Abstract
Vibrio parahaemolyticus is one of the most important seafood-borne bacterial in recent years and is the leading causal agent of human acute gastroenteritis, primarily following the consumption of raw, undercooked or mishandled marine products. Until 1996, infections caused by V. parahaemolyticus were generally associated with diverse serovars. However, in February 1996, a unique serovar (O3:K6) of V. parahaemolyticus with specific genetic markers (tdh, toxRS/New and/or orf8) appeared abruptly in Kolkata, India. In subsequent years, O3:K6 isolates similar to those isolated in Kolkata have been reported from food borne outbreaks in Southeast Asia, as well as in the Atlantic and Gulf coasts of the United States (U.S). More recently, there have been reports in Europe, Africa and Central and South America. Specifically, in the American continent, some countries have reported cases of gastroenteritis due to the pandemic O3:K6 strain and its serovariants; the pandemic strain was first detected in Peru (1996, >100 cases), subsequently spreading to Chile in 1998 (>16,804 human cases), to the U.S. in 1998 (>700 cases), to Brazil in 2001 (>18 cases) and to Mexico in 2004 (>1200 cases). The arrival of the pandemic clone on the American continent may have resulted in a significant shift on the epidemic dynamics of V. parahaemolyticus. However, although O3:K6 is the predominant serovar of the recognized clinical strains in some countries in the Americas, a decrease in clinical cases caused by O3:K6 and an increase in cases associated with a new serotype (O3:K59, Chile) have been recently reported. The emergence and worldwide dissemination of O3:K6 and other pandemic strains since 1996 have come to represent a threat to public health and should concern health authorities. This review focuses on the presence, distribution and virulence factors of the V. parahaemolyticus O3:K6 pandemic clone and its serovariants in clinical and environmental strains on the American continent.
Keywords: American continent, pandemic clone, serotypes, Vibrio parahaemolyticus, biosurveillance, public health
Introduction
Vibrio parahaemolyticus is a Gram-negative halophilic, mesophilic, non-spore forming, curved rod-shaped bacterium that naturally inhabits marine and estuarine environments worldwide (Baumann et al., 1984). It is commonly found free swimming, attached to underwater surfaces, and commensally associated with various species of shellfish (McCarter, 1999). V. parahaemolyticus has recently been recognized as one of the most important foodborne pathogens and as the leading causal agent of human acute gastroenteritis, primarily following the consumption of raw, undercooked or mishandled seafood and marine products (Okuda et al., 1997; Bag et al., 1999; Depaola et al., 2000; Wong et al., 2000; Ansaruzzaman et al., 2005; Martinez-Urtaza et al., 2005; Cabanillas-Beltran et al., 2006; Su and Liu, 2007; Pal and Das, 2010; Velazquez-Roman et al., 2012). The halophilic bacteria was first identified as a cause of food-borne illness during the fall of 1950 within the southern suburbs of Osaka, Japan, where an outbreak of acute gastroenteritis following the consumption of semidried juvenile sardines sickened 272 and killed 20 individuals (Fujino et al., 1953). V. parahaemolyticus causes two additional major clinical pathologies wound infections and septicemia. The most common syndrome is gastroenteritis, the symptoms of which include diarrhea with abdominal cramps, nausea, vomiting, headache, chills and low-grade fever (Honda and Iida, 1993). The associated diarrhea is occasionally bloody, characterized by reddish watery stools (Qadri et al., 2003) that are unlike those observed in cases of dysentery caused by Shigella species or in cases of amoebiasis. The mean incubation period for V. parahaemolyticus infection is 15 h (range: 4–96 h) (Joseph et al., 1982). The illness is self-limiting and of moderate severity, lasting an average of 3 days in immunocompetent patients (Yeung and Boor, 2004). Because of its self-limiting nature, most cases of infection by V. parahaemolyticus can be can be treated by oral rehydration, alone. Occasionally, treatment with antibiotics such as doxycycline, ciprofloxacin, or erythromycin is necessary (Qadri et al., 2003). The infection can be fatal for immunocompromised patients or for those with a preexisting medical condition, such as liver disease or diabetes. Although the overall pathogenetic mechanism of marine Vibrio species such as V. parahaemolyticus is not completely understood, these pathogens are known to produce various extracellular products, some of which are known pathogenicity factors (Hasegawa and Hase, 2009). Potentially virulent strains are commonly distinguished from virulent strains by the presence of two hemolysins, namely the thermostable direct hemolysin (tdh) and the Tdh-related hemolysin (trh), which exhibits 68% homology with the tdh gene (Zhang and Austin, 2005), and plays a significant role in precipitating the disease (Takeda, 1983; Shirai et al., 1990; Bej et al., 1999). Pathogenic V. parahaemolyticus can produce either TDH, TRH, or both (Honda and Iida, 1993; Nishibuchi and Kaper, 1995). An isolate that produces TDH is referred to as Kanagawa phenomenon (KP)-positive and can be identified by β hemolysis on a special agar known as Wagatsuma blood agar (Pal and Das, 2010; Canizalez-Roman et al., 2011; Velazquez-Roman et al., 2012). PCR analysis targeting the tdh gene encoding TDH hemolysin and/or the trh gene encoding TRH hemolysin have been widely used for the identification of pathogenic isolates of V. parahaemolyticus (Shirai et al., 1990; Yoh et al., 1995).
Pathogenic V. parahaemolyticus
Although the presence of V. parahaemolyticus is extensive in marine, estuarine and riverine environments, not all strains of V. parahaemolyticus are considered pathogenic. Vibrios cultured from environmental samples commonly lack the genes coding for proteins with associated pathogenic functions in humans and marine animals (e.g., tdh in V. parahaemolyticus) (Deepanjali et al., 2005; Canizalez-Roman et al., 2011; Gutierrez West et al., 2013). However, few studies have reported the presence of the tdh and trh genes in V. parahaemolyticus strains of environmental origin; only 0–6% of the samples analyzed from the coasts of the U. S. (Kaysner et al., 1990; Depaola et al., 2000), Europe (Hervio-Heath et al., 2002), and Asia contained the tdh and/or trh genes (Vuddhakul et al., 2000; Wong et al., 2000; Alam et al., 2002). Most of the environmental strains are known to be KP-negative, and only 1–2% of the samples are KP-positive (Joseph et al., 1982; Nishibuchi and Kaper, 1995). In molecular epidemiological studies, up to 90% of the isolated clinical strains have been found to possess the tdh and/or trh gene (the serotype O3:K6 strain has increased in prominence) (Okuda et al., 1997; Garcia et al., 2009; Chao et al., 2010; Velazquez-Roman et al., 2012), whereas their presence in environmental isolates is rare (Shirai et al., 1990; Depaola et al., 1992; Nishibuchi and Kaper, 1995; Islam et al., 2004; Yeung and Boor, 2004; Nair et al., 2007; Chao et al., 2010). However, in limited areas of Mexico and the U.S., increased proportions (48–52%) of strains with pathogenic markers (tdh and/or trh) have been detected in environmental isolates (Velazquez-Roman et al., 2012; Gutierrez West et al., 2013). The increased rate of detection of pathogenic V. parahaemolyticus within the marine environment might be attributed to a decreased prevalence of total V. parahaemolyticus rather than to an increased abundance of pathogenic strains (Velazquez-Roman et al., 2012).
Currently, a comprehensive understanding of the ability of this organism to cause disease remains incomplete. Although the isolates that do not contain tdh or those that exhibit a deletion in tdh are still cytotoxic to host cells, the overall mechanism underlying the pathogenesis of these tdh-negative strains remains unclear (Boyd et al., 2008). Isolates TDH positive has been shown to have hemolytic, enterotoxic, cardiotoxic, and cytotoxic activities (Nishibuchi et al., 1992; Raimondi et al., 2000). These cellular events are caused by bacterial effector proteins, include cytolysins, proteases, lipases, siderophores, exopolysaccharides, and effectors which are delivered into intestinal cells (enterotoxic) that directly modulate the activities of host cell proteins and are secreted and translocated into host cells via type III secretion systems (T3SS) (Hiyoshi et al., 2010, 2011; Matsuda et al., 2012) and recently type VI secretion systems (T6SS) (Yu et al., 2012; Salomon et al., 2013).
The complex interactions between pathogens and the hosts that they infect have long been believed to represent the primary driving forces that determine the strategies used by bacteria to counter host defenses. However, new evidence suggests that the external environment, including other hosts, might play a greater role in the evolution of certain pathogens than previously expected (Wilson and Salyers, 2003). Thus, the pandemic strains that exhibit certain biological characteristics, such as elevated toxin production or the ability to survive within the natural environment, might provide further insights into the mechanisms underlying the emergence and spread of these strains (Wong et al., 2000). Factors that affect the incidence and distribution of V. parahaemolyticus in the environment are known to include the water temperature, salt and oxygen concentrations, interaction with plankton, presence of sediment, organic matter in suspension, fish, and seafood, as well as the incorporation and tidal action of estuarine waters (Cabrera-Garcia et al., 2004). However, how these strains disseminate worldwide remains unknown.
vibrio parahaemolyticus O3:K6 in the world
Until 1996, infections caused by V. parahaemolyticus were usually associated with diverse serovars (for example, O1:K38, O3:K29, O4:K8, O3:K6, O2:K3, O4:K8, and other serotypes) (Okuda et al., 1997; Wong et al., 2000) and exhibited a localized distribution, emerging in different areas of the world only during the warmer months of the year. However, the epidemiology of V. parahaemolyticus changed drastically in February 1996, when an atypical increase in V. parahaemolyticus infections occurred in Kolkata, a city in the northeastern part of India, resulting in hospitalized patients with diarrheal infections. These infections were linked to strains that belonged to a clonal group of the O3:K6 serotype, which exhibited the specific genetic markers tdh, toxRS/New and orf8. These strains had not been previously isolated, but subsequent surveillance in Kolkata identified this serotype as responsible for 50–80% of the infections during the following months. This clone rapidly spread throughout the majority of Southeast Asian countries within a single year (Okuda et al., 1997; Chowdhury et al., 2000). During subsequent years, O3:K6 isolates similar to those isolated in Kolkata were reported from foodborne outbreaks and from sporadic cases in the Atlantic and Gulf coasts of the U.S. (Okuda et al., 1997; Chowdhury et al., 2000; Matsumoto et al., 2000). Recently, there have been similar reports in Europe (Martinez-Urtaza et al., 2005), Africa (Ansaruzzaman et al., 2005), North, Central and South America (Daniels et al., 2000a; Gonzalez-Escalona et al., 2005; Velazquez-Roman et al., 2012). The widespread occurrence of this single V. parahaemolyticus serotype had not previously been reported; thus, it became evident that a pandemic strain had emerged.
Currently several serovariants of the pandemic strains of O3:K6, included 14 different serotypes (O1:K25, O1:K41, O1:K56, O1:KUT, O3:K6, O3:K58, O3:K68, O3:K75, O4:K8, O4:K12, O4:K68, O4:KUT, O5:KUT, and OUT:K6), although only O3:K6, O4:K68, O1:K25, O1:26, and O1:KUT have been recognized as the predominant group responsible for most outbreaks since 1996. These serotypes have been identified as pandemic clones (Okuda et al., 1997; Matsumoto et al., 2000; Okura et al., 2003; Chowdhury et al., 2004a; Ansaruzzaman et al., 2005; Mahmud et al., 2007; Nair et al., 2007; Velazquez-Roman et al., 2012). The acquisition of additional serotypes of the pandemic strain may be a selective response to host immunological pressure (Chowdhury et al., 2004b). Molecular studies based on pulsed-field gel electrophoresis and arbitrarily primed PCR (APPCR) have indicated that the pandemic strains exhibit almost identical fragment patterns (Depaola et al., 2000; Parvathi et al., 2006). Notably, since 1996, a filamentous phage (f237) (Nasu et al., 2000; Iida et al., 2001) has been reported to be associated with the pandemic isolates of V. parahaemolyticus (Nasu et al., 2000; Iida et al., 2001). Therefore, from a single O3:K6 serotype, other serotypes that exhibit identical genotypes and molecular profiles to the O3:K6 isolates have emerged. These new serotypes have been collectively called the “serovariants” of O3:K6 isolates (Matsumoto et al., 2000). Generally, pandemic strains belong to the O3:K6 serotype and contain the orf8 gene. The orf8 gene is believed to encode an adherence protein that increases the ability of V. parahaemolyticus to adhere to host intestinal cells or to the surfaces of marine plankton (Nasu et al., 2000). Furthermore, several studies have reported that the toxRS operon of the pandemic strains contains a unique sequence (toxRS/new) that encodes for transmembrane proteins involved in the regulation of virulence-associated genes (Chowdhury et al., 2000; Matsumoto et al., 2000; Okura et al., 2003). Characteristics of the O3:K6 pandemic clone isolates include the O3:K6 antigens, a distinctive toxRS sequence (toxRS/new) (Matsumoto et al., 2000), orf8 (Nasu et al., 2000) and tdh genes, and the absence of the trh gene that is found in some pathogenic strains. Although several polymerase chain reaction (PCR)-based methods targeting toxRS/new, group-specific PCR (GS-PCR) or orf8—have been developed, the presence of the toxRS/new gene is necessary but not always sufficient, whereas the orf8 gene is sufficient but not always necessary for the detection of pandemic strains (Chowdhury et al., 2000; Okura et al., 2003). In general, however, based on the presence or absence of virulence genes, an isolate possessing both tdh and toxRS/new genes can be considered a pandemic strain (Okura et al., 2003). One of the other virulence genes, trh, is not specific to pandemic strains and is rarely present in environmental strains compared to clinical strains (Depaola et al., 2000; Parvathi et al., 2006). Pathogenic strains feature tdh- and/or trh-positivity, whereas non-pathogenic strains are characteristically tdh- and trh-negative. Some clinical isolates have been found to contain both tdh and trh genes, whereas most environmental isolates possess neither (Shirai et al., 1990; Baba et al., 1991; Kishishita et al., 1992; Xu et al., 1994).
Pandemic O3:K6 clone on the american continent
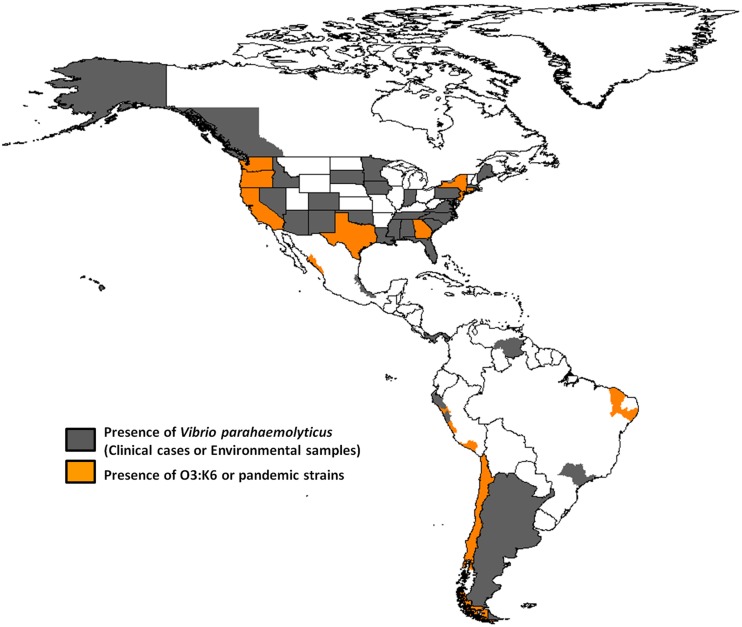
The routes and mechanisms resulting in the dissemination of the pandemic strain are still unclear. Moreover, the epidemiology of V. parahaemolyticus infections remains poorly understood. Recently, there have been occurrences of other extraordinary events regarding pathogenic Vibrio species. The transmission and epidemiology of V. parahaemolyticus infections in places such as Kolkata, India, and Bangladesh are entirely distinct from the rest of the world because seafood is never eaten raw, and freshwater fish are preferred over seawater fish by the local population (Sarkar et al., 1985). However, in February 1996, the O3:K6 serotype emerged in Kolkata with identical genotypes (tdh positive and trh negative) and profiles that were indistinguishable by molecular subtyping techniques (Okuda et al., 1997). In subsequent years, isolates similar to those from Kolkata were reported from foodborne outbreaks and from sporadic cases in several sites worldwide (Daniels et al., 2000a; Matsumoto et al., 2000; Ansaruzzaman et al., 2005; Gonzalez-Escalona et al., 2005; Martinez-Urtaza et al., 2005; Leal et al., 2008; Velazquez-Roman et al., 2012), particularly during the warmer months of the year. Subsequently, the pandemic clone began global dissemination, with many outbreaks. In particular, some countries of the American continent have reported cases of gastroenteritis due to the pandemic O3:K6 strain and its serovariants, where the pandemic strain was first detected in Peru (>100 cases at approximately the same time that it caused an outbreak in Calcutta in February 1996) (Gil et al., 2007). The O3:K6 strain subsequently spread to Chile in 1998 (>16,804 human cases), to the U.S. in 1998 (>700 cases), to Brazil in 2001 (>18 cases) and to Mexico in 2004 (>1200 cases) (Figure 1 and Table 1).
Figure 1.
Map showing the presence of Vibrio parahaemolyticus and the dissemination of pandemic O3:K6 clone throughout the American continent.
Table 1.
Presence of Vibrio parahaemolyticus and the pandemic O3:K6 clone in clinical cases and the environment on the American continent.
| Country (year of isolation) | Source of isolates and serovars collected (origin type) | Reference | |||||
|---|---|---|---|---|---|---|---|
| (No. Cases) Clinical | Pandemic ()* | Other serotypes | (No. Samples) Environmental | Pandemic ()* | Other serotypes | ||
| Peru (1993–2007) | (424) Gastroenteritis | O3:K6 (1996) | O1:K33, O2:K3, O3:K30, O3:K58 | Fish | O3:K6 (1996) | No data available | Gil et al., 2007 |
| O3:K68 | O3:K68, O4:K8, O4:K12, O4:K55 | ||||||
| O3:K58 | O6:K18, O6:K46, O11:K15, O1:KUT, O2:KUT | Shrimp | Gavilan et al., 2013 | ||||
| OUT:K6 | O5:KUT, O6:KUT, O11:KUT, OUT:K3 | ||||||
| OUT:K6, OUT:K8, OUT:K46, OUT:KUT | |||||||
| Chile (1998–2013) | (>16000) Gastroenteritis | O3:K6 (1998) | O1:K56, O3:K6, O3:K59, O4:K12 | Shellfish | O3:K6 (1998) | O3:KUT | Gonzalez-Escalona et al., 2005 |
| O10:K20, O1:KUT, O3:KUT, OUT:KUT | Garcia et al., 2009 | ||||||
| Harth et al., 2009 | |||||||
| United States of America (1973–2012) | 6159 | O3:K6 (1998) | O1:K20, O1:K25, O1:K33, O1:K43 | Shellfish | No data available | Daniels et al., 2000a,b | |
| Gastroenteritis | O1:K56, O3:K39, O3:K5, O3:K56 | Depaola et al., 2000 | |||||
| Wound infections Septicemia | O4:K4, O4:K8, O4:K9, O4:K12 | McLaughlin et al., 2005 | |||||
| O4:K13, O4:K34, O4:K37, O4:K42 | Newton et al., 2012 | ||||||
| O4:K53, O4:K63, O5:K17, O5:K30 | Fish | ||||||
| O5:K47,O6:K18, O8:K41, O8:K70 | |||||||
| O1:KUT, O3:KUT, O4:KUT, O5:KUT | |||||||
| O6:KUT, O8:KUT, O10:KUT, O11:KUT | |||||||
| Brazil (1989–2002) | (39) Gastroenteritis | O3:K6 (2002) | O1:K56, O3:K5 O3:K6, O3:K58 | (23) Shellfish | No data available | Hofer, 1983 | |
| O3:KUT | O3:KUT, O4:K4, O4:K10, O4:K12 O4:K53, O4:KUT, O5:KUT, O10:KUT | Magalhaes et al., 1991a,b, 1992 | |||||
| Leal et al., 2008 | |||||||
| Sobrinho Pde et al., 2010 | |||||||
| Mexico (2001–2010) | (>1379) Gastroenteritis | O3:K6 (2004) | O1:KUT, O1:K9, O1:K33, O3:K58 | (190) Shellfish | O3:K6 (2004) | O1:K25, O1:K26, O1:K32, O1:K33 | Cabrera-Garcia et al., 2004 |
| O3:KUT | O3:K68, O4:KUT, O4:K8, O4:K10 | Fish | O3:KUT | O1:K41, O1:K56, O2:K28, O3:k4 | Cabanillas-Beltran et al., 2006 | ||
| O4:K12, O4:K63, O5:K15, O5:K17 | Seawater | O10:KUT | O3:K6, O3:K29, O3:K32, O3:K33 | ||||
| O6:K46, O8:K20, O10:K52, O1:KUT | O3:K58, O3:K59, O3:K68, O4:K10 | Velazquez-Roman et al., 2012 | |||||
| O4:K12, O4:K53, O4:K68, O5:K15, | |||||||
| O5:K17, O5:K68, O6:K6, O6:K46 | |||||||
| O4:KUT, O5:KUT, O8:KUT, O11:KUT | Sediment | O8:K8, O8:K20, O8:K21, O10:K52, | |||||
| OUT:KUT | O1:KUT, O2:KUT, O3:KUT, O4:KUT | ||||||
| O5:KUT, O6:KUT, O7:KUT, O10:KUT | |||||||
| O11:KUT, OUT:K8, OUT:KUT | |||||||
| Canada (1997–2006) | (>200) Gastroenteritis | No data available | Shellfish | No data available | FAO/WHO, 2011 | ||
| Khaira and Galanis, 2007 | |||||||
| Costa Rica (1985–2002) | (77) Shellfish | No data available | Garcia Cortes and Antillon, 1990 | ||||
| Sediment | Díaz, 2002 | ||||||
| Puerto Rico (1986–1988) | Shellfish | No data available | Rivera Susan et al., 1989 | ||||
| Panama (1974–1975) | (>1) Gastroenteritis | No data available | Shrimp | No data available | Kourany et al., 1974; Kourany and Vasquez, 1975 | ||
| Venezuela (2011) | Shellfish | No data available | Grau et al., 2004 | ||||
| Prawn | Muñoz et al., 2008 | ||||||
| Coral | Garcia-Amado et al., 2011 | ||||||
| Argentina (1977) | Shellfish | No data available | Casellas et al., 1977 | ||||
Year of isolation of O3:K6 pandemic clone.
Peru
Peru was the first country where the V. parahaemolyticus O3:K6 clone was found in marine food products (raw fish or seafood marinated with lemon juice, popularly called “cebiche”) (Guevara Duncan and Del Carpio, 1989) and is where the strain was first associated with gastroenteritis on the American continent (Ibarra and Alvarado, 1999; Gil et al., 2007). Case records from 1994–1996 reflect occasional scattered infections restricted to the warmest months of the year (Martinez-Urtaza et al., 2008). Gil et al. found that the first O3:K6 strains were isolated in February 1996 in Trujillo city, indicating that the pandemic strain was present in Peru at approximately the same time that it caused the outbreak in Calcutta in February 1996 (Gil et al., 2007). In 1997, a sudden increase in the number of O3:K6 V. parahaemolyticus cases was observed in Chiclayo, Department of Lambayeque (in northern Peru) in July, which lasted for 10 months with 2 peaks. Infections were observed along the entire coastline of Peru and spread more than 1500 km within a 4-month period (Martinez-Urtaza et al., 2008). The infections spread in a constant southward direction, affecting the Department of Cajamarca in August, La Libertad in September, and Huaraz in October, finally reaching the southern Peruvian border in November—at the same time as the emergence of V. parahaemolyticus illnesses within the northern Chilean city of Antofagasta (Gonzalez-Escalona et al., 2005; Martinez-Urtaza et al., 2008). In a retrospective analysis of data on V. parahaemolyticus in Peru, Gil et al., indicated that although the first O3:K6 strain was isolated in February 1996 in Trujillo city, the first cluster of cases caused by the pandemic serotype occurred in Lima in 1998, when 27 strains were isolated from hospitalized cases in a short span of time in one hospital (Gil et al., 2007).
Interestingly, the non-pandemic O4:K8 and O4:K55, and O5:KUT serotypes predominated in 1994 and 1996 in Trujillo, and the infections were characteristically related to self-limiting outbreaks that were detected along the coastal regions during the summer months (Martinez-Urtaza et al., 2008). The serovar dominance, particularly that of O4:K8, abruptly changed during the winter of 1997, with the emergence of infections caused by strains belonging to the pandemic clone (Gavilan et al., 2013). Additional strains belonging to the O3:K68, O3:K58, and OUT:K6 serotypes were isolated in Lima and Arequipa in 1997 and found to be positive for Orf8 and GS-PCR, indicating that they were also pandemic strains (Gil et al., 2007). From 1998 to 2002, pandemic strains (O3:K6 and others) were detected in Lima and Trujillo (Gil et al., 2007). Due to the environmental nature of V. parahaemolyticus, this type of change in population dominance and the subsequent population admixture might be expected to lead to an unpredictable impact on the populations of pathogenic V. parahaemolyticus in Peru (Gavilan et al., 2013). Finally, an undefined pattern of serovar dominance was detected after the emergence of the pandemic clone in 1997–1998. In this post-pandemic period, pandemic strains were identified together with O4:K8 strains, as well as multiple serovars that had not been previously detected (O1:K33, O1:KUT, O3:K30, O3:K58, O3:KUT, O5:KUT, O6:KUT, and OUT:KUT) (Gavilan et al., 2013). One particular feature of the epidemiology of V. parahaemolyticus in Peru after the emergence of the pandemic clone was the sudden appearance of diverse serovars that were not previously detected (Gavilan et al., 2013). Recently, in 2009, an outbreak of diarrhea attributed to infection by V. parahaemolyticus of the O3:K59 serotype with pandemic traits was reported in Peru in the Lambayeque, Piura and Lima regions (Zamudio et al., 2011) (Figure 1 and Table 1).
The epidemic dissemination of the O3:K6 pandemic clone along the coast of Peru appears to correspond to the expansion and dynamics of the poleward propagation and receding of tropical waters associated with the 1997 El Niño event (Martinez-Urtaza et al., 2008; Ansede-Bermejo et al., 2010). The arrival of the pandemic clone to Peru in 1997 resulted in a significant shift in the epidemic dynamics of V. parahaemolyticus within the region, replacing the seasonal and local self-limited infections attributed to native genetic groups with the generalization of infections exclusively caused by the pandemic strains across the country and possibly on the American continent (Gil et al., 2007; Martinez-Urtaza et al., 2008; Ansede-Bermejo et al., 2010; Velazquez-Roman et al., 2012). Recently, the Instituto Nacional de Salud del Perú (INS) became a member of the WHO Global Foodborne Infections Network (WHO-GFN) and of the PulseNet Latin American and Caribbean Net (PN-AL & C), with whom it shares the genetic profiles of the isolated pathogenic strains, making it possible to compare the genotypes of similar strains found in different countries and to identify the occurrence of epidemic outbreaks within the region (Zamudio et al., 2011).
Chile
Over the past decade, trends in V. parahaemolyticus illnesses associated with molluscan shellfish have taken unexpected and unprecedented turns. Pandemic V. parahaemolyticus has now spread to at least 5 continents and has caused repeated annual outbreaks in Chile, where V. parahaemolyticus was not even considered a problem when this risk assessment began (Gonzalez-Escalona et al., 2005). The spread of the pandemic clonal O3:K6 complex reached the Southern Hemisphere as early as 1998, two years after the strain abruptly appeared in Calcutta, India, in 1996 (Gonzalez-Escalona et al., 2005). An outbreak that occurred primarily within the northern city of Antofagasta in Chile from November 1997 to March 1998 caused >300 clinical cases (Cordova et al., 2002). This city is near the border with Peru. A second outbreak affecting approximately 1500 individuals occurred from January to March 2004, predominantly in Puerto Montt, a region that is known for cold coastal waters. The outbreak in Puerto Montt was likely triggered by higher-than-normal temperatures during the summer months within the region, which is generally cool during all seasons (Gonzalez-Escalona et al., 2005). This last outbreak had important economic and social repercussions because this region is one of the main shellfish-producing areas in Chile (Gonzalez-Escalona et al., 2005). The specific infection dynamics found in Chile, where infections associated with the pandemic clone started in 1997–1998 and subsequently in 2004 and where the pandemic clone has been predominant among clinical isolates, clearly contrasts with the epidemiological trend of arrival and the rapid decline of pandemic V. parahaemolyticus infections in Peru.
During the summer of 2005, one of the outbreaks in Chile was high-magnitude, causing >11000 cases. This is the largest outbreak documented for this pathogen, most of which have been associated with consumption of clams and mussels but not oysters, which reflects the pattern of shellfish consumption in Chile (Gonzalez-Escalona et al., 2005; Fuenzalida et al., 2006; Cabello et al., 2007). The large epidemics of diarrhea observed during the last two austral summers in Puerto Montt, Chile, appears to be directly related to the introduction of the O3:K6 serovar pandemic strain to this region (Gonzalez-Escalona et al., 2005; Fuenzalida et al., 2006). Until 2006, every analyzed case in Chile was caused by the serovar O3:K6 pandemic strain (Olea et al., 2005; MINSAL, 2013). In the summer of 2007, among the 477 cases reported, 73% corresponded to the pandemic complex (Olea et al., 2005; MINSAL, 2013), among which 40% of the clinical cases were associated with the O3:K59 serotype and 27% were unrelated to the pandemic strain (Garcia et al., 2009; Harth et al., 2009). In the summer of 2008, there was an unexpected increase from 477 to 1143 clinical cases that were associated with the pandemic strain serotype O3:K6 (98%) (Garcia et al., 2009; Harth et al., 2009). According to data from the Ministery of Health of Chile, more than 3640 cases of gastroenteritis were reported in 2008, although the reports do not specify the serotype of the V. parahaemolyticus isolates (MINSAL, 2013). In 2009, clinical cases decreased to 441, and only 64% were related to the pandemic strain; the remaining cases were related to non-pandemic tdh- and trh-negative strains (Garcia et al., 2009). Gradually, outbreaks have decreased to fewer than 10 cases in 2010 and 2011 (Garcia et al., 2013). In Chile, the pandemic strain had become a relatively stable bacterial subpopulation of the diverse V. parahaemolyticus population that is present in shellfish (Garcia et al., 2009).
This cycle of proliferation and the disappearance of pathogenic strains might be the result of a combination of biological and physical environmental factors, including temperature, bacterial solar exposure and interaction with bacteriophages, which has been demonstrated in other regions of the world (Kaneko and Colwell, 1973; Kaneko, 1977; Kaysner et al., 1990; Garcia et al., 2013). However, the significant mechanisms of transmission remain poorly defined.
United states of america
In the U.S., Vibrio infections result in an estimated 80000 illnesses, 500 hospitalizations, and 100 deaths every year (Baba et al., 1991). The United States has been prone to V. parahaemolyticus infections since the identification of this pathogen in 1971 in Maryland, where steamed crabs and crabmeat were implicated in the outbreaks (Dadisman et al., 1972). A total of forty V. parahaemolyticus outbreaks associated with consumption of raw or cooked seafood cross-contamination occurred in the U.S. between 1973 and 1998 (Daniels et al., 2000a). One such report was of sporadic Vibrio infections in two coastal areas of Louisiana and Texas in the U.S. between 1992–1993, when crayfish consumption was reported by 50% of the individuals infected by V. parahaemolyticus (Bean et al., 1998). Furthermore, three large multistate outbreaks occurred in 1997 and 1998. An outbreak linked to the consumption of raw or undercooked shellfish harvested from the waters off the coasts of California, Oregon, Washington, and British Columbia resulted in 1200 illnesses in 7 states and Canada (CDC, 1998), between July to August 1997. The most common V. parahaemolyticus serotypes isolated from patients involved in this outbreak were O4:K12 and O1:K56. The O4:K12 serotype exhibited the highest prevalence among tested clinical V. parahaemolyticus isolates from the U.S. Pacific Coast between 1979 and 1995 (Depaola et al., 2003).
Between May and June 1998, 416 individuals in 13 states reported experiencing gastroenteritis after having eaten raw oysters harvested from Galveston Bay, Tex. All of the 28 available stool samples yielded V. parahaemolyticus O3:K6 isolates, which closely resembled the pandemic Asian O3:K6 isolates by PFGE (Daniels et al., 2000a). During July to September 1998, a smaller outbreak of O3:K6 V. parahaemolyticus associated with the consumption of oysters and clams harvested from Long Island Sound occurred among residents of Connecticut, New Jersey, and New York (CDC, 1999a,b). Interestingly, prior to 1998, outbreaks of the O3:K6 V. parahaemolyticus serotype had never been reported in the U.S. (Daniels et al., 2000a). V. parahaemolyticus has also been detected as far north as Alaska (Vasconcelos et al., 1975). Active surveillance identified a total of 62 patients with gastroenteritis; the outbreak was associated with the O6:K18 serotype, which was responsible for a 30% infection rate among passengers consuming one to six oysters on three cruises over a two-week period in July 2004 (McLaughlin et al., 2005). The O6:K18 isolates from the Alaskan outbreak were indistinguishable by PFGE from those isolated in the sporadic cases from Pacific Coast states over the previous decade. From July to October of 2004, 96 environmental samples (e.g., oysters, water, and sediment) were collected from 17 Alaskan oyster farms, and 31 samples (32 percent) tested positive for V. parahaemolyticus. The most frequently occurring serotypes were O1:K9, O4:K63, and O6:K18 (Newton et al., 2012) (Figure 1 and Table 1).
Clinical isolates in the U.S., especially from the Pacific Northwest also encode trh in addition to tdh (Paranjpye et al., 2012). An increasing proportion of clinical isolates possessed neither tdh nor trh genes, and these were associated with the most severe cases that required hospitalization (FAO/WHO, 2011).
Due to elevated prevalence of Vibrio infection, the following two national surveillance systems have been created to monitor the cases of infection: the national Cholera and Other Vibrio Illness Surveillance (COVIS) system and the Foodborne Diseases Active Surveillance Network (Food-Net). COVIS was established in 1988 by the CDC; the Gulf Coast states of Alabama, Florida, Louisiana, and Texas (states with high incidences of vibriosis); and the US Food and Drug Administration (FDA) to conduct surveillance of illnesses caused by Vibrio species (CDC, 2013a). FoodNet is a collaborative project that includes the CDC, 10 participating state health departments, the Food Safety and Inspection Service of the US Department of Agriculture, and the FDA. FoodNet conducts active, population-based surveillance for all laboratory-confirmed Vibrio infections, as well as other enteric infections that are commonly transmitted through food (CDC, 2013b).
Brazil
In relation to Brazil, there are few references to V. parahaemolyticus in cases of intestinal infections of human isolates (Hofer, 1983; Magalhaes et al., 1991a,b, 1992) in the colonization of cutaneous wounds (Rodrigues et al., 2001). In 1991 in Recife, a tropical city situated in Northeast Brazil, V. parahaemolyticus cultures were isolated from 21 cases of gastroenteritis associated with the consumption of different types of seafood. The serotypes detected belonged to diverse serovars, including O1:K56, O3:K5, O3:K58, O3:KUT, O4:K4,O4:K10, O4:K12, O4:K53, O4:KUT, O5:KUT, and O10:KUT (Magalhaes et al., 1991a). In 1992, V. parahaemolyticus strains were identified in 18 human isolates and found to carry either the tdh, trh, or both genes (Magalhaes et al., 1992). Until 2001–2002, the presence of strains with the characteristics of the pandemic clone (O3:K6) was detected in two outbreaks of gastroenteritis, suggesting that this strain is disseminated in certain regions (Ceara and Alagoas) of northeast Brazil. Nonetheless, the O3:KUT serotype of V. parahaemolyticus was detected in this region (Pernambuco and Alagoas) at the same time (Magalhaes et al., 1992) (Figure 1 and Table 1).
In 2010, studies on oysters harvested in the Southern coastal areas of Sao Paulo, Brazil found that the 99.2–100% contained V. parahaemolyticus (Sobrinho Pde et al., 2010, 2011). The results of these studies indicated that the levels of V. parahaemolyticus in retail oysters are frequently above 10,000 cfu/g, but the pathogenic strains are infrequent (none of the isolates contained the tdh and/or trh genes) (Sobrinho Pde et al., 2010, 2011). This corroborates the limited number of reported outbreaks, as only three V. parahaemolyticus outbreaks were reported between 1999 and 2009, two in 2003 and one in 2006, affecting 39 patients (Sobrinho Pde et al., 2010, 2011). However, because of the virulence potential of the pandemic clone detected at northeast Brazil in 2001–2002, increased seawater temperature and the consumption of seafood (oysters and mussels) represent factors that are extremely favorable for the dissemination of this microorganism.
Mexico
In 1993, the prevalence of V. parahaemolyticus in fresh seafood sold in a region of Mexico was 45.6%, of which 71.4% was fish, 44% was oysters and 27.6% was shrimp. Of the total seafood sold, a larger number of positive samples was observed during the warmer months (Torres-Vitela and Fernandez-Escartin, 1993). Between September and October of 2004, more than 1230 cases of gastroenteritis were reported in the relatively small geographical area of the southern part of Sinaloa, a state located in Northwest Mexico.
These cases were associated with the consumption of raw or undercooked shrimp that were collected in a lagoon system. In Mexico, this was the first outbreak of gastroenteritis that was caused by the pandemic strain of O3:K6 V. parahaemolyticus (Cabanillas-Beltran et al., 2006; Velazquez-Roman et al., 2012). Prior to 2004, there had been no reports of outbreaks caused by V. parahaemolyticus in Mexico (Cabanillas-Beltran et al., 2006; Velazquez-Roman et al., 2012) and few reports of environmental tdh-positive strains had been isolated from water, fish, sediment and shrimp (Cabrera-Garcia et al., 2004; Velazquez-Roman et al., 2012). Since 2004, recurrent sporadic cases have been reported in this state. During recent years, new cases have arisen in different areas of Sinaloa (including the southern and northern areas). Data analyzed over 6 years (from September 2004 to December 2010) in clinical cases and environmental samples along the Pacific coast of Sinaloa detected the presence of pandemic and pathogenic strains belonging to different serotypes of Vibrio parahaemolyticus (Figure 1 and Table 1). The pandemic O3:K6 clone was the most prevalent serotype isolated from clinical samples during every year of the study. The pandemic strain was also detected in environmental samples (seawater, sediment, and shrimp), suggesting that it is endemic to this setting (Velazquez-Roman et al., 2012).
To date, a shift in the epidemiology of outbreaks caused by pandemic V. parahaemolyticus O3:K6 has not been established in Mexico, where 80.5% of clinical cases were caused by this pathogen. However, the data from 2010/2011 in Chile showed that the pandemic O3:K6 strain has practically disappeared and that this condition is associated with a change in serotype of many pandemic isolates to O3:K59, suggesting the emergence of new clinical strains (Harth et al., 2009; Garcia et al., 2013). Thus, on the American continent, the last report of the O3:K6 pandemic clone associated with clinical cases was observed in Mexico.
vibrio parahaemolyticus in countries without the pandemic american continent clone
Canada
In 1997–2006, 212 cases of gastroenteritis were reported in British Columbia and were based predominantly on laboratory confirmation (Vibrio parahaemolyticus), with a minority of cases based on an association of clinical symptoms and food history (Figure 1 and Table 1). From 2001 to 2006, V. parahaemolyticus was the most commonly reported Vibrio species, and the majority of V. parahaemolyticus infections are believed to have resulted from the consumption of raw shellfish (in particular, raw oysters) (CDC, 2008). The majority of V. parahaemolyticus infections occurred during the summer months when ocean temperatures are warmer (PHAC, 2013). Regardless of the underlying reason, these findings suggest the need to increase awareness of the risk of Vibrio infection from shellfish (PHAC, 2013). Considering the increased rate of V. parahaemolyticus infection in 2006, the BC Center for Disease Control, BC health authorities, Health Canada, the CFIA (2007), the BC Shellfish Growers Association and the Canadian Council of Grocery Distributors have developed an education and communication plan to increase awareness for the risks associated with consuming raw shellfish (Khaira and Galanis, 2007).
Costa rica
From November 1985 to October 1987, Vibrio parahaemolyticus and other Vibrios were detected in 36 sediment samples and 41 bivalve samples obtained from 3 collection sites in the Gulf of Nicoya, Costa Rica (Garcia Cortes and Antillon, 1990). However, the first reported case of diarrhea attributed to V. parahaemolyticus was isolated in 1998 in a 30-year-old man in Costa Rica (Díaz, 2002), after consuming “pianguas” (Anadara tuberculosa), he presented severe and watery diarrhea, vomiting and abdominal cramps. No additional data are available for this pathogen in Costa Rica (Figure 1 and Table 1).
Puerto rico
In 1988, water and shellfish samples collected from estuaries, mangroves, and beaches along the coast of Puerto Rico were examined for V. vulnificus and V. parahaemolvticus (Rivera Susan Lugo and Hazen, 1989) (Table 1). That study demonstrated a significant positive correlation between fecal coliform levels and the density of V. parahaemolyticus in the water column, although no other reports are available for this bacterium.
Panama
In 1974, the presence of V. parahaemolyticus was detected in seawater off the Pacific coast of Panama (Kourany et al., 1974), and in 1975 (Kourany and Vasquez, 1975) the first confirmed case of gastroenteritis attributed to Vibrio parahaemolyticus infection in Panama was reported. Beside, Vibrio parahaemolyticus was recovered from fresh shrimp used in preparing seafood dishes (Kourany and Vasquez, 1975) (Figure 1 and Table 1). However, no other data regarding the isolates of V. parahaemolyticus to date have been reported.
Venezuela
In Venezuela, V. parahaemolyticus has been isolated from bivalves (Arca zebra and Perna perna) in an area near the Cariaco Basin (Grau et al., 2004; Muñoz et al., 2008). In 2011, Vibrio parahaemolyticus was detected in the prawns and corals of this system (Garcia-Amado et al., 2011) (Figure 1 and Table 1). However, additional data are unavailable for this pathogen.
Argentina
The only accessible data regarding the detection of V. parahemolyticus in this country is a report in 1977, when V. parahemolyticus was isolated from mussel varieties (Casellas et al., 1977) (Figure 1 and Table 1). However, no additional data are available for this region.
The dissemination of vibrio parahaemolyticus O3:K6 on the american continent
How and when the pandemic strain arrived in these countries and regions of the American continent and why it caused the outbreaks during those years remains a matter for speculation. The routes and mechanisms underlying the dissemination of the pandemic O3:K6 clone have been controversial from their emergence, although biological invasion by Vibrio populations can be mediated by human activities, such as ballast water discharges, or induced by natural events, such as the movement of oceanic waters.
Until the emergence of Vibrio epidemics in South America, the infections had spread predominantly westward, consistent with the prevailing westward movement of water associated with the Indonesian throughflow (a system of currents flowing from the Pacific Ocean to the Indian Ocean through the Indonesian Sea) (Gordon et al., 2003). The infections first surfaced in the north of the country and then spread southwards along more than 1500 km within a 4-month period until they reached the Chilean city of Antofagasta (Gonzalez-Escalona et al., 2005). The origins and routes of dissemination of pandemic V. parahaemolyticus from its arrival in South America remain unknown. A recent revision of the oceanographic conditions during this period revealed that the emergence and dissemination of the pandemic clone in Peru correlated with the dynamics of the progression and recession of the 1997 El Niño waters (Martinez-Urtaza et al., 2008). According to this study, the 1997 El Niño episode might have provided an extraordinary corridor for the displacement of Asian Vibrio populations to America, causing a general disruption of the environmental conditions of coastal areas and the southward displacement of native species (Ansede-Bermejo et al., 2010). In addition to promoting natural range expansions, warming temperatures could facilitate the establishment and spread of deliberately or accidentally introduced species (Carlton, 2000; Stachowicz et al., 2002). The chance of success of a biological invasion positively correlates with the level of ecological disturbance of the invaded environment (Lockwood et al., 2005).
One of the most recurrent explanations has been based on the discharge of ballast waters from ships traveling from areas of V. parahaemolyticus endemicity (Ansede-Bermejo et al., 2010). Ballast water discharges have been considered one of the major vehicles for the worldwide dissemination of marine species and biological invasions (Niimi, 2004), and they have been identified as a reliable mechanism for the propagation of pathogenic Vibrios (Depaola et al., 1992; McCarthy and Khambaty, 1994; Ruiz et al., 2000). Discharge of ballast waters should be related to a low propagule pressure or introductory effort as follows: a unique introduction event of reduced genetic variation with low chances of success of establishment within the invaded area (Lockwood et al., 2005). Ballast water-mediated invasions have been proposed for the introduction of the O3:K6 strains into Texas in 1998 (Daniels et al., 2000b). The infections caused a unique epidemic outbreak in areas close to ports, although after the outbreaks, the strains belonging to the O3:K6 clone were never detected in the clinical cases or the environment (Depaola et al., 2000). Bacterial dispersal through ballast water, however, fails to consistently and comprehensively explain the emergence of some epidemic episodes of V. parahaemolyticus (Ansede-Bermejo et al., 2010). This is particularly true where infections have emerged and spread rapidly over hundreds of kilometers of coastline, as in the case of the arrival, dissemination and establishment of the O3:K6 clone populations along the Pacific Coast of South America (Peru) since 1996 (Leal et al., 2008; Martinez-Urtaza et al., 2008).
The importance of water temperature in the epidemiology of infections is reflected by the fact that most outbreaks have occurred during the warmer months of the year. Thus, the spread or arrival of O3:K6 to Brazil in 2001 may be attributed to the increased seawater temperature, a factor that is extremely favorable for the dissemination of this microorganism. However, how this bacterium spread to Brazil remains to be determined. Warmer temperatures also appear to extend the geographical range of V. parahaemolyticus into areas such as Alaska (McLaughlin et al., 2005) and Chile (Gonzalez-Escalona et al., 2005), whereas the restoration of cold waters into these areas may have begun to reduce the O3:K6 populations toward the tropical areas located in northern Peru, where they remain endemic to date (Martinez-Urtaza et al., 2008). Alaska is located 1000 km further north than any previously reported V. parahaemolyticus illnesses. The unprecedentedly high prevalence of pathogenic strains of V. parahaemolyticus among the population of Alaskan oysters, and perhaps their increased virulence compared to other pathogenic strains, was particularly unexpected and remains unexplained (McLaughlin et al., 2005). Although oysters are the most common food associated with Vibrio infection in certain countries (Hlady and Klontz, 1996), there have been reports of V. parahaemolyticus infections associated with other types of seafood (Velazquez-Roman et al., 2012). Climate warming can increase pathogen development and survival rates, disease transmission and host vulnerability, although a subset of pathogens might decrease with warming, releasing hosts from disease (Harvell et al., 2002).
Climate change might also influence the selection of different modes of transmission and virulence (Marcogliese, 2001). Recently, analysis of the arrival of the O3:K6 clone at the Pacific coasts of South America has provided novel insights linking its origin to an invasion in 1996 from the Asian (Indian) populations and describing the successful establishment of O3:K6 populations, first in Peru and subsequently in the south of Chile (1998) (Ansede-Bermejo et al., 2010). The Peruvian foothold of the O3:K6 clone may be the origin of the arrival of this strain to the coasts of Northwestern Mexico (Sinaloa) in 2003 to 2004, traveling more than 8000 Km due to either ballast water discharge or climate change (Velazquez-Roman et al., 2012). However, the actual dissemination dynamics of the environmental human pathogen Vibrio parahaemolyticus remain uncertain. El Niño provides a partial explanation for the spectrum of marine biotoxins (red tides) on the coast of Mexico (Ochoa, 2003).Notwithstanding, the absence of environmental information about the impact of the arrival of El Niño waters on the native Vibrio community severely limits a comprehensive overview of the role of El Niño episodes in the introduction of new Vibrio pathogens into these remote regions. All of these factors, including climate change and human activities, may have influenced and favored the dissemination of the O3:K6 pandemic clone and its serovariants to American countries.
Nevertheless, pathogenic subpopulations of Vibrios are potential agents of disease outbreaks and pandemics (Siddique et al., 1994; CDC, 1999b; Depaola et al., 2000; Lee et al., 2001; Gonzalez-Escalona et al., 2005; McLaughlin et al., 2005; Noriea et al., 2010). The origin and subsequent spread of the O3:K6 isolates of V. parahaemolyticus must be the consequence of coincidental events that varied in magnitude in developing countries, particularly on the American continent, where the consumption of raw or undercooked shellfish is common (Wong et al., 1999; Chen et al., 2004; Velazquez-Roman et al., 2012) but varies according to prevailing levels of health education, sanitation, risk factors, response from the community at large and climate change (Ansede-Bermejo et al., 2010).
Conclusions
On the American continent, only five countries have reported the presence of the pandemic Vibrio parahaemolyticus clone O3:K6 and its serovariants in clinical cases and/or the environment. The pandemic strain was first observed in Peru (1996), subsequently spread to Chile in 1998, to the U.S. in 1998, to Brazil in 2001 and to Mexico in 2004. However, the presence of pandemic clones and the number of reported outbreaks in the countries of the American continent may be underestimated due to non-reporting. The American continent has a diversified climate. The scientific communities are coming to the conclusion that ballast discharge, global trade and climate change represent the major underlying mechanisms for the global spread of pandemic V. parahaemolyticus, particularly clone O3:K6. Moreover, many American countries require active reporting, diagnosis, and surveillance of cases and contacts, food handlers, and sanitation, as well as health education programs for the prevention of gastroenteritis due to V. parahaemolyticus.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from PROFAPI-UAS 2012/032 to Adrian Canizalez-Roman. Lucio de Jesus Hernández-Díaz was supported by a scholarship from CONACYT No. 290225. We thank Dr. Secundino Muro for critical reading.
References
- Alam M. J., Tomochika K. I., Miyoshi S. I., Shinoda S. (2002). Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol. Lett. 208, 83–87 10.1111/j.1574-6968.2002.tb11064.x [DOI] [PubMed] [Google Scholar]
- Ansaruzzaman M., Lucas M., Deen J. L., Bhuiyan N. A., Wang X. Y., Safa A., et al. (2005). Pandemic serovars (O3:K6 and O4:K68) of Vibrio parahaemolyticus associated with diarrhea in Mozambique: spread of the pandemic into the African continent. J. Clin. Microbiol. 43, 2559–2562 10.1128/JCM.43.6.2559-2562.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansede-Bermejo J., Gavilan R. G., Trinanes J., Espejo R. T., Martinez-Urtaza J. (2010). Origins and colonization history of pandemic Vibrio parahaemolyticus in South America. Mol. Ecol. 19, 3924–3937 10.1111/j.1365-294X.2010.04782.x [DOI] [PubMed] [Google Scholar]
- Baba K., Shirai H., Terai A., Takeda Y., Nishibuchi M. (1991). Analysis of the tdh gene cloned from a tdh gene- and trh gene-positive strain of Vibrio parahaemolyticus. Microbiol. Immunol. 35, 253–258 10.1111/j.1348-0421.1991.tb01554.x [DOI] [PubMed] [Google Scholar]
- Bag P. K., Nandi S., Bhadra R. K., Ramamurthy T., Bhattacharya S. K., Nishibuchi M., et al. (1999). Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J. Clin. Microbiol. 37, 2354–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., Furniss A. l., Lee J. V. (1984). Genus I. Vibrio, in Bergey's Manual of Systematic Bacteriology, Vol. 1 eds Murray R. G. E., Brenner D. J., Bryant M. P. (Baltimore, MD: Williams and Wilkins; ), 518. [Google Scholar]
- Bean N. H., Maloney E. K., Potter M. E., Korazemo P., Ray B., Taylor J. P., et al. (1998). Crayfish: a newly recognized vehicle for vibrio infections. Epidemiol. Infect. 121, 269–273 10.1017/S0950268898001381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej A. K., Patterson D. P., Brasher C. W., Vickery M. C., Jones D. D., Kaysner C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36, 215–225 10.1016/S0167-7012(99)00037-8 [DOI] [PubMed] [Google Scholar]
- Boyd E. F., Cohen A. L., Naughton L. M., Ussery D. W., Binnewies T. T., Stine O. C., et al. (2008). Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8, 110 10.1186/1471-2180-8-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanillas-Beltran H., LLausás-Magaña E., Romero R., Espinoza A., Garcia-Gasca A., Nishibuchi M., et al. (2006). Outbreak of gastroenteritis caused by the pandemic Vibrio parahaemolyticus O3:K6 in Mexico. FEMS Microbiol. Lett. 265, 76–80 10.1111/j.1574-6968.2006.00475.x [DOI] [PubMed] [Google Scholar]
- Cabello F. C., Espejo R. T., Hernandez M. C., Rioseco M. L., Ulloa J., Vergara J. A. (2007). Vibrio parahaemolyticus O3:K6 epidemic diarrhea, Chile, 2005. Emerging Infect. Dis. 13, 655–656 10.3201/eid1304.06-1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Garcia M. E., Vazquez-Salinas C., Quinones-Ramirez E. I. (2004). Serologic and molecular characterization of Vibrio parahaemolyticus strains isolated from seawater and fish products of the Gulf of Mexico. Appl. Environ. Microbiol. 70, 6401–6406 10.1128/AEM.70.11.6401-6406.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canadian Food Inspection Agency. (2007). Available online at: http://www.phac-aspc.gc.ca/publicat/ccdr-rmtc/07vol33/dr3311b-eng.php
- Canizalez-Roman A., Flores-Villasenor H., Zazueta-Beltran J., Muro-Amador S., Leon-Sicairos N. (2011). Comparative evaluation of a chromogenic agar medium-PCR protocol with a conventional method for isolation of Vibrio parahaemolyticus strains from environmental and clinical samples. Can. J. Microbiol. 57, 136–142 10.1139/W10-108 [DOI] [PubMed] [Google Scholar]
- Carlton J. T. (2000). Global change and biological invasions in the oceans, in Invasive Species in a Changing World, eds Mooneyand H. A., Hobbs R. J. (Covelo, CA: Island Press; ), 31–53 [Google Scholar]
- Casellas J. M., Caría M. A., Gerghi M. E. (1977). Isolation of Vibrio parahemolyticus from mussel varieties in Argentina. Rev. Asoc. Argent. Microbiol. 9, 41–53 [PubMed] [Google Scholar]
- CDC. (1998). Outbreak of Vibrio parahaemolyticus infections associated with eating raw oysters–Pacific Northwest, 1997. MMWR Morb. Mortal. Wkly. Rep. 47, 457–462 [PubMed] [Google Scholar]
- CDC. (1999a). From the centers for disease control and prevention. outbreak of vibrio parahaemolyticus infection associated with eating raw oysters and clams harvested from Long Island Sound—Connecticut. New Jersey, and New York, 1998. JAMA 281, 603–604 [PubMed] [Google Scholar]
- CDC. (1999b). Outbreak of Vibrio parahaemolyticus infection associated with eating raw oysters and clams harvested from Long Island Sound–Connecticut, New Jersey, and New York, 1998. MMWR Morb. Mortal. Wkly. Rep. 48, 48–51 10.1001/jama.281.7.603 [DOI] [PubMed] [Google Scholar]
- CDC. (2008). Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food–10 states, 2007. MMWR Morb. Mortal. Wkly. Rep. 57, 366–370 [PubMed] [Google Scholar]
- CDC. (2013a). Cholera and Other Vibrio Illness Surveillance System. [Accessed September 9 2013] Centers for Disease Control and Prevention. Department of Health and Human Services.Available online at: http://www.cdc.gov/nationalsurveillance/cholera_vibrio_surveillance.html
- CDC. (2013b). Foodborne Diseases Active Surveillance Network. Foodborne Diseases Active Surveillance Network (FoodNet). [Accessed September 9]. Available online at: http://www.cdc.gov/foodnet/
- Chao G., Jiao X., Zhou X., Wang F., Yang Z., Huang J., et al. (2010). Distribution of genes encoding four pathogenicity islands (VPaIs), T6SS, biofilm, and type I pilus in food and clinical strains of Vibrio parahaemolyticus in China. Foodborne Pathog. Dis. 7, 649–658 10.1089/fpd.2009.0441 [DOI] [PubMed] [Google Scholar]
- Chen C. H., Shimada T., Elhadi N., Radu S., Nishibuchi M. (2004). Phenotypic and genotypic characteristics and epidemiological significance of ctx+ strains of Vibrio cholerae isolated from seafood in Malaysia. Appl. Environ. Microbiol. 70, 1964–1972 10.1128/AEM.70.4.1964-1972.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A., Ishibashi M., Thiem V. D., Tuyet D. T., Tung T. V., Chien B. T., et al. (2004a). Emergence and serovar transition of Vibrio parahaemolyticus pandemic strains isolated during a diarrhea outbreak in Vietnam between 1997 and 1999. Microbiol. Immunol. 48, 319–327 10.1111/j.1348-0421.2004.tb03513.x [DOI] [PubMed] [Google Scholar]
- Chowdhury N. R., Stine O. C., Morris J. G., Nair G. B. (2004b). Assessment of evolution of pandemic Vibrio parahaemolyticus by multilocus sequence typing. J. Clin. Microbiol. 42, 1280–1282 10.1128/JCM.42.3.1280-1282.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury N. R., Chakraborty S., Ramamurthy T., Nishibuchi M., Yamasaki S., Takeda Y., et al. (2000). Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerging Infect. Dis. 6, 631–636 10.3201/eid0606.000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova J. L., Astorga J., Silva W., Riquelme C. (2002). Characterization by PCR of Vibrio parahaemolyticus isolates collected during the 1997-1998 Chilean outbreak. Biol. Res. 35, 433–440 10.4067/S0716-97602002000300017 [DOI] [PubMed] [Google Scholar]
- Dadisman T. A., Jr., Nelson R., Molenda J. R., Garber H. J. (1972). Vibrio parahaemolyticus gastroenteritis in Maryland. I. Clinical and epidemiologic aspects. Am. J. Epidemiol. 96, 414–426 [DOI] [PubMed] [Google Scholar]
- Daniels N. A., Mackinnon L., Bishop R., Altekruse S., Ray B., Hammond R. M., et al. (2000a). Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181, 1661–1666 10.1086/315459 [DOI] [PubMed] [Google Scholar]
- Daniels N. A., Ray B., Easton A., Marano N., Kahn E., McShan A. L., et al. (2000b). Emergence of a new Vibrio parahaemolyticus serotype in raw oysters: a prevention quandary. JAMA 284, 1541–1545 10.1001/jama.284.12.1541 [DOI] [PubMed] [Google Scholar]
- Deepanjali A., Kumar H. S., Karunasagar I. (2005). Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl. Environ. Microbiol. 71, 3575–3580 10.1128/AEM.71.7.3575-3580.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaola A., Capers G. M., Motes M. L., Olsvik O., Fields P. I., Wells J., et al. (1992). Isolation of Latin American epidemic strain of Vibrio cholerae O1 from US Gulf Coast. Lancet 339, 624 10.1016/0140-6736(92)90917-R [DOI] [PubMed] [Google Scholar]
- Depaola A., Kaysner C. A., Bowers J., Cook D. W. (2000). Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66, 4649–4654 10.1128/AEM.66.11.4649-4654.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaola A., Ulaszek J., Kaysner C. A., Tenge B. J., Nordstrom J. L., Wells J., et al. (2003). Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 69, 3999–4005 10.1128/AEM.69.7.3999-4005.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz J. M. E. (2002). Diarrea por Vibrio parahaemolyticus: primer caso reportado en Costa Rica. Revista Médica del Hospital Nacional de Niños Dr. Carlos Sáenz Herrera 37, 15–17 [Google Scholar]
- FAO/WHO. (2011). Risk assessment of Vibrio parahaemolyticus in seafood: Interpretative summary and Technical report. food and agriculture organization of the united nations/world health organization. Microbiol. Risk Assess. 16, 193 [Google Scholar]
- Fuenzalida L., Hernandez C., Toro J., Rioseco M. L., Romero J., Espejo R. T. (2006). Vibrio parahaemolyticus in shellfish and clinical samples during two large epidemics of diarrhoea in southern Chile. Environ. Microbiol. 8, 675–683 10.1111/j.1462-2920.2005.00946.x [DOI] [PubMed] [Google Scholar]
- Fujino T., Okuno Y., Nakada D., Aoyama A., Fukai K., Mukai T., et al. (1953). On the bacteriological examination of Shirasu food poisoning. Med. J. Osaka Univ. 4, 299–304 [Google Scholar]
- Garcia Cortes V., Antillon F. (1990). [Isolation of enteropathogenic Vibrio in bivalves and mud from the Nicoya Gulf, Costa Rica]. Rev. Biol. Trop. 38, 437–440 [PubMed] [Google Scholar]
- Garcia K., Bastias R., Higuera G., Torres R., Mellado A., Uribe P., et al. (2013). Rise and fall of pandemic Vibrio parahaemolyticus serotype O3:K6 in southern Chile. Environ. Microbiol. 15, 527–534 10.1111/j.1462-2920.2012.02883.x [DOI] [PubMed] [Google Scholar]
- Garcia K., Torres R., Uribe P., Hernandez C., Rioseco M. L., Romero J., et al. (2009). Dynamics of clinical and environmental Vibrio parahaemolyticus strains during seafood-related summer diarrhea outbreaks in southern Chile. Appl. Environ. Microbiol. 75, 7482–7487 10.1128/AEM.01662-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Amado M. A., Bozo-Hurtado L., Astor Y., Suarez P., Chistoserdov A. (2011). Denaturing gradient gel electrophoresis analyses of the vertical distribution and diversity of Vibrio spp. populations in the Cariaco Basin. FEMS Microbiol. Ecol. 77, 347–356 10.1111/j.1574-6941.2011.01116.x [DOI] [PubMed] [Google Scholar]
- Gavilan R. G., Zamudio M. L., Martinez-Urtaza J. (2013). Molecular epidemiology and genetic variation of pathogenic Vibrio parahaemolyticus in Peru. PLoS Negl. Trop. Dis. 7:e2210 10.1371/journal.pntd.0002210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A. I., Miranda H., Lanata C. F., Prada A., Hall E. R., Barreno C. M., et al. (2007). O3:K6 serotype of Vibrio parahaemolyticus identical to the global pandemic clone associated with diarrhea in Peru. Int. J. Infect. Dis. 11, 324–328 10.1016/j.ijid.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escalona N., Cachicas V., Acevedo C., Rioseco M. L., Vergara J. A., Cabello F., et al. (2005). Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerging Infect. Dis. 11, 129–131 10.3201/eid1101.040762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A. L., Susanto R. D., Vranes K. (2003). Cool Indonesian throughflow as a consequence of restricted surface layer flow. Nature 425, 824–828 10.1038/nature02038 [DOI] [PubMed] [Google Scholar]
- Grau C. L. B., Zerpa A., Silva S., Gallardo O. (2004). Aislamiento de Vibrio spp. y evaluacion de la condicion sanitaria de los moluscos bivalvos Arca zebra y Perna perna procedentes de la costa nororiental del Edo. Sucre. Venezuela. Revista Cientımathfica FCV-LUZ, 14, 513–521 [Google Scholar]
- Guevara Duncan J. P., Del Carpio E. (1989). Vibrio parahaemolyticus in ‘cebiches’ sold by hucksters in Lima Peru. Diagnostico (in Spanish). 24, 23–26 [Google Scholar]
- Gutierrez West C. K., Klein S. L., Lovell C. R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 79, 2247–2252 10.1128/AEM.03792-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth E., Matsuda L., Hernandez C., Rioseco M. L., Romero J., Gonzalez-Escalona N., et al. (2009). Epidemiology of Vibrio parahaemolyticus outbreaks, southern Chile. Emerging Infect. Dis. 15, 163–168 10.3201/eid1502.071269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvell C. D., Mitchell C. E., Ward J. R., Altizer S., Dobson A. P., Ostfeld R. S., et al. (2002). Climate warming and disease risks for terrestrial and marine biota. Science 296, 2158–2162 10.1126/science.1063699 [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Hase C. C. (2009). The extracellular metalloprotease of Vibrio tubiashii directly inhibits its extracellular haemolysin. Microbiology 155, 2296–2305 10.1099/mic.0.028605-0 [DOI] [PubMed] [Google Scholar]
- Hervio-Heath D., Colwell R. R., Derrien A., Robert-Pillot A., Fournier J. M., Pommepuy M. (2002). Occurrence of pathogenic vibrios in coastal areas of France. J. Appl. Microbiol. 92, 1123–1135 10.1046/j.1365-2672.2002.01663.x [DOI] [PubMed] [Google Scholar]
- Hiyoshi H., Kodama T., Iida T., Honda T. (2010). Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 78, 1772–1780 10.1128/IAI.01051-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyoshi H., Kodama T., Saito K., Gotoh K., Matsuda S., Akeda Y., et al. (2011). VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe 10, 401–409 10.1016/j.chom.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Hlady W. G., Klontz K. C. (1996). The epidemiology of Vibrio infections in Florida, 1981–1993. J. Infect. Dis. 173, 1176–1183 10.1093/infdis/173.5.1176 [DOI] [PubMed] [Google Scholar]
- Hofer E. (1983). Primeiro isolamento e identificac,a~o de Vibrio parahaemolyticus no Brasil de infecc,a~o gastrointestinal humana. Rev. Microbiol. Sa~o Paulo 4, 174–175 [Google Scholar]
- Honda T., Iida T. (1993). The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct hemolysin and related hemolysins. Rev. Med. Microbiol. 4, 106–113 10.1097/00013542-199304000-00006 [DOI] [Google Scholar]
- Ibarra J. D., A., Alvarado D. (1999). Non-epdemic vibrios and Vibrio cholerae O1 associated with acute diarrhea from cases given during “El Niño” Southern Oscillation (ENSO) 1998. Hospital Nacional Dos de Mayo. An Fac Med (Peru) (in Spanish) 60, 251–256 [Google Scholar]
- Iida T., Hattori A., Tagomori K., Nasu H., Naim R., Honda T. (2001). Filamentous phage associated with recent pandemic strains of Vibrio parahaemolyticus. Emerging Infect. Dis. 7, 477–478 10.3201/eid0703.010325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. S., Tasmin R., Khan S. I., Bakht H. B., Mahmood Z. H., Rahman M. Z., et al. (2004). Pandemic strains of O3:K6 Vibrio parahaemolyticus in the aquatic environment of Bangladesh. Can. J. Microbiol. 50, 827–834 10.1139/w04-072 [DOI] [PubMed] [Google Scholar]
- Joseph S. W., Colwell R. R., Kaper J. B. (1982). Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 10, 77–124 10.3109/10408418209113506 [DOI] [PubMed] [Google Scholar]
- Kaneko T., Colwell R. R. (1973). Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J. Bacteriol. 113, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T. C., R. R. (1977). The annual cycle of Vibrio parahaemolyticus in Chesapeake Bay. Microb. Ecol. 4, 135–155 10.1007/BF02014284 [DOI] [PubMed] [Google Scholar]
- Kaysner C. A., Abeyta C., Jr., Stott R. F., Lilja J. L., Wekell M. M. (1990). Incidence of urea-hydrolyzing Vibrio parahaemolyticus in Willapa Bay, Washington. Appl. Environ. Microbiol. 56, 904–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaira G., Galanis E. (2007). Descriptive epidemiology of Vibrio parahaemolyticus and other Vibrio species infections in British Columbia: 2001-2006. Can. Commun. Dis. Rep. 33, 12–22 [PubMed] [Google Scholar]
- Kishishita M., Matsuoka N., Kumagai K., Yamasaki S., Takeda Y., Nishibuchi M. (1992). Sequence variation in the thermostable direct hemolys in related hemolysin (trh) gene of Vibrio parahaemolyticus Appl. Environ. Microbiol. 58, 2449–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourany M., Kinney R. J., Vasquez M. A. (1974). Vibrio parahaemolyticus in seawater off the Pacific coast of Panama. Am. J. Trop. Med. Hyg. 23, 714–715 [DOI] [PubMed] [Google Scholar]
- Kourany M., Vasquez M. A. (1975). The first reported case from Panama of acute gastroenteritis caused by Vibrio parahaemolyticus. Am. J. Trop. Med. Hyg. 24, 638–640 [DOI] [PubMed] [Google Scholar]
- Leal N. C., Da Silva S. C., Cavalcanti V. O., Figueiroa A. C., Nunes V. V., Miralles I. S., et al. (2008). Vibrio parahaemolyticus serovar O3:K6 gastroenteritis in northeast Brazil. J. Appl. Microbiol. 105, 691–697 10.1111/j.1365-2672.2008.03782.x [DOI] [PubMed] [Google Scholar]
- Lee W. C., Lee M. J., Kim J. S., Park S. Y. (2001). Foodborne illness outbreaks in Korea and Japan studied retrospectively. J. Food Prot. 64, 899–902 [DOI] [PubMed] [Google Scholar]
- Lockwood J. L., Cassey P., Blackburn T. (2005). The role of propagule pressure in explaining species invasions. Trends Ecol. Evol. 20, 223–228 10.1016/j.tree.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Magalhaes M., Magalhaes V., Antas M. G., Tateno S. (1991a). Isolation of urease-positive Vibrio parahaemolyticus from diarrheal patients in northeast Brazil. Rev. Inst. Med. Trop. Sao Paulo 33, 263–265 10.1590/S0036-46651991000400004 [DOI] [PubMed] [Google Scholar]
- Magalhaes V., Lima R. A., Tateno S., Magalhaes M. (1991b). Human gastroenteritis associated with Vibrio parahaemolyticus in Recife, Brazil. Rev. Inst. Med. Trop. Sao Paulo 33, 64–68 10.1590/S0036-46651991000100012 [DOI] [PubMed] [Google Scholar]
- Magalhaes M., Takeda Y., Magalhaes V., Tateno S. (1992). Brazilian urease-positive strains of Vibrio parahaemolyticus carry genetic potential to produce the TDH-related hemolysin. Mem. Inst. Oswaldo Cruz 87, 167–168 10.1590/S0074-02761992000100027 [DOI] [PubMed] [Google Scholar]
- Mahmud Z. H., Neogi S. B., Kassu A., Wada T., Islam M. S., Nair G. B., et al. (2007). Seaweeds as a reservoir for diverse Vibrio parahaemolyticus populations in Japan. Int. J. Food Microbiol. 118, 92–96 10.1016/j.ijfoodmicro.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Marcogliese D. J. (2001). Implications of climate change for parasitism of animals in the aquatic environment. Can. J. Zool. 79, 1331–1352 10.1139/z01-067 [DOI] [Google Scholar]
- Martinez-Urtaza J., Huapaya B., Gavilan R. G., Blanco-Abad V., Ansede-Bermejo J., Cadarso-Suarez C., et al. (2008). Emergence of asiatic vibrio diseases in south america in phase with El Nino. Epidemiology 19, 829–837 10.1097/EDE.0b013e3181883d43 [DOI] [PubMed] [Google Scholar]
- Martinez-Urtaza J., Simental L., Velasco D., Depaola A., Ishibashi M., Nakaguchi Y., et al. (2005). Pandemic Vibrio parahaemolyticus O3:K6, Europe. Emerging Infect. Dis. 11, 1319–1320 10.3201/eid1108.050322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S., Okada N., Kodama T., Honda T., Iida T. (2012). A cytotoxic type III secretion effector of Vibrio parahaemolyticus targets vacuolar H+-ATPase subunit c and ruptures host cell lysosomes. PLoS Pathog. 8:e1002803 10.1371/journal.ppat.1002803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto C., Okuda J., Ishibashi M., Iwanaga M., Garg P., Rammamurthy T., et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38, 578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. (1999). The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1, 51–57 [PubMed] [Google Scholar]
- McCarthy S. A., Khambaty F. M. (1994). International dissemination of epidemic Vibrio cholerae by cargo ship ballast and other nonpotable waters. Appl. Environ. Microbiol. 60, 2597–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. B., Depaola A., Bopp C. A., Martinek K. A., Napolilli N. P., Allison C. G., et al. (2005). Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 353, 1463–1470 10.1056/NEJMoa051594 [DOI] [PubMed] [Google Scholar]
- MINSAL (2013). Vibrio parahaemolyticus. [Accessed 4 September 2013]. Departamento de Epidemiologia. Ministerio de Salud. (Departamento de Epidemiología-DIPLAS-MINSAL). Vibrio parahaemolyticus. Available online at: http://epi.minsal.cl/vigilancia-epidemiologica/enfermedades-de-notificacion-obligatoria/vibrio-parahaemolyticus/
- Muñoz D., Crucita G. D. M., Martımathnez C., Marjal H., Zerpa A. (2008). Prevalencia de Staphylococcus aureus, Vibrio spp. y enterobacterias en carne de pepitona, Arca zebra, comercializada en Cumana, Venezuela. Zoot. Trop. 26, 505–513 [Google Scholar]
- Nair G. B., Ramamurthy T., Bhattacharya S. K., Dutta B., Takeda Y., Sack D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48 10.1128/CMR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu H., Iida T., Sugahara T., Yamaichi Y., Park K. S., Yokoyama K., et al. (2000). A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38, 2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A., Kendall M., Vugia D. J., Henao O. L., Mahon B. E. (2012). Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin. Infect. Dis. 54(Suppl. 5), S391–S395 10.1093/cid/cis243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi A. J. (2004). Role of container vessels in the introduction of exotic species. Mar. Pollut. Bull. 49, 778–782 10.1016/j.marpolbul.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Nishibuchi M., Kaper J. B. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63, 2093–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M., Fasano A., Russell R. G., Kaper J. B. (1992). Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60, 3539–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriea N. F., 3rd., Johnson C. N., Griffitt K. J., Grimes D. J. (2010). Distribution of type III secretion systems in Vibrio parahaemolyticus from the northern Gulf of Mexico. J. Appl. Microbiol. 109, 953–962 10.1111/j.1365-2672.2010.04722.x [DOI] [PubMed] [Google Scholar]
- Ochoa J. L. (2003). ENSO phenomenon and toxic red tides in Mexico. Geofísica Int. 42, 505–515 [Google Scholar]
- Okuda J., Ishibashi M., Hayakawa E., Nishino T., Takeda Y., Mukhopadhyay A. K., et al. (1997). Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35, 3150–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura M., Osawa R., Iguchi A., Arakawa E., Terajima J., Watanabe H. (2003). Genotypic analyses of Vibrio parahaemolyticus and development of a pandemic group-specific multiplex PCR assay. J. Clin. Microbiol. 41, 4676–4682 10.1128/JCM.41.10.4676-4682.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olea A. M., González C., Chiu M., Vallebuona C., Labraña M., Martiniello F. (2005). Brote de gastroenteritis por Vibrio parahaemolyticus en Chile. Rev. Chile. Salud Pública 9, 51–53 [Google Scholar]
- Pal D., Das N. (2010). Isolation, identification and molecular characterization of Vibrio parahaemolyticus from fish samples in Kolkata. Eur. Rev. Med. Pharmacol. Sci. 14, 545–549 [PubMed] [Google Scholar]
- Paranjpye R., Hamel O. S., Stojanovski A., Liermann M. (2012). Genetic diversity of clinical and environmental Vibrio parahaemolyticus strains from the Pacific Northwest. Appl. Environ. Microbiol. 78, 8631–8638 10.1128/AEM.01531-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathi A., Kumar H. S., Bhanumathi A., Ishibashi M., Nishibuchi M., Karunasagar I. (2006). Molecular characterization of thermostable direct haemolysin-related haemolysin (TRH)-positive Vibrio parahaemolyticus from oysters in Mangalore, India. Environ. Microbiol. 8, 997–1004 10.1111/j.1462-2920.2006.00990.x [DOI] [PubMed] [Google Scholar]
- PHAC. (2013). Descriptive epidemiology of vibrio parahaemolyticus and other vibrio species infections in british columbia: 2001–2006. 33, 1–7 [Accessed 4 September 2013]. [PubMed] [Google Scholar]
- Qadri F., Alam M. S., Nishibuchi M., Rahman T., Alam N. H., Chisti J., et al. (2003). Adaptive and inflammatory immune responses in patients infected with strains of Vibrio parahaemolyticus. J. Infect. Dis. 187, 1085–1096 10.1086/368257 [DOI] [PubMed] [Google Scholar]
- Raimondi F., Kao J. P., Fiorentini C., Fabbri A., Donelli G., Gasparini N., et al. (2000). Enterotoxicity and cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin in in vitro systems. Infect. Immun. 68, 3180–3185 10.1128/IAI.68.6.3180-3185.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera Susan, Lugo T., Hazen T. C. (1989). Autecology of Vibrio vulnificus and Vibrio parahaemolyticus in tropical waters. Wat. Res. 23, 923–931 10.1016/0043-1354(89)90018-3 [DOI] [Google Scholar]
- Rodrigues S. M., Goncalves E. G., Mello D. M., Oliveira E. G., Hofer E. (2001). [Identification of Vibrio spp bacteria on skin lesions of fisherman in the county of Raposa-MA]. Rev. Soc. Bras. Med. Trop. 34, 407–411 [DOI] [PubMed] [Google Scholar]
- Ruiz G. M., Rawlings T. K., Dobbs F. C., Drake L. A., Mullady T., Huq A., et al. (2000). Global spread of microorganisms by ships. Nature 408, 49–50 10.1038/35040695 [DOI] [PubMed] [Google Scholar]
- Salomon D., Gonzalez H., Updegraff B. L., Orth K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE 8:e61086 10.1371/journal.pone.0061086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar B. L., Nair G. B., Banerjee A. K., Pal S. C. (1985). Seasonal distribution of Vibrio parahaemolyticus in freshwater environs and in association with freshwater fishes in Calcutta. Appl. Environ. Microbiol. 49, 132–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai H., Ito H., Hirayama T., Nakamoto Y., Nakabayashi N., Kumagai K., et al. (1990). Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58, 3568–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique A. K., Zaman K., Akram K., Mutsuddy P., Eusof A., Sack R. B. (1994). Emergence of a new epidemic strain of Vibrio cholerae in Bangladesh. An epidemiological study. Trop. Geogr. Med. 46, 147–150 [PubMed] [Google Scholar]
- Sobrinho Pde S., Destro M. T., Franco B. D., Landgraf M. (2010). Correlation between environmental factors and prevalence of Vibrio parahaemolyticus in oysters harvested in the southern coastal area of Sao Paulo State, Brazil. Appl. Environ. Microbiol. 76, 1290–1293 10.1128/AEM.00861-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrinho Pde S., Destro M. T., Franco B. D., Landgraf M. (2011). Occurrence and distribution of Vibrio parahaemolyticus in retail oysters in Sao Paulo State, Brazil. Food Microbiol. 28, 137–140 10.1016/j.fm.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Stachowicz J. J., Terwin J. R., Whitlatch R. B., Osman R. W. (2002). Linking climate change and biological invasions: ocean warming facilitates nonindigenous species invasions. Proc. Natl. Acad. Sci. U.S.A. 99, 15497–15500 10.1073/pnas.242437499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. C., Liu C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558 10.1016/j.fm.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Takeda Y. (1983). Thermostable direct hemolysin of Vibrio parahaemolyticus. Pharmacol. Ther. 19, 123–146 10.1016/0163-7258(82)90044-4 [DOI] [PubMed] [Google Scholar]
- Torres-Vitela M. R., Fernandez-Escartin E. (1993). Incidence of V. parahaemolyticus in raw fish, oysters and shrimps. Rev. Latinoam. Microbiol. 35, 276–272 [PubMed] [Google Scholar]
- Vasconcelos G. J., Stang W. J., Laidlaw R. H. (1975). Isolation of Vibrio parahaemolyticus and Vibrio alginolyticus from estuarine areas of Southeastern Alaska. Appl. Microbiol. 29, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Roman J., Leon-Sicairos N., Flores-Villasenor H., Villafana-Rauda S., Canizalez-Roman A. (2012). Association of pandemic Vibrio parahaemolyticus O3:K6 present in the coastal environment of Northwest Mexico with cases of recurrent diarrhea between 2004 and 2010. Appl. Environ. Microbiol. 78, 1794–1803 10.1128/AEM.06953-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuddhakul V., Chowdhury A., Laohaprertthisan V., Pungrasamee P., Patararungrong N., Thianmontri P. (2000). Isolation of a pandemic O3:K6 clone of a Vibrio parahaemolyticus strain from environmental and clinical sources in Thailand. Appl. Environ. Microbiol. 66, 2685–2689 10.1128/AEM.66.6.2685-2689.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. A., Salyers A. A. (2003). Is the evolution of bacterial pathogens an out-of-body experience? Trends Microbiol. 11, 347–350 10.1016/S0966-842X(03)00179-3 [DOI] [PubMed] [Google Scholar]
- Wong H. C., Chen M. C., Liu S. H., Liu D. P. (1999). Incidence of highly genetically diversified Vibrio parahaemolyticus in seafood imported from Asian countries. Int. J. Food Microbiol. 52, 181–188 10.1016/S0168-1605(99)00143-9 [DOI] [PubMed] [Google Scholar]
- Wong H. C., Liu S. H., Wang T. K., Lee C. L., Chiou C. S., Liu D. P., et al. (2000). Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Appl. Environ. Microbiol. 66, 3981–3986 10.1128/AEM.66.9.3981-3986.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Iida T., Yamamoto K., Takarada Y., Miwatani T., Honda T. (1994). Demonstration and characterization of simultaneous production of a thermostable direct hemolysin (TDH/I) and a TDH-related hemolysin (TRHx) by a clinically isolated Vibrio parahaemolyticus strain, TH3766. Infect. Immun. 62, 166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung P. S., Boor K. J. (2004). Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog. Dis. 1, 74–88 10.1089/153531404323143594 [DOI] [PubMed] [Google Scholar]
- Yoh M., Kawakami N., Funakoshi Y., Okada K., Honda T. (1995). Evaluation of two assay kits for thermostable direct hemolysin (TDH) as an indicator of TDH-related hemolysin (TRH) produced by Vibrio parahaemolyticus. Microbiol. Immunol. 39, 157–159 10.1111/j.1348-0421.1995.tb02183.x [DOI] [PubMed] [Google Scholar]
- Yu Y., Yang H., Li J., Zhang P., Wu B., Zhu B., et al. (2012). Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch. Microbiol. 194, 827–835 10.1007/s00203-012-0816-z [DOI] [PubMed] [Google Scholar]
- Zamudio M. L., Meza A., Bailon H., Martinez-Urtaza J., Campos J. (2011). [Experiences in the epidemiological surveillance of foodborne pathogens by pulsed field gel electrophoresis (PFGE) in Peru]. Rev. Peru. Med. Exp. Salud Publica 28, 128–135 10.1590/S1726-46342011000100020 [DOI] [PubMed] [Google Scholar]
- Zhang X. H., Austin B. (2005). Haemolysins in Vibrio species. J. Appl. Microbiol. 98, 1011–1019 10.1111/j.1365-2672.2005.02583.x [DOI] [PubMed] [Google Scholar]