Abstract
Drug abuse alone has been shown to cause epigenetic changes in brain tissue that have been shown to play roles in addictive behaviors. In conjunction with HIV-1 infection, it can cause epigenetic changes at the viral promoter that can result in altered gene expression, and exacerbate disease progression overall. This review entails an in-depth look at research conducted on the epigenetic effects of three of the most widely abused drugs (cannabinoids, opioids, and cocaine), with a particular focus on the mechanisms through which these drugs interact with HIV-1 infection at the viral promoter. Here we discuss the impact of this interplay on disease progression from the point of view of the nature of gene regulation at the level of chromatin accessibility, chromatin remodeling, and nucleosome repositioning. Given the importance of chromatin remodeling and DNA methylation in controlling the retroviral promoter, and the high susceptibility of the drug abusing population of individuals to HIV infection, it would be beneficial to understand the way in which the host genome is modified and regulated by drugs of abuse.
Keywords: drug abuse, epigenetics, chromatin remodeling, HIV-1, LTR
Introduction
Drug abuse is a prevalent public health problem in the United States, affecting the abusing individuals as well as their families and the community at large. Surveys taken from 2010 to 2011 showed that roughly 12.6% of Americans over the age of twelve reported using marijuana in the past year and that 4.6% of Americans between the ages of 18 and 25 reported cocaine use in the past year (SAMHSA, 2012). Additionally, drug abuse and addiction, excluding alcohol and tobacco, cost the United States approximately $181 billion per year in health care and loss of productivity due to crime and other factors (NIDA, 2008).
Drugs of abuse such as cannabinoids, opioids, and cocaine are known to produce numerous physical effects, including addiction. While they can cause seemingly benign symptoms such as euphoria and heightened sensations, they can also lead to harmful long-term effects, especially in terms of cognitive function, such as physical dependence, withdrawal, neurological changes, organ damage, (Vale A, 2012), and even epigenetic changes (Renthal W, 2009). A summary of the mechanisms and side effects of the drugs discussed in this paper can be found in Table 1 (Ameri A, 1999; Molina PE, 2011; Solum D, 2008). As indicated in table 1, cannabinoids, opiates, and cocaine differ in their mechanisms of action, but are all known to produce deleterious effects on the immune system, often by causing an imbalance in Th1 pro- and Th2 anti-inflammatory cytokines, leading to greater susceptibility to infection (Cabral GA, 2006).
Table 1.
Summary of drugs of abuse
| Drug | Mode of Action | Side Effects | Statistics |
|---|---|---|---|
| Cannabinoids | Δ9-THC disrupts cellular membranes in a way that affects membrane-associated enzymes and ion channels. Also binds to CB1 (brain) and CB2 (peripheral) receptors |
Euphoria Enhancement of sensory perception Tachycardia Antinociception Difficulty concentrating Memory impairment |
Most commonly used illicit drug in the United States (approximately 14.6 million current users) |
| Opioids | Primarily act on mu opioid receptor Also act on dopamine, gamma-amino-butyric acid (GABA), and glutamate |
Analgesia Euphoria |
Approximately 4% of North American adults report opiate use. |
| Cocaine | Blocks dopamine, norepinephrine, and serotonin uptake transporters Results in increased dopamine concentrations in critical brain sites |
Increased heart rate, blood pressure, and arousal Greater vigilance, alertness, and self-confidence Euphoria |
Approximately 5.8 million current users in the United States |
| Alcohol | Disturbs balance between excitation and inhibition in the brain through agonistic action on the GABA receptors | Disinhibition Ataxia Sedation |
30% of American adults report having abused alcohol at least once. |
Illegal drug use is considered the second most common route of human immunodeficiency virus type 1 (HIV-1) infection (Pandhare J, 2011), occurring via sharing of needles and unsafe sexual intercourse, and continued abuse has also been found to impact the progression of the disease. According to National Institute on Drug Abuse (NIDA), 8% of new HIV-1 cases were caused by injection drug use, 3% by male-to-male sexual contact and injection drug use, and the remainder by sexual contact. Approximately 40% of infected white women contracted HIV from injection drug use. Other social risk factors include race and ethnicity as 46% of newly infected individuals in 2010 were African-American. In addition, almost a quarter of HIV patients are coinfected with hepatitis C, and 50–80% of intravenous drug users carry both viruses. Active cocaine use is significantly associated with lower adherence to therapeutic regimens. Adherence declined 41% among active cocaine users, which was associated with failure to maintain viral suppression (Arnsten et al., 2002). In addition to an observed association with CD4 T-cell count (Duncan R, 2007), crack cocaine was significantly associated with a higher level of HIV-1 RNA (Baum et al., 2009), and more AIDS-defining illnesses (Nacher et al., 2009). Additionally, exposure of an individual to drugs of abuse, viral infection, and a multitude of other factors can result in epigenetic modifications to their genes and integrated viral genes, prompting further study into the interaction between these pathologies. The relationship between cannabis, opioids, and cocaine, three common drugs of abuse that share a high correlation with HIV-1 infection (Chao C, 2008), and HIV-1 gene expression and epigenetics will be explored in this review.
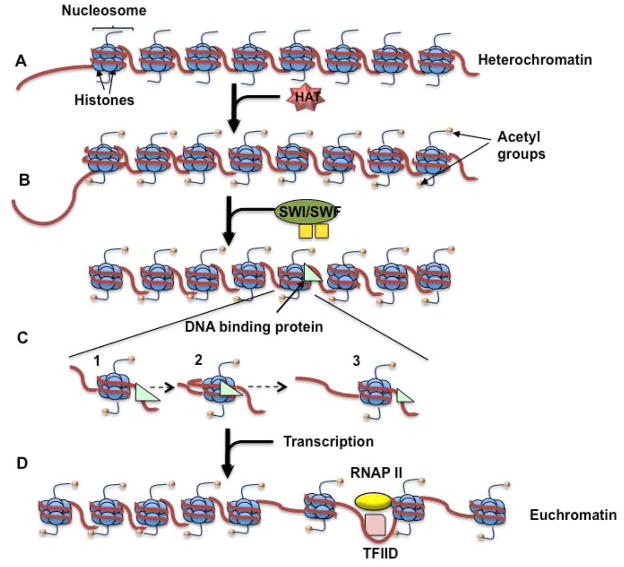
Epigenetics are the chemical and physical code written over the genome’s DNA sequences that involve the regulation of gene expression without any changes to the DNA sequence itself (Arimondo PB, 2012). One of the primary components of epigenetics is chromatin accessibility. Chromatin and its components, nucleosomes and histones, help to condense DNA and are critically important in gene regulatory control and to ensure correct gene expression (Shah S, 2010). It can be categorized into euchromatin, which consists of regions of active, accessible chromatin, and heterochromatin, which consists of inactive, condensed chromatin. Each type of chromatin has its own markers based on the methylation of its DNA and acetylation patterns of its histones (Fig. 1). DNA methylation, which occurs on the cytosine residue of CpG dinucleotides in the 5′ regulatory regions of DNA, is an important epigenetic alteration that can result in silenced genes (Takizawa T, 2008). During DNA methylation, a methyl group is added to the 5th position of cytosine residues (C5) by DNA methyltransferases (DNMTs). This leads to a repression in gene transcription through recruitment of co-repressor complexes such as histone deacetylases (HDACs) and DNA methyl-binding domain proteins (MBDs) (Robison AJ, 2011). Methyl-CpG-binding protein 2 (MeCP2) recruits HDACs to methylated DNA, resulting in the removal of acetyl groups from the histones, preventing transcription (Bergman Y, 2009).
Fig. 1. Steps involved in histone modifications.
(A) The HAT is recruited by a specific DNA sequence and acetylates the histone tails. (B) The bromodomain in the SWI/SNF complex is attracted to the acetylated histone tails. The complex then disrupts the DNA’s rotational phasing, altering the contact between the DNA and the histone. (C) (1) ATPase/translocase binds to the DNA, (2) pulling the linker DNA towards the histone. (3) This forms a DNA loop that spreads through the DNA strand. (D) TFIID is able to bind to the promoter. This results in bending of the DNA strand, causing the modified nucleosome to slide to the next position.
The acetylation of histones is carried out by several histone-modifying enzymes, namely histone acetyltransferases (HATs). There is a clear link between histone acetylation and chromatin remodeling, an ATP-dependent process in which a remodeling complex disrupts the contact between the DNA and the histones by altering its rotational phasing, allowing DNA-binding proteins to access the DNA (Vignali M, 2000). SWI/SNF is one of the widely studied remodeling complexes, which contains a bromodomain in the C-terminal region that is attracted to the histones’ acetylated lysines (Step C) (Zeng L, 2002). While the mechanism by which the complex remodels chromatin is still being explored, it is believed that remodeler ATPases anchor the nucleosome while an ATPase domain involved in translocation binds to the DNA. This ATPase/translocase domain then pulls DNA from the linker towards the histones (Step C1), leaving a loop of DNA around the nucleosome (Step C2). The transient DNA loop disseminates around the nucleosome and disrupts the histone-DNA contacts, eventually replacing them on the edge of the loop (Step C3) (Clapier CR, 2009). This process results in nucleosome repositioning leaving the DNA sequences accessible for transcription (Chatterjee N, 2011; Segal E, 2009).
Epigenetic changes are especially important during infection with retroviruses, such as human T-cell leukemia virus type 1 (HTLV-1) and HIV-1, because they remain integrated in the host’s genome. The activity of both the HTLV-1 and HIV-1 promoters, called long terminal repeat (LTR) are affected by epigenetic changes. The processes of viral attachment to a susceptible cell, uncoating, reverse transcription, and integration for HIV-1 and HTLV-1 are similar in that once they bind to a susceptible cell and the RNA virus is released into the cytoplasm, reverse transcriptase acts to convert the RNA virus into a DNA provirus. These proviruses, like eukaryotic DNA, are coiled with histone proteins to form a nucleosome (Elgin, 1990), and are packaged in a pre-integration complex consisting of integrase, host and viral proteins and transcription factors, and thus is capable of functioning as a surrogate cellular transcriptional unit (Shah S, 2010). Gene expression following proviral integration is a crucial stage in the viral life cycle, which depends on the transactivating proteins Tax in the case of HTLV-1 (Kiyokawa et al., 1984; Sodroski et al., 1984), or Tat in the case of HIV-1, and specific transcription factors (TFs) (Matsuoka and Jeang, 2007; Pandya et al., 2007). Studies by others and us highlight the importance of cellular TFs in Tat- or Tax-mediated LTR activation (Adya and Giam, 1995; Bantignies et al., 1996; Bodor et al., 1995; Kashanchi et al., 1998; Kwok et al., 1996; Li L, 2011; Rahman S, 2012), and the ability of Tat/Tax to interact with these factors independently (Berkhout B, 1992; Deng L, 2001; Harrod et al., 2000; Jiang et al., 1999; Macian F, 1999; Okada and Jeang, 2002; Scoggin et al., 2001). Our previous review (Aliya N, 2012) provides a comprehensive account of the epigenetics of HTLV-1 infection. Thus far there is no report about the effect of any drugs of abuse on HTLV-1 promoter; therefore, we will base our discussion on the HIV-1 LTR (Rafati H, 2011; Treand C, 2006), which is known to be affected by drugs of abuse (Molina PE, 2011; Reynolds JL, 2006). We will also elaborate on the epigenetic effects among overlapping populations of virus infected- and individual-drug abusers.
Epigenetics of HIV-1 infection
The integration of HIV-1 into the host’s genome is controlled by several epigenetic factors, and therefore is more likely to occur in active transcription units (Lewinski MK, 2005). Histone 3 (H3) acetylation, H4 acetylation, and H3 K4 methylation have been shown to result in higher instances of HIV-1 integration, while H3 K27 trimethylation and DNA CpG methylation prevent integration through transcription inhibition. However, further investigation into the biochemistry of these phenomena is needed (Pandhare J, 2011).
As stated previously, once integrated, HIV-1 can remain transcriptionally quiescent in the host’s resting CD4+ cells during highly active anti-retroviral therapy (HAART), preventing the virus from being fully eliminated from this reservoir. This latent stage is achieved through transcriptional shutdown and the formation of restrictive chromatin at the LTR. The transcription initiation factor NF-κB is necessary to reactivate HIV-1, although other methods have been discovered (reviewed in (Shah S, 2010)). CpG methylation of the viral promoter at the 5′ LTR is known to silence many retroviruses, such as HIV-1 (Kauder SE, 2009). Briefly, in HIV-1, MBD2 is recruited to the methylated CpG island on the promoter, which aids in transcriptional silencing through the recruitment of other factors, such as the nucleosome remodeling and histone deacetylation (NuRD) complex. Blocking methylation results in decreased recruitment of MBD2 and HDAC2, and thus, greater gene expression. Selective hypermethylation observed in CpG sites of the 5′ LTR implies that these sites are involved in activation and deactivation of the virus (Kauder SE, 2009). While demethylation of the 5′ LTR has been observed at specific CpG sites during tumor necrosis factor-α (TNF-α)-induced reactivation, CpG methylation at 3′ LTR remains unaffected in presence of TNF-α (Ishida T, 2006). Therefore, CpG methylation at specific sites on the LTR helps the virus maintain its latency by hindering its reactivation (Blazkova J, 2009) and potentially inducing viral latency (Pandhare J, 2011). This understanding has lead to the use of a DNA methyltransferase inhibitor, 5-aza-2′deoxycytidine, that interacts with TNF-α thereby allowing reactivation of latent HIV and increasing viral clearance during highly active antiretroviral therapy (Deng L, 2001).
Important epigenetic changes occur during the virus’s integration into the host genome as a result of the chromatin remodeling that occurs in order to facilitate the efficient viral transcript production of RNA Polymerase II (Easley R, 2009). Access to the LTR is controlled by five nucleosomes, nuc-0 to nuc-4, located on the 5′ LTR that help to regulate gene expression of the virus. In a condensed chromatin state, nucleosome-1 (nuc-1), located at the −2 to +140 position, inhibits the promoter when HIV-1 is silent. Therefore, it is necessary for the nucleosome to be remodeled before transcription can occur. There exists a nucleosome-free region from −255 to −3, which is extended in the presence of TNF-α and certain HDAC inhibitors. This is further shown by the histone acetylation that occurs at nucleosome-1 when treated with Trichostatin A (TSA), a histone deacetylase inhibitor (Easley R, 2009).
Another mechanism by which HIV-1 maintains latency within host cells is through the actions of HDACs. HDACs are recruited to the LTR by cellular DNA binding proteins and are then able to deacetylate lysine residues of core histones, repressing transcription through this covalent modification. Transcriptionally inactive proviruses are also more likely to contain the histone methyltransferase Suv39H1, histone H3 methylated on K9, repressive heterochromatin protein 1 (HP1) proteins, HP1-α (a methylated histone binding protein), and trimethylated histone H3 with modification on lysines 9 and 27. Suv39H1 also interacts with COUP-TF interacting protein 2 (CTIP2) to increase H3K9 methylation (Imai K, 2010). In addition, CTIP2 aids in the recruitment of a chromatin-modifying complex that leaves the promoter in a heterochromatic state within microglial cells, preventing viral transcription. This occurs through interaction with the transcription factor, Sp1, and with Tat, a viral trans-activator protein that is then relocated to the areas of the chromatin that are transcriptionally inactive. The aforementioned epigenetic changes are thought to block the initiation of HIV transcription, stopping the Tat-mediated regulatory circuit (Pearson R, 2008).
There are regulatory nucleosomes situated around the HIV transcription start site (TSS) that are acetylated when the promoter is activated. While Tat recruits ATP-dependent chromatin remodeling complexes and HATs to the LTR when the virus is active, the chromatin serves as a barrier to polymerase progression in the latent state. Latent infection is often observed when the provirus is integrated into regions that are rich in heterochromatin. Examples include proviruses that are integrated into centromeric alphoid repeats or gene deserts (Lewinski MK, 2005). Silent HIV promoters are also found in the presence of HDACs, heterochromatin proteins, CpG methylation, and methyltransferases, further proving the effect that epigenetic factors have on HIV expression. In fact, latent HIV has a tendency to be depressed in the absence of cotranscriptional chromatin reassembly factors (CRFs). The absence of CRFs results in greater chromatin accessibility due to inadequate nucleosome architecture (Gallastegui E, 2011).
The HIV LTR is generally located in a strategic position relative to certain regulatory elements. The virus has been found to preferentially integrate into euchromatic regions where genes are transcriptionally active (Lewinski MK, 2005), allowing for greater accessibility to necessary transcription factors prior to tat-mediated transcription and RNA polymerase. The integrated viral genome is subject to the same regulatory controls that effect human genes and therefore, if the HIV-1 promoter is activated, the nucleosomes associated with the LTR are quickly disrupted allowing the necessary factors access to the LTR, and vice versa when the promoter is silenced. One region of the LTR promoter that binds to the transcription factor, USF, tends not to contain any nucleosomes despite the fact that nucleosomes would be able to fit in that space. This is most likely due to the presence of USF, which is known to cause promoter bending. The DNA bending leaves the region inhospitable to nucleosomes (Marzio G, 1999). Exposure to drugs that prevent HDAC activity results in viral activation due to the resulting increase in histone acetylation, illustrating the extent to which epigenetic effects dictate the lifecycle of the virus.
Epigenetics of drug abuse and its impact on HIV-1 infection
As the lifespan of the average HIV-sufferer increases, so does the prevalence of HIV-associated neurocognitive disorders (HAND). They are believed to be caused by the secretion of viral proteins that interact with neurons or by the tendency of infected glia to secrete inflammatory molecules (Regan PM, 2012). Given the fact that drug abuse is known to cause an increase in the incidence of these symptoms (Regan PM, 2012), it would be worth considering whether or not these drugs are causing an increase in the neurons’ susceptibility to the disease or the immune cells’ tendency to release inflammatory factors in the brain. In addition to impact of infection at the gene level, drug addiction is in itself responsible for epigenetic changes that involve transcriptional irregularities, such as overexpression of ΔFOSB in the brain reward regions, rendering individuals more prone to drug addiction and modifying neuronal plasticity (Robison AJ, 2011; Wong CCY, 2011). Drug addiction is also believed to activate the brain’s reward circuitry, with greater dopaminergic transmission between the ventral tegmental area (VTA) and nucleus accumbens (NAc), leaving individuals with a decreased sensitivity to pleasure in the absence of the drug. Specifically, regulation of DNA methyltransferases (to be discussed later) aid in the regulation of hippocampal plasticity (Wong CCY, 2011). Given the complexity of these effects, a more in depth look at the role of epigenetics that is further impacted upon by drug use in the development of HAND is necessary. A detailed discussion on the significance of this phenomenon among the most commonly abused drugs is provided below.
Cannabinoids
Cannabis sativa, also known as marijuana, is one of the most commonly used drugs of abuse. It comprises over sixty cannabinoids, including cannabidiol, cannabinol, and Δ-9 tetrahydrocannabinol (Δ-9-THC), which is the primary psychoactive ingredient. The mediator of the neuro-behavioral effects observed with the use of cannabis is the CB1 receptor with operates primarily in the brain. The homologous CB2 receptor functions in the periphery and is found on the surface of immune cells, including T-cells and B-cells. Thus, HIV-infected patients using cannabis often experience epigenetic effects from immunomodulation, contributing to changes in HIV-1 progression (Molina PE, 2011).
Epigenetic mechanisms associated with cannabinoid use
The transcription factor ΔFosB, which is largely accepted as being an important component in the development of drug addiction, is generally activated in the presence of drugs of abuse, although the extent and location of its activity depends on the drug. Studies conducted on the effect of Δ-9-THC on ΔFosB activity involved the twice-daily injection of Δ-9-THC into mice (Perrotti LI, 2008), which resulted in ΔFosB induction prominently in the prefrontal cortex (Miller EK, 2000), the region of the brain known for decision-making, planning, and impulse control, implying that cognitive performance can most likely be affected. Besides this, no significant increase in ΔFosB activity was observed in the nuclear accumbens shell or dorsal striatum, which are instrumental in impulsivity and the ability to learn responses, respectively (Basar K, 2010). Interestingly, increases were observed in these areas as a result of other drugs of abuse, namely cocaine, morphine, and ethanol.
Studies on the impact of cannabinoids on epigenetic changes in fertility have been conducted, where cannabinoid receptor 1 (CNR1) null mutant mice displayed higher histone retention in germ cells compared to the wild type. CNR1 is important in spermiogenesis and helps to control the chromatin quality in the resulting sperm. The activation of this receptor mediates chromatin remodeling in spermatids by regulating transition protein 2 (Tnp2) levels or by increasing the level of histone displacement. Due to the fact that the use of exocannabinoids such as marijuana has been shown to cause lower fertility (Chioccarelli T, 2010), it can be assumed that these can have the same observed effect as the endocannabinoids described in this study. Marijuana use is believed to deregulate the endogenous cannabinoid receptor system, which could potentially inhibit histone displacement during spermiogenesis, resulting in poor sperm quality, demonstrating the epigenetic ramifications of inactivating this receptor (Chioccarelli T, 2010).
Interactions between cannabinoids and HIV-1
Cannabis use is common within the HIV-infected population because it is often considered a relatively low-risk drug useful for appetite stimulation in AIDS patients. In fact, Dronabinol, a synthetic form of Δ-9-THC, has been approved by the FDA to treat HIV-associated anorexia (ElSohly MA, 2001). In addition, one longitudinal study found no adverse effects of cannabis use on the CD4+ T-cell counts of HIV-positive men over the course of eleven years (Chao C, 2008).
The potential effects of cannabis use in HIV-1 infection have led to several studies albeit with conflicting observations. Cannabinoids have been shown to have an impact on cytokine production as well as lymphocyte function and survival (Molina PE, 2011). They also utilize a CB2 receptor-dependent pathway to regulate the balance and activation of human Th1/Th2 (T-helper) cells (Yuan M, 2002). HIV replication leads to the activation of the immune system, which causes an increase in pro-inflammatory cytokines, lymphocyte proliferation, and lymphocyte apoptosis (Meyaard L, 1992). Molina et al. explored the effects of a month’s worth of regular injection of Δ-9-THC in rhesus macaques prior to simian immunodeficiency virus (SIV) infection (Molina PE, 2010), one of the best animal models for HIV studies. When plasma Δ-9-THC levels of the monkeys were maintained at 22 ng/mL, it resulted in decreased plasma and cerebrospinal fluid viral load as well as a lower level of tissue inflammation leading to a greater survival rate upon SIV infection. The ability of activation of the cannabinoid receptor to help suppress infection can be further seen in microglia where viral replication was inhibited by a synthetic nonselective CB1/CB2 receptor agonist called WIN55,212-2 (Peterson PK, 2004; Rock RB, 2007). In yet another study using SIV-infected rhesus macaques treated with Δ-9-THC, lower plasma and brain viral loads were observed compared to controls (Molina PE, 2011). These Δ-9-THC treated SIV-infected animals had lower levels of episomal SIV 2-long terminal repeat (2-LTR) DNA circles. These animals also displayed changes in the expression of 153 genes with several genes implicated in inflammation and HIV infectivity modulation. The decreases in tissue inflammation that have been observed with cannabis use in these studies, seemingly contribute to the decreases in virus replication.
In contrast to these studies, it has been reported that cannabinoid receptor stimulation during HIV-1 infection can contribute to disease spread and pathogenesis. One study observed an increase in CCR5 and CXCR4 expression in peripheral blood leukocytes (PBLs) after ten days of Δ-9-THC administration (10 mg/kg) in immunodeficient mice along with an increase in HIV-infected cells (Roth MD, 2005). It has also been shown that when MT-2 cells were cultured with cannabinoid agonists and infected with HIV, there is observably greater syncytia formation compared to cells exposed to HIV alone, implying that stimulation of the CB2 increased HIV-1 cytopathology and infection (Noe SN, 1998). Thus, regardless of consensus on the role of cannabinoid receptor stimulation on HIV-1 infection, it is certain that epigenetic changes translate into apparent changes in disease progression as described above.
Evidence of cannabinoids possessing the ability to elicit antiviral responses that manifest through DNA methylation is also present. DNA methylation, an epigenetic mechanism that occurs as a result of exposure to cannabinoids, has been studied by a few groups, but the full mechanism has not yet been established. DNA methyltransferase (DNMT) activity has been shown to be increased when the p38 and p42/44 mitogen-activated protein kinase (MAPK) pathways are activated by the CB1 receptor (Paradisi A, 2008). This results in high levels of DNA methylation and, consequently, the inhibition of gene expression. Studies conducted on Δ-9-THC- exposed, SIV-infected animals showed that at least 50% of the differentially expressed genes that were found are involved in DNA methylation (Molina PE, 2011). Some of these hypermethylated genes, such as CXCR4 and HIV-1 enhancer binding protein (HIVEP1), play a role in the entry, production, and integration of the virus. These genes also affect inflammation and viral kinetics in the brain through the hypermethylation of CCAAT enhancer binding protein delta (C/EBPδ), which is involved in the mediation of IL-1 activation of NF-κB. Δ-9-THC exposure also affected the expression of miRNA in SIV-infected animals. Differentially expressed miRNA was found in CD4+ T-cells, intestinal mucosa, and the brain. The miR-21, which is generally upregulated during inflammatory conditions, was found to be downregulated in the brains of Δ-9-THC exposed, SIV-infected animals (Molina PE, 2011).
Opioids
Opioids are a class of drugs that act on the opioid receptors and induce symptoms such as respiratory depression, vomiting, depression of consciousness, and low blood pressure and pulse rate (Bateman DN, 2012). Opioid users traditionally administer the drugs intravenously or through inhalation due to the rapid high that results. Heroin, a very common street drug that raises the risk of HIV transmission due to the possibility of sharing infected needles, breaks down into the active metabolite morphine in the body. Other examples of opioids that are abused include codeine, oxycodone, and methadone (Bateman DN, 2012). Due to the variety of opiates that an infected individual can take, it is important to study any epigenetic mechanisms that may affect disease progression in this group of drug users.
Epigenetic mechanisms associated with opioid use
Among all opioids, morphine has been the best studied with respect to its potential epigenetic effects. The μ-opioid receptor (MOR), which is primarily controlled by its proximal promoter during neuronal expression, recruits HDAC1 and HDAC2. Additionally, MOR gene transcription increases in the presence of TSA, an HDAC inhibitor. This is thought to be due to the fact that TSA acetylates the local nucleosomes, allowing access to the necessary transcription factors (Lin Y, 2008). Some studies have been performed regarding the role that HDACs play in morphine-induced conditioned place preference (CPP), which is the tendency for addicted animals to prefer locations associated with drug administration. It was found that HDAC inhibition can aid in morphine-induced CPP extinction and attenuate reinstatement behavior, emphasizing the role that HDAC activity plays in the development of addiction (Wang R, 2010).
Interactions between opioids and HIV-1
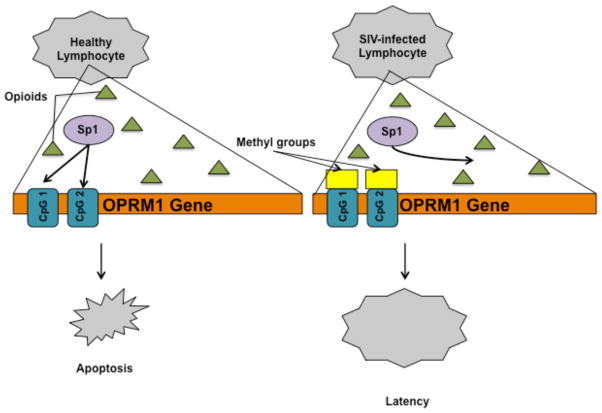
Opioid abuse has long been observed to exacerbate HIV-1 infection, particularly increasing the chances of neurological symptoms, due to its immunosuppressive effects, and constitutive production of proinflammatory cytokines that are known to alter blood-brain barrier (BBB) permeability, as well as HIV-1-opioid interactions that cause inflammation and glial dysfunction (Strazza M, 2011). More specifically, it is thought that opioids amplify the effects of HIV-1 neurotoxicity by promoting apoptosis through the MOR (Hauser KF, 2006). The OPRM1 (opioid receptor, mu 1) gene regulates MOR and is subject to many epigenetic changes. One such epigenetic mechanism is regulation of OPRM1 by Methyl-CpG-binding protein 2 (MeCP2) using Brg1, a chromatin-remodeling factor, and DNA methyltransferase 1 (DNMT1) (Hwang CK, 2009). The promoter’s CpG islands are methylated, allowing for MeCP2 binding. This leads to the recruitment of repressor proteins that facilitate the deacetylation of the relevant histones. It has already been established that neuronal cells that do not express the OPRM1 promoter are methylated, while OPRM1-expressing neuronal cells are not, confirming the importance of DNA methylation in the activation of this gene (Andria ML, 1999). In addition, former heroin addicts have been found to display hypermethylation of the OPRM1 promoter in the peripheral lymphocytes, regardless of whether or not they were HIV-infected, illustrating the epigenetic ramifications of long-term use (Nielsen DA, 2009). SIV is further known to affect the OPRM1 promoter through DNA methylation in lymphocytes, although the exact mechanism is poorly understood (Liu H, 2009). SIV infection increases DNMT activity, resulting in a methylated promoter, which blocks the initiation of the gene by preventing the Specificity protein 1 (Sp1) transcription factor from binding to the promoter at CpG island 1 and CpG island 2 (Figure 2). It is believed that this activity increases the viability of the infected lymphocytes in the presence of morphine exposure. This phenomenon has only been observed in the early stages of infection, and it is believed that this allows the virus to remain latent in the cells for a longer period of time. Histone acetylation also plays an important role in the epigenetic effects of opioid use as HDAC1 and HDAC2 interact with the OPRM1 promoter in neuronal NMB (neuroblastoma) cells. Additionally, TSA has also been found to impact OPRM1 gene expression, opening avenues for research on the effects that opioids have on chromatin accessibility (Lin Y, 2008).
Fig. 2. Epigenetic changes to the OPRM1 promoter.<.
br>DNMT activity increases upon the early stages of SIV infection, methylating the OPRM1 promoter at CpG island 1 and CpG island 2. This prevents the Sp1 transcription factor from initiating the gene, allowing the virus to remain latent in the lymphocytes in the presence of morphine.
Chronic morphine use has been shown to inhibit IL-2 secretion by activated T-cells via histone acetylation and trimethylation (Wang J, 2007) and reducing CpG dinucleotide demethylation in the IL-2 promoter, thereby suggesting a strong link between morphine and changes in immune function. IL-2 therapy has been shown to aid in the survival of CD4+ T-cells during HIV infection (Kovacs JA, 2005), meaning that a reduction in IL-2 would have detrimental effects on these patients. An increase in cAMP levels due to morphine exposure, leading to the upregulation of inducible cAMP early repressor (ICER) and cAMP response element modulator (CREM), has been shown to result in down regulation of CREB in activated T-cells (Wang J, 2007). This causes CPB/p300, a chromatin-remodeling complex, to uncouple and prevent IL-2 transcription. A decrease in acetylated H3 and H4 binding, as well as reduced trimethylated H3K4 binding with regard to the IL-2 promoter has also been observed (Kovacs JA, 2005).
Cocaine
Cocaine is an extremely addictive psychomotor stimulant that acts by inhibiting monoamine re-uptake by the presynaptic terminals. This extends the duration of their local anesthetic and vasoconstricting effects. Side effects include euphoria, stroke, ventricular arrhythmias, acute myocardial infarction, and renal and intestinal infarctions. It also hinders heat perception, leading to thermoregulatory adjustment impairment. Additionally, chronic use can lead the nasal septum to perforate. Cocaine can be taken intranasally, orally, or intravenously as it is a water-soluble powder (Vale A, 2007). Intravenous injection leaves the cocaine users vulnerable to HIV infection due to the risk of using unsterile needles.
Epigenetic mechanisms associated with cocaine use
Drug-induced alterations in the levels of histone acetylation in the brain are thought to play a role in drug addiction. While cocaine use often leads to greater H3 and H4 acetylation in the nucleus accumbens (NAc), some genes display less histone acetylation after chronic cocaine exposure (Robison AJ, 2011). The reason for this discrepancy is unknown, and further research is required. Various isoforms of HDAC have been shown to yield different responses in drug-exposed mice.
In fact, when animals were injected with Trichostatin A (TSA), a pan-HDAC inhibitor, in the NAc shell and then exposed to cocaine, they displayed elevated self-administration rates and thus a greater level of addiction (Wang L, 2010). It was also found that these animals were willing to work harder for higher injection doses compared to the control animals. However, these results were unique to the NAc shell and were not observed when TSA was injected in to the NAc core or the medial prefrontal cortex. Further investigation confirmed that these observations were a result of increased H3 and H4 histone acetylation leading to greater expression in the genes related to addictive behavior.
Chromatin immunoprecipitation studies were conducted on the BDNF (brain-derived neurotrophic factor) and Cdk5 (cyclin-dependent kinase 5) genes in order to determine what effect cocaine use has on their chromatin accessibility (Kumar A, 2005). BDNF plays an important role in synaptic plasticity, increasing the tendency to respond to cues that are related to natural rewards. Its continued expression during periods of withdrawal is thought to be related to the persistence of the cravings that define addiction. Both genes are known to be activated in response to chronic cocaine exposure. In this study, with chronic cocaine exposure, striking increases in H3 acetylation of the BDNF promoter were observed and persisted for a week after withdrawal. This is consistent with previous findings that BDNF protein levels increase during cocaine withdrawal (Grimm JW, 2003). Although Cdk5 did not display the same persistence, another study has shown Cdk5 to play an important role in the mediation of dendritic arborization outgrowth in striatal neurons in response to chronic cocaine exposure (Kumar A, 2005). Chromatin immunoprecipitation assays confirmed chronic cocaine exposure increases ΔFOSB/Cdk5 gene promoter association, elucidating the previously unknown mechanism behind this observed phenomenon. Since the ΔFOSB promoter is already known to impact brain plasticity during addiction (Kumar A, 2005), the association of Cdk5 with ΔFOSB strengthens the finding that Cdk5 affects the striatal neurons in this manner.
Besides BDNF, tyrosine hydroxylase has been implicated as an important factor in cocaine-induced reward behavior. The mRNA levels of tyrosine hydroxylase were found to increase in the ventral midbrain upon exposure to a D1 agonist and the HDAC inhibitor, sodium butyrate, in cocaine-treated mice. This was determined to be the result of chromatin remodeling through H3 acetylation at the tyrosine hydroxylase promoter and the BDNF promoter of these regions (Schroeder FA, 2008).
One of the most widely studied transcription factors implicated in the epigenetic changes caused by drug abuse is ΔFOSB, the expression of which is increased in the chronic presence of drugs of abuse. This increase is most noticeable in the NAc and dorsal striatum in D1 dopamine receptor expressing medium spiny neurons (Nestler EJ, 2008). The overexpression of ΔFOSB has been shown to cause drug self-administration and increased impulsive behavior during withdrawal in mice. This is due to the large impact that ΔFOSB has on neuronal plasticity as a result of its regulation of various proteins associated with dendritic spine architecture, such as microtubule associated proteins, actin-related proteins, synaptotagmin, activity-regulated cytoskeleton-associated protein, kinesin, and cyclin-dependent kinase 5. ΔFOSB is also involved in the regulation of proteins that are vital to glutamatergic synaptic function and plasticity, providing further evidence that this transcription factor has a large effect on the plasticity that is thought to contribute to drug addiction (Robison AJ, 2011). One known mechanism by which ΔFOSB can have such a wide range of effects is by controlling nuclear factor-κB (NF-κB) levels in the NAc. NF-κB levels have been seen to increase in the presence of cocaine, contributing to the increase in medium spiny neuron dendritic spine density that sensitizes the subject to the drug’s effects (Russo SJ, 2009).
Histone modifications at the cFos and FosB promoters have been studied due to their established responses to cocaine intake. Chromatin immunoprecipitation studies have shown that acute cocaine use results in H4 acetylation and H3 phosphoacetylation at the cFos promoter (Kumar A, 2005). These effects were observed 30 minutes after exposure and subsided after three hours with no relapse. However, chronic exposure to cocaine yielded different results. No changes in H4 acetylation or H3 phosphoacetylation were observed, showing that the induction of the cFos promoter is desensitized to the drug. While the response of the FosB promoter to acute cocaine exposure was very similar to that of the cFos promoter, its response to chronic cocaine ingestion differed due to an increase in H3 acetylation. This difference was expected because FosB does not display the same level of desensitization that cFos does in protein studies. The same results were found when the rats being tested were allowed to self-administer the drug, showing that these epigenetic changes play a role in cocaine addiction.
FosB accumulation has been established in chronic cocaine exposure implicating its role in addiction-related behavior and the sensitization of animals to the drug response. CREB-binding protein (CBP) is a HAT that acetylates certain lysine residues and is recruited by CREB during cAMP-dependent PKA phosphorylation (Levine AA, 2005). CBP recruitment to the FosB promoter and H4 acetylation were found to increase in rodents that were injected with cocaine (Levine AA, 2005). In addition, suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor, was shown to induce an increase in FosB expression after cocaine exposure. No such effect was observed in animals that were not treated with cocaine, reflecting the lack of FosB accumulation. The increase in FosB observed in the cocaine-treated animals can be explained by SAHA’s inhibition of the deacetylation that would normally partially negate the effects of CBP.
Chronic cocaine use also suppresses striatal multiple myocyte-specific enhancer factor 2 (MEF-2) activity by way of D1 receptor cAMP dependent inhibition of calcineurin. MEF-2 binds to DNA recruiting coactivators such as p300 or corepressors including HDACs. It has recently been shown to regulate excitatory synapses suggesting the possibility for its role in addiction. MEF-2 is a substrate of Cdk5, which displays increased activity after chronic cocaine use. However, the exact role of MEF-2 in addiction is ambiguous because it causes an increase in medium spiny neuron dendritic spine numbers while also decreasing the subject’s behavioral sensitization to cocaine in some cases (Pulipparachuravil, 2008; Robison AJ, 2011). Recently, we identified a novel role for MEF-2 in the regulation of HTLV-1 LTR and subsequent viral infection (unpublished observations).
DNA methylation also seems to play an important role in cocaine addiction. Methyl CpG binding protein 2 (MeCP2) displays increased expression in the NAc and dorsal striatum in the presence of cocaine (Robison AJ, 2011). Both acute and chronic cocaine exposure have been found to cause hypermethylation of the PP1c (protein phosphatase-1 catalytic subunit) promoter and hypomethylation at the fosB promoter. This was found in conjunction with increased DNMT3A and DNMT3B expression in the NAc. It is believed that MeCP2 aids in the recruitment of HDACs and histone methyltransferases to methylated PP1c promoters, resulting in increased CBP recruitment, and therefore, histone acetylation (Anier K, 2010).
Interactions between cocaine and HIV-1
Cocaine use plays a role in the neuropathogenesis of HIV-1 and can accelerate the development of HIVE (Dhillon NK, 2007). Cocaine increases the release of cytokines in immunoeffector cells and impairs macrophage and CD4+ T-cell function. The increased expression of inflammatory cytokines and endothelial adhesion molecules caused by cocaine leads to greater viral invasion and a higher incidence of cerebrovascular complications. Cocaine also increases interleukin-10 (IL-10) expression, which in some cases has been shown to promote HIV-1 replication in monocyte-derived macrophages in the presence of TNF-α (Strazza M, 2011), while in other studies IL-10 has been linked with inhibiting HIV-1 transcription (Kauder SE, 2009). Cocaine use is known to enhance leukocyte migration as evidenced by the increased infiltration of mononuclear blood cells into the blood vessels of the brain (Gan X, 1999). Cocaine promotes the activation of the virus in latently infected promonocytic U1 cells, as evidenced by elevated levels of viral p24 protein. It activates the LTR through the mediation of NF-κB, which is essential for the transcription of the virus. The interplay between cocaine and HIV-1 is reviewed recently (Buch S, 2012), providing insight into the myriad of ways that the abuse of this drug aids in the progression of the disease.
HIV-1 is known to affect normal human astrocytes (NHA) when it enters the central nervous system (CNS), which usually occurs during the few days following infection. HIV replication in the CNS primarily occurs in the macrophage and microglial cells, although infection of the astrocytes has also been recorded. This leads to neuroimmunopathogenesis as the infected astrocytes spread the infection and cause damage to the surrounding cells. Cocaine use has been shown to enhance HIV-1 replication in the NHAs through the upregulated HIV-LTR-R/U5 region (Reynolds JL, 2006). Cocaine-exposed, HIV-infected NHAs also exhibited upregulated p24 antigen levels, indicative of high viral protein content. It is worth noting that viral latency has been found to be correlated with cognitive defects, neuroinflammatory changes, and neurodegenerative alterations, indicating that viral load does not play as large a role in the development of neuroAIDS as previously believed (Desplats P, 2013). The puzzling prevalence of HAND in aviremic patients can be explained by a deregulation of BCLL1B, a transcriptional regulator involved in provirus silencing. It is proposed that this protein increases HIV latency while also impacting inflammation-related gene expression. BCLL1B recruits a multienzymatic complex to the LTR resulting in a heterochromatic region. The effect that drugs of abuse have on this process have not yet been studied, but given the clear impact that cocaine and other drugs have on neuroinflammation, such research could yield fruitful results.
Cocaine exposure does affect HIV-1 replication in CD4+ T-cells and in peripheral blood mononuclear cells (Mantri CK, 2012). It is believed that it impacts the entry of the virus, as well as the post entry steps that direct the replication of the virus. The proposed mechanism involves the downregulation of miR-125b, a microRNA involved in the differentiation of CD4+ T-cells, by cocaine, which enhances HIV replication. However, there is more work to be done in order to determine if there are any epigenetic mechanisms underlying these observed phenomena.
Other miscellaneous drugs and their impact on epigenetics and/or HIV-1
Alcoholism affects millions of Americans and their families. 30% of adults have engaged in alcohol abuse at least once, and the economic burden of alcoholism is around $400 billion annually (Alcoholism, 2011). Alcohol abuse symptoms often have epigenetic causes. The anxiolytic effects of acute alcohol ingestion occur in conjunction with the downregulation of HDAC activity and upregulation of H3/H4 acetylation and CBP. This process results in greater expression of Neuropeptide Y (NPY), an endogenous anxiolytic compound that has been implicated in the effects of ethanol, in the amygdala. The withdrawal effects that occur following chronic alcohol use have the opposite effect. Exposure to TSA following withdrawal from chronic alcohol use prevents these effects, leaving the amygdala with close to normal levels of acetylation (Pandey SC, 2008). Alcohol use is common in the HIV-infected population, as evidenced by the fact that they are twice as likely to have a history of alcohol use than the general population and that almost half of HIV carriers report a history of alcohol abuse (Lefevre F, 1995). Chronic alcohol consumption has been found to increase viral replication and upregulate proinflammatory cytokine production in HIV-positive patients (Amedee AM, 2010). Individuals afflicted with both HIV and alcoholism show far greater brain abnormalities compared to those who are victim to one condition alone (Pfefferbaum A, 2010). Cognitive deficiencies are also observed to occur more frequently within this population. However, not much is known about the epigenetic components of these observed phenomena.
Amphetamines are another type of commonly abused drug and are considered to be stimulants, much like cocaine, but they remain in the CNS longer, increasing the duration of their effects (Drug Enforcement Administration, 2011). Amphetamines are also known to affect Th1/Th2 production in the murine model of AIDS (Cabral GA, 2006). They impact epigenetics by mechanisms similar to those employed by cocaine. Acute exposure of amphetamines causes the induction of the c-fos and fosb genes in the NAc, which results in increased H4 acetylation on those gene promoters (Renthal W, 2009). Recently, a connection between methamphetamine use, high-risk sexual behavior, and HIV transmission has been reported (Kim W, 2012). Treatment with increasing doses of this drug correlated with altered glutamatergic function in the dorsal striatum of rats (Kauer JA, 2007; Marban C, 2007). Epigenetic effects included an increase in recruitment of corepressor of RE1 silencing transcription (CoREST), HDAC2, MeCP2, and sirtuin 2 (SIRT2) causing a downregulation of GluA1 and GluA2 mRNA levels upon methamphetamine treatment. Methamphetamine-induced increases in expression of genes such as Crf (major regulator of the pituitary-adrenal axis) in the NAc are associated with increases in histone H4K5 and H4K8 acetylation whereas decreases in gene expression (ex. CCK; gene that encodes cholecystokinin neurotransmitter) correlate with hypoacetylation of histones (Yao H, 2012). These changes are not effected immediately; rather they occur secondarily to methamphetamine-induced increases in ATF2 expression, resulting in altered gene expression in the NAc. In an HIV-1 transgenic rat model, oxidative stress caused by the combined and individual effects of HIV viral protein and low dose repeated methamphetamine exposure showed alterations in glutathion (GSH; the most abundant antioxidant in the brain) content in both the thalamus and striatum in the brain depending on the type of exposure (du Chene I, 2007). In addition to this concerted effect by viral infection and drug use, exposure to both methamphetamine and supernatant from HIV infected monocyte-derived macrophages (MDM), dopaminergic neurons showed an increase in microRNA-9, -124 and let-7d, making the CNS neurons susceptible to drug tolerance and addiction (Tyagi M, 2007). Individuals infected with HIV who use methamphetamines exhibit greater interneuron loss due to HIVE (Krebs FC, 2002), as well as higher levels of reverse transcriptase activity in macrophages (Alexaki A, 2007). Thus, the abuse of alcohol and/or amphetamines leave HIV-infected individuals vulnerable to exacerbated symptoms, many of which can likely be owed to epigenetic changes to the retroviral promoter, although the precise mechanisms are still being studied.
Impact of other comorbidities
HIV-infected individuals with a history of drug abuse display a high rate of hepatitis C virus (HCV) coinfection (Letendre S, 2007). HCV on its own is associated with cognitive decline, but when paired with HIV, the symptoms can be even more extreme. For example, HCV exacerbates the cognitive deficits observed in HIV-infected individuals who also use methamphetamines, proving that the viruses do interact in the CNS. One suggested mechanism involves the blocking of astroglial glutamate clearance, causing excitotoxicity. Although the epigenetic effects of HCV in the brain have not been studies in depth, HCV is known to induce epigenetic changes that results in the development of hepatocellular carcinoma (Feng Q, 2013). It is possible that HCV causes other changes at the histone level that impact the development of HAND in a manner similar to various drugs of abuse.
Mental illness is another risk factor for HIV as approximately 3% of mentally ill individuals are infected with HIV compared to 0.3% of the general US population (Hammond E, 2007). According to data from HIV clinic at Johns Hopkins, 54% of patients had an Axis I disorder, and 74% presented with a substance use disorder. Neuropsychiatric disorders such as schizophrenia, anxiety disorders, and depression are well-established contributor to HAND (Power, 2009) and are also known to be associated with various epigenetic changes. Depression is associated with increased H3K27 methylation, while schizophrenia is associated with hypermethylation in GABA-containing neurons (Tsankova N, 2007). Such psychiatric disorders may also cause changes at the chromatin level that eventually contribute to the development of HAND.
Concluding notes
There is still a great deal of work to be done in order to elucidate the epigenetic interactions between the previously discussed drugs of abuse and the LTR promoter. The need for this research is obvious given the high incidence of drug use within the HIV-infected population. Extensive work has been done on the epigenetics of drug abuse within the context of addiction, proving that these drugs do in fact affect chromatin remodeling and DNA methylation. The next step is to investigate the extent to which these effects extend to the LTR, given that it is well known that there are many epigenetic factors that dictate the progression of HIV.
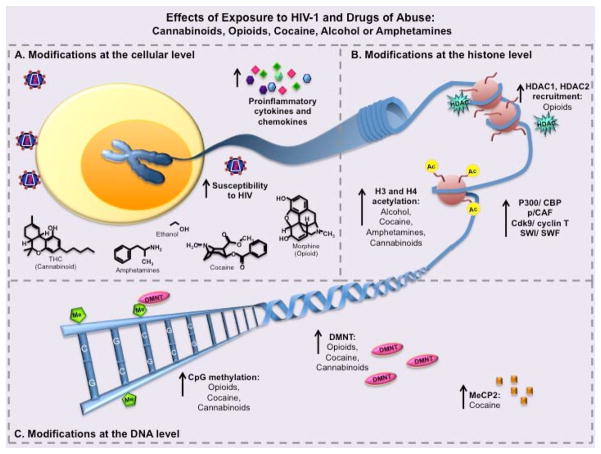
Drugs of abuse interact with HIV-1 in a variety of ways, many of them leading to similar end-point actions such as increased HIV-1 susceptibility and proinflammatory cytokines to increases in CpG methylation and histone modifications (summarized in Figure 3). Although these interactions are widely documented, there is uncertainty with respect to the epigenetic interactions between the drugs and the LTR. Given the heritability of certain epigenetic changes, it is possible that some of the modifications caused by the drug abuse in infected individuals could be passed on to the next generation, increasing their susceptibility to drug addiction. The relevance of this research is clear, and there is a great deal of evidence suggesting that there may be a larger link than previously believed. Studies conducted both in vitro and in vivo have confirmed that cannabinoids have an effect on HIV-1 in terms of DNA methylation and miRNA expression, highlighting the need for further investigation into the precise interactions that are occurring, especially with regard to the retroviral promoter. The implications of OPRM1 methylation during opioid abuse and its influence on HIV latency are vast and raise a lot of questions about whether there is direct effect on the viral promoter LTR as well. Given the extensive research on the epigenetic effects of cocaine use, it makes sense to explore the impact that these effects may have on the retroviral promoter, especially since cocaine use is known to spur HIV replication. Applying an animal model similar those used in cannabinoid studies to cocaine studies is sure to elucidate the nature of the epigenetic interactions between the drug and the virus, giving way to refining treatment strategies for drug abusing, HIV-infected populations. Treatment for drug abuse has long been considered a viable method of HIV prevention based on observations that users who are undergoing treatment experience lower rates of infection (Metzger DS, 1998). Understanding the mechanisms that cause changes in gene expression at the epigenetic level could pave the way for HMT inhibitors that could be used along with current treatments in order to combat the effects of drug abuse on the disease. The use of HMT inhibitors in HIV treatment has already been suggested (Imai K, 2010), but few studies have been conducted on the potential of this therapeutic option in the context of drug abuse. It may be beneficial to counter the deleterious effects of drug abuse on patients’ prognosis in addition to administering traditional HAART in the hopes that the severity of the disease could be reduced.
Fig. 3. Epigenetic effects of exposure to HIV-1 and drugs of abuse.
Cells in the brain or periphery that are exposed to HIV-1 and drugs of abuse (DoA) undergo a variety of epigenetic changes. Despite some of the differences in the pathways, many different DoA lead to similar end-point results in terms of epigenetic modifications. (A) Exposure of cells to HIV-1 and various drugs of abuse such as cannabinoids, opioids, cocaine, alcohol, or amphetamines result in increased susceptibility to HIV-1 viral infection, and increases in proinflammatory cytokines, which can also exacerbate disease. (B) This exposure can also lead to epigenetic changes at the histone level allowing for DNA and genes of interest to be more or less available to host transcription factors and transcriptional machinery. The interaction of HIV-1 and DoA generally lead to an increase in chromatin modifying factors such as P300/CBP, p/CAF, and SWI/SWF. Additionally, use of alcohol, cocaine, amphetamines, or cannabinoids can lead to histone H3 and H4 acetylation through various pathways. In contrast, signaling through MOR (μ-opioid receptor) by use of opioids can lead to HDAC1 and HDAC2 recruitment to the histones preventing gene transcription. (C) These same DoA can also cause epigenetic changes at the DNA level potentially resulting in increases in MeCP2 and DMNT (DNA methyltransferase) recruitment to the DNA, depending on cell type, and leading to CpG methylation and prevention of specific gene expression.
Using drugs such as cocaine, cannabinoids, and methamphetamine is known to exacerbate neurological deficiencies by increasing dopamine in the brain (Purohit V, 2011). It has also been suggested that the increased dopamine levels potentiate oxidative stress, contributing further to neuronal degeneration. However, there are many other risk factors that may cause an individual to develop HAND, such as coinfection with HCV, which can lead to greater neuropsychological impairment (Annoni J, 2011). Traditional therapies generally do not prevent the onset of HAND as up to 69% of HIV patients who have an undetectable plasma load still display mild cognitive defects. This is believed to be caused by continued neuroinflammation or neurodegenerative mechanisms similar to those observed in Parkinson’s and Alzheimer’s diseases. This shows that any epigenetic changes observed in these patients may be the result of any number of factors. However, it is still worth pursuing the impact that drugs of abuse have this pathology given the clear correlation between drug use and cognitive decline.
There is no consensus on the epigenetic effects of drug abuse in any context as no definitive causal relationships have been shown (Nielsen DA, 2013). As more studies are conducted, a correlation is almost sure to be confirmed. What is not clear is whether the observed epigenetic changes are caused by the drugs themselves or whether they simply signify a predisposition for drug use. This merely illustrates the need for further research on the matter. Even without knowing the causality, there are still observed epigenetic differences between users and nonusers that bear investigation. It is clear that drug abuse imparts epigenetic changes to the genome, and it is likely that similar effects are taking place on the HIV-1 LTR given the well-documented incidence of altered HIV replication in response to these drugs.
Acknowledgments
We wish to acknowledge United States Public Health Service/National Institutes of Health Grants wherein PJ was funded through NIAID R01 AI077414 and NCI R01 CA054559 while ZKK was funded through NIAID R21 AI 093172-01. In addition, we thank the Philadelphia NeuroAIDS Training Grant: T32 MH079785 and internal funds from the Department of Microbiology & Immunology, Drexel University College of Medicine to P. J.
List of Abbreviations
- Δ-9-THC
Δ-9 tetrahydrocannabinol
- BDNF
brain-derived neurotrophic factor
- C/EBPD
CCAAT/enhancer binding protein
- cAMP
adenosine monophosphate
- CB1/CB2
cannabinoid receptor type 1/type 2
- CBP
CREB-binding protein
- CCR5
C-C chemokine receptor type 5
- Cdk5
cyclin-dependent kinase 5
- CpG
Cytosine-phosphate-Guanine
- CPP
conditioned place preference
- CREB
cAMP response element-binding
- CREM
cAMP response element modulator
- CRF
chromatin assembly factors
- CSF
cerebrospinal fluid
- CTIP2
COUP-TF interacting protein 2
- CXCR4
C-X-C chemokine receptor type 4
- DNMT
DNA methyltransferase
- HAND
HIV-associated neurocognitive disorders
- HAT
histone acetyl transferase
- HCV
hepatitis C virus
- HDAC
histone deacetylase
- HIVEP1
HIV-1 enhancer binding protein
- HMT
histone methyltransferase
- HP1-α
heterochromatin protein 1-α
- ICER
inducible cAMP early repressor
- IL-2/4
interleukin-2/4
- LTR
long terminal repeat
- MAPK
mitogen-activated protein kinase
- MBD
methyl binding domain
- MeCP2
methyl CpG binding protein 2
- MEF-2
myocyte-specific enhancer factor 2
- MOR
mu-opioid receptor
- NAc
nucleus accumbens
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B-cells
- NHA
normal human astrocytes
- NPY
Neuropeptide Y
- NuRD
nucleosome remodeling and histone deacetylation
- OPRM1
opioid receptor, mu 1
- SAHA
suberoylanilide hydroxamic acid
- Sp1
Specificity protein 1
- Th
T helper cells
- TNF-α
tumor necrosis factor-α
- Tnp2
transition protein 2
- TSA
trichostatin A
Footnotes
Competing Interests
The authors declare no competing interests.
Authors’ Contributions
JS, DS, ZK, and PJ contributed to the conception and design of the paper. JS and DS contributed to the literature search. JS and PJ drafted the manuscript. JS and SS designed the figures. SS, MRN, BW, ZK and PJ revised it critically for important intellectual content and gave final approval of the version to be published. All authors read and approved the final manuscript.
References
- [Accessed on Oct. 1, 2013]; Available at http://www.drugabuse.gov/publications/hivaids/who-risk-hiv-infection-which-populations-are-most-affected.
- Adya N, Giam CZ. Distinct regions in human T-cell lymphotropic virus type I tax mediate interactions with activator protein CREB and basal transcription factors. J Virol. 1995;69:1834–1841. doi: 10.1128/jvi.69.3.1834-1841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcoholism, R.S.o. Impact of Alcoholism and Alcohol INduced Disease on America. 2011. [Google Scholar]
- Alexaki A, QS, Liu Y, Irish B, Kilareski E, Nonnemacher MR, Wigdahl B. PMA-induced differentiation of a bone marrow progenitor cell line activates HIV-1 LTR-driven transcription. DNA and Cell Biology. 2007;26:387–394. doi: 10.1089/dna.2006.0542. [DOI] [PubMed] [Google Scholar]
- Aliya N, RS, Khan Z, Jain P. Cotranscriptional Chromatin Remodeling by Small RNA Species: An HTLV-1 Perspective. Leuk Res Treatment. 2012 doi: 10.1155/2012/984754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedee AM, BG, Happel KI, Nelson S, Pandrea I. Alcohol’s role in HIV transmission and disease progression. Alcohol Research and Health. 2010;33:203–218. [PMC free article] [PubMed] [Google Scholar]
- Ameri A. The effects of cannabinoids on the brain. Progress in Neurobiology. 1999;58:315–348. doi: 10.1016/s0301-0082(98)00087-2. [DOI] [PubMed] [Google Scholar]
- Andria ML, SE Localization of promoter elements in the human mu-opioid receptor gene and regulation by DNA methylation. Mol Brain Res. 1999;70:54–65. doi: 10.1016/s0169-328x(99)00126-6. [DOI] [PubMed] [Google Scholar]
- Anier K, MK, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA Methylation Regulates Cocaine-Induced Behavioral Sensitization in Mice. Neuropsychopharmacology. 2010;35:2450–2461. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annoni J, CM, Du Pasquier RA, Hirschel B, Simioni S. HIV-associated neurocognitive disorders: a changing pattern. Future Neurology. 2011;6:81. [Google Scholar]
- Arimondo PB, EG, Tost J. Epigenetics. Biochimie. 2012;94:2191–2192. doi: 10.1016/j.biochi.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Arnsten JH, Demas PA, Grant RW, Gourevitch MN, Farzadegan H, Howard AA, Schoenbaum EE. Impact of active drug use on antiretroviral therapy adherence and viral suppression in HIV-infected drug users. J Gen Intern Med. 2002;17:377–381. doi: 10.1046/j.1525-1497.2002.10644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Rousset R, Desbois C, Jalinot P. Genetic characterization of transactivation of the human T-cell leukemia virus type 1 promoter: Binding of Tax to Tax-responsive element 1 is mediated by the cyclic AMP-responsive members of the CREB/ATF family of transcription factors. Mol Cell Biol. 1996;16:2174–2182. doi: 10.1128/mcb.16.5.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basar K, ST, Groenewegen H, Steinbusch HWM, Visser-Vandewalle V, Ternel Y. Nucleus accumbens and impulsivity. Progress in Neurobiology. 2010;92:533–557. doi: 10.1016/j.pneurobio.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Bateman DN. Opioids. Medicine. 2012;40:141–143. [Google Scholar]
- Baum MK, Rafie C, Lai S, Sales S, Page B, Campa A. Crack-cocaine use accelerates HIV disease progression in a cohort of HIV-positive drug users. J Acquir Immune Defic Syndr. 2009;50:93–99. doi: 10.1097/QAI.0b013e3181900129. [DOI] [PubMed] [Google Scholar]
- Bergman Y, CH Linking DNA methylation and histone modification: patterns and paradigms. Nature Reviews Genetics. 2009;10:295. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Berkhout B, JK Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency virus type 1 long terminal repeat. Journal of Virology. 1992;66:139–149. doi: 10.1128/jvi.66.1.139-149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazkova J, TK, Gondois-Rey F, Halfon P, Philibert P, Guigen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I. CpG Methylation Control Reactivation of HIV from Latency. PLoS Pathog. 2009;5:e1000554. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor J, Walker W, Flemington E, Spetz AL, Habener JF. Modulation of Tax and PKA-mediated expression of HTLV-I promoter via cAMP response element binding and modulator proteins CREB and CREM. FEBS Lett. 1995;377:413–418. doi: 10.1016/0014-5793(95)01299-0. [DOI] [PubMed] [Google Scholar]
- Buch S, YH, Guo M, Mori T, Su T, Wang J. Cocaine and HIV-1 Interplay: Molecular Mechanisms of Action and Addiction. J Neuroimmune Pharmacol. 2012;6:503–515. doi: 10.1007/s11481-011-9297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral GA. Drugs of Abuse, Immune Modulation, and AIDS. Journal of Neuroimmune Pharmacology. 2006;1:280–295. doi: 10.1007/s11481-006-9023-5. [DOI] [PubMed] [Google Scholar]
- Chao C, JL, Tashkin D, Martinez-Maza O, Roth MD, Margolick JB, Chmiel JS, Rinaldo C, Zhang Z, Detels R. Recreational drug use and T lymphocyte subpopulations in HIV-uninfected and HIV-infected men. Drug and Alcohol Dependence. 2008;94:165–171. doi: 10.1016/j.drugalcdep.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, SD, Lemma-Dechassa M, Tan S, Shogren-Knaak MA, Bartholomew B. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Research. 2011;39:8378–8391. doi: 10.1093/nar/gkr535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioccarelli T, CG, Altucci L, Lewis SEM, Simon L, Ricci G, Ledent C, Meccariello R, Fasano S, Pierantoni R, Cobellis G. Cannabinoid Receptor 1 Influences Chromatin Remodeling in Mouse Spermatids by Affecting Content of Transition Protein 2 mRNA and Histone Displacement. Endocrinology. 2010;151:5017–5029. doi: 10.1210/en.2010-0133. [DOI] [PubMed] [Google Scholar]
- Clapier CR, CB The Biology of Chromatin Remodeling Complexes. Annual Review of Biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Deng L, KF, Wang D, Wang L, de la Fuente C, Li H. Enhancement of the p300 HAT Activity by HIV-1 Tat on Chromatin DNA. Virology. 2001;289:312–326. doi: 10.1006/viro.2001.1129. [DOI] [PubMed] [Google Scholar]
- Desplats P, DW, Smith D, Adame A, Everall I, Letendre S, Ellis R, Cherner M, Grant I, Masliah E. Molecular and pathologic insights from latent HIV-1 infection in the human brain. Neurology. 2013;80:1415–1423. doi: 10.1212/WNL.0b013e31828c2e9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon NKWR, Peng F, Tsai Y, Dhillon S, Nicolay B, Gadgil M, Kumar A, Buch SJ. Cocaine-mediated enhancement of virus replication in macrophages: Implications for human immunodeficiency virus–associated dementia. Journal of Neurovirology. 2007;13:483–495. doi: 10.1080/13550280701528684. [DOI] [PubMed] [Google Scholar]
- Drug Enforcement Administration, U.D.o.J. A DEA Resource Guide. 2011. Drugs of Abuse, 2011 Edition. [Google Scholar]
- du Chene IBE, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, Bertrand E, Marcello A, Emiliani S, Kiernan R, Benkirane M. Suv39H1 and HP1γ are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO. 2007;26:424–435. doi: 10.1038/sj.emboj.7601517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan RSP, Page JB. Crack cocaine: effect modifier of RNA viral load and CD4 count in HIV infected African American women. Front Biosci. 2007;12:488–495. doi: 10.2741/2162. [DOI] [PubMed] [Google Scholar]
- Easley RVDR, Coley W, Guendel I, Dadgar S, Kehn-Hall K, Kashanchi F. Chromatin dynamics associated with HIV-1 Tat activated transcription. Biochim Biophys Acta. 2009;1799:275–285. doi: 10.1016/j.bbagrm.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SC. Chromatin structure and gene activity. Curr Opin Cell Biol. 1990;2:437–445. doi: 10.1016/0955-0674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- ElSohly MAdH, Wachtel SR, Feng S, Murphy TP. Delta9-tetrahydrocannabiviran as a marker for the ingestion of marijuana versus Marinol: reslts of a clinical study. J Anal Toxicol. 2001;25:565–571. doi: 10.1093/jat/25.7.565. [DOI] [PubMed] [Google Scholar]
- Feng QHS, Das T, Huang J, Feng Z. Epigenetic mechanisms in hepatitis C virus-associated hepatocellular carcinoma: a potential new link between stem cells, virology and cancer. American Medical Journal. 2013;4:21–35. [Google Scholar]
- Gallastegui EM-ZG, Terme J, Chavez S, Jordan A. Chromatin Reassembly Factors Are Involved in Transcriptional Interference Promoting HIV Latency. J Virol. 2011;85:3187–3202. doi: 10.1128/JVI.01920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan XZL, Berger O, Stin MF, Way D, Taub DT, Chang SL, Kim KS, House SD, Weinand M, Witte M, Graves MC, Fiala M. Cocaine Enhances Brain Endothelial Adhesion Molecules and Leukocyte Migration. Clinical Immunology. 1999;91:68–76. doi: 10.1006/clim.1998.4683. [DOI] [PubMed] [Google Scholar]
- Grimm JW, LL, Hayashi T, Hope BT, Su T, Shaham Y. Time-Dependent Increases in Brain-Derived Neurotrophic Factor Protein Levels within the Mesolimbic Dopamine System after Withdrawal from Cocaine: Implications for Incubation of Cocaine Craving. Journal of Neuroscience. 2003;23:742–747. doi: 10.1523/JNEUROSCI.23-03-00742.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond ETG. HIV and Psychiatric Illness. Psychiatric Times. 2007;24:57. [Google Scholar]
- Harrod R, Kuo YL, Tang Y, Yao Y, Vassilev A, Nakatani Y, Giam CZ. p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J Biol Chem. 2000;275:11852–11857. doi: 10.1074/jbc.275.16.11852. [DOI] [PubMed] [Google Scholar]
- Hauser KF, E-HN, Buch S, Nath A, Tyor WR, Bruce-Keller AJ, Knapp PE. Impact of opiate-HIV-1 interactions on neurotoxic signaling. J Neuroimmune Pharmacol. 2006;1:98–105. doi: 10.1007/s11481-005-9000-4. [DOI] [PubMed] [Google Scholar]
- Hwang CK, SK, Kim CS, Choi HS, Guo X-H, Law P-Y, Wei L-N, Loh HH. Epigenetic programming of μ-opioid receptor gene in mouse brain is regulated by MeCP2 and Brg1 chromatin remodelling factor. J Cell Mol Med. 2009;13:3591–3615. doi: 10.1111/j.1582-4934.2008.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, TH, Okamoto T. Involvement of Histone H3 Lysine 9 (H3K9) Methyltransferase G9a in the Maintenance of HIV-1 Latency and Its Reactivation by BIX01294. Journal of Biological Chemistry. 2010;285:16538–16545. doi: 10.1074/jbc.M110.103531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, HA, Koiwa T, Watanabe T. 5′ long terminal repeat (LTR)- selective methylation of laently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology. 2006;3:69. doi: 10.1186/1742-4690-3-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lu H, Schiltz RL, Pise-Masison CA, Ogryzko VV, Nakatani Y, Brady JN. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol Cell Biol. 1999;19:8136–8145. doi: 10.1128/mcb.19.12.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashanchi F, Duvall JF, Kwok RP, Lundblad JR, Goodman RH, Brady JN. The coactivator CBP stimulates human T-cell lymphotrophic virus type I Tax transactivation in vitro. J Biol Chem. 1998;273:34646–34652. doi: 10.1074/jbc.273.51.34646. [DOI] [PubMed] [Google Scholar]
- Kauder SE, BA, Lindqvist A, Planelles V, Verdin E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PloS Pathogens. 2009;5:e1000495. doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer JA, MR Synaptic plasticity and addiction. Nature Reviews Neuroscience. 2007;8:844–858. doi: 10.1038/nrn2234. [DOI] [PubMed] [Google Scholar]
- Kim W, KR, Park G, Park JW, Kim JE. Deficiency of H3K79 Histone Methyltransferase Dot1-like Protein (DOT1L) Inhibits Cell Proliferation. J Biol Chem. 2012;287:5588–5599. doi: 10.1074/jbc.M111.328138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyokawa T, Seiki M, Imagawa K, Shimizu F, Yoshida M. Identification of a protein (p40x) encoded by a unique sequence pX of human T-cell leukemia virus type I. Gann. 1984;75:747–751. [PubMed] [Google Scholar]
- Kovacs JA, LR, Sidorov IA, Adelsberger JW, Sereti I, Sachau W, Kelly G, Metcalf JA, Davey RT, Falloon J, Polis MA, Tavel J, Stevens R, Lambert L, Hosack DA, Bosche M, Issaq HJ, Fox SD, Leitman S, Baseler MW, Masur H, Di Mascio M, Dimitrov DS, Lane HC. Induction of prolonged survival of CD4+ T lymphocytes by intermittent IL-2 therapy in HIV-infected patients. J Clin Invest. 2005;115:2139–2148. doi: 10.1172/JCI23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs FCHT, Quiterio S, Gartner S, Wigdahl B. Lentiviral LTR-directed expression, sequence variation, and disease pathogenesis. Los Alamos National Laboratory HIV Sequence Compendium; 2002. [Google Scholar]
- Kumar A, CK, Renthal W, Tsankova NM, Theobald DEH, Truong H, Russo SJ, LaPlant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ. Chromatin Remodeling is a Key Mechanism Underlying Cocaine-Induced Plasticity in Striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Kwok RP, Laurance ME, Lundblad JR, Goldman PS, Shih H, Connor LM, Marriott SJ, Goodman RH. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- Lefevre F, OLB, Moran M, Mossar M, Yarnold PR, Martin GJ, Lassroth GJ. Alcohl consumption among HIV-infected patients. J Gen Intern Med. 1995;10:458–460. doi: 10.1007/BF02599920. [DOI] [PubMed] [Google Scholar]
- Letendre S, PA, Rockenstein E, Adame A, Crews L, Cherner M, Heaton R, Ellis R, Everall IP, Grant I, Masliah E. Pathogenesis of Hepatitis C Virus Coinfection in the Brains of Patients Infected with HIV. Journal of Infectious Diseases. 2007;196:361–370. doi: 10.1086/519285. [DOI] [PubMed] [Google Scholar]
- Levine AA, GZ, Barco A, Xu S, Kandel ER, Schwartz JH. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc Natl Acad Sci. 2005;102:19186–19191. doi: 10.1073/pnas.0509735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, BD, Shinn P, Chen H, Hoffman C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD. Genome-Wide Analysis of Chromosomal Features Repressing Human Immunodeficiency Virus Transcription. Journal of Virology. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, AB, Pirrone V, Nonnemacher MR, Wojno A, Passic S, Flaig K, Kilareski E, Blakey B, Ku J, Parikh N, Shah R, Martin-Garcia J, Moldover B, Servance L, Downie D, Lewis S, Jacobson JM, Kolson D, Wigdahl B. Development of co-selected single nucleotide polymorphisms in the viral promoter precedes the onset of human immunodeficiency virus type 1-associated neurocognitive impairment. Journal of Neurovirology. 2011;17:92–109. doi: 10.1007/s13365-010-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, FK, Cook RJ, Hunkele AJ, Loh HH, Ko JL. Effects of Trichostatin A on Neuronal mu-Opioid Receptor Gene Expression. Brain Research. 2008;1246:1–10. doi: 10.1016/j.brainres.2008.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, LH, Guo L, Li C, Li M, Jiang W, Liu X, McNutt M, Li G. The mechanism involved in the repression of the μ opioid receptor gene expression in CEM ×174 cells infected by simian immunodeficiency virus. Journal of Leukocyte Biology. 2009;85:684–691. doi: 10.1189/jlb.0908543. [DOI] [PubMed] [Google Scholar]
- Macian F, RA Reciprocal Modulatory Interaction between Human Immunodeficiency Virus Type 1 Tat and Transcription Factor NFAT1. Molecular and Cellular Biology. 1999;19:3645–3653. doi: 10.1128/mcb.19.5.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantri CK, DJ, Mantri JV, Dash CCV. Cocaine Enhances HIV-1 Replication in CD4+ T Cells by Down-Regulating MiR-125b. PLoS ONE. 2012;7:e51387. doi: 10.1371/journal.pone.0051387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marban C, SS, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO. 2007;26:412–423. doi: 10.1038/sj.emboj.7601516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G, GM Chromatin control of HIV-1 gene expression. Genetica. 1999;106:125–130. doi: 10.1023/a:1003797332379. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- Metzger DS, NH, Woody GE. Drug Abuse Treatment as AIDS Prevention. Public Health Reports. 1998;113:97–106. [PMC free article] [PubMed] [Google Scholar]
- Meyaard L, OS, Jonker RR, Mijnster MJ, Keet RP, Miedema F. Programmed death of T cells in HIV-1 infection. Science. 1992;257:217–219. doi: 10.1126/science.1352911. [DOI] [PubMed] [Google Scholar]
- Miller EK. The Prefrontal Cortex and Cognitive Control. Nature Reviews Neuroscience. 2000;1:59–65. doi: 10.1038/35036228. [DOI] [PubMed] [Google Scholar]
- Molina PE, AA, LeCapitaine NJ, Zabaleta J, Mohan M, Winsauer P, Vande Stouwe C. Cannabinoid Neuroimmune Modulation of SIV Disease. J Neuroimmune Pharmacol. 2011;6:516–527. doi: 10.1007/s11481-011-9301-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina PE, WP, Zhang P, Walker E, Birke L, Amedee A, Stouwe CV, Troxclair D, McGoey R, Varner K, Byerley L, Lamotte L. Cannabinoid Administration Attenuates the Progression of Simian Immunodeficiency Virus. AIDS Res Hum Retrovir. 2010;27:585–592. doi: 10.1089/aid.2010.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacher M, Adenis A, Hanf M, Adriouch L, Vantilcke V, El Guedj M, Vaz T, Dufour J, Couppie P. Crack cocaine use increases the incidence of AIDS-defining events in French Guiana. AIDS. 2009;23:2223–2226. doi: 10.1097/QAD.0b013e32833147c2. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Transcriptional mechanism of addiction: role of DeltaFosB. Phil Trans R Soc Lond B. 2008;363:3245–3255. doi: 10.1098/rstb.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIDA. Addiction Science: From Molecules to Managed Care 2008 [Google Scholar]
- Nielsen DA, UA, Reyes JA, Simons DD, Kosten TR. Epigenetics of drug abuse: predisposition or response. Pharmacogenomics. 2013;13:1149–1160. doi: 10.2217/pgs.12.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DA, YV, Hamon S, Jackson C, Ho A, Ott J, Kreek MJ. Increased OPRM1 DNA methylation in lymphocytes of methadone-maintained former heroin addicts. Neuropsychopharmacology. 2009;34:867–873. doi: 10.1038/npp.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe SN, NS, Ugen K, Friedman H, Klein TW. Cannabinoid receptor agonists enhance syncytia formation in MT-2 cells infectd with cell free HIV-1MN. Adv Exp Med Biol. 1998;437:223–229. doi: 10.1007/978-1-4615-5347-2_25. [DOI] [PubMed] [Google Scholar]
- Okada M, Jeang KT. Differential requirements for activation of integrated and transiently transfected human T-cell leukemia virus type 1 long terminal repeat. J Virol. 2002;76:12564–12573. doi: 10.1128/JVI.76.24.12564-12573.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, UR, Zhang H, Tang L, Prakash A. Brain Chromatin Remodeling: A Novel Mechanism of Alcoholism. Journal of Neuroscience. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandhare J, DC A prospective on drug abuse-associated epigenetics and HIV-1 replication. Life Sciences. 2011;88:995–999. doi: 10.1016/j.lfs.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya D, Rahman S, Wigdahl B, Khan ZK, Jain P. New insights into the pathogenesis, diagnosis and treatment of human T-cell leukemia virus type 1-induced disease. Future Virology. 2007;2:481–493. [Google Scholar]
- Paradisi A, PN, Barcaroli D, Maccarrone M. Anandamide regulates keratinocyte differentiation by inducing DNA methylation in a CB1 receptor-dependent manner. J Biol Chem. 2008;283:6005–6012. doi: 10.1074/jbc.M707964200. [DOI] [PubMed] [Google Scholar]
- Pearson R, KY, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. Epigenetic Silencing of Human Immunodeficiency Virus (HIV) Transcription by Formation of Restrictive Chromatin Structures at the Viral Long Terminal Repeat Drives the Progressive Entry of HIV into Latency. J Virol. 2008;82:12291–12303. doi: 10.1128/JVI.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti LI, WR, Robison B, Renthal W, Maze I, Yazdani S, Elmore RG, Knapp DJ, Selley DE, Martin BR, Sim-Selley L, Bachtell RK, Self DW, Nestler EJ. Distinct Patterns of ΔFosB Induction in Brain by Drugs of Abuse. Synapse. 2008;62:358–369. doi: 10.1002/syn.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson PK, GG, Hu S, Cabral G, Lokensgard JR. Cannabinoids and morphine differentially affect HIV-1 expression in CD4+ lymphocyte and microglial cell cultures. J Immunol. 2004;147:123–126. doi: 10.1016/j.jneuroim.2003.10.026. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, RM, Sullivan EV. Focus on the brain: HIV infection and alcoholism: comorbidity effects on brain structure and function. Alcohol Research and Health. 2010;33:247. [PMC free article] [PubMed] [Google Scholar]
- Power C, Boisse L, Rourke S, Gill MJ. NeuroAIDS: An Evolving Epidemic. Canadian Journal of Neurological Sciences. 2009;36:285–295. doi: 10.1017/s0317167100007009. [DOI] [PubMed] [Google Scholar]
- Pulipparachuravil S, Renthal W, Hale CF, Taniguchi M, Guanghua X, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, Greengard P, Naim AC, Nestler EJ, Cowan CW. Cocaine Regulates MEF2 to Control Synaptic and Behaviral Plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit V, RR, Shurtleff D. Drugs of Abuse, Dopamine, and HIV-Associated Neurocognitive Disorders/HIV-Associated Dementia. Molecular Neurobiology. 2011;44:102–110. doi: 10.1007/s12035-011-8195-z. [DOI] [PubMed] [Google Scholar]
- Rafati H, PM, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. Repressive LTR Nucleosome Positioning by the BAF Complex Is Required for HIV Latency. PLoS Biol. 2011:9. doi: 10.1371/journal.pbio.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, QK, Pandya D, Singh S, Khan Z, Jain P. HTLV-1 Tax Mediated Downregulation of miRNAs Associated with Chromatin Remodeling Factors in T Cells with Stably Integrated Viral Promoter. PLoS. 2012;7:13. doi: 10.1371/journal.pone.0034490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan PM, DR, Datta PK, Khalili K. Epigenetics of μ-opioid receptors: Intersection with HIV-1 infection of the central nervous system. Journal of Cellular Physiology. 2012;227:2832–2841. doi: 10.1002/jcp.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, NE Histone acetylation in drug addiction. Seminars in Cell & Developmental Biology. 2009;20:387–394. doi: 10.1016/j.semcdb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JL, MS, Bindukumar B, Sykes D, Schwartz SA, Nair MPN. Proteomic analysis of the effects of cocaine on the enhancement of HIV-1 replication in normal human astrocytes (NHA) Brain Research. 2006;1123:226–236. doi: 10.1016/j.brainres.2006.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, NE Transcriptional and epigenetic mechanisms of addiction. Nature Reviews Neuroscience. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock RB, GG, Hu S, Shend WS, Cabral GA, Martin BR, Peterson PK. WIN55,212-2-mediated inhibition of HIV-1 expression in microglial cells: involvement of cannabinoid receptors. J Neuroimmune Pharmacol. 2007;2:178–183. doi: 10.1007/s11481-006-9040-4. [DOI] [PubMed] [Google Scholar]
- Roth MD, TD, Whittaker KM, Choi R, Baldwin GC. Tetrahydrocannabinol suppresses immune function and enhances HIV replication in the huPBL-SCID mouse. Life Sciences. 2005;77:1711–1722. doi: 10.1016/j.lfs.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Russo SJ, WM, Mazei-Robison MS, Dietz DM, Maze I, Krishnan V, Renthal W, Graham A, Birnbaum SG, Green TA, Robison B, Lesselyong A, Perrotti LI, Bolanos CA, Kumar A, Clark MS, Neumaier JF, Leve RL, Bhakar AL, Barker PA, Nestler EJ. Nuclear Factor kB Signaling Regulates Neuronal Morphology and Cocaine Reward. Journal of Neuroscience. 2009;29:3529–3537. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMHSA. 2010–2011 National Survey on Drug Use and Health Model-Based Estimates (50 States and the District of Columbia) 2012. [Google Scholar]
- Schroeder FA, PK, Matevossian A, Jones SR, Konradi C, Tapper AR, Akbarian S. Drug-Induced Activation of Dopamine D1 Receptor Signaling and Inhibition of Class I/II Histone Deacetylase Induce Chromatin Remodeling in Reward Circuitry and Modulate Cocaine-Related Behaviors. Neuropsychopharmacology. 2008;22:2981–2992. doi: 10.1038/npp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoggin KE, Ulloa A, Nyborg JK. The oncoprotein Tax binds the SRC-1-interacting domain of CBP/p300 to mediate transcriptional activation. Mol Cell Biol. 2001;21:5520–5530. doi: 10.1128/MCB.21.16.5520-5530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, WJ What controls nucleosome positions? Trends in Genetics. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, NM, Pirrone V, Wigdahl B. Innate and Adaptive Factors Regulating Human Immunodeficiency Virus Type 1 Genomic Activation. Journal of Neuroimmune Pharmacology. 2010;5:278–293. doi: 10.1007/s11481-010-9207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski JG, Rosen CA, Haseltine WA. Trans-acting transcriptional activation of the long terminal repeat of human T lymphotropic viruses in infected cells. Science. 1984;225:381–385. doi: 10.1126/science.6330891. [DOI] [PubMed] [Google Scholar]
- Solum D. Substance abuse. Psychopharmacology Educational Updates (PsychEd Up) 2008:4. [Google Scholar]
- Strazza M, PV, Nonnemacher MR. Breaking Down the Barrier: The effects of HIV-1 on the Blood-Brain Barrier. Brain Research. 2011;1399:96–115. doi: 10.1016/j.brainres.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, ME Chromatin and nuclear architecture in the nervous system. Trends in Neurosciences. 2008;31:343–352. doi: 10.1016/j.tins.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Treand C, dCI, Bres V, Kiernan R, Benarous R, Benkirane M, Emiliani S. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO. 2006;25:1690–1699. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, RW, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nature Reviews Neuroscience. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tyagi M, KJ CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO. 2007;26:4985–4995. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale A. Cocaine. Medicine. 2007;35:607. [Google Scholar]
- Vale A. Drugs of abuse (amfetamines, BZP, cannabis, cocaine, GHB, LSD) Poisoning. 2012;40:84–87. [Google Scholar]
- Vignali M, HA, Neely KE, Workman JL. ATP-Dependent Chromatin-Remodeling Complexes. Molecular and Cellular Biology. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, BR, Roy S. Transcriptional and Epigenetic Regulation of Interleukin-2 Gene in Activated T Cells by Morphine. Journal of Biological Chemistry. 2007;282:7164–7171. doi: 10.1074/jbc.M604367200. [DOI] [PubMed] [Google Scholar]
- Wang L, LZ, Hu Z, Sheng J, Hui B, Sun J, Ma L. Chronic Cocaine-Induced H3 Acetylation and Trascriptional Activation of CaMKIIα in the Nucleus Accumbens Is Critical for Motivation for Drug Reinforcement. Neuropsychopharmacology. 2010;35:913–928. doi: 10.1038/npp.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, ZY, Qing H, Liu M, Yang P. The extinction of morphine-induced conditioned place preference by histone deacetylase inhibition. Neuroscience Letters. 2010;483:137–142. doi: 10.1016/j.neulet.2010.07.080. [DOI] [PubMed] [Google Scholar]
- Wong CCY, MJ, Fernandes C. Drugs and addiction: an introduction to epigenetics. Addiction. 2011;106:480–489. doi: 10.1111/j.1360-0443.2010.03321.x. [DOI] [PubMed] [Google Scholar]
- Yao H, KK, Duan M, Hayashi T, Guo M, Morgello S, Prat A, Wang J, Su T, Buch S. Cocaine hijacks sigma-1 receptor to initiate induction of ALCAM: Implication for increased monocyte adhesion and migration in the central nervous system. Journal of Neuroscience. 2012;31:5942–5955. doi: 10.1523/JNEUROSCI.5618-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, KS, Cheng Q, Zoumalan R, Tashkin DP, Roth MD. Delta 9-Tetrahydrocannabinol regulates Th1/Th2 cytokine balance in activated human T cells. J Neuroimmunol. 2002;133:124–131. doi: 10.1016/s0165-5728(02)00370-3. [DOI] [PubMed] [Google Scholar]
- Zeng L, ZM Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]