between 1969-1978. Offentl Gesundheitswes 42(Suppl. 2):75-128. (Authors' translation.) [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1999. Salmonella surveillance: annual tabulation summary, 1998. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Ga.
- 7.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garaizar, J., S. Porwollik, A. Echeita, A. Rementeria, S. Herrera, R. M. Wong, J. Frye, M. A. Usera, and M. McClelland. 2002. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40:2074-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geoffrey, E., S. Gaines, M. Landy, W. D. Tigertt, H. Sprintz, R.-J. Trapani, A. D. Mandel, and A. S. Benenson. 1960. Studies on infection and immunity in experimental typhoid fever: typhoid fever in chimpanzees orally infected with Salmonella typhosa. J. Exp. Med. 112:143-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guard-Petter, J., D. J. Henzler, M. M. Rahman, and R. W. Carlson. 1997. On-farm monitoring of mouse-invasive Salmonella enterica serovar Enteritidis and a model for its association with the production of contaminated eggs. Appl. Environ. Microbiol. 63:1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henzler, D. J., and H. M. Opitz. 1992. The role of mice in the epizootiology of Salmonella enteritidis infection on chicken layer farms. Avian Dis. 36:625-631. [PubMed] [Google Scholar]
- 12.Herzberg, M., P. Nash, and S. Hino. 1972. Degree of immunity induced by killed vaccines to experimental salmonellosis in mice. Infect. Immun. 5:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelterborn, E. 1967. Salmonella species. First isolations, names and occurrence. S. Hirzel Verlag Leipzig, Karl-Marx-Stadt, Germany.
- 14.Kinde, H., D. H. Read, A. Ardans, R. E. Breitmeyer, D. Willoughby, H. E. Little, D. Kerr, R. Gireesh, and K. V. Nagaraja. 1996. Sewage effluent: likely source of Salmonella enteritidis, phage type 4 infection in a commercial chicken layer flock in southern California. Avian Dis. 40:672-676. [PubMed] [Google Scholar]
- 15.Kingsley, R. A., A. D. Humphries, E. H. Weening, M. R. De Zoete, S. Winter, A. Papaconstantinopoulou, G. Dougan, and A. J. Baumler. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect. Immun. 71:629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krabisch, P., and P. Dorn. 1980. Epidemiologic significance of live vectors in the transmission of salmonella infections in broiler flocks. Berl. Munch. Tierarztl. Wochenschr. 93:232-235. [PubMed] [Google Scholar]
- 17.Kudva, I. T., P. S. Evans, N. T. Perna, T. J. Barrett, F. M. Ausubel, F. R. Blattner, and S. B. Calderwood. 2002. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol. 184:1873-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilleengen, K. 1948. Typing of Salmonella typhimurium by means of bacteriophage. Acta Pathol. Microbiol. Scand. Suppl. 77:2-125. [Google Scholar]
- 19.Liu, G. R., K. Edwards, A. Eisenstark, Y. M. Fu, W. Q. Liu, K. E. Sanderson, R. N. Johnston, and S. L. Liu. 2003. Genomic diversification among archival strains of Salmonella enterica serovar Typhimurium LT7. J. Bacteriol. 185:2131-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 21.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgenroth, A., and J. P. Duguid. 1968. Demonstration of different mutational sites controlling rhamnose fermentation in FIRN and non-FIRN rha-strains of Salmonella typhimurium: an essay in bacterial archaeology. Genet. Res. 11:151-169. [DOI] [PubMed] [Google Scholar]
- 23.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 24.Pasmans, F., F. Van Immerseel, M. Heyndrickx, A. Martel, C. Godard, C. Wildemauwe, R. Ducatelle, and F. Haesebrouck. 2003. Host adaptation of pigeon isolates of Salmonella enterica subsp. enterica serovar Typhimurium variant Copenhagen phage type 99 is associated with enhanced macrophage cytotoxicity. Infect. Immun. 71:6068-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelludat, C., S. Mirold, and W. D. Hardt. 2003. The SopEPhi phage integrates into the ssrA gene of Salmonella enterica serovar Typhimurium A36 and is closely related to the Fels-2 prophage. J. Bacteriol. 185:5182-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabsch, W., H. L. Andrews, R. A. Kingsley, R. Prager, H. Tschape, L. G. Adams, and A. J. Baumler. 2002. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect. Immun. 70:2249-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roudier, C., M. Krause, J. Fierer, and D. G. Guiney. 1990. Correlation between the presence of sequences homologous to the vir region of Salmonella dublin plasmid pSDL2 and the virulence of twenty-two Salmonella serotypes in mice. Infect. Immun. 58:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos, R. L., R. M. Tsolis, S. Zhang, T. A. Ficht, A. J. Baumler, and L. G. Adams. 2001. Salmonella-induced cell death is not required for enteritis in calves. Infect. Immun. 69:4610-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato, G., T. Miyamae, and S. Miura. 1970. A long term epizootiological study of chicken salmonellosis on a farm with reference to elimination of paratyphoid infection by cloacal swab culture. Jpn. J. Vet. Res. 18:47-62. [PubMed] [Google Scholar]
- 31.Schneider, H. A., and N. D. Zinder. 1956. Nutrition of the host and natural resistance to infection. V. An improved assay employing genetic markers in the double strain inoculation test. J. Exp. Med. 103:207-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scholtens, R. T., and G. Caroli. 1971. Role of pigeons in the spread of salmonellosis: incidence of different types of Salmonella typhi-murium var. copenhagen in pigeons, man, and other animals. Antonie Leeuwenhoek 37:473-476. [DOI] [PubMed] [Google Scholar]
- 33.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swords, W. E., B. M. Cannon, and W. H. Benjamin, Jr. 1997. Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect. Immun. 65:2451-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Oye, E., and J. Borghijs. 1979. Do pigeons play a part in human infections with Salmonella typhimurium var. Copenhagen? Dtsch. Tierarztl. Wochenschr. 89:306-307. (Authors' translation.) [PubMed] [Google Scholar]
- 36.Wilmes-Riesenberg, M. R., J. W. Foster, and R. Curtiss III. 1997. An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect. Immun. 65:203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuthe, H. H., H. Brandis, L. Andries, and S. Wuthe. 1975. Phage typing and biotyping of Salmonella typhi-murium. Zentralbl. Bakteriol. 230:172-185. (Authors' translation.) [PubMed] [Google Scholar]