Abstract
In North America, tick-borne relapsing fever (TBRF) is caused by the spirochete species Borrelia hermsii, Borrelia parkeri, and Borrelia turicatae. We previously demonstrated that some isolates of B. hermsii and B. parkeri are capable of binding factor H and that cell-bound factor H can participate in the factor I-mediated cleavage of C3b. Isolates that bound factor H expressed a factor H-binding protein (FHBP) that we estimated to be approximately 19 to 20 kDa in size and thus, pending further characterization, temporarily designated FHBP19. Until this report, none of the FHBPs of the TBRF spirochetes had been characterized. Here we have recovered the gene encoding the FHBP of B. hermsii YOR from a lambda ZAP II library and determined its sequence. The gene encodes a full-length protein of 22.7 kDa, which after processing is predicted to be 20.5 kDa. This protein, which we redesignate factor H-binding protein A (FhbA), is unique to B. hermsii. Two-dimensional pulsed-field gel electrophoresis and hybridization analyses revealed that the B. hermsii gene encoding FhbA is a single genetic locus that maps to a linear plasmid of approximately 220 kb. The general properties of FhbA were also assessed. The protein was found to be surface exposed and lipidated. Analysis of the antibody response to FhbA in infected mice revealed that it is antigenic during infection, indicating expression during infection. The identification and characterization of FhbA provides further insight into the molecular mechanisms of pathogenesis of the relapsing fever spirochetes.
Tick-borne relapsing fever (TBRF) is characterized by an undulating fever. In North America, relapsing fever is caused by three species of Borrelia: B. hermsii, B. turicatae, and B. parkeri (4). Several other Borrelia species are associated with TBRF in other parts of the world. In some regions, TBRF represents a serious human health concern. In the Mvumi and Mwanza districts of Tanzania, the incidence of TBRF is remarkably high (11). In these regions, the incidence of TBRF in children under 1 year of age is 384 per 1,000. While the incidence of TBRF in North America is significantly lower, the actual number of cases each year is unclear since it is usually treated prior to a definitive diagnosis. TBRF is endemic in the western and northwestern regions of the United States, with outbreaks having been reported in Colorado, Montana, Washington, New Mexico, and California (28). TBRF is transmitted to humans through the bite of infected Ornithordoros and Argasid ticks. The relapsing fever ticks are primarily nocturnal feeders and can complete the intake of a blood meal in minutes. In the bloodstreams of infected animals, relapsing fever spirochetes can achieve remarkable cell densities of 106 spirochetes ml−1. It is well-established that antigenic variation, mediated by the Vmp proteins, represents an important pathogenic mechanism for the relapsing fever spirochetes (7). The dominant molecular mechanism associated with Vmp antigenic variation is gene conversion (2). The TBRF spirochete B. hermsii carries approximately 40 different partial vmp gene cassettes and a single vmp expression locus. The gene cassettes lack a promoter and are not expressed. The vmp gene that resides at the expression locus is expressed at high levels and represents the dominant surface antigen. During infection, this antigen is the primary target of the humoral immune response, which is effective in clearing most of the spirochete population. However, at a frequency of 10−4 a gene conversion event results in the replacement of the expressed vmp gene with one of the silent vmp cassettes. This results in the production of an antigenically distinct Vmp protein variant that is not recognized by existing anti-Vmp antibodies. As this antigenically distinct population of spirochetes emerges, antibodies to the new Vmp variant are produced. Once again the antibody response leads to the elimination of most of the bacterial population, but as described above, a new antigenic variant will emerge, and the cycle will be repeated numerous times. In this manner the bacteria evade the humoral immune system and induce the symptoms of relapsing fever.
It stands to reason that the TBRF spirochetes would possess mechanisms which allow for evasion of the innate arm of the immune system. We recently demonstrated that several TBRF isolates are able to bind factor H and that cell-bound factor H is competent to participate as a cofactor for the factor I-mediated cleavage of C3b (24). In the closely related Lyme disease spirochetes, factor H binding has also been shown to play a role in innate immune system evasion (1, 15, 18, 25, 26). McDowell et al. have demonstrated a strong correlation between the species identity of a Lyme disease isolate and its ability to bind factor H (25). Borrelia garinii, a species which has been frequently isolated from the central nervous system (CNS), generally does not bind factor H, leading to the hypothesis that its tendency to localize in the CNS is a reflection of its inability to thwart complement attack through the binding of factor H. The CNS, which is devoid of complement, would provide a protected niche for the spirochetes. It is noteworthy that in our earlier analysis of factor H binding by the relapsing fever spirochetes we found that B. turicatae, which tends to localize to the CNS, cannot bind factor H (24), suggesting that this species might also seek the protective environment of the CNS. However, it is premature to draw a correlation between species identity and factor H binding for the relapsing fever spirochetes because only a single B. turicatae isolate was available for analysis.
The goals of this study were to recover and sequence the gene (fhbA) encoding the factor H-binding protein (FHBP) of B. hermsii from a genomic DNA library, characterize the protein and the immune response to it during infection, and determine the molecular basis of the factor H binding phenotype.
MATERIALS AND METHODS
Bacterial cultivation under different environmental conditions.
Table 1 lists and describes the Borrelia isolates analyzed in this study. All isolates were cultivated in BSK-H complete medium supplemented with rabbit serum (Sigma-Aldrich, St. Louis, Mo.) to 12% at 33°C. Bacteria were harvested by centrifugation and washed with phosphate-buffered saline (PBS).
TABLE 1.
Borrelia isolates employed in this study
| Species and isolate | Origin | Factor H binding phenotype | Molecular mass(es) (kDa) of FHBP(s) |
|---|---|---|---|
| B. hermsii | |||
| YOR | Human blood, California | + | 20 |
| MAN | Human blood, California | + | 20 |
| HSI | Ornithordoros hermsii, Washington | + | 20 |
| CON | Human blood, California | − | NAa |
| FRO | Human blood, Washington | − | NA |
| DAH | Human blood, Washington | − | NA |
| B. parkeri RML | Ornithordoros parkeri, RML collection | + | 20, 27 |
NA, not applicable.
Construction and screening of a lambda ZAP II library.
To recover the gene encoding the FHBP of B. hermsii YOR, a lambda ZAP II library was constructed. DNA from B. hermsii YOR was isolated as previously described and digested with Sau3A (New England Biolabs). Library construction was as described by the manufacturer (Stratagene), using BamHI-predigested lambda arms. Once the appropriate titer for the phage library was determined, the phage were plated with Escherichia coli and incubated overnight at 42°C. The plates were removed, and then plaque lifts were performed as follows. Nitrocellulose disks were soaked in 100 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 min at room temperature, allowed to dry, placed on the NZY plates, and incubated at 37°C for 4 to 6 h. The nitrocellulose membranes were removed and washed with PBS containing 0.02% Tween 20. To identify plaques that were expressing FHBPs, the blots were incubated with human factor H (Calbiochem) (10 ng μl−1 in PBS; room temperature; 4 h) and washed with PBS-0.02% Tween 20 to remove unbound factor H, and then factor H binding to phage-expressed proteins was detected by using anti-human factor H antiserum and chemiluminescence. Positive plaques were cored from the plates, placed in SM buffer (50 mM Tris-HCl,10 mM MgSO4, 0.001% gelatin, 100 mM NaCl [pH 7.5]) with chloroform (40% vol/vol), vortexed, and incubated for 2 h at room temperature. Phagemids were then generated by using the ExAssist helper phage as described by the manufacturer (Stratagene).
DNA sequence analysis, PCR, ligase-independent cloning, and production of recombinant protein.
To determine the gene sequence of the putative FHBP-encoding gene, a 4.1-kb subfragment of a 6.5-kb phagemid insert was sequenced on a fee-for-service basis by using automated methods (MWG Biotech). The determined sequence was translated to identify potential reading frames by using the TRANSLATE program.
To generate recombinant protein for the putative FHBP, primers were designed to amplify the entire coding sequence (minus the leader peptide). The primers were designed with sequences that allow for annealing of the PCR product into the pET32-Ek/LIC vector in a ligase-independent fashion (Novagen). The primers FhbA-R-LIC (5′-GAG GAG AAG CCC GGT CAA CTT AAG TTT TTA AAT ATT CC) and FhbA-F-LIC (5′-GAC GAC GAC AAG ATT AGC TGT GAT TTA TTC AAT AAA AAC) were employed. PCR was performed with the Expand high-fidelity PCR system as described by the manufacturer (Roche). The PCR product was then treated with T4 DNA polymerase to generate single-stranded overhangs and annealed into the pET32-Ek/LIC vector as instructed by the supplier (Novagen). The recombinant plasmid was transformed into and propagated in E. coli NOVABlue cells. Plasmid purified from the NOVABlue cells was then transformed into E. coli BL21(DE3) cells. Colonies found to be carrying the desired insert through PCR screening were cultivated overnight at 37°C in LB with ampicillin (50 μg ml−1). IPTG induction was performed as previously described (23). All recombinant proteins expressed from pET32-Ek/LIC were generated with an N-terminal fusion that contains both S and His tags. The N-terminal fusion adds approximately 17 kDa to the molecular mass of the recombinant protein.
Two-dimensional PFGE and Southern hybridization analysis.
To conduct pulsed-field gel electrophoresis (PFGE), plugs from 50-ml cultures were prepared as previously described (8). Electrophoresis was conducted with the Bio-Rad contour-clamped homogenous electric field mapper system with 1% GTG-agarose gels in 0.5× Tris-acetate-EDTA buffer at 14°C. The autoalgorithim function was used to determine electrophoresis parameters, using a separation size range of 10 to 400 kb. The gel was then rotated 90° and electrophoresed (second dimension) under constant voltage. After staining with ethidium bromide, the DNA was transferred onto a Hybond-N membrane by using the VacuGene vacuum blotting system (Pharmacia) as previously described (19). The DNA was fixed to the membrane by UV irradiation with the Stratagene UV cross-linker. The membrane was then incubated for 2 h in hybridization buffer (0.2% bovine serum albumin, 0.2% polyvinylpyrrolidine, 0.2% Ficoll, 50 mM Tris-HCl, 0.1% sodium pyrophosphate, 1% sodium dodecyl sulfate [SDS], 10% dextran sulfate, 100 μg of herring sperm DNA μl−1, 50% formamide) at 50°C, and then the buffer was replaced with fresh buffer containing the radioactively labeled probe. The probe was generated by PCR amplification and labeled by using the Prime-A-Gene labeling system (Promega) with [α-32P]dATP (6,000 Ci mmol−1). The probe was incubated with the membrane overnight at 42°C. The blot was washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS for 10 min at 37°C and then once with 0.2 × SSC-0.1% SDS for 2 h at 37°C. The membranes were wrapped in saran and exposed to film at −70°C with intensifying screens for 3 h.
Determination of the number of fhbA-related alleles in the B. hermsii genome.
To determine if there are multiple fhbA alleles or paralogs in B. hermsii, DNA was isolated from the B. hermsii isolates YOR and MAN as previously described (5), digested with HaeIII (New England Biolabs) under standard conditions, and fractionated by electrophoresis in a 0.8% agarose gel in Tris-acetate-EDTA buffer. The DNA was visualized by staining with ethidium bromide, transferred onto a Hybond-N membrane by vacuum blotting, and fixed to the membrane by UV cross-linking. Southern hybridization was performed with a random-prime-labeled PCR probe. All methods were as described in preceding sections.
Northern hybridization analysis.
Total cellular RNA was isolated from B. hermsii YOR grown at 33°C in BSK-H medium as previously described (19). Ten micrograms of RNA was fractionated in 1.2% agarose-formaldehyde gels and transferred onto Hybond-N+ membranes by vacuum blotting as previously described (21). Hybridization was performed exactly as described above for the Southern analyses. The hybridization probe was generated by PCR and then labeled internally by random priming as described above.
Experimental infection of mice and generation of infection sera.
C3h-HeJ mice were infected with B. hermsii YOR and B. parkeri RML by intradermal injection of 103 spirochetes in PBS between the shoulder blades as previously described (9). The establishment of infection was confirmed by dark-field examination of blood smears at days 3, 4, and 7 postinoculation. Blood was collected from the mice at 1 month postinoculation and at the time of sacrifice by cardiac puncture. Prior to use of the sera (henceforth referred to as infection sera), nonspecific antibodies were absorbed by using cell lysates of E. coli as previously described (23).
Generation of anti-FhbA antiserum.
To generate antiserum to FhbA, production of the recombinant protein in E. coli BL21(DE3) cells was induced with IPTG (Novagen), and inclusion bodies were isolated as instructed by the supplier of the vector. Antiserum was generated in C3h-HeJ mice as previously described (26), using inclusion bodies as the inoculum. Prior to its use in immunoblots, antibodies that target the N-terminal S-tag fusion of the protein were removed by preabsorption with an irrelevant S-tag, His-tag protein as described by McDowell et al. (22).
Proteinase K digestion assays and Triton X-114 extraction and phase partitioning.
To determine whether the FHBPs of the relapsing fever spirochetes are surface exposed, proteinase K digestion assays were performed as described previously (6). Briefly, exponential-phase cells at an optical density at 600 nm of 0.5 were diluted 1:1 with PBS, recovered by centrifugation, resuspended in proteinase K solution (0.2 μg ml−1 in PBS), and incubated at 20°C for 40 min. Phenylmethylsulfonyl fluoride (Fisher), prepared as a 50-mg ml−1 stock solution in isopropanol, was added to a final concentration of 0.5 μg μl−1, and the cells were harvested by centrifugation (11,228 × g; 4°C), washed with PBS, and immediately resuspended in SDS solubilizing solution containing PMSF (0.5 μg μl−1).
Triton X-114 extraction and phase partitioning was performed as described by Cunningham et al. (10). Briefly, exponential-phase cells at an optical density at 600 nm of 0.5 were harvested by centrifugation, resuspended in 1% Triton X-114 (in PBS), and incubated at 4°C overnight with gentle rocking. The detergent-insoluble phase was collected by centrifugation at 4°C at 15,000 × g. The supernatant was incubated at 37°C for 15 min and then centrifuged (15,000 × g) at room temperature to separate the aqueous and detergent-soluble phases. Each sample was extracted twice to ensure complete partitioning. The resulting samples were prepared for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) by solubilization in SDS solubilizing solution.
Immunoblot analysis and ALBI assays.
SDS-PAGE was performed with Bio-Rad 12% Ready Gels, and the proteins were transferred onto Immobilon-P membranes (Millipore) by electroblotting as previously described (23). Factor H affinity ligand binding immunoblot assays (ALBI assays) were performed as previously described (24, 26). Briefly, immunoblots of E. coli BL21(DE3) cells that had been induced to express recombinant protein were incubated with purified human factor H (Calbiochem), and then the formation of factor H-protein complexes was detected with goat anti-human factor H antisera (1:800) (Calbiochem). Rabbit anti-goat immunoglobulin G antibody (1:40,000) served as the secondary antibody. As a control, an immunoblot was screened with S-protein-horseradish peroxidase (HRP) conjugate to verify expression of the recombinant proteins as instructed by the manufacturer (Novagen). In all cases the Pierce Super Signal substrate was employed for chemiluminescent detection of antibody binding.
RESULTS
Recovery of FhbA from a genomic library and DNA sequence analysis.
Screening of plaque lifts from a B. hermsii YOR library with human factor H and anti-human factor H antiserum led to the recovery of a phagemid expressing a putative FHBP. Analysis of the phagemid revealed that it carried a 6.5-kb insert. The phagemid was then digested with BglII and subcloned to identify the smallest DNA fragment that still allowed for production of a FHBP. A subclone harboring a 4.1-kb insert conveyed the factor H binding phenotype in E. coli. The insert contained a putative open reading frame of 576 bp with an appropriately spaced ribosomal binding site (Fig. 1). The deduced protein is 192 amino acids with a calculated molecular mass of 22.7 kDa. After cleavage of the leader peptide, the predicted molecular mass is 20.5 kDa. Computer analysis of the physical properties of the deduced processed protein revealed it to be highly hydrophilic, with no hydrophobic or transmembrane-spanning domains. Computer analyses also revealed that the protein has a high predictive probability of having coiled-coil domains. A BLAST search did not detect any proteins with significant homology, indicating that this protein is unique to B. hermsii. Henceforth, we designate this protein FhbA (factor H-binding protein A) to reflect its functional activity and its genetic uniqueness with respect to other Borrelia proteins.
FIG. 1.
DNA and deduced amino acid sequences of FhbA of B. hermsii YOR. The gene encoding FhbA was recovered from a lambda ZAP II library and sequenced. The deduced amino acid sequence is presented, with each amino acid aligned above its corresponding codon. The ribosomal binding site is underlined, and the translational start codon is indicated by uppercase letters.
Demonstration of factor H binding by recombinant FhbA.
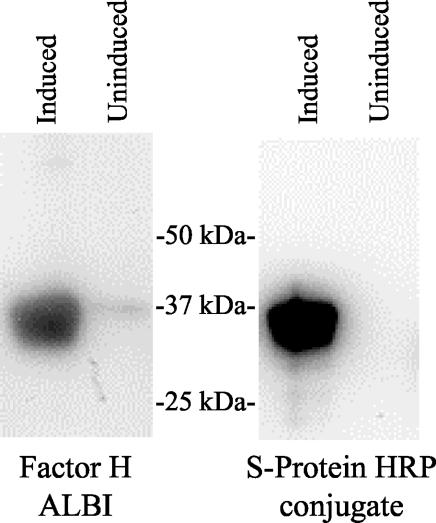
To confirm the ability of recombinant FhbA to bind factor H, the gene (without its leader peptide) was PCR amplified, cloned by using ligase-independent cloning (LIC) methodologies, and expressed as an S-tag, His-tag fusion protein as previously described (23). Lysates of E. coli BL21(DE3) carrying the recombinant plasmid that had been induced with IPTG were immunoblotted and used in factor H ALBI assay analysis (Fig. 2). The induced cultures expressed a 37-kDa FHBP, consistent with the size expected for the FhbA fusion protein (17 kDa is derived from the N-terminal S-tag sequence).
FIG. 2.
Demonstration of factor H binding by recombinant FhbA. The gene encoding FhbA was amplified with primers containing tails that allow for LIC cloning and expression as an N-terminal S-tag fusion protein. The PCR fragment was annealed with pretreated pET32-Ek/LIC and propagated in NOVABlue cells, and the plasmid was isolated. The plasmid was transformed into E. coli BL21(DE3) cells, and colonies carrying the appropriate recombinant plasmids were grown in Luria-Bertani broth with ampicillin (50 μg ml−1). The cultures were induced with IPTG (or not induced), fractionated by SDS-PAGE, immunoblotted, and then tested for factor H binding by using the factor H ALBI assay or screened with S-protein-HRP conjugate (to confirm expression). For the factor H ALBI assay, the blots were incubated with purified human factor H, and then bound factor H was detected by using anti-human factor H antiserum and chemiluminescence.
Identification of the genomic element carrying the fhbA gene.
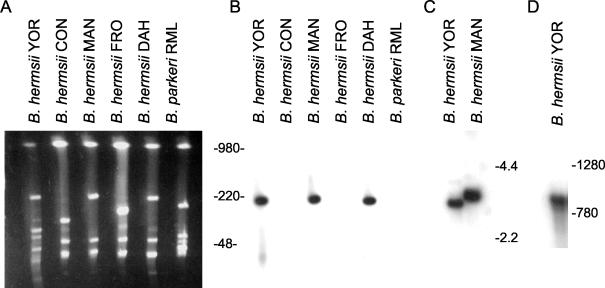
To identify the genetic element carrying the fhbA gene, PFGE was performed, followed by electrophoresis in the second dimension under constant voltage. This approach allows for the differentiation of linear and circular plasmids, because the circular plasmids exhibit retarded mobility relative to the linear plasmids upon electrophoresis in the second dimension. A Southern blot of the fractionated DNA was screened with a PCR-generated, full-length fhbA probe labeled by random priming (Fig. 3A and B). The probe hybridized exclusively with a linear plasmid of approximately 220 kb (lp220) in B. hermsii isolates YOR, MAN, and DAH. The B. hermsii isolates CON and FRO, which we previously demonstrated do not bind factor H (24), lack this plasmid and were hybridization negative. B. parkeri RML, which carries a linear plasmid of approximately 180 kb, was also hybridization negative. This isolate produces two FHBPs. The negative hybridization data suggest that the genes encoding these proteins are divergent from fhbA of B. hermsii.
FIG. 3.
Demonstration that fhbA is a single genetic locus carried by a 220-kb linear plasmid and expressed as a monocistronic mRNA. DNAs from several TBRF isolates were fractionated in the first dimension by PFGE. The gel was rotated 90°, and then electrophoresis in the second dimension was performed, followed by ethidium bromide staining (A). The DNA was transferred onto membranes for hybridization analysis (B). To determine whether fhbA is a single genetic locus or a member of a paralogous gene family, hybridization analyses were performed with HaeIII-digested DNAs from B. hermsii isolates YOR and MAN. The DNA was fractionated in a 0.8% agarose gel and transferred onto membranes for hybridization analyses (C). To determine the size of the fhbA transcript, Northern blot analysis was performed (D.) All blots were hybridized with an internally labeled, PCR-generated fhbA probe. All methods are described in the text. DNA size markers (in kilobases) are indicated for panels A, B, and C. RNA size markers (in bases) were used for panel D.
Demonstration that fhbA is a single-copy locus.
To determine if there are multiple fhbA alleles in the B. hermsii genome, HaeIII-restricted DNA was screened by Southern hybridization with an fhbA PCR-generated probe. The probe hybridized with fragments of ∼3.5 and ∼3.0 kb in B. hermsii isolates MAN and YOR, respectively (Fig. 3C). This, coupled with the sequence analysis of the flanking sequence of fhbA, which demonstrated that the gene is not tandemly duplicated, indicates that fhbA represents a single genetic locus with no apparent paralogous genes.
Determination of the size of the fhbA transcript.
To determine the size of the fhbA transcript, Northern blot hybridization analysis was performed. RNA was isolated from B. hermsii YOR that had been cultivated at 33°C. A transcript approximately 900 bases in length was readily detected, indicating that fhbA is most likely expressed as a monocistronic message (Fig. 3D). A similarly sized transcript was detected in B. hermsii MAN (data not shown).
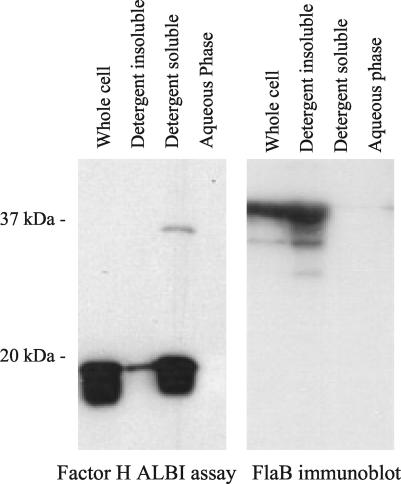
Demonstration of the surface exposure of FhbA.
To determine if FhbA is surface exposed, several relapsing fever isolates were treated with proteinase K to digest surface proteins or not treated. The cells were fractionated by SDS-PAGE and immunoblotted. The factor H ALBI assay was used to determine whether proteinase K treatment inhibited factor H binding. Treatment with proteinase K resulted in the complete elimination of factor H binding by both B. hermsii and B. parkeri (Fig. 4). As a negative control, an immunoblot of the samples was also screened with anti-Fla antiserum. As expected, in view of the periplasmic localization of the FlaB protein in the Borrelia, FlaB was not degraded by proteinase K treatment. These analyses demonstrate that FhbA is a surface-exposed protein and thus is potentially available in vivo to interact with factor H.
FIG. 4.
Demonstration of the surface exposure of FhbA. To assess surface exposure, spirochetes were cultivated in BSK-H medium at 33°C, and then aliquots of each culture were incubated with (+) or without (−) proteinase K as described in the text. The blot on the left was used in a factor H ALBI to determine whether proteinase K treatment abolished factor H binding. As a control, the immunoblot on the right was screened with anti-Fla antiserum to detect the FlaB proteins.
Analysis of the subcellular localization of FhbA.
To assess the subcellular localization and other properties of FhbA, Triton X-114 extraction and phase partitioning were performed. As described above, we assayed for FhbA in the subcellular fractions by using the factor H ALBI assay. The FHBP partitioned almost exclusively to the detergent phase (Fig. 5). This observation, coupled with its demonstrated surface exposure, indicates that FhbA is an outer membrane lipoprotein. As a control for these analyses, an identical immunoblot was screened with anti-FlaB, and as expected, FlaB was detected only in the whole-cell lysate and detergent-insoluble phase.
FIG. 5.
Demonstration that FhbA is a lipidated outer membrane protein. To assess possible lipidation of FhbA, cultures of B. hermsii YOR were subjected to Triton X-114 phase partitioning and extraction as described in the text. The individual fractions (indicated above each lane) were separated by SDS-PAGE and immunoblotted, and a factor H ALBI assay was performed. As a control, an identical immunoblot was screened with anti-Fla antiserum.
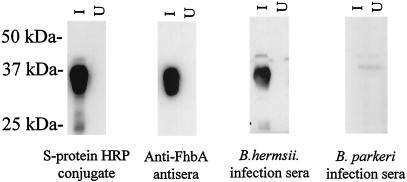
Analysis of the immune response to FhbA during experimental infection of mice.
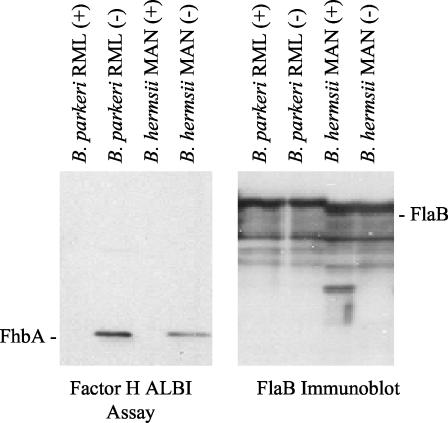
To determine whether FhbA elicits an antibody response during infection and is thus expressed during infection, mice were experimentally infected with B. hermsii YOR and B. parkeri RML. Serum was collected 1 month after the establishment of infection. We refer to these sera as infection sera. After preabsorption of the infection sera with E. coli expressing an irrelevant S-tag recombinant protein, the sera were used to screen immunoblots of E. coli expressing recombinant FhbA (Fig. 6). The serum from a mouse infected with B. hermsii YOR reacted with recombinant FhbA, while the serum from a mouse infected with B. parkeri RML did not. The presence of antibodies recognizing FhbA in the infection serum indicates that B. hermsii expresses FhbA during infection. The absence of antibodies in the serum derived from the B. parkeri-infected mouse suggests that the FHBP of B. parkeri is antigenically distinct from FhbA.
FIG. 6.
Analysis of the antibody response to FhbA in experimentally infected mice. Cell lysates of E. coli were induced with IPTG (lanes I) or uninduced (lanes U), fractionated by SDS-PAGE, and immunoblotted. The immunoblots were screened with sera collected from mice infected with either B. hermsii YOR or B. parkeri RML (as indicated) that had been preabsorbed with E. coli expressing an irrelevant S-tag recombinant protein. As a control, the migration position of recombinant FhbA was determined by using S-protein-HRP conjugate, and the identity of the expressed protein was verified by screening with anti-FhbA antiserum. All methods are described in the text. Molecular mass markers are indicated.
DISCUSSION
A unique feature of the relapsing fever spirochetes is their ability to reach high cell densities in the blood and to persist in the blood of infected mammals. We recently demonstrated that the TBRF spirochetes can bind the serum complement regulatory protein factor H and that factor H bound to B. hermsii is competent to participate in the factor I-mediated cleavage of C3b (24). The primary objective of this study was to identify and characterize the FHBP of B. hermsii.
The gene encoding the B. hermsii FHBP was recovered from a lambda ZAP II genomic library by screening of plaque lifts for the factor H binding phenotype by using the factor H ALBI assay approach. Sequence analysis of the gene revealed that it encodes a mature protein of 20.5 kDa. Database analyses indicate that the protein is unique to B. hermsii, as it exhibits no significant homology to other known FHBPs or to any proteins encoded by the Borrelia burgdorferi genome. We originally designated this protein FHBP19 (24), but here we redesignate it FhbA. A variety of conflicting nomenclatures have been assigned to the FHBPs of Borrelia species associated with Lyme disease (16, 17, 25, 29). Many of these nomenclatures were devised prior to the determination that these proteins bind factor H. Most recently, the FHBPs of the Lyme disease spirochetes have been designated complement regulator-acquiring surface proteins (CRASPs) (16, 17). However, since there are numerous complement regulatory proteins that are not bound by factor H-binding proteins such as FhbA, and since the CRASP designation is not consistent with recommended bacterial gene nomenclature schemes, we opted not to employ this designation for the FHBPs of the relapsing fever spirochetes. Here we propose the three-letter, functionally based Fhb (for factor H binding) designation, and since this gene is the first of its functional class to be identified in the relapsing fever spirochetes, its product is referred to as FhbA.
The Borrelia genome is segmented and composed of a linear chromosome and a series of linear and circular minichromosomes or plasmids (3, 27). To determine where the fhbA gene resides in the B. hermsii genome, two-dimensional PFGE and Southern blotting were performed. This technique has been previously employed to differentiate the linear and circular plasmids of the Borrelia species (19, 21). In this study, the fhbA gene was localized to an approximately 220-kb linear plasmid that is present only in B. hermsii isolates exhibiting the factor H binding phenotype. The origin of the fhbA gene remains to be determined. The presence of this gene on a plasmid raises the possibility that the factor H binding phenotype could be potentially transferable and acquired via lateral transfer. Lateral transfer of the ospD gene, which is carried on a linear plasmid of the Lyme disease spirochetes, has been postulated (20). However, direct evidence of transfer of Borrelia linear plasmids has not yet been obtained. In contrast, Eggers et al. have directly demonstrated that lateral transfer of the 32-kb circular plasmids can occur. These plasmids are packaged and transferred as bacteriophages (12-14).
Analysis of the general properties of FhbA revealed it to be surface exposed and lipidated. Surface exposure was demonstrated through the treatment of intact cells with proteinase K and lipidation through Triton X-114 extraction and phase partitioning. The surface exposure of FhbA and the previously demonstrated ability of cell surface-bound factor H to mediate C3b cleavage (24) provide strong support for the assertion that FhbA, if expressed during infection, may play an important role in evasion of the innate immune system. The detection of anti-FhbA antibodies in B. hermsii infection serum indicates that FhbA is produced during experimental infection in mice. Interestingly, serum from a mouse infected with B. parkeri, which produces a similarly sized FHBP, did not harbor antibodies that recognized the recombinant B. hermsii FhbA. This observation suggests that the B. parkeri FHBP is antigenically distinct from FhbA. This suggestion is consistent with the hybridization analyses in which a PCR-generated fhbA probe did not hybridize with B. parkeri DNA.
It has been postulated that the FHBPs of the Lyme disease spirochetes possess specific linear sequence elements that are involved in factor H binding (17). Sequences with homology to these putative binding determinants were not found in FhbA. An earlier study of the OspE protein by Metts et al. (26) provided evidence that factor H binding is not strictly dependent on a specific-sequence element but rather is dependent on the formation of a discontinuous binding site or possibly a conformationally defined binding site. It is interesting that while FhbA exhibits no discernible homology with other FHBPs of the Lyme disease spirochetes, computer analyses indicate that all FHBPs of the Borrelia species have a high predictive probability of forming coiled-coil motifs. These motifs are thought to be important in inter- and intramolecular interactions and could be an important determinant in factor H binding. The potential importance of structural elements in factor H binding is supported by an earlier analysis that demonstrated that antibody and factor H binding to OspE requires both the N and C termini of the protein (26). This suggests that either structural determinants or discontinuous binding sites are required for factor H binding.
The ability of the TBRF spirochetes to persist in the blood is clearly a multifactorial process, with antigenic variation allowing for evasion of the humoral immune response and factor H binding possibly contributing to evasion of the innate immune system. It is important to note, however, that not all TBRF isolates exhibit the factor H binding phenotype. It remains to be determined whether factor H binding allows TBRF spirochete isolates with the factor H binding-positive phenotype to persist longer in infected animals or to achieve higher cell densities in the blood. In conclusion, the characterization of FhbA represents an important step forward in our understanding of the molecular mechanisms of pathogenesis of the TBRF spirochetes.
REFERENCES
- 1.Alitalo, A., T. Meri, L. Rämö, T. S. Jokiranta, T. Heikkilä, I. J. T. Seppälä, J. Oksi, M. Viljanen, and S. Meri. 2001. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 69:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, A. G., C. J. Carter, N. Burman, C. S. Freitag, C. F. Garon, and S. Bergstom. 1991. Tandem insertion sequence-like elements define the expression site for variable antigen genes of Borrelia hermsii. Infect. Immun. 59:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour, A. G., and C. F. Garon. 1987. Linear plasmids of the bacterium, Borrelia burgdorferi, have covalently closed ends. Science 237:409-411. [DOI] [PubMed] [Google Scholar]
- 4.Barbour, A. G., and S. F. Hayes. 1986. Biology of Borrelia species. Microbiol. Rev. 50:381-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour, A. G., R. A. Heiland, and T. R. Howe. 1985. Heterogeneity of major proteins in Lyme disease borreliae: a molecular analysis of North American and European isolates. J. Infect. Dis. 152:478-484. [DOI] [PubMed] [Google Scholar]
- 6.Barbour, A. G., S. L. Tessier, and S. F. Hayes. 1984. Variation in a major surface protein of Lyme disease spirochetes. Infect. Immun. 45:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour, A. G., S. L. Tessier, and H. G. Stoenner. 1982. Variable major proteins of Borrelia hermsii. J. Exp. Med. 156:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlyon, J. A., and R. T. Marconi. 1998. Cloning and molecular characterization of a multicopy, linear plasmid-carried, repeat motif-containing gene from Borrelia turicatae, a causative agent of relapsing fever. J. Bacteriol. 180:4974-4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlyon, J. A., D. M. Roberts, M. Theisen, C. Sadler, and R. T. Marconi. 2000. Molecular and immunological analyses of the B. turicatae Bdr protein family. Infect. Immun. 68:2369-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham, T. M., E. M. Walker, J. N. Miller, and M. A. Lovett. 1988. Selective release of the Treponema pallidum outer membrane and associated polypeptides with Triton X-114. J. Bacteriol. 170:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cutler, S., and A. Talbert. 2003. Tick-borne relapsing fever in Tanzania—a forgotten problem? ASM News 69:542-543. [Google Scholar]
- 12.Eggers, C. H., and D. S. Samuels. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eggers, C. H., J. L. Bono, A. Elias, P. Rosa, and D. S. Samuels. 2001. Transduction of an antibiotic-resistance marker by BB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggers, C. H., S. Casjens, S. F. Hayes, C. F. Garon, C. J. Damman, D. B. Oliver, and D. S. Samuels. 2000. Bacteriophages of spirochetes. J. Mol. Microbiol. Biotechnol. 4:365-373. [PubMed] [Google Scholar]
- 15.Hellwage, J., T. Meri, T. Heikkila, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppala, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 16.Kraiczy, P., J. Hellwage, C. Skerka, H. Becker, M. Kirschfink, M. S. Simon, V. Brade, P. F. Zipfel, and R. Wallich. 2003. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 279:2421-2429. [DOI] [PubMed] [Google Scholar]
- 17.Kraiczy, P., J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, and R. Wallich. 2003. Immune evasion of Borrelia burgdorferi: mapping of a complement inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 33:697-707. [DOI] [PubMed] [Google Scholar]
- 18.Kraiczy, P., C. Skerka, M. Kirschfink, V. Brade, and P. F. Zipfel. 2001. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and factor H. Eur. J. Immunol. 31:1674-1684. [DOI] [PubMed] [Google Scholar]
- 19.Marconi, R. T., D. S. Samuels, and C. F. Garon. 1993. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J. Bacteriol. 175:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marconi, R. T., D. S. Samuels, R. K. Landry, and C. F. Garon. 1994. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J. Bacteriol. 176:4572-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marconi, R. T., D. S. Samuels, T. G. Schwan, and C. F. Garon. 1993. Identification of a protein in several Borrelia species which is related to OspC of the Lyme disease spirochetes. J. Clin. Microbiol. 31:2577-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDowell, J. V., S. Y. Sung, L. T. Hu, and R. T. Marconi. 2002. Evidence that the variable regions of the central domain of VlsE are antigenic during infection with the Lyme disease spirochetes. Infect. Immun. 70:4196-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDowell, J. V., S.-Y. Sung, G. Price, and R. T. Marconi. 2001. Demonstration of the genetic stability and temporal expression of select members of the Lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 69:4831-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDowell, J. V., E. Tran, D. Hamilton, J. Wolfgang, K. Miller, and R. T. Marconi. 2003. Analysis of the ability of spirochete species associated with relapsing fever, avian borreliosis, and eptizootic bovine abortion to bind factor H and cleave C3b. J. Clin. Microbiol. 41:3905-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDowell, J. V., J. Wolfgang, E. Tran, M. S. Metts, D. Hamilton, and R. T. Marconi. 2003. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 71:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metts, S., J. V. McDowell, M. Theisen, P. R. Hansen, and R. T. Marconi. 2003. Analysis of the OspE determinants involved in the binding of factor H and OspE targeting antibodies elicited during infection in mice. Infect. Immun. 71:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plasterk, R. H. A., M. I. Simon, and A. G. Barbour. 1985. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium B. hermsii. Nature 318:257-263. [DOI] [PubMed] [Google Scholar]
- 28.Schwan, T. G., P. F. Policastro, Z. Miller, R. L. Thompson, T. Damrow, and J. E. Keirans. 2003. Tick-borne relapsing fever caused by Borrelia hermsii in Montana. Emerg. Infect. Dis. 9:1151-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]