Abstract
In Buchnera aphidicola strains associated with the aphid subfamilies Thelaxinae, Lachninae, Pterocommatinae, and Aphidinae, the four leucine genes (leuA, -B, -C, and -D) are located on a plasmid. However, these genes are located on the main chromosome in B. aphidicola strains associated with the subfamilies Pemphiginae and Chaitophorinae. The sequence of the chromosomal fragment containing the leucine cluster and flanking genes has different positions in the chromosome in B. aphidicola strains associated with three tribes of the subfamily Pemphiginae and one tribe of the subfamily Chaitophorinae. Due to the extreme gene order conservation of the B. aphidicola genomes, the variability in the position of the leucine cluster in the chromosome may be interpreted as resulting from independent insertions from an ancestral plasmid-borne leucine gene. These findings do not support a chromosomal origin for the leucine genes in the ancestral B. aphidicola and do support a back transfer evolutionary scenario from a plasmid to the main chromosome.
Aphids (family Aphididae, sensu Remaudière and Remaudière [32]) are plant sap-feeding insects that maintain an endosymbiotic association with the bacterium Buchnera aphidicola, a member of the γ3 group of the Proteobacteria (2, 28). After their association, which started at least 150 million years ago, host and symbiont lineages have subsequently diverged strictly in parallel, by maternal transmission of the symbiont to eggs or embryos at blastoderm stage (24). The major role of B. aphidicola in the symbiosis is the provision of amino acids, which are lacking from the phloem sap diet (8).
In past years, the discovery of plasmids in B. aphidicola that carry both the rate-limiting genes for biosynthesis of tryptophan (trpEG) and the genes for biosynthesis of leucine (leuABCD) was considered evidence of the overproduction of these essential amino acids, thus supporting the nutrient-provisioning role of B. aphidicola in aphid symbiosis (3, 4, 6, 17, 33, 40, 45, 47, 48). The main B. aphidicola chromosome is also present in multicopy in each cell (15), and in some cases B. aphidicola has fewer leucine and tryptophan plasmid copies than chromosome copies in each cell (30). The discovery that ratios of plasmid-borne trpEG and leuABCD copies to chromosomal gene copies vary, both within and between species (30, 44), casts doubt on the idea that plasmid location is a means of leucine and tryptophan overproduction that leads to a quick response to changes in demand for these amino acids (25). The evolutionary history of the plasmids is puzzling, due to the fact that not all of the lineages of aphids carry plasmids and not all plasmids have the same gene content and/or gene order. B. aphidicola strains associated with aphids of the subfamily Aphidinae and some tribes of the subfamily Pemphiginae contain tryptophan plasmids (17, 33, 47), ranging in size from 3.0 to 12.8 kb, which contain the two first genes of the tryptophan pathway (trpEG). The variability in size is due mainly to variability in the number of tandem repeats of these genes or pseudogenes.
In the case of leucine plasmids, only a single replicon, named repA1, has been found, but the gene content and/or gene order is different in different lineages, indicating a great plasticity of the leucine plasmids throughout B. aphidicola evolution. In fact, up to seven plasmids that are different in both gene order and gene content have been found (45). The first leucine plasmid, pRPE (renamed pBRp), was described for B. aphidicola strains associated with Rhopalosiphum padi (6), a member of the subfamily Aphidinae. It contains the genes encoding key enzymes in the pathway leading to leucine, in the same order as in Escherichia coli (leuABCD). The other genes of the leucine plasmid are two copies of repA, which code for plasmid replicases, and open reading frame 1 (ORF1) (renamed yqhA), encoding a putative integral membrane protein. The same gene content in the same order was found in strains associated with other species of the Aphidinae subfamily (3, 41). The leucine genes have also been located in plasmids in B. aphidicola strains associated with members of the subfamilies Pterocommatinae, Thelaxinae, and Lachninae, with each lineage showing special features (40, 45, 48). Finally, in strains associated with the subfamily Pemphiginae, cryptic plasmids have been found. They are phylogenetically related to the leucine plasmids but do not have the structural leucine genes. These plasmids contain only the origin of replication and one or two copies of the repA gene, plus one or two more genes (ibp or yqhA), depending on the different tribe within the subfamily (Pemphigini, Eriosomatini, or Fordini). It was suggested that they probably represent the ancestral replicon, related to the IncFII plasmids in which the other genes were relocated (48).
During the past 3 years, the whole genomes of three B. aphidicola strains have been completely sequenced (37, 42, 46): B. aphidicola BAp and BSg, associated with the aphids Acyrthosiphon pisum and Schizaphis graminum, respectively, which belong to the same aphid subfamily (Aphidinae) but to different tribes (Macrosiphini and Aphidini, respectively), and B. aphidicola BBp, associated with the aphid Baizongia pistaciae, a member of the subfamily Pemphiginae (tribe Fordini). A comparison of BAp and BSg, with an estimated divergence time of 50 to 70 million years, revealed an extreme conservation of the genome order, with neither chromosomal rearrangements (translocations, inversions, or duplications) nor gene acquisition by horizontal gene transfer, thus being the most extreme case of genome stability to date (42). The comparison with BBp revealed nearly perfect gene order conservation, with only four minor rearrangements (two inversions and two translocations involving the leucine and tryptophan plasmid-carried genes) in the BBp strain. Since the Aphidinae and Pemphiginae lineages diverged about 80 to 150 million years, van Ham et al. (46) suggested that B. aphidicola can be considered a “gene order fossil” and that the onset of genomic stasis coincided with the establishment of the symbiosis. However, the gene contents are different in the three lineages, indicating that independent gene losses have occurred from the last common symbiotic ancestor (LCSA) of B. aphidicola (38).
In the case of BBp, the leucine cluster is located in the chromosome, flanked by the genes yqgF and yggS. However, in a B. aphidicola strain (BPs) associated with the aphid Pemphigus spyrothecae, which is also a member of the Pemphiginae but belongs to a different tribe (Pemphigini), the cluster is also located in the chromosome but is flanked by the genes trxA and rep (34).
In the present work we have characterized the four leucine genes, as well as the flanking regions, that are located in the chromosome in B. aphidicola strains associated with two new species: Tetraneura caerulescens, a member of the tribe Eriosomatini (subfamily Pemphiginae), and Chaitophorus populeti, a member of the tribe Chaitophorini (subfamily Chaitophorinae). These data, together with the two previous leucine cluster chromosomal locations in BBp and BPs, are consistent with four independent insertions of the leucine plasmid throughout B. aphidicola evolution from an ancestral plasmid present in the LCSA.
MATERIALS AND METHODS
Aphid material and DNA extraction.
C. populeti aphids were collected from leaves of white poplar trees (Populus alba) in Benifaió (Valencia, Spain). T. caerulescens galls were collected from elm trees (Ulmus minor) in Bugarra (Valencia, Spain). The total aphid DNA (tDNA) was isolated as previously described (18). Genomic DNA of B. aphidicola was obtained by isolating symbiotic bacteria (13) by the cetyltrimethylammonium bromide-NaCl protocol (36, 48).
Location of leucine cluster.
To determine the location of the leucine genes, either amplified in a plasmid or in the bacterial chromosome, as well as the gene order of the four leucine genes, we followed the procedure outlined by van Ham et al. (48), based on structural PCRs, restriction maps, and hybridization with probes from the pBRp plasmid as described previously (34).
Amplification, cloning, and sequencing of the leucine cluster and flanking regions.
A strategy based on overlapping PCR fragments was used to obtain the sequences of the chromosomal regions containing the leucine genes in the two species. All PCR products were purified and cloned into T-pBluescript (19) or into pGEM-TEasy (Promega). Table 1 lists the specific primers used in the amplifications.
TABLE 1.
Primers used to amplify the leucine cluster and flanking regions in this study
| Primer | Sequence (5′→3′)a | 5′ position in BAp genomeb | PCR | Refer- ence |
|---|---|---|---|---|
| Leucine general primers | ||||
| leuA.du2 | (CGGATCCTGCAG)GAT GAT GTW GAA TTT TCW TGY GAR GAY GC | 3431 (pLeu) | ilPCR | 48 |
| leuA.dl3 | (CGGATCCGTCGAC)AR ACT WGC TTG WAR WGC TTG TTC WCC ATC | 3099 (pLeu) | ilPCR | 48 |
| leuD.du2 | (CCCATCCTGCAG)GGW TGT GGW TCW TCW AGA GAR CAT GC | 7400 (pLeu) | Regular PCR | 48 |
| leuA-R2 | (GGAATTC)WG TAT AWC CWA CWG TAT CWG G | 3556 (pLeu) | Regular PCR | TSc |
| BCp-specific primers | ||||
| gnd-lo1 | CAA GCT CAA AGA GAT TAT TTT GGA GCT C | 112951 | ilPCR | TS |
| gnd-up1 | CTG CTC TAA TGA TAC TAC CTG CAC G | 112711 | ilPCR | TS |
| dcd-dF1 | WGT GAY AVA GAY ATH GAR TGG | 113245 | Regular PCR | TS |
| dcd-dR1 | ATK CCA WCC WGG RTC DAT NCK RTG | 113632 | Regular PCR | TS |
| dcd-CloR1 | TAA AGA AGA ACG ACC ATC TAA CCA TCC | 113574 | Regular PCR | TS |
| leuC-CloR1 | TCC TCC TCT ACC TTG CCT GCC TTC | 7063 (pLeu) | Regular PCR | TS |
| BTc-specific primers | ||||
| TcleuC-Fi | (GGAATTCCTCTAGA)A ATA TGG CTA TTG AAA TGG GAG CTA AAT CAG | 6405 (pLeu) | ilPCR | TS |
| TcleuB-Ri | (GGAATTCGCTGCAG)ATT TCA AAA GCA AAA TTC GCT ATC CTA C | 5201 (pLeu) | ilPCR | TS |
| TcleuD-Ri | (GGAATTCGCTGCAG)CGG AAG CCA TAA TCT AAA ATA GCC CAA AC | 7427 (pLeu) | ilPCR | TS |
| TcleuA-Fi | (GGAATTCCTCTAGA)G ATA CAA CAT TAA GAG ATG GTG AAC AAG C | 3056 (pLeu) | Regular PCR | TS |
| TcleuAi-F2 | (GGAATTC)AA ATT TTC GAG AAA CTG TTG ATC TAG C | 3371 (pLeu) | Regular PCR | TS |
| mrcB900d-UR2 | ATA TAA WAG WGC WCC TTT GAC CAT ACC WAC | 217163 | Regular PCR | TS |
| mrcB310d-UR1 | ATC WGG WAA ATC AAA WGS ACG WCG | 216574 | Regular PCR | TS |
Sequences in parentheses correspond to restriction enzyme sites for cloning purposes.
Main chromosome or pLeu plasmid.
TS, this study.
(i) B. aphidicola strain BCp, associated with C. populeti.
The restriction map showed that the enzyme HindIII yielded a 3.5-kb fragment that contained the genes leuA, leuC, and leuD as well as flanking regions. tDNA was cut with this enzyme and used as a template for an inverse long PCR (ilPCR), performed with a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer), using the Expand long-template PCR system (Roche) and the outwardly oriented primers leuA.dl3 and leuA.du2, located 331 nucleotides apart within leuA (48). A 3.2-kb fragment was amplified and sequenced. It contained the gene leuD; the partial genes leuC, leuA, and gnd; and the corresponding intergenic regions igleuA-gnd, igleuD-leuA, and igleuC-leuD (Fig. 1). The region downstream of leuA was obtained by ilPCR with two specific primers designed on gnd (gnd-lo1 and gnd-up1) that contained two complete ORFs (hisI and hisF) and three partial ORFs (gnd, hisA, and leuA). The 5′ end of the cluster containing the leuB gene and flanking regions was obtained by using a different strategy. As the gene dcd is contiguous to gnd in the three sequenced B. aphidicola strains, and based on the B. aphidicola conserved gene order (see the introduction), two degenerate primers on dcd (dcd-dF1 and dcd-dR1) were designed that amplified a 500-bp fragment, which, after sequencing, enabled the design of specific primers. PCR with primers dcd-cloR1 and leuC-CloR1, based on the 3.2-kb fragment previously sequenced, amplified a 3-kb fragment.
FIG. 1.
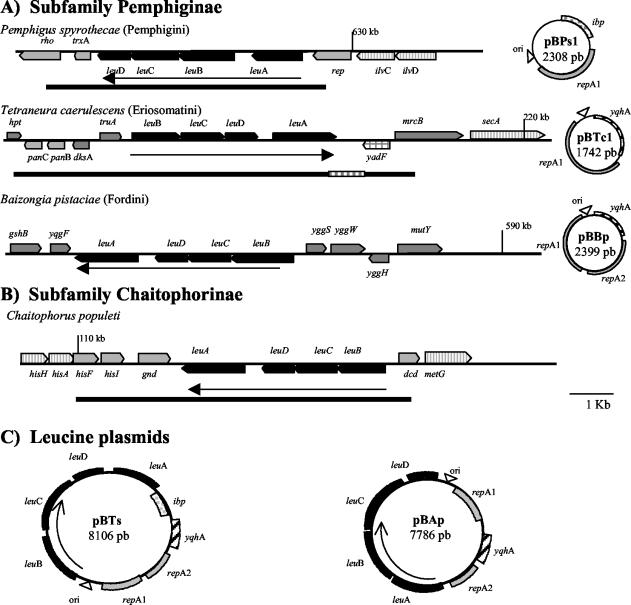
Gene order and flanking regions of the leucine cluster in B. aphidicola strains. (A) Members of the subfamily Pemphiginae, showing the chromosomal regions with their corresponding cryptic leucine-related repA plasmids. (B) Subfamily Chaitophorinae. (C) Leucine plasmids in pBTs (subfamily Thelaxinae) and BAp (subfamily Aphidinae; plasmids from strains BSg, BDn, and BRp are present the same gene content and order) (see Table 2 for strain designations). Black bars indicate the sequenced regions (reference 34 and this study). Arrows are oriented according to the coding strand. Number in kilobases indicate the position of the gene in the BAp chromosome (37).
(ii) B. aphidicola strain BTc, associated with T. caerulescens.
A medium PCR was performed with primers leuA.dl3 and leuD.du2 (48) to determine the partial gene order and to sequence some fragments in order to design new specific BTc primers. B. aphidicola tDNA was digested with EcoRI and run in a 0.8% agarose gel, and the smear was cut between the 5.5-to-5.0-kb and 1.6-to-1.2-kb ranges (restriction map analysis showed that the complete region containing the leucine cluster and flanking regions was contained within two EcoRI fragments of 1.4 kb (leuBC) and 5.2 kb (leuCDA and downstream) (Fig. 1). DNA was recovered from the agarose by using GeneClean II (Bio 101), and ligated to λZAP II-EcoRI (Stratagene) according to the manufacturer's instructions. Four recombinants of the 1.6- to 1.2-kb partial EcoRI library were in vivo excised to plasmid with helper phage ExAssistant (Stratagene) according to the manufacturer's instructions. Specific primers (TcleuC-Fi, TcleuB-Ri, TcleuD-Ri, and TcleuA-Fi) were designed outwardly to amplify the upstream region of the cluster by ilPCR. The other BTc primers (Table 1) were used to finish sequencing of the region.
PCR mixtures contained 40 or 15 pmol of each degenerate or specific primer, respectively; 500 nM deoxynucleoside triphosphates; 1× buffer system 3; and 0.75 μl of Taq polymerase mix in a 50-μl final reaction volume. The amplification profile was 92°C for 2 min; 10 cycles of 92°C for 10 s, 52°C (62°C for iPCR) for 30 s, and 68°C for 1 min (10 min for ilPCR); 20 more cycles with an autoextension of 20 s/cycle at 68°C; and a final extension at 68°C for 7 min. The annealing temperature varied, depending on the primer pair, from 62 to 52°C.
The ilPCR with outwardly oriented primers within leuA lacks 389 nucleotides of the original leuA (48). For completion of the cluster fragment, one leuA degenerate primer (leuA-R2 [Table 1]), was designed and used in PCR in combination with leuD.du2 to obtain a 500-nucleotide fragment containing the missing leuA fragment in both species.
The sequencing of all of the clones (in both directions) was carried out in a PE/ABI 377, 310, or 3100 instrument with a dRhodamine or BigDye version 1.0 dye terminator cycle sequencing kit (Perkin-Elmer). Universal primers T3, T7, UNI17-mer, and UNIrev as well as specific primers were also used.
Computer and phylogenetic analysis.
DNA sequence data were assembled with the program Sequencher version 4.0 (Genecodes Co.). Blastx version 2.2.1 (http://www.ncbi.nlm.nih.gov/BLAST) was used to identify the ORFs and for gene assignment.
For comparative analysis we chose representative B. aphidicola strains that had the leucine cluster, either in a plasmid or in the main chromosome, completely sequenced. Table 2 summarizes the main features of the clusters (gene order and location) as well as the GenBank/EMBL nucleotide sequence accession numbers. We classified the aphids as described previously (32).
TABLE 2.
Taxonomic status, location, and gene order of the leucine cluster in the aphid species (family Aphididae) analyzed in this study.
| Subfamily | Tribe | Species | B. aphidicda plasmid or strain | Localization of leucine cluster | Gene order | Accession no. (reference) |
|---|---|---|---|---|---|---|
| Aphidinae | Rhopalosiphini | Rhopalosiphum padi | pBRp | Plasmid | leuABCD | X71612 (6) |
| Schizaphis graminum | pBSg | Plasmid | leuABCD | AF041836 (3) | ||
| Macrosiphini | Acyrthosiphum pisum | pBAp | Plasmid | leuABCD | AJ006878 (41) | |
| Diuraphis noxia | pBDn | Plasmid | leuABCD | AF041837 (3) | ||
| Chaitophorinae | Chaitophorini | Chaitophorus populeti | BCp | Chromosome | leuBCDA | AY375291 (this study) |
| Thelaxinae | Thelaxes suberi | pBTs | Plasmid | leuBCDA | Y11966 (48) | |
| Pemphiginae | Pemphigini | Pemphigus spyrothecae | BPs | Chromosome | leuABCD | AJ426489 (34) |
| Fordini | Baizongia pistaciae | BBp | Chromosome | leuBCDA | NC004545 (46) | |
| Eriosomatini | Tetraneura caerulescens | BTc | Chromosome | leuBCDA | AY375290 (this study) |
The phylogeny of the leucine cluster was obtained by using a concatenated alignment of the four leucine genes (leuB, leuC, leuD, and leuA), with the previous removal of the 5′ and 3′ nonconserved ends of each gene, resulting in 4,152 nucleotide positions. The phylogenetic tree was obtained with the neighbor-joining algorithm (35) implemented in the program MEGA (16) with the following parameters: second codon positions, complete deletion, and Tamura-Nei model of nucleotide substitution (43). The reliability of the different branches was evaluated by bootstrapping (1,000 replicates). Outgroup species and accession numbers are given in the legend to Fig. 2.
FIG. 2.
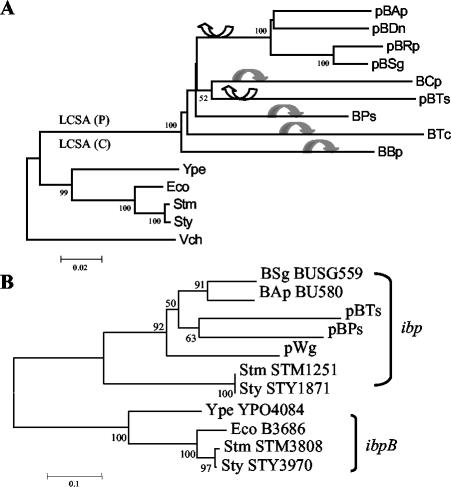
Phylogenetic trees obtained with the neighbor-joining algorithm for the leucine cluster (A) and the ibp genes (B). In panel A, representative events according to the proposed scenarios for leucine cluster evolution are shown: (i) (back transfer) LCSA (P), cluster present in an ancestral plasmid, and then four insertions in the chromosome (black arrows); (ii) LCSA (C), cluster present in the ancestral chromosome, and then two transfers into plasmids (white arrows). Abbreviations for B. aphidicola strains and accession numbers are in Table 2. Other γ-proteobacteria are Escherichia coli K-12 MG1655 (Eco) (accession number U00096), Salmonella enterica subsp. enterica serovar Typhi CT18 (STY) (AL513382), S. enterica subsp. enterica serovar Typhimurium LT2 (STM) (AE006468), Yersinia pestis strain CO92 (Ype) (AL590842), and Vibrio cholerae strain N16961 (Vch) (AE003852). The same species were used in the ibp gene phylogeny, except for V. cholerae and the gene of W. glossinidia that was present in the plasmid pWb1 (NC 003425). Bootstrap values of below 50% were not reported.
The phylogeny of the ibp genes was obtained after removing the 5′ and 3′ nonconserved ends of each gene, which gave 528 nucleotide positions. The first and second positions of the alignment were used. The remaining parameters were the same as for the leucine cluster phylogeny. Species and accession numbers are given in the legend to Fig. 2.
RESULTS
Structure of the leucine cluster in BCp.
A chromosomal fragment of 8.5 kb, which contains nine complete ORFs and two partial ones, was amplified (Fig. 1). The gene content is as follows: the leucine cluster, with the gene order leuB, leuC, leuD, and leuA, flanked by the genes gnd and dcd; two complete genes (hisI and hisF) and a partial one (hisA) upstream of gnd; and the partial gene dcd downstream of leuB. The leucine genes are expressed in a strand different from that of the rest of the genes. It is worth noting that in the three B. aphidicola sequenced genomes, the chromosome structure of the region (both gene order and transcriptional direction) is the same as that found in BCp but lacks the four leucine genes. The corresponding positions on the B. aphidicola chromosome are from positions 109133, 109588, and 108962 (hisA) to 113817, 113513, and 113517 (dcd) in BAp, BSg, and BBp, respectively (37, 42, 46). The search for regulatory regions has shown inverted repeats that could act as transcription terminators between leuA and gnd (data not shown). Putative −35 and −10 promoter sequences, similar to those found for other B. aphidicola genes (34, 48), were not found upstream of leuA. Table 3 shows the intergenic regions found between the four leucine genes and the two flanking genes. The position of leuA, together with the large intergenic region between leuD and leuA, indicates that there probably is not an operon structure, as occurs in free living enterobacteria such as E. coli.
TABLE 3.
Sizes of the intergenic regions between the four leucine genes and the corresponding flanking genes of the four B. aphidicola strains with the genes in a chromosomal location and two leucine plasmids
| Strain or plasmid | Intergenic region | Size (nucleotides) |
|---|---|---|
| BCp | leuB-leuC | 3 |
| leuC-leuD | −3a | |
| leuD-leuA | 123 | |
| leuA-gnd | 158 | |
| dcd-leuB | 304 | |
| BTc | leuB-leuC | 4 |
| leuC-leuD | 2 | |
| leuD-leuA | 290 | |
| leuA-yadF | —b | |
| truA-leuB | 36 | |
| BBp | leuB-leuC | −3a |
| leuC-leuD | 24 | |
| leuD-leuA | 235 | |
| leuA-yqgF | 322 | |
| yggS-leuB | 449 | |
| BPs | leuB-leuC | 2 |
| leuC-leuD | 13 | |
| leuD-trxA | 36 | |
| leuA-leuB | 465 | |
| rep-leuA | 467 | |
| pBRpc | leuB-leuC | 2 |
| leuC-leuD | 3 | |
| leuD-repA1 | 374 | |
| leuA-leuB | 25 | |
| repA2-leuA | 146 | |
| pBTs | leuB-leuC | 3 |
| leuC-leuD | 37 | |
| leuD-leuA | 127 | |
| leuA-hspA | 227 | |
| repA1-leuB | 462 |
Overlapped nucleotides.
Not known.
pBRp was chosen as a representative of the B. aphidicola plasmid from the subfamily Aphidinae.
Structure of the leucine cluster in BTc.
A chromosomal fragment of 7.3 kb containing nine complete and two partial ORFs was amplified (Fig. 1). The gene content is as follows: the leucine cluster, with the gene order leuB, leuC, leuD, and leuA; the genes hpt (partial), panC, panB, dksA (in the opposite strand), and truA upstream of leuB; and an ORF followed by partial mrcB gene downstream of leuA. The genes hpt, panC, panB, dksA, truA, and mrcB are contiguous in the BAp chromosome (positions 212355 [hpt] and 218544 [mrcB]), whereas in BSg mrcB is a pseudogene. In BBp the genes hpt, panC, and panB have been lost, whereas dksA, truA, and mrcB are also contiguous (positions 209657 [dskA] and 213749 [mrcB]). The product of the ORF found upstream of mrcB was similar to the enzyme carbonic anhydrase (EC 4.2.1.1.) that is encoded by the gene yadF in E. coli (5). This gene is not present in any of the three B. aphidicola sequenced genomes, so it must have been lost in these lineages. The lengths of the three intergenic regions between truA and mrcB are 72, 78, and 249 nucleotides in BAp, BSg, and BBp, respectively (Table 4). According to the process of gene disintegration, which is postulated to be active during the evolution of the B. aphidicola genome (38), these sizes would indicate that the gene has recently been lost in the lineage to BBp and that the length of the intergenic region (249 nucleotides) is due to the remnant DNA (see Discussion).
TABLE 4.
Sizes of intergenic regions between contiguous genes of the three sequenced B. aphidicola genomes where the leucine cluster has been inserted in the four chromosomal versions
| Strain | Size (nucleotides)
|
|||
|---|---|---|---|---|
| truA-mrcB (BTc) | gnd-dcd (BCp) | trxA-rep (BPs) | yqgF-yggS (BBp) | |
| BAp | 72 | 163 | 202 | 118 |
| BSg | 78 | 162 | 159 | 32 |
| BBp | 249 | 305 | 149 | Leucine cluster |
In the search for regulatory regions, two inverted repeats (one imperfect) were found in the intergenic region between leuD and leuA, followed by a thymine-rich region, which is a necessary element for a rho-independent terminator (data not shown). This hairpin is different from the short inverted repeats found in the leucine plasmids (40). Two putative promoter regions were found, both upstream of leuA and leuB, thus suggesting that the leucine cluster in BTc is transcribed in two different transcripts leuBCD and leuA, which is supported by the sizes of the intergenic regions between the genes (Table 3).
Potential ribosome-binding sites.
The search for putative ribosome-binding sites upstream of the four leucine genes in the four B. aphidicola strains with the chromosomal location (Table 5) revealed that in four cases this regulatory sequence seems to be absent. The apparent absence of regulatory sequences similar to the eubacterial consensus sequence is a recurrent observation in studies of B. aphidicola DNA (27, 48). Evidence of this is demonstrated by the difficulty of finding a −35 sequence in the promoter of the genes, and it is mainly due to the high A+T content of the B. aphidicola genome (around 75%).
TABLE 5.
Potential ribosome-binding sites of the four leucine genes in the species that contain the leucine cluster in the chromosome
| Gene | Sequencea in:
|
|||
|---|---|---|---|---|
| C. populeti | T. caerulescens | P. spyrothecae | B. pistaciae | |
| leuA | ATTTGATATAAGAAAAAAATG | GTTAAGAGATATTTATATATG | AATTTGTAGGAAAATTTAATG | CATTTTTTGGACAATGAAATG |
| leuD | TCGTGAGAATAATCTTGTATG | AAATAAAATATTTTAAGGATG | ATTTAAAATAGGAAAAGAATG | TAATTAATAAGGATAAAAATG |
| leuC | TATTGAGGAACTATAAATATG | AACATGAGGAATAGTTTAATG | TAAAAAAGAGAATATAAAATG | TAATTAATAAGGATAAAAATG |
| leuB | AACTATATAGAAAAAAATATG | TTTTTGTCGAGGTTTAAAATG | ATTTAGAGTTATTTTATTATG | GACTTAAAATAATAACTTATG |
Underlining indicates potential ribosome-binding sites; boldface indicates start codons.
Chromosomal locations of the leucine cluster in four B. aphidicola strains.
Knowledge about the flanking genes of the leucine cluster in each species allows us to locate the position on the B. aphidicola ancestral chromosome. As mentioned above, the three sequenced genomes have a very well conserved gene order. More precisely, the pairs of genes trxA-rep, truA-mrcB, yqdF-yggS, and gnd-dcd are contiguous in the three sequenced B. aphidicola genomes, with the only exception being BBp, where the leuB, -C, -D, and -A genes are between yqgF and yggS. Table 4 shows the intergenic sizes for each pair of genes in the three sequenced genomes, where the four leucine genes have been located, and Fig. 1 shows the four different chromosomal structures of the regions containing the leucine cluster in the chromosome, as well as the gene order of the two leucine plasmids (Table 2) and the cryptic repA plasmids.
Phylogenetic analysis of leucine and ibp genes.
Two different phylogenetic analyses were carried out to learn more about the origin of the different chromosomal locations of the leucine gene and also to asses whether the plasmid versus chromosomal location had any influence on the phylogenetic relationship of the B. aphidicola strains.
Figure 2A shows the phylogenetic reconstruction obtained with the four concatenated leucine genes in the nine B. aphidicola strains (see Table 2 and Materials and Methods), four closely related free-living bacteria, and Vibrio cholerae, a distantly related species that was used as an outgroup. As it can be seen, all of the B. aphidicola strains cluster together, thus corroborating their monophyletic origin. The branch lengths show the evolutionary acceleration that B. aphidicola has undergone compared to its free-living relatives, as has already been stated in several previous works (see, e.g., reference 22). Regarding the relationship of the B. aphidicola strains, there is a clear monophyletic group formed by those associated with the Aphidinae subfamily (bootstrap value, 100). The remaining strains give a group formed by strains associated with the Thelaxinae and Chaitophorinae and, finally, the three strains of the Pemphiginae. This topology agrees with the one proposed by Heie (14), although the low support for some of the branches confirms the difficulty in obtaining a consistent phylogeny of the main B. aphidicola aphid lineages, as already pointed out (20, 29). Regarding the present work, the most relevant aspect of this phylogeny is that the four leucine genes seem to have evolved independently of their position, in either a plasmid or a chromosome, as shown by the cluster formed by BCp and pBTs. The group formed by strains associated with the Chaitophorinae and Thelaxinae has previously been obtained with other genes, such as those encoding GroEL (9).
The genome of Wigglesworthia glossinidia, the primary endosymbiont of the tse-tse fly (1), contains a small plasmid (pWig1) carrying eight genes. The W. glossinidia plasmid does not contain the leucine genes but does contain the gene ibp, the same as has been found in the plasmid pBTs and in the cryptic plasmid pBPs1 (Fig. 1). The similarity between the ibp gene from B. aphidicola and that from W. glossinidia was high (52 to 55%).
The ibp gene encodes a small heat shock protein belonging to the HSP20 gene family. This gene is present in several copies in some gamma-proteobacterial species. For example, Salmonella species contain three different copies, while E. coli contains only two (ibpA and ibpB). An analysis of orthology revealed that neither of the two E. coli genes was an ortholog of the Buchnera ibp genes. In Fig. 2B, a phylogenetic tree with two (ibp and ibpB) of the three paralogous genes is shown. The orthologous ibp group includes the four B. aphidicola ibp genes, located in either the plasmid or the chromosome; the plasmid-located Wigglesworthia gene; and the chromosomally located Salmonella genes STM1251 and STY1871. The close phylogenetic relationship between W. glossinidia and B. aphidicola was expected, according to a recent phylogenetic reconstruction (12). This topology suggests that an ibp gene carried by a plasmid was present in the ancestor of the endosymbiont species and that this gene was transferred to the main chromosome in an ancestor of B. aphidicola associated with the Aphidinae.
DISCUSSION
The sequencing of the two first B. aphidicola genomes, those of BAp (37) and BSg (42), showed complete conservation of the gene order, in spite of the 50 to 70 million years of divergence between the two strains (7). This perfect gene order conservation led Tamas et al. (42) to conclude that it was an extreme case of genome stability. As the strains showed differences in the presence of some genes and/or pseudogenes in the chromosome, the only evolutionary processes that could have changed the structure of the genome were gene loss and gene disintegration, thus supporting the reductive evolutionary process postulated for B. aphidicola evolution (21, 23, 38). Tamas et al. (42) proposed that both losses of repeated elements and losses of important genes involved in recombination, such as recA or recF, could account for the genomic stability in the B. aphidicola strains. The publication of the genome of the B. aphidicola endosymbiont (BBp) of B. pistaciae, belonging to the Pemphiginae (46), a lineage that diverged from the Aphidinae 80 to 150 million years ago (49), showed almost perfect gene order conservation of the chromosome, with only two small inversions and two translocations. Thus, the previously reported perfect chromosomal synteny, or genomic stasis, was also found in the BBp genome. Taking into account the data for the three genomes, and considering that the Pemphiginae could be the most basal branching among all present B. aphidicola, van Ham et al. (46) proposed that B. aphidicola possesses a “gene order fossil” that has remained practically unchanged since the origin of aphid infection. Following this reasoning, it was possible to estimate that 638 genes, which is the total number of genes found in all the strains, would be the minimum gene content of the LCSA of the three B. aphidicola lineages. Since then, 164 independent gene losses would have occurred in the lineages, leading to the three present genomes (39).
Finally, taking into account that horizontal gene transfer is a very rare phenomenon in B. aphidicola (but see reference 45), together with the observed synteny, it was possible to infer that gene loss in B. aphidicola is an ongoing process in all of the B. aphidicola lineages. This fact is corroborated by the finding of some B. aphidicola strains associated with the Lachininae subfamily with chromosomal genome sizes of 450 to 470 kb (11). In fact, we can consider the genome of a B. aphidicola strain associated with the aphid Cinara cedri to be the smallest known bacterial genome reported so far (450 kb). The evolution of B. aphidicola would be a case of degenerated, rather than adaptive, genome evolution. Genetic isolation and small effective population size may be main determinants of this degenerative process (22). According to van Ham et al. (46), prolonged genomic stasis could be unsustainable in the long term and could be a symptom of genome degeneracy, despite the strength of compensatory processes such as the stabilizing effect of chaperones on cellular proteins (10). It has also been stated that B. aphidicola was essential to the success of aphids in the initial radiation but is no longer a source of ecological innovation for its host, because the ecological diversification of aphids cannot be attributed to the current genetic diversity of B. aphidicola (26, 42).
However, the results obtained in the present work cannot be explained under the genomic stasis hypothesis. Regarding the leucine genes, up to seven different repA plasmids (6, 40, 45, 48) and four different leucine gene chromosomal positions have been found (references 34 and 45 and the present work).
Since van Ham et al. (48) discovered different locations of the leucine gene cluster, either on the chromosome or on a plasmid, two possible scenarios have been proposed, as follows.
(i) The leucine cluster, probably an operon as in E. coli (50), was located in the chromosome of the B. aphidicola LCSA that predated the symbiosis about 200 million years ago. This bacterium would have carried a cryptic plasmid with at least a repA gene. After establishing symbiosis, the leucine genes were transferred to plasmids independently in several B. aphidicola lineages, resulting in leucine plasmids, with a different gene order and gene content. The minimum number of transfer events would have depended on the phylogenetic relationship used (two in the relationship shown in Fig. 2A). This was the scenario first proposed by our group, for the evolution of both the leucine cluster (48) and the trpEG genes (47). Accordingly, the four different locations of the leucine cluster in the B. aphidicola chromosome would be due to intrachromosomal rearrangements.
(ii) Alternatively, the transfer of the leucine cluster to a repA plasmid took place only once in the common ancestor of B. aphidicola. The different locations of the leucine genes in the B. aphidicola chromosome were due to independent back transfers to the main chromosome throughout B. aphidicola evolution.
The first leucine chromosomal cluster and its flanking genes, from a B. aphidicola strain (BPs) associated with a member of the Pemphiginae, were sequenced by Sabater-Muñoz et al. (34) (Table 2 and Fig. 1). In that work it was postulated that a leucine plasmid was present in the B. aphidicola LCSA that preceded the diversification of all the endosymbionts and that the chromosomal location of the leucine genes observed in some B. aphidicola strains arose by a transfer of such genes from a plasmid to the main chromosome. A three-step back transfer scenario was then postulated, supported by the large sizes of the intergenic regions between leuB and leuA (Table 3) and between the genes ibp and repA1 in the cryptic plasmid (858 bp) of BPs (Fig. 1).
The sequencing of the genome of BBp, another strain associated with the Pemphiginae, showed that the leucine cluster was flanked by different genes (yqgF and yggS) and had the gene order leuBCDA (Fig. 1). A striking fact was that yqgF and yggS were adjacent in BAp and BSg, while the leucine cluster flanking genes trx and rep were contiguous in BBp chromosome (Table 4). These results suggest that the chromosomal positions of the leucine cluster in BBp and BPs were due to two independent insertion events in the ancestral LCSA chromosome, even though the two aphid species belong to the same subfamily.
The sequencing of two new chromosomal leucine clusters carried out in the present work (those of BTc, another strain associated with Pemphiginae from a third tribe, and of BCp, associated with the subfamily Chaitophorinae) also supports the second scenario. Moreover, given the overall data, the four chromosomal locations found in three strains associated with the subfamily Pemphiginae and one associated with the Chaitophorinae can be explained only by four independent insertions (Fig. 2A). The cryptic repA plasmids found in the subfamily Pemphiginae would be the remnant of the ancestral leucine plasmid, as previously postulated (34).
In BTc, the cluster is inserted between the genes truA and yadF (followed by mrcB). In the three sequenced genomes, the gene yadF is absent (Table 4). As mentioned above, a possible explanation is that yadF was present in the LCSA but was convergently lost and disintegrated both in the lineage of B. pistaciae and in the Aphidinae. In the case of the Pemphiginae family, the loss would have taken place after the divergence of the Eriosomatini from the Fordini tribe. The large intergenic region in BBp (249 bp [Table 4]) would indicate that, in fact, this process of disintegration began recently in the Fordini tribe. In the case of BSg and BAp, the small intergenic spaces would indicate that the yadF gene started its disintegration in the ancestor of the Aphidinae subfamily.
In BCp the insertion occurred between the genes gnd and dcd. These two genes are also contiguous in the three sequenced genomes (Table 4), thus indicating that the insertion must have occurred in the lineage leading to the Chaitophorinae. We know that, at least in Chaitophorus leucomelas, another species of the same genus, the leucine cluster is inserted in the same position (data not shown). More data are needed to know whether the back transfer predates the divergence of the Chaitophorinae lineage from the rest of Drepanosiphine group (31).
It has been postulated that the absence of essential genes involved in recombination and repair processes would explain, at least in part, the gene order conservation. We postulate that as recA is absent from the B. aphidicola genome, the possible insertions would have been mediated by the recBCD system that has been retained in the three sequenced genomes and in the B. aphidicola strain associated with C. cedri, which is currently being sequenced (data not shown). Thus, in the absence of recA, recBCD may serve as a general exonuclease repair enzyme functioning as a substitute for recombinational repair.
The ancestral LCSA plasmid may contain the four leucine genes and at least one repA gene, plus two additional genes, ibp and yqhA. The ibp gene is present in two of the seven types of B. aphidicola repA plasmids, and an orthologous gene was detected in the small plasmid present in W. glossinidia (Fig. 2B). The presence of ibp in the chromosome in BAp and BSg probably indicates a back transfer event. A very intriguing fact is that the ibp gene is absent in the BBp genome, while the trpEG genes in the BBp genome are located in the place where the ibp gene is located in the BAp and BSg genomes. The possibility that the trpEG genes also contained in plasmid in some B. aphidicola lineages are in some way related to the repA plasmid is a question that deserves more intense study.
Regarding the yqhA gene, it is present in five of the seven types of B. aphidicola repA plasmids, in strains associated with the subfamilies Aphidinae, Pterocommatinae, Thelaxinae, and Pemphiginae. This wider distribution suggests that the gene may be present in the ancestral plasmid, and it could have been transferred to the main chromosome in the other four types of repA plasmids.
Finally, the reason why some lineages transferred the plasmid back to the main chromosome is an open and unsolved question but is probably related to a nutritional basis (25) and/or to the chromosome and plasmid copy numbers. The presence of the leucine genes in a plasmid in the LCSA was probably advantageous, due to the large number of plasmid copies. However, the loss of genes involved in the control of chromosome replication and segregation led to polyploidy of the bacterial cell. Thus, in many strains, the number of chromosomal copies could be higher than the number of plasmid copies (44). In these circumstances, the presence of the leucine genes in the chromosome could have turned out to be more advantageous for the stability and expression of these genes.
Acknowledgments
Financial support was provided by a fellowship (FP97-9194430) to B.S.-M. and by projects BMC2003-00305 from Ministerio de Ciencia y Tecnología (Spain) and GRVPOS203/204 from Generalitat Valenciana (Spain).
We thank J. M. Michelena and P. González for aphid species identification and F. Barraclough for help with the English. We also acknowledge Servicio de Secuenciación de Ácidos Nucléicos y Proteínas and Servicio de Bioinformática at SCSIE (Universitat de València) for sequencing and bioinformatics support, respectively.
REFERENCES
- 1.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, L., P. Baumann, C. Y. Lai, D. Rouhbakhsh, N. A. Moran, and M. A. Clark. 1995. Genetics, physiology, and evolutionary relationships of the genus B. aphidicola: intracellular symbionts of aphids. Annu. Rev. Microbiol. 49:55-94. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, L., P. Baumann, N. A. Moran, J. Sandström, and M. L. Thao. 1999. Genetic characterization of plasmids containing genes encoding enzymes of leucine biosynthesis in endosymbionts (B. aphidicola) of aphids. J. Mol. Evol. 48:77-85. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, P., N. A. Moran, and L. Baumann. 1997. The evolution and genetics of aphid endosymbiotics. Bioscience 47:12-20. [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Bracho, A. M., D. Martínez-Torres, A. Moya, and A. Latorre. 1995. Discovery and molecular characterization of a plasmid localized in B. aphidicola sp., bacterial endosymbiont of the aphid Rhopalosiphum padi. J. Mol. Evol. 41:67-73. [DOI] [PubMed] [Google Scholar]
- 7.Clark, M. A., N. A. Moran, and P. Baumann. 1999. Sequence evolution in bacterial endosymbionts having extreme base composition. Mol. Biol. Evol. 16:1586-1598. [DOI] [PubMed] [Google Scholar]
- 8.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria B. aphidicola. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 9.Fares, M. A., E. Barrio, B. Sabater-Muñoz, and A. Moya. 2002. The evolution of the heat-shock protein GroEL from Buchnera, the primary endosymbiont of aphids, is governed by positive selection. Mol. Biol. Evol. 19:1162-1170. [DOI] [PubMed] [Google Scholar]
- 10.Fares, M. A., M. X. Ruiz-González, A. Moya, S. F. Elena, and E. Barrio. 2002. Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature 417:398. [DOI] [PubMed] [Google Scholar]
- 11.Gil, R., B. Sabater-Muñoz, A. Latorre, F. J. Silva, and A. Moya. 2002. Extreme genome reduction in Buchnera spp.: toward the minimal genome needed for symbiotic life. Proc. Natl. Acad. Sci. USA 99:4454-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil, R., F. J. Silva, E. Zientz, F. Delmotte, F. González-Candelas, A. Latorre, C. Rausell, J. Kamerbeek, J. Gadau, B. Hölldobler, R. C. H. J. van Ham, R. Gross, and A. Moya. 2003. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc. Natl. Acad. Sci. USA 100:9388-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison, C. P., A. E. Douglas, and A. F. G. Dixon. 1989. A rapid method to isolate symbiotic bacteria from aphids. J. Invertebr. Pathol. 53:427-428. [Google Scholar]
- 14.Heie, O. E. 1987. Paleontology and phylogeny, p. 367-391. In A. K. Minks and P. Harrewijn (ed.), World crop pests, vol. 2A. Aphids: their biology, natural enemies and control. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 15.Komaki, K., and H. Ishikawa. 1999. Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J. Mol. Evol. 48:717-722. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, I. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetic analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 17.Lai, C. Y., L. Baumann, and P. Baumann. 1994. Amplification of trpEG: adaptation of B. aphidicola to an endosymbiotic association with aphids. Proc. Natl. Acad. Sci. USA 91:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latorre, A., A. Moya, and F. J. Ayala. 1986. Evolution of mitochondrial DNA in Drosophila subobscura. Proc. Natl. Acad. Sci. USA 83:8649-8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchuck, D., M. Drumm, A. Saulino, and F. S. Collins. 1992. Construction of T-vectors, a rapid general system for direct cloning of unmodified PCR products. Nucleic Acid Res. 19:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez-Torres, D., C. Buades, A. Latorre, and A. Moya. 2001. Molecular systematics of aphids and their primary endosymbionts. Mol. Phylogenet. Evol. 20:437-449. [DOI] [PubMed] [Google Scholar]
- 21.Mira, A., H. Ochman, and M. A. 2001. Deletion bias and the evolution of bacterial genomes. Trends Genet. 17:589-596. [DOI] [PubMed] [Google Scholar]
- 22.Moran, N. A. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran, N. A., and A. Mira. 2001. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2:0054.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran, N. A., M. A. Munson, P. Baumann, and H. Ishikawa. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect host. Proc. R. Soc. London B 253:167-171. [Google Scholar]
- 25.Moran, N. A., G. R. Plague, J. P. Sandström, and J. L. Wilcox. 2003. A genomic perspective on nutrients provisioning by bacterial symbionts of insects. Proc. Natl. Acad. Sci. USA 100:14543-14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran, N. A., and J. J. Wernegreen. 2000. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 15:321-326. [DOI] [PubMed] [Google Scholar]
- 27.Munson, M. A., and P. Baumann. 1993. Molecular cloning and nucleotide sequence of a putative trpDC(F)BA operon in Buchnera aphidicola (endosymbiont of the aphid Schizaphis graminum). J. Bacteriol. 175:6426-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munson, M. A., M. A. Clark, L. Baumann, N. A. Moran, D. J. Voegtlin, and B. C. Campbell. 1991. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J. Bacteriol. 173:6321-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz-Rivas, B., A. Moya, and D. Martínez-Torres. 2003. Molecular systematics of aphids (Homoptera: Aphididae): new insights from the long-wavelength opsin gene. Mol. Phylogenet. Evol. 30:24-37. [DOI] [PubMed] [Google Scholar]
- 30.Plague, G. R., C. Dale, and N. A. Moran. 2003. Low and homogeneous copy number of plasmid-borne symbiont genes affecting host nutrition in Buchnera aphidicola of the aphid Uroleucon ambrosiae. Mol. Ecol. 12:1095-1100. [DOI] [PubMed] [Google Scholar]
- 31.Quednau, F. W. 1999. Atlas of the Drepanosiphine aphids of the world, part I. Panaphidini Oestlund, 1922-Myzocallidina Börner, 1942 (Hemiptera: Aphididae: Calaphidinae). Contrib. Am. Entomol. Inst. 31:4-70. [Google Scholar]
- 32.Remaudière, G., and M. Remaudière. 1997. Catalogue des Aphididae du monde: Homptera, Aphidoidea. Institut National de la Recherche Agronomique, Paris, France.
- 33.Rouhbakhsh, D., C. Y. Lai, C. von Dohlen, M. A. Clark, L. Baumann, P. Baumnn, N. A. Moran, and D. J. Voegtlin. 1996. The tryptophan biosynthesis pathway of aphid endosymbionts (Buchnera): genetics and evolution of plasmid-associated anthranilate synthase (trpEG) within the Aphididae. J. Mol. Evol. 42:414-421. [DOI] [PubMed] [Google Scholar]
- 34.Sabater-Muñoz, B., L. Gómez-Valero, R. C. H. J. van Ham, F. J. Silva, and A. Latorre. 2002. Molecular characterization of the leucine cluster in B. aphidicola sp. strain PSY, a primary endosymbiont of the aphid Pemphigus spyrothecae. Appl. Environ. Microbiol. 68:2572-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids B. aphidicola sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 38.Silva, F. J., A. Latorre, and A. Moya. 2001. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 17:615-618. [DOI] [PubMed] [Google Scholar]
- 39.Silva, F. J., A. Latorre, and A. Moya. 2003. Why are the genomes of endosymbiotic bacteria so stable?. Trends Genet. 19:176-180. [DOI] [PubMed] [Google Scholar]
- 40.Silva, F. J., R. C. H. J. van Ham, B. Sabater, and A. Latorre. 1998. Structure and evolution of the leucine plasmids carried by the endosymbiont (Buchnera aphidicola) from aphids of the family Aphididae. FEMS Microbiol. Lett. 168:43-49. [DOI] [PubMed] [Google Scholar]
- 41.Soler, T., A. Latorre, B. Sabater, and F. J. Silva. 2000. Molecular characterization of the leucine plasmid from B. aphidicola, primary endosymbiont of the aphid Acyrtosiphon pisum. Curr. Microbiol. 40:264-268. [DOI] [PubMed] [Google Scholar]
- 42.Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 43.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 44.Thao, M. L., L. Baumann, P. Bauman, and N. A. Moran. 1998. Endosymbionts (Buchnera) from the aphids Schizaphis graminum and Diuraphis noxia have different copy numbers of the plasmid containing the leucine biosynthetic genes. Curr. Microbiol. 36:238-240. [DOI] [PubMed] [Google Scholar]
- 45.van Ham, R. C. H. J., F. González-Candelas, F. J. Silva, B. Sabater, A. Moya, and A. Latorre. 2000. Postsymbiotic plasmid acquisition and evolution of the repA1-replicon in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 97:10855-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Ham, R. C. H. J., J. Kamerbeek, C. Palacios, C. Rausell, F. Abascal, U. Bastolla, J. M. Fernández, L. Jiménez, M. Postigo, F. J. Silva, J. Tamames, E. Viguera, A. Latorre, F. Morán, and A. Moya. 2003. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 100:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Ham, R. C. H. J., D. Martínez-Torres, A. Moya, and A. Latorre. 1999. Plasmid-encoded anthranilate synthase (trpEG) in Buchnera aphidicola from the family Pemphigidae. Appl. Environ. Microbiol. 65:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Ham, R. C. H. J., A. Moya, and A. Latorre. 1997. Putative evolutionary origin of plasmids carrying the genes involved in leucine biosynthesis in Buchnera aphidicola (endosymbiont of aphids). J. Bacteriol. 179:4768-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Dohlen, C. D., and N. A. Moran. 2000. Molecular data support a rapid radiation of aphids in the Cretaceous and multiple origins of host alternation. Biol. J. Linn. Soc. 71:689-717. [Google Scholar]
- 50.Wessler, S. R., and J. M. Calvo. 1981. Control of leu operon expression in Escherichia coli by a transcription attenuation. J. Mol. Biol. 149:579-597. [DOI] [PubMed] [Google Scholar]