Abstract
The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon at two levels. H-NS binds upstream of the promoter, represses transcription initiation, and binds downstream within the coding region of the first gene, where it induces polarity of transcription elongation. In hns mutants, silencing of the bgl operon is completely relieved. Various screens for mutants in which silencing of bgl is reduced have yielded mutations in hns and in genes encoding the transcription factors LeuO and BglJ. In order to identify additional factors that regulate bgl, we performed a transposon mutagenesis screen for mutants in which silencing of the operon is strengthened. This screen yielded mutants with mutations in cyaA, hfq, lon, and pgi, encoding adenylate cyclase, RNA-binding protein Hfq, protease Lon, and phosphoglucose isomerase, respectively. In cyaA mutants, the cyclic AMP receptor protein-dependent promoter is presumably inactive. The specific effect of the pgi mutants on bgl is low. Interestingly, in the hfq and lon mutants, the downstream silencing of bgl by H-NS (i.e., the induction of polarity) is more efficient, while the silencing of the promoter by H-NS is unaffected. Furthermore, in an hns mutant, Hfq has no significant effect and the effect of Lon is reduced. These data provide evidence that the specific repression by H-NS can (directly or indirectly) be modulated and controlled by other pleiotropic regulators.
The abundant histone-like nucleoid structuring protein H-NS is a key regulator in the adaptation of Escherichia coli to its environment. H-NS directly or indirectly affects the expression of ∼5% of the genes in E. coli K-12, including pathogen determinants, several motility and adhesion systems, and proteins of the osmotic and acid stress responses, many of which are controlled by environmental signals (17). H-NS binds nonspecifically to DNA with a preference for bent and AT-rich DNA sequences, represses the transcription of most loci, and is present at very high cellular levels (∼20,000 to 60,000 molecules per cell) (47). To date, little is known about how H-NS activity is modulated. Expression of the hns gene is growth rate regulated and autorepressed (6, 11). At the translational level, hns is repressed upon overproduction of the 87-nucleotide regulatory RNA DsrA. This repression depends on Hfq (25, 26, 49). In addition, H-NS activity may be modulated by its homologue, StpA, with which it can form heterodimers (18, 19, 53).
Among the H-NS-controlled loci, the repression of the E. coli bgl operon and the proU operon by H-NS is exceptionally specific. The proU operon encoding an uptake system for the osmoprotectants glycine and betaine is induced by osmotic stress. The bgl operon encodes the gene products for the fermentation of aryl-β,d-glucoside. Its gene products are the positive regulator and antiterminator BglG, the β-glucoside-specific permease EIIBgl (or BglF), and the phospho-β,d-glucosidase BglB (see Fig. 2). The bgl operon is silent under all laboratory growth conditions (35, 39). Both the proU and bgl operons are repressed ∼50- to 100-fold by H-NS, and in both systems, regulatory elements located upstream and downstream of the promoter are required for the H-NS-mediated repression (9, 20, 21, 43, 48). We have shown that repression of the bgl operon by H-NS occurs at two levels (Fig. 1). H-NS represses transcription initiation at the cyclic AMP (cAMP) receptor protein (CRP)-dependent promoter by binding to an AT-rich and presumably bent upstream silencer sequence (32, 43, 46, 48). In addition, H-NS binds to a downstream silencer located within the coding region of the first gene, bglG, ∼600 to 700 bp downstream of the transcription initiation site, where it induces a Rho-dependent polarity of transcription (9).
FIG. 2.
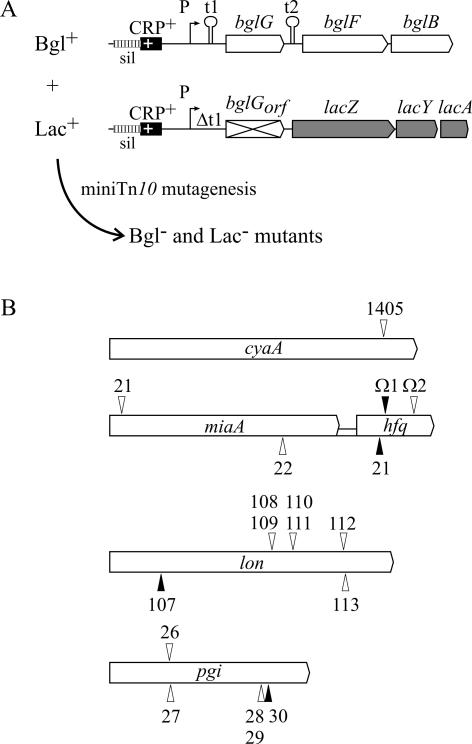
Mutagenesis screen for mutants with reduced expression of the bgl operon. (A) Mutagenesis strategy to isolate in trans mutations affecting expression of the bgl operon. Strain S594 carries an activated bgl operon and a fusion of the bgl regulatory region to the lac operon. In both the bgl operon and the bgl-lac fusion, the bgl promoter is activated by an improved CRP-binding site, and thus strain S594 is Bgl+ Lac+. Expression of the bgl-lac fusion is independent of BglG-mediated antitermination at terminator t1, due to a deletion (Δt1) of the terminator (including positions +55 to +120 relative to the transcription start site). In addition, translation of bglG was excluded by mutation of the translation start codon and codon 3 to GCG (bglGorf). After mini-Tn10 mutagenesis, mutants with a double-phenotype change to Bgl− Lac− were isolated. (B) Schematic representation of the mini-Tn10 insertion mutants obtained in the screen. The insertion sites were sequenced using a Tn10-specific primer (Materials and Methods and Table 1). Arrowheads above the mutated gene depict insertions in orientation I, while arrowheads underneath the gene were used for insertions in orientation II. Solid arrowheads represent the alleles used throughout the study. The hfq alleles Ω1 and Ω2 (50) were also used.
FIG. 1.
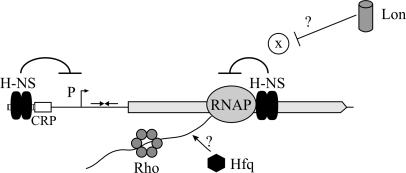
Model of the H-NS-mediated repression of the bgl operon at two levels. H-NS binds upstream of the promoter and represses transcription initiation. In addition, H-NS binds within the coding region of the first gene, approximately 600 to 700 bp downstream of the transcription initiation site, where it induces a Rho-dependent polarity (9). Hfq and Lon reduce the H-NS-induced polarity (this work).
In addition to H-NS, which is essential for bgl operon silencing, the operon is affected by other pleiotropic regulators. Constitutive expression of leuO and bglJ relieves silencing of bgl (14, 51). The leuO gene encodes a pleiotropic transcription factor that controls, e.g., dsrA (37), while bglJ encodes a putative transcription factor of the LuxR family of unknown function. Furthermore, RpoS contributes to repression of the operon (8, 33, 44), while the H-NS homologue StpA and the RNA chaperone Hfq are involved in regulation of bgl in an hns mutant that expresses a truncated H-NS protein (12, 50). Silencing of the bgl operon can also be relieved by in cis mutations that disrupt the upstream silencer or make the CRP-binding site more similar to the consensus CRP-binding site (40, 45, 48).
In this work, we report that the RNA chaperone Hfq and the protease Lon reduce silencing of the bgl operon. While in all screens to date, mutants were isolated in which silencing of bgl is relieved, these factors were characterized in a reverse screen for mutants in which silencing of bgl is strengthened. Further characterization of the hfq and lon mutants revealed that the H-NS-mediated silencing of bgl via the downstream silencer was more efficient, while silencing of the bgl promoter was unchanged. Furthermore, in an hns null mutant background, mutations in hfq and lon had little effect on bgl, which suggests that Hfq and Lon directly or indirectly control H-NS to induce polarity at this specific locus.
MATERIALS AND METHODS
Strains and plasmids.
The genotypes of the E. coli strains used in this study are given in Table 1. All experiments were performed by using isogenic strains derived from E. coli K-12 CSH50 [ara Δ(gpt-lac) ara thi] (27). Mutations were transduced by using phage T4GT7 (54). Integration of bgl-lacZ reporter gene fusions into the chromosomal phage λ attachment site attB was performed as described previously (7, 8). Strains S581 and S594 carry replacements of the lac promoter by the bgl promoter, including the upstream and downstream regulatory elements. They were constructed by site-specific recombination using plasmids pKES50 and pKES51, respectively, according to Dabert and Smith (4). These plasmids carry a fragment encompassing the lacI gene, followed by an Ω spectinomycin resistance cassette (36), the bgl regulatory region, and the lacZ gene. Within the bgl fragment terminator, t1 was deleted (from positions +55 to +120 relative to the transcription start) and translation of bglG was excluded by mutation of the translation start codon and codon 3 of bglG (ATG to GCG). The lacI-bgl-lacZ cassette is flanked by chi sites (5′GCTGGTGG) in the proper orientation to enhance site-specific recombination (4). Recombinants were selected on spectinomycin plates, and correct replacement was tested by PCR.
TABLE 1.
Characteristics of the E. coli K-12 strains used in this study
| Strain | Genotypea | Constructionb or reference |
|---|---|---|
| AM111 | MC4100 hfq1::Ω(Kmr) | 31, 50 |
| AM112 | MC4100 hfq2::Ω(Kmr) | 31, 50 |
| CSH50 | bglo Δ(lac-pro) ara thi | 27 |
| CY307 | zcb-222::Tn10 pyrD34 relA1 spoT1 metB1-Hfr (argF→lac) | CGSC#6428 |
| JT4000 | Pro+ Tcs Δlon-510 | Susan Gottesman |
| KL788 | λ−Thr-1 Δ(gpt-lac)5 tsx-35 sulA3 el4−Rac-0? rfbD1? mgl-51 recA441 (Ts) relA1? rpsL31 (strR) kdgK51? mtl-1 spoT1? thi-1? lexA71::Tn5 creC510? | CGSC#6218 |
| M182 | 55 | |
| stpA::Tcr | ||
| PD32 | MC4100 hns-206::Apr Strr | 6 |
| RH90 | RH90 = MC4100 rpoS359::Tn10 | 24 |
| SG1039 | Δlac proC mutant zaj-403::tet | Susan Gottesman |
| W3110 | λ− F− IN(rrnD-rrnE) | CGSC#4474 (2) |
| S486 | CSH50 bglo (Bgl−) (gpt-lac)+ | 8 |
| S539 | CSH50 Δbgl-AC11 (gpt-lac)+ | 8 |
| S541 | CSH50 Δbgl-AC11 (gpt-lac)+ ΔlacZ-Y217 | 8 |
| S544 | CSH50 bgl-CRP+ (Bgl+) (gpt-lac)+ ΔlacZ-Y217 | 8 |
| S572 | S544 (gpt-lac)+ | ×− T4GT7(W3110), Lac+ Pro+ |
| S581 | S486 bglo ΔlacOP::(Spcr Pbgl Δ(+55 to 120) bglGorf) | × pKES50 |
| S594 | S572 bgl-CRP+ ΔlacOP::(Spcrbgl-CRP+ PbglΔ(+55 to 120) bglGorf) | × pKES51 |
| S690 | S539 hns-206::Apr | × T4GT7(PD32), Apr |
| S706 | S594 hns-206::Apr | × T4GT7(PD32), Apr |
| S749 | S594 lon-110::mTn10 (orientation I, position lon:1537) | This work |
| S750 | S594 miaA21::mTn10 (I, miaA:50) | This work |
| S751 | S594 pgi-30::mTn10 (II, pgi:1325) | This work |
| S752 | S594 pgi-28::mTn10 (II, pgi:1270) | This work |
| S753 | S594 pgi-29::mTn10 (identical to pgi-28::mTn10) | This work |
| S754 | S594 lon-113::mTn10 (II, lon:1976) | This work |
| S755 | S594 pgi-27::mTn10 (II, pgi:509) | This work |
| S756 | S594 cyaA1405::mTn10 (I, cyaA:2296) | This work |
| S757 | S594 hfq-21::mTn10 (I, hfq:96) | This work |
| S759 | S594 lon-108::mTn10 (I, lon:1360) | This work |
| S760 | S594 lon-112::mTn10 (I, lon:1960) | This work |
| S762 | S594 lon-111::mTn10 (same as S749) | This work |
| S763 | S594 miaA22::mTn10 (II, miaA:733) | This work |
| S764 | S594 lon-107::mTn10 (II, lon:430) | This work |
| S765 | S594 pgi-26::mTn10 (I, pgi:500) | This work |
| S766 | S594 lon-109::mTn10 (as lon-108::mTn10) | This work |
| S778 | S539 hfq-21::mTn10 | × T4GT7 (S757), Tcr |
| S794 | S539 lon-107::mTn10 | × T4GT7(S764), Tcr |
| S1048 | S594 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1097 | S541 attB::[SpcrlacUV5 t1-L bglG lacZ] | 8 |
| S1105 | S539 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1132 | S1079 rpoS359::Tn10 | × T4GT7(RH90), Tcr |
| S1182 | S541 zcb-222::Tn10 pyrD34 | × T4GT7(S1179), Tcr |
| S1185 | S1182 sulA3 pyrD+ (a lon-107::mTn10 derivative of this strain is nitrofurantoin resistant) (13) | × T4GT7(CY307), Ura+ |
| S1189 | S541 attB::[SpcrlacUV5 bgl-t1L bglGorf lacZ] | 9 |
| S1193 | S541 attB::[SpcrlacUV5 (+95)bglG lacZ] | 9 |
| S1195 | S541 attB::[SpcrlacUV5 (+95)bglGorf lacZ] | 9 |
| S1211 | S541 attB::[Spcr Pbgl(−76 to +25) lacZ] | 9 |
| S1213 | S541 attB::[Spcr Pbgl(+25) lacZ] | 9 |
| S1215 | S541 attB::[Spcrbgl-CRP+ Pbgl(+25) lacZ] | 9 |
| S1218 | S1185 proC mutant zaj::Tn10 | × T4GT7(SG1039), Tcr |
| S1219 | S1189 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1223 | S1193 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1225 | S1195 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1258 | S1195 hns::Apr | 9 |
| S1311 | S1097 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1371 | S1195 stpA::Tcr | × T4GT7(M182 stpA::Tcr) |
| S1379 | S541 Δlon-510 sulA3 clpP::CmrstpA::TcrattB::[SpcrlacUV5 (+95)bglGorf lacZ] | × T4GT7(M182 stpA::Tcr) |
| S1416 | S1132 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1422 | S539 pgi-30::mTn10 | × T4GT7(S751), Tcr |
| S1424 | S539 pgi-26::mTn10 | × T4GT7(S765), Tcr |
| S1432 | S541 Δlon-510 sulA3 clpP::Cmrhns-206::AprattB::[SpcrlacUV5 (+95)bglGorf lacZ] | × pKESD49 |
| S1553 | S1218 Δlon-510 proC+ | × T4GT7(JT4000), Pro+ Tcs |
| S1554 | S1553 attB::[Spcr Pbgl(−76 to +25) lacZ] | × pKEKB25 |
| S1556 | S1553 attB::[Spcr Pbgl(+25) lacZ] | × pKEKB30 |
| S1558 | S1553 attB::[Spcrbgl-CRP+ Pbgl(+25) lacZ] | × pKEYK1 |
| S1562 | S1553 attB::[SpcrlacUV5 (+95)bglG lacZ] | × pKESD48 |
| S1564 | S1553 attB::[SpcrlacUV5 (+95)bglGorf lacZ] | × pKESD49 |
| S1582 | S1213 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1584 | S1211 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1586 | S1215 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1588 | S1217 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1624 | S541 attB::[SpcrlacUV5 bgl(1-173)t1-L lacZ] | × pKESD44 |
| S1642 | S1553 attB::[SpcrlacUV5 bgl(1-173)t1-L lacZ] | × pKESD44 |
| S1654 | S594 hfq2::Ω(Kmr) | × T4GT7(AM112), Kmr |
| S1656 | S539 hfq2::Ω(Kmr) | × T4GT7(AM112), Kmr |
| S1676 | S1624 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
| S1680 | S1553 attB::[SpcrlacUV5 t1-L bglG lacZ] | × pKESD20 |
| S1684 | S1553 attB::[SpcrlacUV5 t1-L bglGorf lacZ] | × pKESD28 |
| S1767 | S1371 hns:amp | × T4GT7(PD32), Apr |
| S1828 | S1258 hfq1::Ω(Kmr) | × T4GT7(AM111), Kmr |
Strains carrying the silent wild-type operon, designated bglo, are Bgl− (for nomenclature see reference 39). bgl-CRP+ (a C-to-T exchange in the CRP binding site at position −66 relative to the transcription start) causes activation of the bgl operon. In the t1-L terminator mutant, 3 bases in the stem-loop structure of the terminator are mutated (8). Pbgl and lacUV5 represent the bgl and lacUV5 promoters, respectively, and bglGorf represents a bglG mutant, in which the translation initiation codon was mutated. The coordinates of cloned fragments are given in brackets, where relevant. For mini-Tn10 mutations, the orientation (I or II) and the position of the insertion site are given relative to the translation start of the mutated gene (e.g., lon:1360). Orientation I indicates that the Tn10 tetA gene is in the same orientation as the mutated gene. Tn10 usually generates a target site duplication of 9 bp. The position of the nucleotide located directly 3′ to mini-Tn10 is given for insertions in orientation I, while the position of the nucleotide located directly 5′ to mini-Tn10 is given for insertions in orientation II. Tcr, tetracycline resistant; Apr ampicillin resistant; Kmr, kanamycin resistant; and Spcr, spectinomycin resistant.
For strains that were obtained from the E. coli Genetic Stock Center, the respective strain number is given as a reference. ×, in strains S581 and S594, the lac promoter was replaced by the bgl regulatory region by site-specific recombination using plasmids pKES44 and pKES51, respectively, according to Dobert and Smith (4). Transductants (using T4GT7) were selected as indicated. In case of the transduction of lon alleles, which map at 9.8 min, phenotype analysis or PCR confirmed that the lac locus, which is located at 7.8 min, was not cotransduced. Integrations into attB were performed as described previously (8).
Plasmids were constructed according to standard techniques (42). Site-specific mutations and fusion of bgl and lac sequences were introduced by PCR. All regions of plasmids that were derived from PCR fragments were sequenced. The relevant structures of the plasmids are schematically shown in the figures. Details of constructions and compiled sequences of the plasmids are available upon request. Media and plates were used as described previously (8). Where necessary, antibiotics were added to final concentrations of 25-μg/ml kanamycin, 50-μg/ml ampicillin, 15-μg/ml chloramphenicol, and 50-μg/ml spectinomycin.
Transposon mutagenesis.
The transposon mutagenesis screen was performed using λ phage NK1323, which carries a mini-Tn10 transposon with a tetracycline resistance marker (23). Due to several mutations, the λ phage does not replicate in wild-type E. coli and it cannot integrate into the genome of E. coli as a prophage (23). In addition, the transposase gene is encoded outside of the mini-Tn10 on the λ phage and thus was lost along with the other λ sequences. Therefore, after infection, single transposition events of the mini-Tn10 transposon into the chromosome can be selected. Transduction experiments and direct sequencing of the chromosomal DNA confirmed that all mutants isolated were due to single transposition events (data not shown).
In the mutagenesis screen, strains S581 and S594, respectively, were infected with λ NK1323 and plated onto MacConkey lactose-tetracycline plates. Mutants with a change in the lactose phenotype were restreaked, and their Bgl phenotype was tested on bromthymol blue-salicin indicator plates (8). For mutants with a double phenotype change, the insertion position of the mini-Tn10 transposon on the chromosome was determined by sequencing of chromosomal DNA using the mini-Tn10-specific primer S156 with the sequence 5′-GATGATAAAAGGCACCTTTGGTCA. To confirm the integration site, the mutated gene fragment carrying the mini-Tn10 insertion of some mutants was amplified with gene-specific primers and the integration site was sequenced using the Tn10-specific primer S156.
Determination of β-galactosidase activities.
For enzyme assays, cells were grown in M9 medium containing 1% (wt/vol) glycerol, 0.66% (wt/vol) Casamino Acids (Difco), and 1-μg/ml vitamin B1 or in NB medium (Difco) as indicated. Cultures were inoculated from fresh overnight cultures grown in the same medium. Cells were harvested after approximately 3 h of growth at 37°C at an optical density at 600 nm (OD600) of 0.5. The β-galactosidase assays were performed as described previously (27). The enzyme activities were determined at least three times from at least two independent transformants or integration derivatives. Standard deviations were <10%.
RESULTS
A mutagenesis screen for identification of factors involved in bgl operon regulation.
In order to identify factors that regulate the bgl operon in addition to H-NS, we performed a transposon mutagenesis screen using a phage λ mini-Tn10 system (23). In one approach, mutants were isolated that cause derepression of the silent wild-type bgl operon. In a second, reverse screen, we screened for mutants in which expression of an active bgl operon is down-regulated. To avoid mutations that map in cis to the operon, a double-phenotype screening strategy was established (Fig. 2). We constructed strains that carry the bgl operon and a fusion of the bgl regulatory region to the lac operon at its natural chromosomal locus. In this fusion, the lac promoter was replaced by the bgl promoter, including the upstream and downstream negative regulatory elements. In the bgl-lac fusion, the terminator gene t1 was deleted (from nucleotides +55 to +120 relative to the transcription start, Δt1) and translation of bglG was excluded by mutation of the translation start codon and an additional ATG (codon 3) to GCG (bglGorf) to render expression independent of BglG-mediated antitermination and to avoid cross talk between expression of the bgl-lac fusion and the bgl operon.
In the screen for mutations that activate the operon, strain S581 was used, in which both the bgl operon and the bgl-lac operon fusion carry the wild-type bgl promoter. This screen yielded six mutations, which were Bgl and Lac positive and which all mapped in hns (data not shown). The reverse screen for mutations reducing expression of bgl was performed with strain S594, in which expression of both the bgl operon and the bgl-lac fusion is activated by the identical point mutation improving the bgl CRP-binding site. This mutation is a C-to-T exchange at position −66 relative to the transcription start, which generates the conserved TGTGA motif in the promoter distal half-site of the CRP-binding site. This strain is therefore Bgl and Lac positive. Sixteen transposon mutants of this strain with a double-phenotypic change to Bgl− Lac− (from a total of more than 50,000 mutants) were isolated and characterized by sequencing of the mini-Tn10 insertion site. One of the mutations carried by these mutants mapped in cyaA, five mapped in pgi, seven mapped in lon, and three mapped in the miaA-hfq locus (Fig. 2).
Gene cyaA codes for adenylate cyclase. Due to a lack of cAMP, the CRP-dependent bgl promoter is likely to be inactive and thus the bgl operon and the bgl-lac fusion are not expressed in the cyaA1405::mini-Tn10 mutant. This mutation was not further analyzed. Lon is a highly conserved ATP-dependent protease that degrades abnormally folded proteins during heat shock and starvation. It also degrades some proteins specifically (15), including protein StpA, which is 67% similar to the H-NS protein (18, 19). Gene pgi codes for the enzyme phosphoglucose isomerase, which catalyzes the isomerization of glucose-6-phosphate to fructose-6-phosphate. The genes miaA and hfq code for the tRNA modification enzyme Δ(2)-isopentenylpyrophosphate transferase (MiaA) and the host factor for replication of RNA phage Qβ (Hfq), respectively. Hfq, a 15-kDa protein with ∼30,000 molecules per cell (1), has an RNA binding/chaperone activity (28, 29, 38, 56). It is involved in the regulation of translation by the regulatory RNAs DsrA and OxyS, and it interferes with translation of the ompA mRNA (31, 49, 52). The MiaA-mediated modification of tRNAs affects translation efficiency and is required for translation of the virulence-related transcriptional regulator VirF (10).
In hfq and lon mutants, expression of the bgl-lacZ fusion is specifically down-regulated.
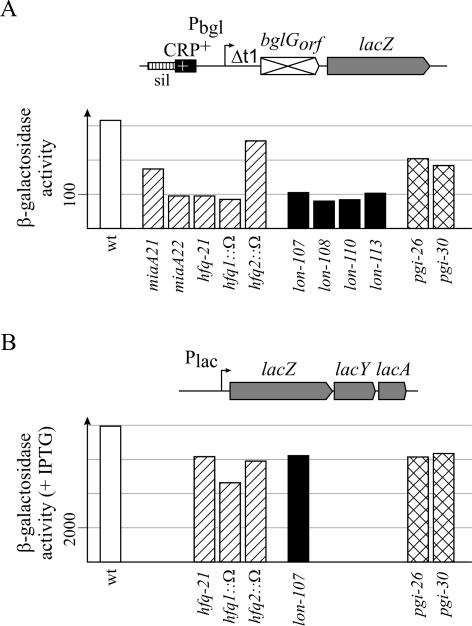
For the quantification of the effect of mutations in the miaA-hfq, lon, and pgi loci, we determined the β-galactosidase expression level directed by the bgl-lac fusion, which carries the bgl promoter derivative activated by the improved CRP-binding site (Fig. 3). As a control, we measured the influence of mutations in miaA-hfq, lon, and pgi on the expression of the wild-type lac operon, which was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (Fig. 3). β-Galactosidase assays were performed with cultures grown to the exponential phase (OD600 of 0.5) in M9 minimal medium containing glycerol and Casamino Acids.
FIG. 3.
Mini-Tn10 mutants of miaA-hfq and lon specifically reduce expression of a bgl-lac fusion. (A) The expression level directed by the bgl-lac fusion (see Fig. 1 for details) was determined in the wild-type (wt) strain (S594) and its mini-Tn10 derivatives (strains S749, S754, S759, S762, S764, S751, S765, S750, S757, S763, S1048, and S1654). (B) As a control, the effect of some of the mutations on the wild-type lac operon was also determined (strains S539, S794, S1422, S1424, S778, S1105, and S1656). Cultures were grown in minimal M9 medium containing glycerol and Casamino Acids to an OD600 of 0.5.
In the lon mutants (alleles 107, 108, 110, 112, and 113), the expression level of the bgl-lac fusion is approximately three- to fourfold lower (between 80 and 105 U) than in the wild-type strain background (315 U) (Fig. 3). The mutation lon-107, which carries the mini-Tn10 insertion closest to the 5′ end of the gene (Fig. 2), was chosen for further analysis. This mutation did not significantly reduce (less than 1.3-fold) expression of the lac operon (Fig. 3). These data show that mutations in lon specifically down-regulated expression of bgl.
The two pgi mutations pgi-26 and pgi-30 caused a weak (1.5- to 1.7-fold) decrease in the expression of the bgl-lac fusion, while the expression level of the lac operon was also weakly (1.3-fold) reduced (Fig. 3). Because of this minimal weak effect on bgl, the pgi mutant was not analyzed further.
The three mutations mapping in the miaA-hfq loci (miaA21, miaA22, and hfq-21) caused a specific decrease in the expression of the bgl-lac fusion (Fig. 3). Genes miaA and hfq are part of the complex amiB-mutL-miaA-hfq-hflX operon, which has several promoters directing expression of the different genes of the operon to various degrees (50). Out of the three mutants isolated in the screen, two mutations (miaA21 and miaA22) map in the miaA gene and one (hfq-21) maps in the hfq gene (Fig. 2). To determine whether these mutations affect bgl expression due to the mutation of miaA or hfq or polar effects on downstream genes, their effect was compared to that caused by the two previously described mutations hfq1::Ω and hfq2::Ω (50). In hfq1::Ω, an Ω cassette carrying a kanamycin resistance marker flanked by Rho-independent transcriptional terminators (36) is inserted towards the 5′ end of the hfq gene and thus causes an Hfq-negative phenotype. In hfq2::Ω the Ω cassette is inserted towards the 3′ end of the hfq gene, which allows expression of an active Hfq protein. Both mutations have the same polar effects on expression of the downstream genes (50). The mutation hfq1::Ω had a negative effect on expression of the bgl-lac fusion very similar to that of hfq-21 (85 and 95 U, respectively, compared to 315 U in the wild-type), while hfq2::Ω had no significant effect on bgl expression (256 U) (Fig. 3), which demonstrates that Hfq reduces bgl silencing. The effect of the mutation miaA22, which maps very close to the hfq gene, was similar to hfq-21 (both 95 U), while the effect of miaA21, which maps at the 5′ end of the miaA gene, was weaker (175 U) (Fig. 3). This suggests that these two mutations cause a down-regulation of bgl expression due to their polar effects on hfq. The expression of the lac operon was also moderately (1.3- to 1.7-fold) reduced in the hfq-21 and hfq1::Ω mutants (Fig. 3). However, in contrast to the bgl-lacZ fusion, the expression of the lac operon was similarly (1.3-fold) reduced in the hfq2::Ω mutant, which expresses a functional Hfq protein (Fig. 3). Taken together, these data suggest that Hfq also reduces silencing of bgl. Further analyses were performed using the hfq-21 and hfq1::Ω alleles. For comparison, expression of the bgl-lac fusion construct that is activated by an improved CRP-binding site was also determined in an hns background. Mutation of the hns gene resulted in an approximately sevenfold increase (to 1,940 U) in the expression level of the bgl-lacZ fusion, while expression of the wild-type lac operon was lowered twofold (to 2,860 U) in the hns mutant. This decrease in lac operon expression may be due to poor growth of the hns mutant in minimal medium. Since the other mutants also showed a reduced growth rate in minimal medium, all further experiments were performed with cultures grown in NB or Luria-Bertani (LB) medium, as indicated.
The bgl promoter is not affected by Hfq and Lon.
Silencing of the bgl operon requires H-NS, which represses the expression at two levels. H-NS hinders transcription initiation, and H-NS binds 600 to 700 bp downstream of the transcription start, where it induces polarity of transcription (9). To further analyze the regulation of the bgl operon by Lon and Hfq, we used a series of chromosomally encoded bgl-lacZ reporter gene fusions, which carry either the promoter and upstream regulatory region (Fig. 4) or the downstream regulatory region (Fig. 5). Expression of the latter is directed by the constitutive lacUV5 promoter (9). The role of Lon in bgl regulation was studied in strains in which a deletion of the lon gene (Δlon-510) was combined with a mutation of sulA (sulA3), encoding the cell division inhibitor SulA. The SOS-induced SulA is normally degraded by Lon. However, in lon mutants, the block of cell division remains and the cells are SOS sensitive (15, 41). In our hands, the growth rate and reproducibility of enzyme assays were significantly improved with the Δlon sulA3 double mutant compared to those with the single lon-107 mutant (data not shown). For the analyses of the role of Hfq in regulation of the bgl operon allele, hfq1::Ω was used.
FIG. 4.
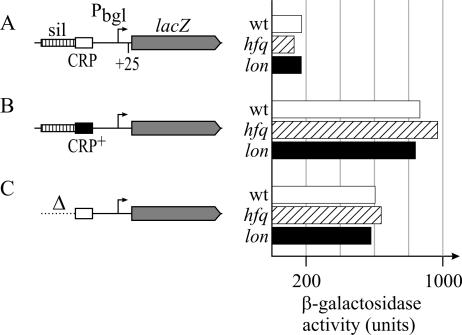
Effect of Hfq and Lon on the bgl promoter. The activity of the wild-type bgl promoter (wt) and activated derivatives, which carry a deletion (Δ, including position −77 and upstream) of the upstream silencer (sil), and an improved CRP-binding site (CRP+; a C-to-T exchange at position −66) were determined in hfq, lon, and pgi mutants by using lacZ gene fusion 25 bp downstream of the transcription start side and compared to the expression level in the wild-type strain tested previously (9). Cultures were grown in NB medium to an OD600 of 0.5. (A) Wild-type bgl promoter. Shown are results for strains S1213 (wild type [wt]), S1582 (hfq1::Ω), and S1556 (Δlon-510 sulA3). (B) bgl promoter activated by an improved CRP-binding site (CRP+). Shown are results for strains S1215 (wild type), S1586 (hfq1::Ω), and S1558 (Δlon-510 sulA3). (C) bgl promoter activated by deletion of the upstream silencer (Δ). Shown are results for strains S1211 (wild type), S1584 (hfq1::Ω), and S1554 (Δlon-510 sulA3).
FIG. 5.
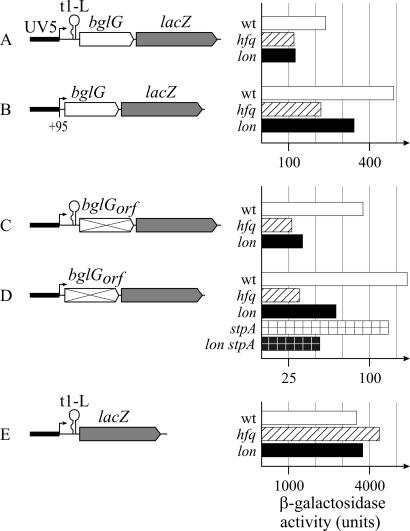
Effect of Hfq and Lon on repression of bgl via the downstream silencer. The expression level directed by the bgl-downstream silencer-lacZ reporter constructs (9) was determined in hfq, lon, and pgi mutants and compared to the expression level in the wild-type strain tested previously (9). The expression of these bgl-lacZ fusions is directed by the constitutive lacUV5 promoter. Cultures were grown in NB medium to an OD600 of 0.5. These constructs carry the following insertions between the lacUV5 promoter and the lacZ gene. (A) The complete downstream region including the leader (with the tl-L terminator mutant) and gene bglG fragment (S1097, wild-type) (9), S1311 (hfq1::Ω), S1680 (Δlon-510 sulA3), S1132 (rpoS359::Tn10), and S1416 (hfq1::Ω, rpoS359::Tn10). (B) The bglG gene fragment (from position +95) (S1193, wild type), S1223 (hfq1::Ω), and S1562 (Δlon-510 sulA3). (C) Same as A, but translation of bglG excluded due to mutations of codons 1, 3, and 27 to GCG (S1189, wild-type) (9), S1219 (hfq1::Ω), and S1684 (Δlon-510 sulA3). (D) Same as B, but translation of bglG excluded due to mutations of codons 1, 3, and 27 to GCG (S1195, wild-type), S1225 (hfq1::Ω), S1564 (Δlon-510 sulA3), S1371 (stpA::Tcr), and S1379 (Δlon-510 sulA3 clp::Cmr stpA::Tcr). (E) bgl leader (from positions +1 to +176) including the tl-L terminator mutant (S1624, wild type), S1676 (hfq1::Ω), and S1642 (Δlon-510 sulA3).
To determine the effect of Hfq and Lon on the bgl promoter, we used bgl promoter-lacZ fusions, in which the lacZ gene was fused at position +25 (relative to the transcription start) (Fig. 4). The expression of three bgl promoter constructs carrying either the wild-type promoter, an allele with a deletion of the upstream silencer, or an allele with the improved CRP-binding site was determined from cultures grown to the exponential phase (OD600 of 0.5) in NB medium. The expression level directed by the wild-type bgl promoter is approximately 3.5-fold lower than that of the promoter that lacks the upstream silencer (175 and 600 U, respectively) (Fig. 4A and C), while the improvement of the CRP-binding site leads to an ∼5-fold increase in the promoter activity (to 865 U) (Fig. 4B) (9). In the hfq::Ω1 and Δlon sulA3 mutants, the expression level directed by the wild-type bgl promoter and its two activated derivatives was not significantly altered (Fig. 4). These results demonstrate that neither Hfq nor Lon affects transcription initiation at the bgl promoter.
Hfq and Lon affect the bgl operon via the downstream silencer.
The second level of repression of the bgl operon by H-NS occurs at a downstream silencer located within the bglG coding region (9). To analyze whether Hfq and Lon (directly or indirectly) affect this level of regulation, a set of bgl-downstream silencer-lacZ reporter constructs were used (9). In these reporter constructs, expression is directed by the constitutive lacUV5 promoter. Between the lacUV5 promoter and the lacZ reporter, fragments were inserted that encompass the complete downstream region, including the leader and the bglG gene (from positions +1 to +971), the bglG gene alone (+95 to +971), or, as a control, the leader only (+1 to +176) (9). To prevent transcription termination at terminator t1 located in the leader, the tl-L terminator mutant was used, in which 3-base exchange in the left stem of the terminator hairpin structure disrupts the terminator (8). In addition, variants of those fusions carrying the bglG gene were used in which the translation of bglG is prevented by mutation of the translation start codon and two additional ATG codons (codons 3 and 27) to GCG. When translation of bglG is excluded, the repression by H-NS via the downstream silencer is more efficient (9).
A construct in which the lacUV5 promoter is followed by the bgl leader with the t1-L terminator mutant, the bglG gene, and the lacZ reporter gene directs 240 U of β-galactosidase activity in the wild-type strain (Fig. 5A) (9). In the hfq1::Ω and lon sulA3 mutants, the expression level of this lacUV5-t1-L-bglG-lacZ fusion decreased approximately twofold to 120 and 125 U, respectively (Fig. 5A). The expression level of a reporter construct carrying a bglG fragment inserted between the lacUV5 promoter and the lacZ gene decreased from 490 U in the wild type to 220 U in the hfq mutant. However, the decrease in the lon mutant (to 370 U) was less significant (Fig. 5B). The effects of Hfq and Lon on the corresponding reporter constructs, in which translation of bglG is excluded, were higher. The expression level of the lacUV5-t1-L-bglGorf-lacZ reporter construct decreased 2.5- to 3-fold in the hfq and lon mutants (Fig. 5C). Ninety-five units of β-galactosidase activity was detected in the wild type, 28 U was detected in the hfq1::Ω mutant, and 44 U was detected in the Δlon-510 sulA3 mutant (Fig. 5C). Likewise the expression level of the lacUV5-bglGorf-lacZ reporter construct decreased two- to threefold from 135 U in the wild type to 35 U in the hfq mutant and 57 U in the lon mutant (Fig. 5D). A control carrying the bgl leader, including the t1-L terminator mutant (positions +1 to +179), which lacks the downstream H-NS binding site, was not affected in the hfq and lon mutants (Fig. 5E). These data show that Hfq and Lon control the expression of the bgl operon via the downstream regulatory region located within bglG. Their effects are stronger when the bglG gene is not translated, which indicates that access to the mRNA is important in this process. Repression by H-NS via the downstream silencer is also more efficient when bglG is not translated (9).
Hfq affects the bgl operon independent of RpoS.
Hfq is required for translation of the rpoS mRNA, and RpoS levels are thus low in hfq mutants (31). In addition, RpoS is known to repress the bgl operon approximately two- to threefold in the exponential phase (8, 33, 44). If Hfq affected bgl indirectly via RpoS, bgl expression should increase in the hfq mutant. However, here we found that the expression of bgl decreases in hfq mutants. In addition, previous data suggest that RpoS affects the bgl promoter (8), while here we found that Hfq acts via the downstream silencer. This suggests that Hfq affects the bgl operon independent of RpoS. To confirm this, the expression level directed by the lacUV5-t1-L-bglG-lacZ construct was also determined in an rpoS mutant and an hfq rpoS double mutant. The results support the assumption that Hfq reduces silencing of bgl independent of RpoS (data not shown).
H-NS is epistatic over Hfq and Lon.
H-NS induces a Rho-dependent polarity of transcription within bglG (9). Here we found that Hfq and Lon also act via the bglG coding region. One possibility is that Hfq and Lon act on a different level of regulation from H-NS. Another possibility is that these proteins directly or indirectly counteract H-NS to induce polarity. For example, Lon and Hfq could reduce the activity or action of H-NS or another protein involved in this process. If this were the case, then the mutation of hfq or lon should have no effect on bgl when tested in an hns mutant. To address this question, the expression level of the downstream silencer reporter construct (lacUV5-bglGorf-lacZ), which carries the nontranslatable bglG coding region, was tested in hfq hns double and lon sulA3 hns triple mutants and compared to the expression level directed in the hns mutant (Fig. 6).
FIG. 6.
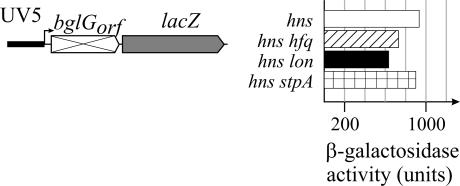
Effect of Hfq and Lon via the downstream silencer in an hns mutant. The expression level directed by the bgl-downstream silencer-lacZ reporter constructs carrying the bglG coding region (Fig. 5D) was determined in hfq hns, lon hns, and stpA hns mutants and compared to the expression level in the hns strain tested previously (9). Cultures were grown in NB medium to an OD600 of 0.5. The hfq hns double mutant was grown in LB medium. β-Galactosidase activity is shown for the following strains from top to bottom: S1258 (hns-206::Apr) (9), S1828 (hfq1::Ω hns-206::Apr), S1432 (Δlon-510 sulA3 hns-206::Apr), and S1767 (stpA::Tcr hns-206::Apr).
The expression level of the lacUV5-bglGorf-lacZ fusion has been shown to increase approximately sevenfold in the hns mutant (from 135 U to 905 U) (9), while it decreased approximately twofold in the lon mutant and approximately fourfold in the hfq mutant. Since the hns hfq double mutant grew very poorly in NB medium, the β-galactosidase activity expressed in this strain was determined from cultures grown in LB medium. The expression levels directed in the wild type and the hns and hfq single mutants were similar when cultures were grown in NB or LB medium (data not shown). In the hns hfq double mutant, 730 U of β-galactosidase activity was determined and in the hns Δlon sulA3 triple mutant 640 U was determined, as compared to 905 U in the hns mutant (Fig. 6). Thus, the effect of Hfq and Lon was reduced in the hns mutant. This suggests that Hfq and Lon control a protein or process that is required for H-NS to induce polarity. In case of Lon, the H-NS homologue StpA is a candidate. StpA is degraded by Lon (18, 19), and StpA plays a role in silencing of bgl by a truncated H-NS variant (12). We therefore analyzed whether StpA is important for the stimulation of bgl expression by Lon and determined the expression level of the downstream silencer reporter lacUV5-bglGorf-lacZ in stpA and stpA lon sulA3 mutants (Fig. 5D). In the stpA mutants, the expression level was unchanged compared to the level of expression in the wild-type and lon sulA3 strains (Fig. 5D), suggesting that StpA is not the protein substrate via which Lon affects bgl. The mutation of stpA in the hns mutant likewise had no effect (Fig. 6).
DISCUSSION
H-NS silences the bgl operon at two levels: it represses transcription initiation, and it induces polarity within the transcription unit. Here we presented evidence that the latter level of silencing by H-NS, the induction of polarity, is reduced by RNA chaperone Hfq and by protease Lon. However, the H-NS-mediated silencing of the bgl promoter is not altered by these proteins. Our results suggest that specificity in the control by the pleiotropic regulator H-NS can be achieved by other pleiotropic factors, which control or modulate the action of H-NS at a specific locus, as shown here for Hfq and Lon, which counteract only the second level of silencing of bgl by H-NS.
The bgl operon is silent under laboratory growth conditions. To date, only mutants in which silencing of the operon is overcome have been isolated. Silencing can be relieved by mutations that map in cis to the promoter and interfere with its repression by H-NS, or they map in the hns gene (5, 39, 40). Constitutive expression of the leuO or bglJ gene, which encode transcription factors, relieves silencing by unknown mechanisms (14, 51). In this work, a reverse transposon mutagenesis screen for mutants was performed, in which expression of a bgl operon derivative was reduced. This screen yielded mutations mapping in the cyaA, miaA-hfq, lon, and pgi loci (Fig. 2). The mutation of cyaA presumably impairs the CRP-dependent promoter. In pgi mutants, the glycolytic pathway is blocked, leading to the accumulation of glucose-6-phosphate. It has been shown that this accumulation stimulates the RNase E-dependent destabilization of the ptsG mRNA (22, 30). The ptsG gene encodes the glucose permease enzyme IICBGlc, which belongs to the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) (34). The use of a bgl-lacZ reporter construct revealed that the specificity of the pgi mutant for bgl is low (Fig. 3), and the mutant was not analyzed further.
Mutation of hfq and lon, respectively, had no effect on the bgl promoter. In these mutants, the second level of bgl silencing by H-NS, the induction of polarity downstream of the promoter, was more efficient. The use of double mutants revealed that in an hns mutant, Hfq plays no role and Lon plays a rather reduced role: i.e., H-NS is epistatic over Hfq and also Lon. What could be the mechanisms by which expression of bgl is reduced in these mutants? H-NS binds approximately 600 to 700 bp downstream of the transcription initiation site, within the coding region of the first gene, bglG, and represses expression beyond bglG. The repression is more efficient when the bglG mRNA is not translated, suggesting that access of an RNA-binding protein, such as, e.g., termination factor Rho, or an RNase to the bglG mRNA is important for H-NS to induce polarity (9). In the hfq and lon mutants, the expression is further reduced, indicating that Hfq and Lon reduce the silencing by H-NS. The highly conserved ATP-dependent protease Lon degrades nonfunctional proteins and some proteins specifically (15). In case of the bgl operon, Lon could degrade a protein required for H-NS to induce polarity. In a lon mutant, this negative regulator protein would be stabilized and accumulate to higher levels, causing low bgl expression. As a candidate, we tested StpA (a protein with 67% similarity to the H-NS protein), which is a specific Lon substrate (18, 19) and which is involved in the repression of bgl by a truncated H-NS protein (12). When we determined the expression level of the bgl-downstream silencer-lacZ reporter construct in stpA and lon sulA3 stpA mutants, no difference compared to the stpA+ strains was observed (Fig. 5). Thus, StpA is likely not to be the specific substrate via which Lon affects bgl.
Hfq is an RNA chaperone that affects the stability and translation of several RNAs (3, 28, 29, 38, 56, 57). For example, Hfq binds to and stabilizes the small regulatory DsrA RNA (3). The dsrA gene is induced at low temperature, allowing its product, the DsrA RNA, to induce translation of the rpoS mRNA in exponentially growing cells under these conditions (49). DsrA, when overproduced, represses translation of hns by an Hfq-dependent mechanism (26, 49). However, it is not clear whether this is relevant under physiological conditions (38). At 30°C, a weak (1.5-fold) relief of the H-NS-mediated repression of the rcsA gene by Hfq/DsrA was observed (49). Whether at 37°C Hfq affects H-NS levels via DsrA is not known. In the case of specific reduction of the H-NS-mediated downstream silencing of bgl by Hfq, several mechanisms seem plausible. First, specific competition experiments suggest that the affinity of H-NS to the upstream silencer is higher than to the downstream silencer. Thus, a small reduction of H-NS amounts in the hfq mutant could result in the specific reduction of downstream silencing by H-NS. Second, Hfq could affect downstream silencing indirectly: e.g., via another small regulatory RNA. Third, Hfq could bind to the bglG mRNA and change the secondary structure of the bglG mRNA and/or reduce access of Rho or an RNase and thus counteract H-NS-mediated polarity.
To date, the biological meaning of bgl operon silencing is not understood. The operon is present in approximately 70% of E. coli isolates, and in all of the strains in which it is present, its silent state is conserved (G. Neelakanta and K. Schnetz, unpublished observations). This and its complex regulation suggest that the expression of the operon is disadvantageous under some conditions, while it is required and induced under certain physiological and environmental conditions. Irrespective of these open questions, the bgl operon is a valuable tool for the understanding of specific repression by H-NS. How specificity in the control by the pleiotropic regulator H-NS is achieved is largely unknown. The bgl operon is controlled at two levels by H-NS. Here we have shown that one of these levels of silencing by H-NS is modulated by the RNA chaperone Hfq and by the protease Lon, while the other level—the silencing of the promoter—is unaffected by these proteins. This finding suggests that the action of H-NS at a specific locus can be modulated by other pleiotropic regulators. Thus, locus-specific control by H-NS may be the result of the combination of H-NS with various sets of pleiotropic factors. A similar combinatorial mechanism has been discussed for the specific regulation by RpoS, the second key regulator of the adaptation response (16).
Acknowledgments
We thank Sandra Kühn for excellent technical assistance.
This work was funded by the Deutsche Forschungsgemeinschaft through the Graduiertenkolleg “Genetik zellulärer Systeme.”
REFERENCES
- 1.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 3.Brescia, C. C., P. J. Mikulecky, A. L. Feig, and D. D. Sledjeski. 2003. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA 9:33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dabert, P., and G. R. Smith. 1997. Gene replacement with linear DNA fragments in wild-type Escherichia coli: enhancement by Chi sites. Genetics 145:877-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Defez, R., and M. de Felice. 1981. Cryptic operon for β-glucoside metabolism in Escherichia coli K12: genetic evidence for a regulatory protein. Genetics 97:11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dersch, P., K. Schmidt, and E. Bremer. 1993. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 8:875-889. [DOI] [PubMed] [Google Scholar]
- 7.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for integration into the lambda attachment site attB of the Escherichia coli chromosome. Plasmid 28:14-24. [DOI] [PubMed] [Google Scholar]
- 8.Dole, S., S. Kühn, and K. Schnetz. 2002. Post-transcriptional enhancement of Escherichia coli bgl operon silencing by limitation of BglG-mediated antitermination at low transcription rates. Mol. Microbiol. 43:217-226. [DOI] [PubMed] [Google Scholar]
- 9.Dole, S., V. Nagarajavel, and K. Schnetz. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol., in press. [DOI] [PubMed]
- 10.Durand, J. M., B. Dagberg, B. E. Uhlin, and G. R. Bjork. 2000. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol. Microbiol. 35:924-935. [DOI] [PubMed] [Google Scholar]
- 11.Free, A., and C. J. Dorman. 1995. Coupling of Escherichia coli hns mRNA levels to DNA synthesis by autoregulation: implications for growth phase control. Mol. Microbiol. 18:101-113. [DOI] [PubMed] [Google Scholar]
- 12.Free, A., M. E. Porter, P. Deighan, and C. J. Dorman. 2001. Requirement for the molecular adapter function of StpA at the Escherichia coli bgl promoter depends upon the level of truncated H-NS protein. Mol. Microbiol. 42:903-918. [DOI] [PubMed] [Google Scholar]
- 13.Gayda, R. C., L. T. Yamamoto, and A. Markovitz. 1976. Second-site mutations in capR (lon) strains of Escherichia coli K-12 that prevent radiation sensitivity and allow bacteriophage lambda to lysogenize. J. Bacteriol. 127:1208-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giel, M., M. Desnoyer, and J. Lopilato. 1996. A mutation in a new gene, bglJ, activates the bgl operon in Escherichia coli K-12. Genetics 143:627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 16.Hengge-Aronis, R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 17.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 18.Johansson, J., S. Eriksson, B. Sondén, S. N. Wai, and B. E. Uhlin. 2001. Heteromeric interactions among nucleoid-associated bacterial proteins: localization of StpA-stabilizing regions in H-NS of Escherichia coli. J. Bacteriol. 183:2343-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson, J., and B. E. Uhlin. 1999. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary-phase survival of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:10776-10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordi, B. J., and C. F. Higgins. 2000. The downstream regulatory element of the proU operon of Salmonella typhimurium inhibits open complex formation by RNA polymerase at a distance. J. Biol. Chem. 275:12123-12128. [DOI] [PubMed] [Google Scholar]
- 21.Jordi, B. J. A. M., A. E. Fielder, C. M. Burns, J. C. D. Hinton, N. Dover, D. W. Ussery, and C. F. Higgins. 1997. DNA binding is not sufficient for H-NS mediated repression of proU expression. J. Biol. Chem. 272:12083-12090. [DOI] [PubMed] [Google Scholar]
- 22.Kimata, K., Y. Tanaka, T. Inada, and H. Aiba. 2001. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 20:3587-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 24.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 25.Lease, R. A., and M. Belfort. 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. USA 97:9919-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lease, R. A., M. E. Cusick, and M. Belfort. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. USA 95:12456-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Moll, I., D. Leitsch, T. Steinhauser, and U. Blasi. 2003. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 4:284-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moller, T., T. Franch, P. Hojrup, D. R. Keene, H. P. Bachinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9:23-30. [DOI] [PubMed] [Google Scholar]
- 30.Morita, T., W. El Kazzaz, Y. Tanaka, T. Inada, and H. Aiba. 2003. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J. Biol. Chem. 278:15608-15614. [DOI] [PubMed] [Google Scholar]
- 31.Muffler, A., D. Fischer, and R. Hengge-Aronis. 1996. The RNA-binding protein HF-1, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10:1143-1151. [DOI] [PubMed] [Google Scholar]
- 32.Mukerji, M., and S. Mahadevan. 1997. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol. Microbiol. 24:617-627. [DOI] [PubMed] [Google Scholar]
- 33.Ohta, T., C. Ueguchi, and T. Mizuno. 1999. rpoS function is essential for bgl silencing caused by C-terminally truncated H-NS in Escherichia coli. J. Bacteriol. 181:6278-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasad, I., and S. Schaefler. 1974. Regulation of the β-glucoside system in Escherichia coli K-12. J. Bacteriol. 120:638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 37.Repoila, F., and S. Gottesman. 2001. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 183:4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Repoila, F., N. Majdalani, and S. Gottesman. 2003. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 48:855-861. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds, A. E., J. Felton, and A. Wright. 1981. Insertion of DNA activates the cryptic bgl operon of E. coli K12. Nature 293:625-629. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds, A. E., S. Mahadevan, S. F. J. LeGrice, and A. Wright. 1986. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J. Mol. Biol. 191:85-95. [DOI] [PubMed] [Google Scholar]
- 41.SaiSree, L., M. Reddy, and J. Gowrishankar. 2000. lon incompatibility associated with mutations causing SOS induction: null uvrD alleles induce an SOS response in Escherichia coli. J. Bacteriol. 182:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schnetz, K. 1995. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 14:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnetz, K. 2002. Silencing of the Escherichia coli bgl operon by RpoS requires Crl. Microbiology 148:2573-2578. [DOI] [PubMed] [Google Scholar]
- 45.Schnetz, K., and B. Rak. 1992. IS5: a mobile enhancer of transcription in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:1244-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schnetz, K., and J. C. Wang. 1996. Silencing of Escherichia coli bgl promoter: effects of template supercoiling and cell extracts on promoter activity in vitro. Nucleic Acids Res. 24:2422-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schröder, O., and R. Wagner. 2002. The bacterial regulatory protein H-NS—a versatile modulator of nucleic acid structures. Biol. Chem. 383:945-960. [DOI] [PubMed] [Google Scholar]
- 48.Singh, J., M. Mukerji, and S. Mahadevan. 1995. Transcriptional activation of the bgl operon of E. coli: negative regulation by DNA structural elements near the promoter. Mol. Microbiol. 17:1085-1092. [DOI] [PubMed] [Google Scholar]
- 49.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsui, H.-C. T., H.-C. E. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13:35-49. [DOI] [PubMed] [Google Scholar]
- 51.Ueguchi, C., T. Ohta, C. Seto, T. Suzuki, and T. Mizuno. 1998. The leuO gene-product has a latent ability to relieve the bgl silencing in Escherichia coli. J. Bacteriol. 180:190-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vytvytska, O., I. Moll, V. R. Kaberdin, A. von Gabain, and U. Blasi. 2000. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 14:1109-1118. [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, R. M., S. Rimsky, and H. Buc. 1996. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 178:4335-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, G. G., K. Y. K. Young, G. J. Edlin, and W. Konigsberg. 1979. High-frequency generalised transduction by bacteriophage T4. Nature 280:80-82. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, A., S. Rimsky, M. E. Reaban, H. Buc, and M. Belfort. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acids dynamics. EMBO J. 15:1340-1349. [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, A., K. M. Wassarman, C. Rosenow, B. C. Tjaden, G. Storz, and S. Gottesman. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50:1111-1124. [DOI] [PubMed] [Google Scholar]