SUMMARY
Increased activation of the serine-glycine biosynthetic pathway is an integral part of cancer metabolism that drives macromolecule synthesis needed for cell proliferation. Whether this pathway is under epigenetic control is unknown. Here we show that the histone H3 lysine 9 (H3K9) methyltransferase G9A is required for maintaining the pathway enzyme genes in an active state marked by H3K9 monomethylation and for the transcriptional activation of this pathway in response to serine deprivation. G9A inactivation depletes serine and its downstream metabolites, triggering cell death with autophagy in cancer cell lines of different tissue origins. Higher G9A expression, which is observed in various cancers and is associated with greater mortality in cancer patients, increases serine production and enhances the proliferation and tumorigenicity of cancer cells. These findings identify a G9A-dependent epigenetic program in the control of cancer metabolism, providing a rationale for G9A inhibition as a therapeutic strategy for cancer.
INTRODUCTION
Histone lysine methylation has a central role in the control of gene transcription. The histone lysine methylation state is controlled by histone lysine methyltransferases (KMTs) and demethylases (KDMs). The enzymatic reactions catalyzed by KMTs and KDMs depend on metabolic coenzymes including S-adenosylmethionine (SAM), flavin adenine dinucleotide (FAD), and α-ketoglutarate (α-KG). KMTs catalyze lysine methylation using SAM as the methyl group donor, whereas LSD (KDM1A and KDM1B) and JmjC domain-containing KDMs (KDM2-KDM8) require FAD and α-KG for demethylation, respectively (Black et al., 2012; Mosammaparast and Shi, 2010). Their dependence on metabolic coenzymes suggests that KMTs and KDMs could reprogram gene expression in response to changes in cellular metabolism. This notion has also led to the provocative hypothesis that KMTs and KDMs may contribute to metabolic control through transcriptional regulation (Teperino et al., 2010).
G9A, also known as EHMT2, is a H3K9 methyltransferase that has a primary role in catalyzing monomethylation and dimethylation of H3K9 (H3K9me1 and H3K9me2) in euchromatin (Peters et al., 2003; Rice et al., 2003; Shinkai and Tachibana, 2011; Tachibana et al., 2002), with H3K9me1 being associated with transcriptional activation and H3K9me2 with transcriptional repression (Black et al., 2012; Mosammaparast and Shi, 2010). Elevated levels of G9A expression have been observed in many types of human cancers, and G9A knockdown has been shown to inhibit the proliferation of cancer cell lines (Chen et al., 2010; Cho et al., 2011; Huang et al., 2010; Kondo et al., 2008). The molecular basis of G9A action in the control of cancer cell proliferation is not well understood. In this study, we identify an essential role of G9A in sustaining cancer cell survival and proliferation by transcriptional activation of the serine-glycine biosynthetic pathway. Our findings provide direct evidence for a G9A-dependent epigenetic program in the control of amino acid production and cancer metabolism.
RESULTS
G9A Is Essential for Sustaining Cancer Cell Proliferation and Survival
We examined the role of G9A in cell survival and proliferation in human cancer cell lines of different tissue origins, including the bladder (J82), bone (U2OS), brain (U251), breast (MCF10A and MCF7), cervix (HeLa), colon (HCT116 and RKO), liver (Hep2G), lung (H1299), and sympathetic nervous system (BE(2)-C, SMS-KCNR, and SHEP1). We treated these cell lines with BIX01294 (BIX), a small molecule inhibitor of G9A (IC50 = 1.7 μM) (Kubicek et al., 2007). BIX at 2–5 μM significantly reduced the global levels of H3K9me1 and H3K9me2 (Figure S1A available online) and completely inhibited the proliferation of all the cancer cell lines examined (Figure S1B for representative cell lines). In addition, we observed a significant decrease in cell survival following BIX treatment (Figure S1C, BIX-5 μM_5d). To confirm that BIX targets G9A to inhibit cell proliferation and survival, we examined the effect of G9A silencing by small hairpin RNA (shRNA). G9A knockdown exerted a pronounced inhibitory effect on cell proliferation and survival (Figures S1D–S1G). Together, these findings indicate an essential role of G9A in sustaining cell proliferation and survival in a wide range of cancer cell lines.
G9A Inhibition or Silencing Induces Autophagy
An early and prominent morphological feature of the cells with G9A inhibition or silencing was the appearance of numerous cytoplasmic vesicles and vacuoles (Figures S1C and S1G) that morphologically resemble autophagosomes, a double-membraned structure that sequesters cellular organelles, proteins, and/or lipids during autophagy. Thus, we examined the possibility that G9A inhibition or silencing might induce autophagy by electron microscopy for ultrastructural morphology, by immunoblotting for detecting the lipidation of LC3 (microtubule-associated protein light chain 3), and by immunofluorescence for monitoring the formation of LC3-positive puncta. LC3 is the mammalian homolog of the yeast autophagy-related protein Atg8 and is proteolytically processed to LC3-I by the Atg4 protease following translation. Upon autophagy induction, LC3-I is converted to the lipidated LC3-II form, which is then incorporated into the autophagosomal membrane, resulting in the redistribution of LC3 from a diffuse pattern to a punctate pattern. Mammalian cells express three LC3 isoforms (LC3A, LC3B, and LC3C), with LC3B-II levels correlating with the steady-state levels of autophagosomes (Klionsky et al., 2008; Mizushima et al., 2010).
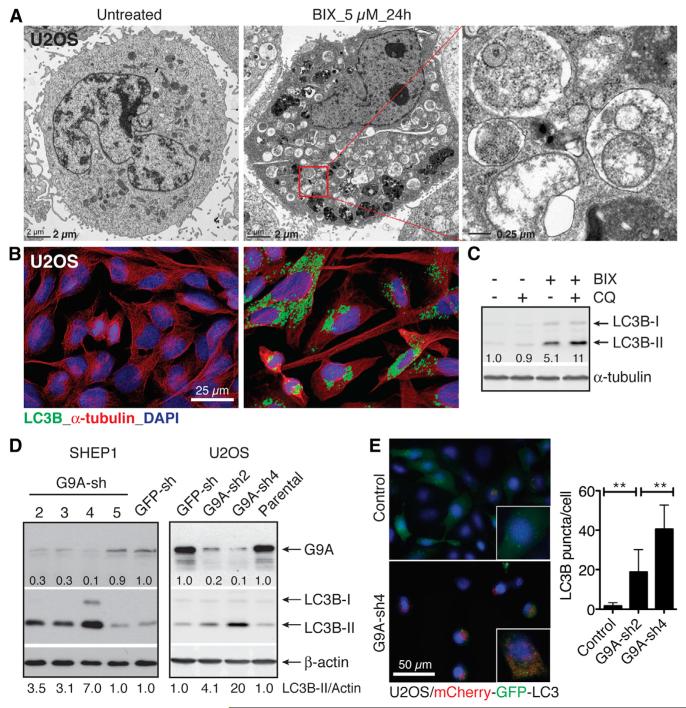
Electron microscopy revealed numerous double-membraned vacuoles in BIX-treated cells that contained fragments of the endoplasmic reticulum and other cytoplasmic components (Figure 1A). Immunofluorescence confirmed that these vesicles and vacuoles were LC3B positive (Figure 1B). To determine whether the accumulation of autophagosomes resulted from an increase in autophagosome formation or from a block in autophagosome turnover, we examined BIX-induced LC3B-II production in the presence or absence of chloroquine (CQ), which blocks lysosomal acidification and thus degradation of autophagosomal components. The presence of CQ further increased LC3B-II levels in BIX-treated cells (Figure 1C), indicating that BIX increased LC3B-II production rather than blocking its degradation.
Figure 1. G9A Inhibition or Silencing Induces Autophagy.
(A) Electron micrographs of U2OS cells with or without 5 μM BIX for 24 hr.
(B) Immunofluorescence of LC3B-positive autophagosomes in U2OS cells with or without 5 μM BIX for 24 hr.
(C) Immunoblotting of LC3B in U2OS cells with or without 5 μM BIX or 10 μM chloroquine (CQ) for 24 hr. LC3B-II levels were quantified against α-tubulin.
(D) Immunoblotting of G9A and LC3B in U2OS and SHEP1 cells expressing GFP shRNA (GFP-sh) or individual G9A-sh sequences. G9A and LC3B-II levels were quantified against β-actin.
(E) Fluorescence and quantification of LC3B-positive autophagosomes in U2OS/mCherry-EGFP-LC3B cells with or without G9A knockdown. Error bars represent SD (n = 100 cells). **p < 0.001. See also Figures S1 and S2.
Dose-titration and time course studies revealed that different tumor cell lines varied in their sensitivity to BIX. For the most sensitive SHEP1 and U2OS cells, BIX as low as 1 μM triggered significant levels of autophagy, as determined by LC3B-II production (Figure S2A). The maximal level of autophagy occurred at 6 hr (SHEP1 and U2OS) or 24 hr (HeLa) following 5 μM BIX treatment (Figure S2B). We further confirmed the ability of BIX to induce autophagy with immortalized Atg5+/+ and Atg5−/− mouse embryonic fibroblasts (MEFs). Atg5 is required for autophagosome formation (Kuma et al., 2004; Mizushima et al., 2001). BIX at the concentrations of 2–5 μM was able to induce LC3B-positive puncta in Atg5+/+, but not in Atg5−/−, MEFs (Figure S2C).
To verify that BIX targets G9A to induce autophagy, we examined the effect of BIX on cells with G9A overexpression. Both SHEP1 and U2OS cells overexpressing G9A were highly resistant to BIX-induced autophagy, as evidenced by a significant reduction in LC3B-II levels following BIX treatment compared to control cells (Figure S2D). Additionally, cells with G9A silencing fully recapitulated the autophagy phenotype induced by BIX, showing a significant increase in the production of LC3B-II (Figure 1D) and LC3B-positive puncta (Figure 1E). Collectively, these results demonstrate that G9A inhibition or silencing induces cell death with autophagy.
G9A Inhibition or Silencing Transcriptionally Represses Serine-Glycine Biosynthesis
To investigate the molecular basis of autophagy induction by G9A inhibition, we examined the kinase activity of the mammalian target of rapamycin complex 1 (mTORC1), which has a major role in the control of autophagy by integrating a wide range of signals including concentrations of nutrients and growth factors (He and Klionsky, 2009; Kim and Guan, 2011; Rabinowitz and White, 2010). mTORC1 inhibits autophagy by suppressing the autophagy-activating kinase ULK1. Nutrient or growth factor deprivation inhibits mTORC1 activity, thereby activating ULK1 and inducing autophagy. Immunoblotting revealed that BIX significantly inhibited mTORC1 kinase activity as demonstrated by a marked decrease in the phosphorylation of the ribosome protein S6 kinase (S6K) (Figure S3A), a key mTORC1 substrate. This finding suggests that G9A inhibition triggers autophagy by interfering with cell growth or survival signals upstream of mTORC1.
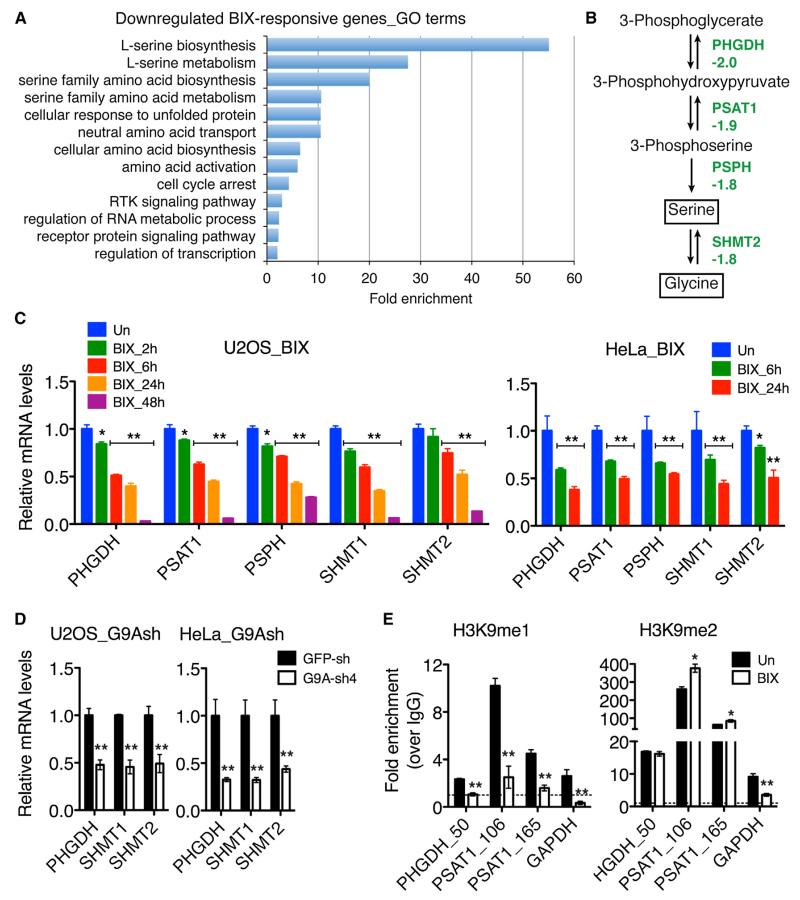
Given the role of G9A in epigenetic control of transcription, we performed microarray profiling to identify potential G9A targets involved in autophagy induction. A total of 615 BIX-responsive genes (≥±1.50-fold, p < 0.01) were identified, with 302 genes being upregulated and 313 downregulated (Table S1). Gene Ontology (GO) analysis revealed that among the genes downregulated by BIX, those within the serine-glycine biosynthetic pathway were significantly enriched (Figure 2A), including phosphoglycerate dehydrogenase (PHGDH), phosphoserine aminotransferase 1 (PSAT1), phosphoserine phosphatase (PSPH), and serine hydroxymethyltransferase 2 (SHMT2) (Figure 2B). We confirmed the ability of BIX to downregulate mRNA expression of these genes by quantitative reverse-transcription PCR (qRT-PCR) in four different cancer cell lines (Figures 2C and S3B). Time course studies revealed significant downregulation of these genes within 2 and 6 hr of BIX treatment in U2OS and HeLa cells, respectively (Figure 2C), which occurred at least several hours before most of the cells underwent autophagy (Figure S2B). We noticed that SHMT1, though downregulated in HeLa, SHEP1, and U2OS cells, was activated in BE(2)-C cells following BIX treatment (Figure S3B). The significance of this apparently cell-type-dependent regulation of SHMT1 expression is currently under investigation. In agreement with the microarray and qRT-PCR data, we observed a significant decrease in PHGDH protein levels in BIX-treated cells (Figure S3C). Similarly, G9A silencing resulted in marked downregulation of the same group of genes at both mRNA and protein levels (Figures 2D, S3D, and S3E).
Figure 2. G9A Inhibition or Silencing Represses Serine-Glycine Biosynthesis.
(A) GO analysis of downregulated BIX-responsive genes (enrichment fold >2.0, p < 0.01).
(B) Serine-glycine biosynthetic pathway with the indicated fold changes in mRNA expression determined by microarray.
(C and D) qRT-PCR analysis of mRNA expression of the pathway enzyme genes in U2OS and HeLa cells with or without 5 μM BIX (C) or G9A silencing (D). Error bars represent SD (n = 3).
(E) ChIP-qPCR analysis of H3K9me1 and H3K9me2 levels at the promoters of PHGDH and PSAT1 in U2OS cells with or without 5 μM BIX for 4 hr. Data on the GAPDH coding region are shown as positive control. Dashed line indicates IgG control. Error bars represent SD of triplicate and are representatives of two independent experiments.
*p < 0.01, **p < 0.001. See also Figure S3, Table S1, and Table S3.
Chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR) assay revealed that BIX treatment significantly reduced the H3K9me1 levels around the transcriptional start sites (TSSs) of PHGDH and PSAT1 (Figure 2E, H3K9me1). By contrast, H3K9me2 levels in the same regions were either unchanged or significantly increased following BIX treatment (Figure 2E, H3K9me2). Together, these results indicate that the serine-glycine biosynthetic pathway is under the direct transcriptional control of G9A, primarily through the regulation of H3K9me1 levels associated with the pathway enzyme genes.
Supplemental Serine Rescues the Cell Death Phenotype of G9A Inhibition
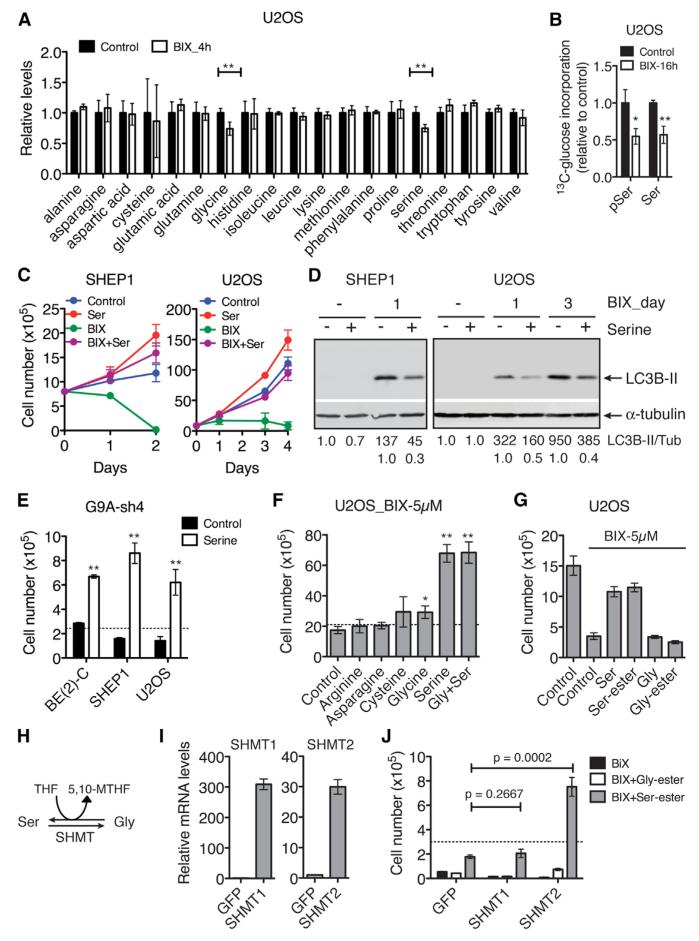
The serine-glycine biosynthetic pathway is an important source of precursors and coenzymes for the biosynthesis of amino acids, nucleotides, and lipids (de Koning et al., 2003; Kalhan and Hanson, 2012) and is crucial for cancer cell survival and proliferation (Locasale, 2013; Locasale et al., 2011; Pollari et al., 2011; Possemato et al., 2011). In addition, amino acid deprivation is a major trigger of autophagy (Kim and Guan, 2011). Thus, we investigated the possibility that suppression of this biosynthetic pathway might be a cause of the cell death phenotype induced by G9A inhibition or silencing. Gas chromatography-mass spectrometry (GC-MS) analysis revealed that only serine and glycine levels were significantly reduced in U2OS cells within 4 hr of BIX treatment (Figure 3A), demonstrating that G9A activity is essential for maintaining the intracellular steady-state levels of serine and glycine. We further assessed the activity of this biosynthetic pathway by [U-13C] glucose flux analysis using liquid chromatography-tandem mass spectrometry (LC-MS/MS). BIX treatment significantly decreased the incorporation of [U-13C] glucose into 3-phosphoserine and serine (Figure 3B). Importantly, addition of serine to the culture medium significantly diminished the effect of BIX (Figures 3C and 3D) or G9A silencing (Figure 3E) on cell proliferation and autophagy in all the cancer cell lines examined. By contrast, supplemental glycine had only a small protective effect on BIX-treated cells, and addition of both serine and glycine was no more effective than adding serine alone (Figures 3F, S4A, and S4B). We confirmed these findings with cell-permeable methyl-serine-ester and methyl-glycine-ester (Figure 3G). Other individual amino acid supplements all failed to prevent cell death with autophagy induced by BIX (Figures 3F, S4A, and S4C), providing further evidence for the specificity of serine action.
Figure 3. Supplemental Serine Rescues the Phenotype of G9A Inhibition or Silencing.
(A) GC-MS analysis of amino acid levels in U2OS cells with or without 5 μM BIX for 4 hr. Error bars represent SD (n = 6).
(B) LC-MS/MS analysis of [U-13C] glucose flux into 3-phosphoserine (pSer) and serine (Ser) biosynthesis in U2OS cells with or without 5 μM BIX for 16 hr. The fraction of labeled to total pSer and Ser, 13C/(13C + 12C), was calculated and normalized to control. Error bars represent SD (n = 3).
(C) Growth assay of SHEP1 and U2OS cells with or without 5 μM BIX or supplemental serine (Ser). Error bars represent SD (n = 4).
(D) Immunoblotting of LC3B in the cell samples from (C). LC3B-II levels were quantified against α-tubulin.
(E) Growth assay of the indicated cell lines with G9A silencing in the presence or absence of supplemental serine for 4 days. Dashed line indicates the number of cells plated at time zero. Error bars represent SD (n = 4).
(F and G) Growth assay of U2OS cells in the presence of 5 μM BIX with or without the indicated supplemental amino acids for 2 days. Gly, glycine; Gly-ester, methyl-glycine-ester; Ser, serine; Ser-ester, methyl-serine-ester. Dashed line (F) indicates the number of cells plated at time zero. Error bars represent SD (n = 4).
(H) Schematic of the interconversion between serine and glycine. THF, tetrahydrofolate; 5,10-MTHF, 5,10-methylenetetrahydrofolate.
(I) qRT-PCR analysis of SHMT mRNA expression in SHEP1 cells overexpressing SHMT1 or SHMT2. Error bars represent SD (n = 3).
(J) Growth assay of SHEP1 cells expressing GFP, SHMT1, or SHMT2 in the presence of 5 μM BIX with or without supplemental Gly-ester or Serester. Dashed line indicates the number of cells plated at time zero. Error bars represent SD (n = 3).
*p < 0.01, **p < 0.001. See also Figure S4.
The observation that serine, but not glycine, was able to rescue the cell death phenotype suggests that the production of 5,10-methylenetetrahydrofolate (5,10-MTHF) might be crucial for cell survival and proliferation. The interconversion between serine and glycine is catalyzed by SHMT: serine conversion to glycine generates 5,10-MTHF, whereas glycine conversion to serine consumes 5,10-MTHF (Figure 3H). To test this idea, we generated SHEP1 cells overexpressing SHMT1, a cytoplasmic enzyme, or SHMT2, a predominantly mitochondrial enzyme (Figure 3I). Their overexpression alone had no significant effect on BIX-induced cell death (Figure 3J). However, overexpression of SHMT2, but not SHMT1, synergized with supplemental serine to enhance cell survival and proliferation in the presence of BIX (Figure 3J). This observation is consistent with our model, since previous studies have suggested that SHMT2 has a major role in the conversion of serine to glycine and 5,10-MTHF, whereas SHMT1 primarily catalyzes the conversion of glycine to serine (Herbig et al., 2002; Narkewicz et al., 1996; Pfendner and Pizer, 1980; Stover et al., 1997; Tibbetts and Appling, 2010; Yoshida and Kikuchi, 1970). Collectively, these results indicate that maintaining the production of serine and its downstream metabolites, including 5,10-MTHF, is a major mechanism by which G9A sustains cancer cell survival and proliferation.
G9A Is a Key Component of the Serine Deprivation Response, Linking Serine Production, Ribosome Biogenesis, and Cell-Cycle Progression
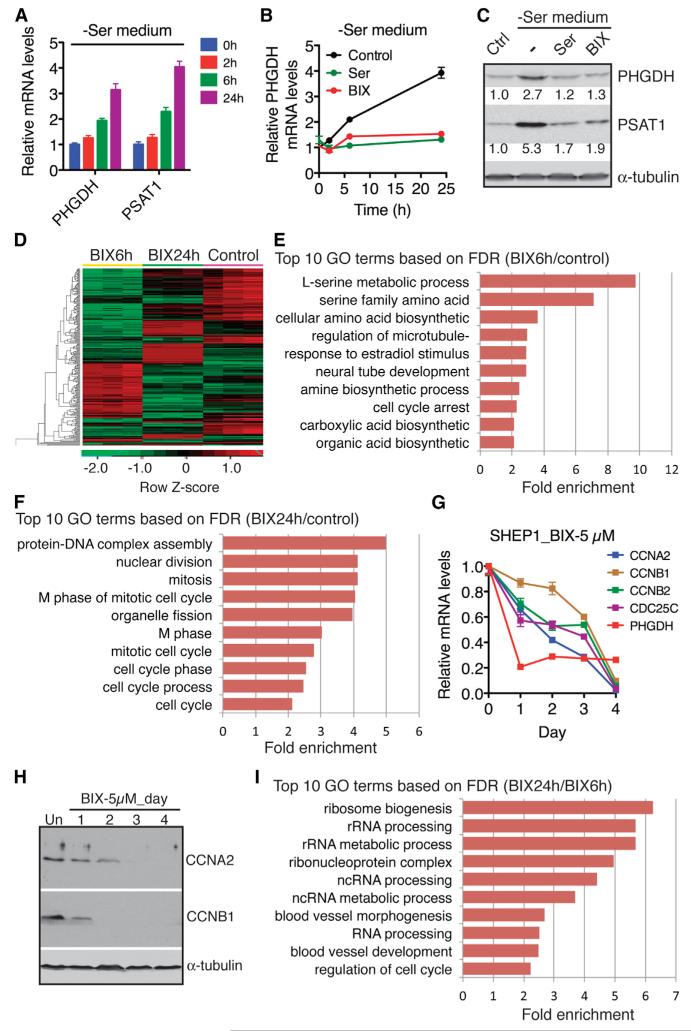
It has been shown recently that serine deprivation leads to transcriptional activation of the serine biosynthetic pathway (Ye et al., 2012; Figure 4A). In light of our findings above, we asked whether G9A has a role in activation of this pathway in response to serine deprivation. Addition of BIX completely abrogated the induction of PHGDH and PSAT1 by serine deprivation (Figures 4B and 4C), supporting a physiological function of G9A in mediating the serine deprivation response.
Figure 4. G9A Links Serine Production, Ribosome Biogenesis, and Cell Proliferation.
(A) qRT-PCR analysis of PHGDH and PSAT1 mRNA levels in SHEP1 cells cultured in serine-deficient (−Ser) medium for 0, 2, 6, and 24 hr. Error bars represent SD (n = 3).
(B) qRT-PCR analysis of PHGDH mRNA levels in SHEP1 cells cultured in −Ser medium with or without 5 μM BIX or supplemental serine for 0, 2, 6, and 24 hr. Error bars represent SD (n = 3).
(C) Immunoblotting of PHGDH and PSAT1 in SHEP1 cells cultured in control (with serine) or −Ser medium with or without 5 μM BIX or supplemental serine for 24 hr. PHGDH and PSAT1 levels were quantified against α-tubulin.
(D) Heatmap showing differential gene expression in U2OS cells without (control) or with 5 μM BIX for 6 (BIX6h) or 24 hr (BIX24h).
(E and F) GO analysis of downregulated BIX-responsive genes at the 6 hr (E) or 24 hr (F) time point. Shown are top ten GO biological process terms.
(G and H) qRT-PCR (G) and immunoblot (H) analyses of cell-cycle genes in SHEP1 cells with or without 5 μM BIX for the indicated time. Error bars in (G) represent SD (n = 3).
(I) GO analysis of the genes downregulated by BIX at 24 hr versus 6 hr. Shown are top ten GO biological process terms.
Amino acid availability controls the rates of protein synthesis and cell growth (Kilberg et al., 2005), and it has been reported recently that G9A is required for activation of rRNA transcription (Yuan et al., 2007), a rate-limiting step in ribosome biogenesis. We therefore asked whether G9A has a role in linking serine metabolism to ribosome biogenesis and cell growth. To test this idea, we performed temporal transcriptome profiling of BIX-responsive genes by RNA sequencing (RNA-seq), which revealed distinctive patterns of gene expression in cells that were treated with BIX for 6 or 24 hr (Figure 4D). In agreement with the microarray data shown above (Figure 2A; Table S1), GO analysis of the genes downregulated by BIX at 6 hr revealed significant enrichment of genes within the serine biosynthetic pathway (Figure 4E; Table S2). However, cells treated with BIX for 24 hr displayed a gene expression pattern characterized by significant downregulation of genes that control cell-cycle progression (Figure 4F), including cyclins A2 and B2, and CDC25C (Table S2). We confirmed the RNA-seq result by qRT-PCR, which showed sequential downregulation of serine synthesis and cell-cycle genes following BIX treatment (Figure 4G). We also confirmed the time-dependent downregulation of cyclins A2 and B1 by immunoblotting (Figure 4H). Importantly, we found marked downregulation of genes involved in ribosome biogenesis in cells treated with BIX for 24 hr compared to those treated for 6 hr (Figure 4I; Table S2), a finding consistent with the reported role of G9A in activation of rRNA transcription (Yuan et al., 2007). Collectively, these results suggest a key role of G9A in coupling serine sensing to the transcription of genes that control protein synthesis and cell proliferation.
G9A Promotes Cell Proliferation and Tumorigenicity
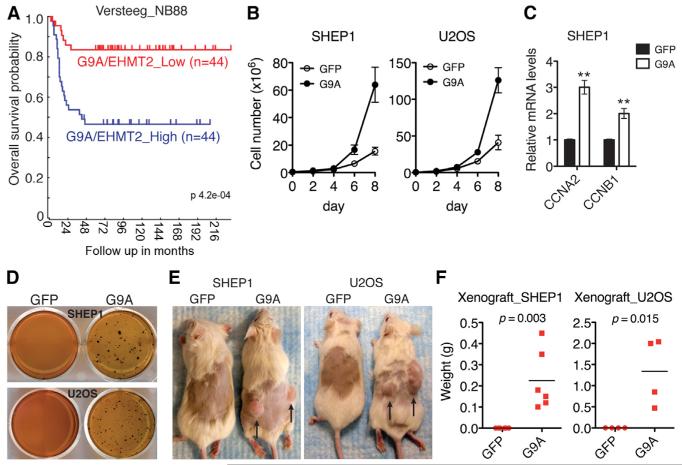
G9A is overexpressed in various human cancers (Chen et al., 2010; Cho et al., 2011; Huang et al., 2010; Kondo et al., 2008), and it has been reported recently that elevated levels of G9A expression positively correlate with disease progression and poor prognosis in lung cancer (Chen et al., 2010). In line with these reports, we found that higher G9A expression was significantly associated with reduced overall survival in neuroblastoma patients (Figure 5A). These observations prompted us to investigate the functional consequence of high G9A expression. Ectopic expression of G9A in SHEP1 and U2OS cells significantly increased cell proliferation (Figure 5B) and the expression of genes that promote cell-cycle progression (Figure 5C), indicating that high G9A expression alone is sufficient to confer a growth advantage to cancer cells. Moreover, G9A overexpression markedly enhanced the anchorage-independent growth (Figure 5D) and tumorigenicity (Figures 5E and 5F) of SHEP1 and U2OS cells, demonstrating that G9A has transforming potential. Together, our results from both human primary tumors and cell lines suggest an oncogenic function of G9A in cancer development.
Figure 5. G9A Promotes Cell Proliferation and Tumorigenicity.
(A) Kaplan-Meier overall survival for G9A/EHMT2 expression in primary neuroblastoma tumors (Versteeg NB88 data set).
(B) Growth assay of SHEP1 and U2OS cells with or without G9A overexpression. Error bars represent SD (n = 4).
(C) qRT-PCR analysis of cyclin A2 and B1 mRNA levels in SHEP1 cells with or without G9A overexpression. Error bars represent SD (n = 3). **p < 0.001.
(D) Soft agar assay of SHEP1 and U2OS cells with or without G9A overexpression. Shown are representatives of three independent experiments.
(E and F) Xenograft assay of SHEP1 and U2OS cells with or without G9A overexpression. Images (E) were taken on day 42 after inoculation. Tumor weight (F) was analyzed by scatterplot with horizontal lines indicating the mean.
Activation of the Serine-Glycine Biosynthetic Pathway Is Essential for the Oncogenic Activity of G9A
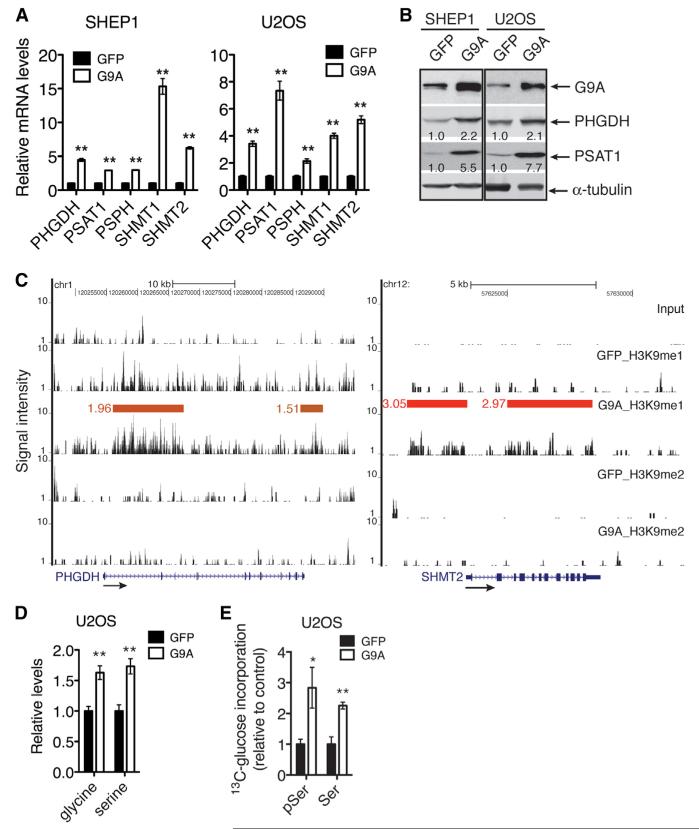
Given the critical role of the serine-glycine biosynthetic pathway in sustaining cancer cell survival and proliferation (Locasale et al., 2011; Pollari et al., 2011; Possemato et al., 2011), we asked whether elevated levels of G9A lead to increased activation of this pathway and whether this is a major mechanism for the oncogenic activity of G9A. Ectopic expression of G9A in SHEP1 and U2OS cells markedly increased the mRNA (Figure 6A) and protein (Figure 6B) levels of the pathway enzyme genes. Moreover, ChIP in combination with DNA sequencing (ChIP-seq) revealed a significant increase in H3K9me1 levels at these gene loci (Figure 6C for PHGDH and SHMT2). We observed no significant change in H3K9me2 levels in the same regions (Figure 6C). These results suggest that G9A activates the expression of the pathway enzyme genes by increasing the H3K9me1 levels associated with their loci. As expected, G9A overexpression significantly increased the intracellular levels of serine and glycine (Figure 6D), and the flux of glucose into the serine biosynthetic pathway (Figure 6E).
Figure 6. G9A Transcriptionally Activates Serine-Glycine Biosynthesis.
(A) qRT-PCR analysis of mRNA levels of serine-glycine biosynthetic enzyme genes in SHEP1 and U2OS cells with or without G9A overexpression. Error bars represent SD (n = 3).
(B) Immunoblotting of PHGDH and PSAT1 in SHEP1 and U2OS cells with or without G9A overexpression. PHGDH and PSAT1 levels were quantified against α-tubulin.
(C) ChIP-seq tag profiles for H3K9me1 and H3K9me2 levels in the PHGDH and SHMT2 loci in U2OS cells with or without G9A overexpression. Horizontal bars represent chromatin regions in which H3K9 methylation levels are significantly changed (G9A/GFP), and the numbers indicate fold enrichment.
(D) GC-MS analysis of serine and glycine levels in U2OS cells with or without G9A overexpression. Error bars represent SD (n = 6).
(E) LC-MS/MS analysis of [U-13C] glucose flux into 3-phosphoserine (pSer) and serine (Ser) biosynthesis in U2OS cells with or without G9A overexpression. The fraction of labeled to total pSer and Ser, 13C/(13C + 12C), was calculated and then normalized to GFP control. Error bars represent SD (n = 3).
*p < 0.05, **p < 0.001. See also Table S3.
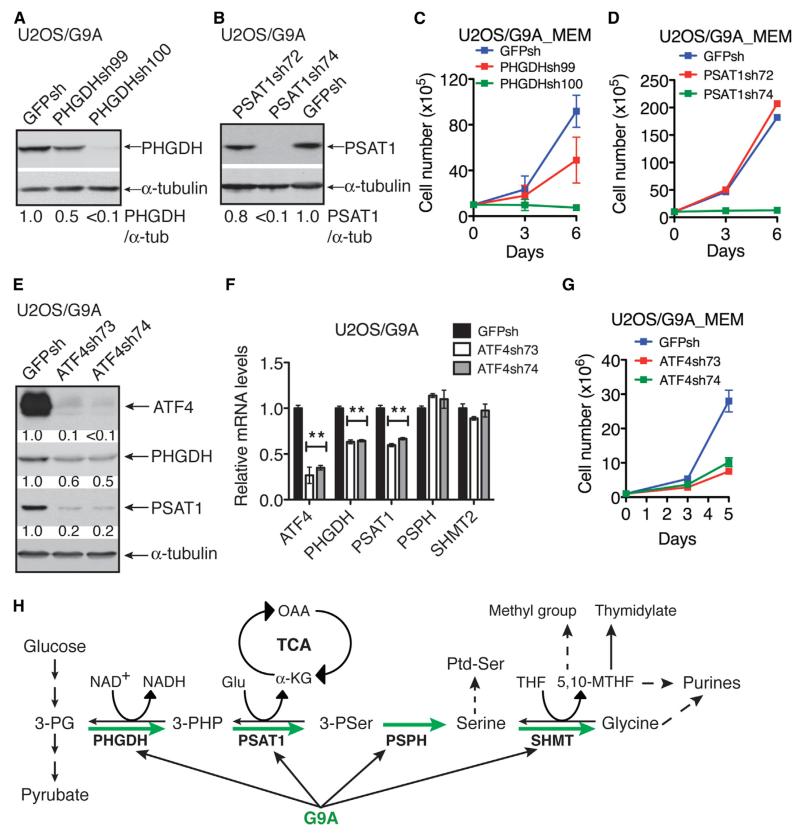
To determine whether the oncogenic function of G9A depends on its ability to activate the serine-glycine pathway, we silenced the expression of PHGDH or PSAT1 in G9A-overexpressing U2OS cells by shRNA (Figures 7A and 7B). Knockdown of either PHGDH or PSAT1 abolished the ability of G9A to promote cell proliferation in a manner dependent on levels of PHGDH or PSAT1 downregulation (Figures 7C and 7D). In fact, G9A-overexpressing cells with more than 90% of PHGDH or PSAT1 knockdown failed to proliferate in the absence of supplemental serine (Figures 7C and 7D). We obtained essentially the same results with G9A-overexpressing SHEP1 (Figures S5A–S5D). Collectively, these data demonstrate that G9A promotes cell proliferation by transcriptionally activating the serine-glycine biosynthetic pathway.
Figure 7. Activation of Serine-Glycine Biosynthesis Is Required for G9A to Promote Cell Proliferation.
(A and B) Immunoblotting of PHGDH and PSAT1 in G9A-overexpressing U2OS cells with PHGDH (A) or PSAT1 (B) knockdown. PHGDH and PSAT1 levels were quantified against α-tubulin.
(C and D) Growth assay of G9A-overexpressing U2OS cells with PHGDH (C) or PSAT1 (D) knockdown. Error bars represent SD (n = 3).
(E and F) Immunoblot (E) and qRT-PCR (F) analysis of the expression of serine-glycine biosynthetic enzyme genes in G9A-overexpressing U2OS cells with ATF4 knockdown. **p < 0.001.
(G) Growth assay of G9A-overexpressing U2OS cells with ATF4 knockdown. Error bars represent SD (n = 3).
(H) Model for G9A regulation of serine metabolism. 3-PG, 3-phosphoglycerate; 3-PHP, 3-phosphohydroxypyruvate; 3-PSer, 3-phosphoserine; OAA, oxaloacetate; Ptd-Ser, phosphatidylserine.
See also Figure S5 and Table S3.
The serine biosynthetic genes are direct transcriptional targets of activating transcription factor 4 (ATF4) (Adams, 2007; Seo et al., 2009; Ye et al., 2012), which is activated in response to amino acid deprivation (Kilberg et al., 2009). It has been shown that ATF4 knockdown or deficiency led to lower expression of serine biosynthetic enzymes (Ye et al., 2012), suggesting an essential role of ATF4 in maintaining transcriptional activation of this pathway. To examine the role of ATF4 in G9A action, we silenced ATF4 expression in G9A-overexpressing U2OS (Figure 7E) or SHEP1 cells (Figures S5E), which resulted in marked downregulation of PHGDH and PSAT1 at both mRNA and protein levels (Figures 7E and 7F and S5E and S5F). ATF4 silencing also significantly reduced the proliferation of G9A-overexpressing U2OS (Figure 7G) or SHEP1 cells (Figure S5G). Together, these results indicate that ATF4 is required for G9A to transcriptionally activate serine biosynthesis and to stimulate cell proliferation.
DISCUSSION
In this study, we show that the histone H3K9 methyltransferase G9A is essential for transcriptional activation of the serineglycine biosynthetic pathway by specifically marking the pathway enzyme genes with H3K9me1, an epigenetic marker associated with active chromatin (Black et al., 2012; Mosammaparast and Shi, 2010). We further present evidence that G9A is a component of the molecular pathway that couples serine sensing to the transcriptional control of serine production, ribosome biogenesis, and cell proliferation. Finally, we provide evidence that G9A has an oncogenic function in cancer development by conferring survival and growth advantages to tumors through increasing the production of serine and its downstream metabolites. Our study uncovers an epigenetic mechanism for the control of amino acid metabolism and provides a molecular explanation for the functional significance of G9A overexpression observed in various human cancers (Chen et al., 2010; Cho et al., 2011; Huang et al., 2010; Kondo et al., 2008).
Cell growth and proliferation depend on protein synthesis, which must in turn match the cellular nutritional status including amino acid availability (Grummt and Ladurner, 2008; Kilberg et al., 2005). The mechanistic connection between these cellular processes remains to be clarified. Our study suggests an epigenetic mechanism for the control of ribosome biogenesis and cell proliferation in response to serine abundance. We show that G9A is essential for the serine deprivation response that leads to transcriptional activation of genes for serine biosynthesis. Moreover, we show that G9A activity or expression levels affect serine production and cell proliferation. Specifically, G9A inhibition initiated a transcriptional program that results in sequential repression of genes required for serine production, ribosome biogenesis, and cell-cycle progression, leading to lower levels of serine biosynthesis and cell growth arrest. Conversely, G9A overexpression activated genes for serine biosynthesis and cell-cycle progression, leading to higher levels of serine production and cell proliferation. These results are of particular interest in the context of recent work showing that G9A is required for activation of rRNA transcription (Yuan et al., 2007). Thus, we suggest that G9A-mediated H3K9 methylation serves as a key link between amino acid sensing and transcriptional control of ribosome biogenesis and cell proliferation.
Also importantly, we show that G9A requires ATF4 for transcriptional activation of the serine biosynthetic pathway and for stimulation of cell proliferation. ATF4 has a key role in the cellular response to amino acid limitation, which leads to increased ATF4 expression primarily at the translational level. ATF4 in turn transcriptionally activates a large number of genes including those for amino acid synthesis and transport (Harding et al., 2003; Kilberg et al., 2009; Ye et al., 2012). Although the mechanistic detail remains to be defined, the observed functional connection between G9A and ATF4 provides further evidence for G9A as a component of the signaling pathway that responds to amino acid abundance.
It is well documented that cancer cells alter cellular metabolism to meet the biosynthetic challenge of growth and proliferation (DeBerardinis et al., 2008). Cancer metabolism is characterized by aerobic glycolysis with a high rate of glucose consumption and lactate production (Warburg, 1956). It is now increasingly recognized that a major function of aerobic glycolysis is to divert glycolic intermediates for the biosynthesis of macromolecules needed for cell growth and proliferation (Cairns et al., 2011; Vander Heiden et al., 2009). Accumulated evidence suggests that increased activation of the serine-glycine biosynthetic pathway is an important part of cancer metabolism (Kalhan and Hanson, 2012; Locasale, 2013). In addition to generating serine and glycine for the biosynthesis of proteins, purines (via one-carbon units), and lipids (via phosphatidylserine), this pathway produces equimolar amounts of reduced nicotinamide adenine dinucleotide (NADH), α-KG, and 5,10-MTHF (Figure 7H). These metabolites have critical roles in the control of cellular metabolism for cell proliferation and survival: NADH participates in ATP production and redox regulation (Corkey and Shirihai, 2012); α-KG supplies carbon to the tricarboxylic acid (TCA) cycle for the generation of many essential biosynthetic precursors (DeBerardinis et al., 2008); and 5,10-MTHF is a coenzyme for the only cellular pathway of de novo thymidylate biosynthesis catalyzed by thymidylate synthase and a major source of one-carbon units for purine synthesis and methyl group biogenesis, such as the generation of SAM, a coenzyme for histone and DNA methyltransferases (Kalhan and Hanson, 2012; Teperino et al., 2010; Tibbetts and Appling, 2010; Touroutoglou and Pazdur, 1996).
Importantly, we present evidence suggesting that the generation of 5,10-MTHF is crucial for the function of the serine-glycine biosynthetic pathway in promoting cancer cell survival and proliferation. We show that supplemental serine, but not glycine, is able to rescue the cell death phenotype induced by G9A inhibition or silencing. In addition, we show that supplemental serine synergizes with SHMT2 overexpression to enhance not only cell survival but also cell proliferation under the condition of G9A inhibition. Since serine and glycine are interconvertible, the most obvious explanation for these observations is that the conversion of serine to glycine generates 5,10-MTHF, a metabolite essential for DNA replication and one-carbon metabolism. This model is also consistent with previous reports that the mitochondrial SHMT2 has a major role in the conversion of serine to glycine and 5,10-MTHF (Herbig et al., 2002; Narkewicz et al., 1996; Pfendner and Pizer, 1980; Stover et al., 1997; Tibbetts and Appling, 2010; Yoshida and Kikuchi, 1970). It should be pointed out that the mitochondrial enzyme glycine cleavage system catabolizes glycine into carbon dioxide, ammonia, and 5,10-MTHF (Tibbetts and Appling, 2010). Therefore, supplemental glycine could be a source of 5,10-MTHF when G9A is inhibited. However, G9A inhibition markedly reduces the steady-state levels of both serine and glycine. The demand for serine production from supplemental glycine would decrease the 5,10-MTHF pool. Thus, under the condition of G9A inhibition, 5,10-MTHF production from the cleavage of supplemental glycine might not be sufficient to sustain DNA synthesis and one-carbon metabolism.
Recent studies have identified a genetic mechanism for activating the serine-glycine biosynthetic pathway in breast cancer and melanoma through amplification of PHGDH (Locasale et al., 2011; Possemato et al., 2011). Our findings, coupled to the observation that G9A is overexpressed in various cancers (Chen et al., 2010; Cho et al., 2011; Huang et al., 2010; Kondo et al., 2008), suggest an epigenetic mechanism for activating this biosynthetic pathway in cancer cells. Thus, by increasing the amount of glycolic carbon diverted into macromolecule biosynthesis, G9A contributes to the generation of a metabolic microenvironment favorable for cancer cell survival and proliferation.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Cells were cultured in DMEM (HyClone SH30022, Thermo Scientific) or MEM (GIBCO 61100-061, Invitrogen) supplemented with 10% dialyzed FBS (GIBCO 26400-036). BIX01294 (B9311), CQ (C6628), and amino acids were purchased from Sigma-Aldrich. Stock solutions of 10 μM were made in DMSO (BIX) or H2O (CQ). In all BIX experiments, DMSO was used as control (untreated). For rescue experiments, amino acids (100 μM stock in H2O) were added to MEM at the final concentration of 0.4 μM (same as in DMEM). For serine deprivation assay, cells were cultured in MEM supplemented with nonessential amino acids (MEM-NEAA) and then transferred to serine-deficient MEM-NEAA. Phase contrast images were captured using an Axio Observer microscope and AxioVision software (Carl Zeiss MicroImaging). Trypan blue exclusion assay was used to assess cell growth and survival.
Immunoblotting
Cells were suspended in SDS sample buffer and protein concentrations determined using a protein assay kit (Bio-Rad). Proteins (20 μg) were separated on SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and probed with primary antibodies detailed in Supplemental Experimental Procedures. Horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit IgG (Santa Cruz Biotechnology) were used as secondary antibodies. Proteins were visualized using a SuperSignal West Pico chemiluminescence kit (Pierce) and quantified with ImageJ (version 1.47d). Films were exposed for various times for protein quantification within linear range of detection. For visualization and quantification with the Odyssey system, goat anti-mouse IRDye 800 or 680 and anti-rabbit IRDye 800 or 680 (LI-COR Biosciences) were used as secondary antibodies.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and costained with rabbit anti-LC3B (3868, Cell Signaling, 1:200) and mouse anti-α-tubulin (B-5-1-2, Sigma-Aldrich, 1:2000). All secondary fluorescence antibodies (Alexa Fluor 488 and 594) were from Molecular Probes and used at 1:1,000 dilution. Nuclei were stained with DAPI (D3571, Invitrogen) or Hoechst 33342 (H3570, Invitrogen). Fluorescent images were captured with an Axio Observer microscope and AxioVision software. For quantification of LC3B-positive puncta per cell, approximately 100 cells were counted from a random selection of four to six 630× fields independently by two investigators.
Microarray
Total RNA was isolated using Trizol (Invitrogen) from three independent samples of BE(2)-C cells with or without 5 μM BIX for 24 hr. Affymetrix microarray was performed using the Human Gene 1.0 ST microarray chip. Data were normalized, significance determined by ANOVA, and fold change calculated with the Partek Genomics Suite. GO analysis was performed with DAVID (Huang et al., 2009) for all differentially expressed genes (≥±1.5-fold, p < 0.01).
RNA-Seq
Total RNA was isolated using Trizol from three independent samples of U2OS cells either untreated or treated with 5 μM BIX for 6 or 24 hr. RNA-seq libraries were generated with an Illumina TruSeq RNA sample preparation kit (RS-122-2001) and sequenced using Illumina high-seq 2000 with a read length of 50 bp with pair ends. RNA-seq reads were mapped to the human genome (hg19) using TopHat (Trapnell et al., 2009). Only those reads mapped to unique genomic locations and with <5% mismatches were analyzed further. We used Cufflinks to measure gene transcripts, and Cuffdiff to identify differentially expressed genes with a RefSeq GTF file downloaded from Illumina iGenomes (Roberts et al., 2011). Deferentially expressed genes were fed into DAVID for functional annotation. Bioconductor (Gentleman et al., 2004) packages were used to generate heatmap.
qRT-PCR
Total RNA was isolated from three independent samples using Trizol. Reverse transcription was performed using SuperScript II Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed using a RT2 SYBR green/Fluorescein PCR master mix (SABiosciences) on an iQ5 real-time PCR system (Bio-Rad) with primers against ATF4, CCNA2, CCNB1, CCNB2, CDC25C, PHGDH, PSAT1, PSPH, SHMT1, SHMT2, and B2M (Table S3). All primer pairs were verified by melting curve analysis following qRT-PCR, with each primer pair showing a single desired amplification peak. All samples were normalized to b2 microglobulin mRNA levels.
ChIP-qPCR and ChIP-Seq
ChIP was performed as described (Lee et al., 2006), using 2 × 107 parental, GFP-overexpressing, or G9A-overexpressing U2OS cells with or without 5 μM BIX for the indicated times. Crosslinked chromatin was sheared through sonication and immunoprecipitated using ChIP grade mouse anti-H3K9me1 (17–680), rabbit anti-H3K9me2 (17–648), or control mouse IgG or rabbit IgG (all from Millipore). For qPCR, two independent ChIP samples were analyzed, and each sample was assayed in triplicate using primers that cover the promoter regions of PHGDH and PSAT1, and the coding region of GAPDH (detailed in Supplemental Experimental Procedures). For ChIP-seq, libraries were generated from ChIP samples using an Illumina TruSeq ChIP Sample Prep kit (IP-202-1012) and sequenced using Illumina high-seq 2000 with a read length of 50 bp with pair ends. Raw Illumina sequencing reads in the FASTQ format were cleaned using in-house scripts by trimming sequencing adaptors and low-quality bases in both ends (Q < 67 in Illumina 1.5). Cleaned sequences were then mapped to the human genome (hg19) using Novoalign v2.07. The reads that mapped uniquely to a single genomic locus were used for peak finding with MACS v1.4 (Zhang et al., 2008), and only those peaks with false discovery rate (FDR) <1% were compared with RefSeq genes in the UCSC genome browser.
Metabolite Analysis
GC-MS metabolomic analysis was performed in the University of Utah Metabolomics Core. Parental, GFP- or G9A-overexpressing U2OS cells were cultured in MEM supplemented with 10% dialyzed FBS and were either untreated or treated with 5 μM BIX for 4 hr. Cells were washed with PBS, harvested by trypsin digestion, transferred to a microfuge tube, and frozen on dry ice. Metabolites were extracted by 80% methanol at −20°C and dried by vacuum centrifugation. Six biological replicate samples (5 × 106 cells/sample) were analyzed for each condition. GC-MS analysis was performed with a Waters GCT Premier mass spectrometer fitted with an Agilent 6890 gas chromatograph and a Gerstel MPS2 autosampler. Data were collected using MassLynx 4.1 software (Waters). Metabolites were identified and their peak area was recorded using QuanLynx. Data were normalized for extraction efficiency and analytical variation by mean centering the area of D4-succinate.
Flux Analysis
U2OS cells with or without overexpression of GFP or G9A were cultured in either glucose-deficient DMEM (11966-025, Invitogen) or MEM supplemented with 10% dialyzed FBS and 25 μM [U-13C] glucose (CDLM-3813-1, Cambridge Isotope Laboratories) with or without 5 μM BIX for 16 hr before metabolite extraction with 80% methanol. Conversion of [U-13C] glucose to other metabolites was analyzed by targeted LC-MS/MS using selected reaction monitoring (SRM) with a 5500 QTRAP mas spectrometer (Yuan et al., 2012). Three biological replicate samples were analyzed for each condition.
Analysis of Tumor Data Sets
The Versteeg NB88 data set contains 88 primary neuroblastoma tumors of all stages (Valentijn et al., 2012). Kaplan-Meier analysis was conducted online (http://r2.amc.nl), and the resulting survival curve and p value (log-rank test) were downloaded.
Statistics
Data are presented as mean ± SD. Statistics were determined with unpaired, two-tailed Student’s t test using GraphPad Prism 6.01 for Mac.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Vadivel Ganapathy for stimulating discussion and Min Yuan for help with metabolomic flux analysis. The work was supported by grants from NIH (R01CA124982), DoD (W81XWH-12-1-0613) and Georgia Cancer Coalition Distinguished Scholar Award to H.-F.D. In part, T.L. and X.W. were supported by grants from the National Natural Science Foundation of China (number 31172268 to T.L. and number 81172443 to X.W.) and J.M.A. by NIH grants (5P30CA006516 and 5 P01CA120964).
Footnotes
ACCESSION NUMBERS
The NCBI Gene Expression Omnibus (GEO) accession number for the microarray data reported in this paper is GSE51512.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures, three tables, and Supplemental Experimental Procedures and can be found with this article at http://dx.doi.org/10.1016/j.cmet.2013.11.004.
REFERENCES
- Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J. Biol. Chem. 2007;282:16744–16753. doi: 10.1074/jbc.M610510200. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Chen M-W, Hua K-T, Kao H-J, Chi C-C, Wei L-H, Johansson G, Shiah S-G, Chen P-S, Jeng Y-M, Cheng T-Y, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- Cho HS, Kelly JD, Hayami S, Toyokawa G, Takawa M, Yoshimatsu M, Tsunoda T, Field HI, Neal DE, Ponder BA, et al. Enhanced expression of EHMT2 is involved in the proliferation of cancer cells through negative regulation of SIAH1. Neoplasia. 2011;13:676–684. doi: 10.1593/neo.11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corkey BE, Shirihai O. Metabolic master regulators: sharing information among multiple systems. Trends Endocrinol. Metab. 2012;23:594–601. doi: 10.1016/j.tem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- de Koning TJ, Snell K, Duran M, Berger R, Poll-The BT, Surtees R. L-serine in disease and development. Biochem. J. 2003;371:653–661. doi: 10.1042/BJ20021785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Ladurner AG. A metabolic throttle regulates the epigenetic state of rDNA. Cell. 2008;133:577–580. doi: 10.1016/j.cell.2008.04.026. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J. Biol. Chem. 2002;277:38381–38389. doi: 10.1074/jbc.M205000200. [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang J, Dorsey J, Chuikov S, Pérez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhan SC, Hanson RW. Resurgence of serine: an often neglected but indispensable amino acid. J. Biol. Chem. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilberg MS, Pan YX, Chen H, Leung-Pineda V. Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guan KL. Amino acid signaling in TOR activation. Annu. Rev. Biochem. 2011;80:1001–1032. doi: 10.1146/annurev-biochem-062209-094414. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR, Issa J-PJ. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS ONE. 2008;3:e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- Narkewicz MR, Sauls SD, Tjoa SS, Teng C, Fennessey PV. Evidence for intracellular partitioning of serine and glycine metabolism in Chinese hamster ovary cells. Biochem. J. 1996;313:991–996. doi: 10.1042/bj3130991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AHFM, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Pfendner W, Pizer LI. The metabolism of serine and glycine in mutant lines of Chinese hamster ovary cells. Arch. Biochem. Biophys. 1980;200:503–512. doi: 10.1016/0003-9861(80)90382-3. [DOI] [PubMed] [Google Scholar]
- Pollari S, Käkönen SM, Edgren H, Wolf M, Kohonen P, Sara H, Guise T, Nees M, Kallioniemi O. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res. Treat. 2011;125:421–430. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Seo J, Fortuno ES, 3rd, Suh JM, Stenesen D, Tang W, Parks EJ, Adams CM, Townes T, Graff JM. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes. 2009;58:2565–2573. doi: 10.2337/db09-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover PJ, Chen LH, Suh JR, Stover DM, Keyomarsi K, Shane B. Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J. Biol. Chem. 1997;272:1842–1848. doi: 10.1074/jbc.272.3.1842. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12:321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- Touroutoglou N, Pazdur R. Thymidylate synthase inhibitors. Clin. Cancer Res. 1996;2:227–243. [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentijn LJ, Koster J, Haneveld F, Aissa RA, van Sluis P, Broekmans ME, Molenaar JJ, van Nes J, Versteeg R. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc. Natl. Acad. Sci. USA. 2012;109:19190–19195. doi: 10.1073/pnas.1208215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, Thompson CB. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc. Natl. Acad. Sci. USA. 2012;109:6904–6909. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Kikuchi G. Major pathways of glycine and serine catabolism in rat liver. Arch. Biochem. Biophys. 1970;139:380–392. doi: 10.1016/0003-9861(70)90490-x. [DOI] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.