Abstract
Background
The genus Micronycteris is a diverse group of phyllostomid bats currently comprising 11 species, with diploid number (2n) ranging from 26 to 40 chromosomes. The karyotypic relationships within Micronycteris and between Micronycteris and other phyllostomids remain poorly understood. The karyotype of Micronycteris hirsuta is of particular interest: three different diploid numbers were reported for this species in South and Central Americas with 2n = 26, 28 and 30 chromosomes. Although current evidence suggests some geographic differentiation among populations of M. hirsuta based on chromosomal, morphological, and nuclear and mitochondrial DNA markers, the recognition of new species or subspecies has been avoided due to the need for additional data, mainly chromosomal data.
Results
We describe two new cytotypes for Micronycteris hirsuta (MHI) (2n = 26 and 25, NF = 32), whose differences in diploid number are interpreted as the products of Robertsonian rearrangements. C-banding revealed a small amount of constitutive heterochromatin at the centromere and the NOR was located in the interstitial portion of the short arm of a second pair, confirmed by FISH. Telomeric probes hybridized to the centromeric regions and weakly to telomeric regions of most chromosomes. The G-banding analysis and chromosome painting with whole chromosome probes from Carollia brevicauda (CBR) and Phyllostomus hastatus (PHA) enabled the establishment of genome-wide homologies between MHI, CBR and PHA.
Conclusions
The karyotypes of Brazilian specimens of Micronycteris hirsuta described here are new to Micronycteris and reinforce that M. hirsuta does not represent a monotypic taxon. Our results corroborate the hypothesis of karyotypic megaevolution within Micronycteris, and strong evidence for this is that the entire chromosome complement of M. hirsuta was shown to be derivative with respect to species compared in this study.
Background
The big-eared bats genus Micronycteris Gray 1866 is an antique and diversified lineage of phyllostomids occurring from Mexico to Paraguay and throughout most parts of South America [1]. This lineage diverged from a sister group, Lampronycteris, approximately 23.2 million years ago (MYA) [2-5]. The Sanborn review [6] recognized 13 species classified in six subgenera: Glyphonycteris, Lampronycteris, Micronycteris, Neonycteris, Trinycteris and Xenoctenes. Since then, Micronycteris lato sensu has undergone considerable taxonomic changes. The monophyly of Micronycteris (sensu Sanborn [6]) was not supported by morphological traits, and the fusion of Xenoctenes and Micronycteris had been proposed [7,8]. Wetterer et al.[3] recommended the elevation to genus status for all the Micronycteris subgenera, and this has been supported by molecular data [9,10]. They recognized six species: Micronycteris megalotis, M. microtis, M. hirsuta, M. minuta, M. sanborni and M. schmidtorum. Simmons et al.[11] recognized two additional species in this genus: M. brosseti and M. homezi. Lately, three new species have been discovered: M. matses[11], M. giovanniae[12] and M. buriri[4], totaling eleven species in Micronycteris (stricto sensu).
From external morphology aspects the big-eared bats (Micronycteris spp.) can be clustered into two distinct groups: the “dark-bellied” (hirsuta, matses, megalotis, microtis, giovanniae and buriri) and “pale-bellied” (brosseti, homezi, minuta, sanborni and schmidtorum) groups [7,11,13]. However, molecular data suggested that “dark-bellied” and “pale-bellied” are not natural groups [13].
In the last three decades, phyllostomid bats have been extensively studied by cytogenetics. Recent advances in molecular cytogenetic methods, that enable genomic mapping and comparison by chromosome painting, have improved greatly our comprehension of karyotypic evolution in mammals. Chromosome painting has been used successfully in the investigation of the evolutionary history of the order Chiroptera. Classical banding techniques together with chromosome painting has allowed the identification of chromosomal rearrangements that have occurred during karyotype evolution of the group and has confirmed the effectiveness of the probes produced for comparative studies of bats [14-25].
Cytogenetic studies in various species of Micronycteris show diploid number ranging from 26–40 [12,26-29]. Despite technical advances, the karyotype of Micronycteris hirsuta has been poorly characterized with conventional staining [12,26], and with C-banding of sex chromosomes [30,31]. These studies reveal three chromosomal races with distinct karyotypes, namely 2n = 26, FN = 30 (Ecuador), 2n = 28, FN = 32 (Trinidad and Tobago) and 2n = 30, FN = 32 (Honduras, Nicaragua and Suriname). According to Baker et al.[26], karyotypes with 2n = 28 and 30 chromosomes differ due to centric fusion events. Molecular data supports high divergence within M. hirsuta, corroborating the karyotypic studies [13]. Therefore, it is not clear if M. hirsuta represents a monotypic taxon.
Chromosome painting is important on the resolution of phylogenetic and cytotaxonomic questions, and is useful to understand the mechanisms of chromosomal differentiation occurred during the evolution of bats [14-23]. Until now, just six species encompassing few Phyllostomidae subfamilies were studied by ZOO-FISH using probes from Phyllostomus hastatus (Phyllostominae) and Carollia brevicauda (Carolliinae) [17]: Desmodus rotundus, Diaemus youngi and Dyphylla ecaudata (Desmodontinae) [32], Artibeus obscurus, Uroderma bilobatum and U. magnirostrum (Stenodermatinae) [18]. Two chromosomes were found entirely preserved in karyotypes of these subfamilies: (CBR7 = PHA11 and CBR9 = PHA14), and probably were present of the ancestral karyotype of Phyllostomidae since they are preserved in different species distant phylogenetically. These results leave no doubt that these paints will be useful together for studying the chromosomal relationships among the Phyllostomidae bats.
In order to improve our understanding of karyotypic diversity of Micronycteris genus and the chromosomal evolution of Phyllostomid bats, we have analyzed the karyotype of Micronycteris hirsuta (Micronycterinae) from the Amazon Forest (Brazil). We used classical banding and comparative genomic mapping with whole chromosome probes from Carollia brevicauda (Carolliinae) and Phyllostomus hastatus (Phyllostominae). The chromosomal homologies observed were used to infer evolutionary relationships among the different subfamilies of Phyllostomidae.
Results
Classic cytogenetic and FISH of telomeric and rDNA 18S probes
All Microncyteris hirsuta samples have a diploid number of 2n = 26, FN = 32, except for the specimen LR-275 that had a 2n = 25 karyotype. In the karyotype with 2n = 26, two chromosomal pairs are metacentric, one pair is submetacentric, one pair is subtelocentric and eight pairs are acrocentric (Figure 1; Table 1). The sex system is simple (XX/XY) and the sex chromosomes are acrocentric, with a small Y. The karyotype with 2n = 25 was heterozygous for a centric fusion rearrangement involving chromosomes 4 and 10 (Figure 2). C-banding detected the presence of small amounts of constitutive heterochromatin in the centromeric region of all chromosomes (Figure 3a).
Figure 1.
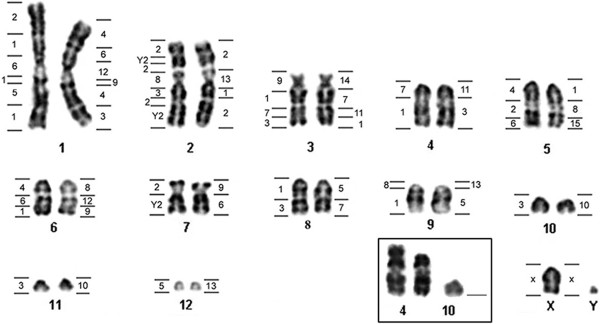
Micronycteris hirsuta G-banding karyotype showing regions homologous to Carollia brevicauda (left) and Phyllostomus hastatus (right). The boxed chromosomes show the centric fusion between pairs 4 and 10.
Table 1.
Karyotypic data of Micronycteris hirsuta
|
Locality |
2n |
NF |
Chromosomal morphology |
Reference |
||
|---|---|---|---|---|---|---|
| Acro | Subtelo | Meta/Sub | ||||
| Itaituba-Brazil |
25 |
32 |
14 |
2 |
7 |
This paper |
| Itaituba-Faro-Brazil |
26 |
32 |
16 |
2 |
6 |
This paper |
| Ecuador |
26 |
30 |
18 |
2 |
4 |
Fonseca et al., 2007 [12] |
| Trinidad and Tobago |
28 |
32 |
20 |
2 |
4 |
Baker et al., 1973 [26] |
| Honduras, Nicaragua and Suriname | 30 | 32 | 24 | 2 | 2 | Baker et al., 1973, 1981; Baker, 1979 [26,27,29] |
Figure 2.
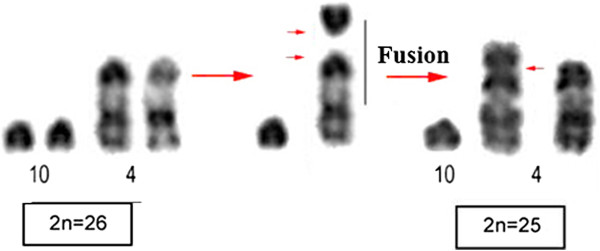
Robertsonian fusion involved in the origin of chromosomes 4 and 10. Small red arrows suggest the break points.
Figure 3.
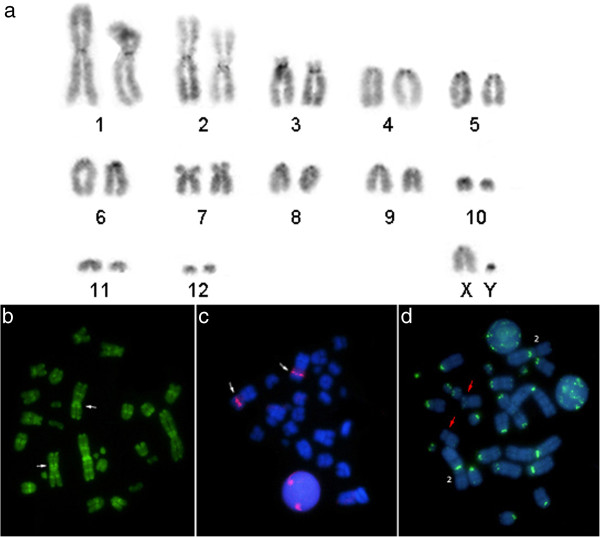
Micronycteris hirsuta karyotype and metaphases. a. C-banding. b. CMA3 staining. c. Partial metaphase of FISH with 18S rDNA probe. d. FISH with telomeric probe. White arrows show NOR bearing chromosome 2, and red arrows indicate the chromosome pair which does not show telomeric signals on its centromeric region.
Staining with AgNO3, CMA3 and FISH with 18S rDNA probes revealed a Nucleolar Organizer Region (NOR) in the middle of the short arm of chromosome 2 (Figure 3c). Telomeric probes hybridized to the centromeric regions of all chromosomes, except the smallest submetacentric. Weak hybridization signals could be visualized on the telomeric region of a few chromosomes (Figure 3d).
Hybridization of Phyllostomus hastatus probes onto Micronycteris hirsuta
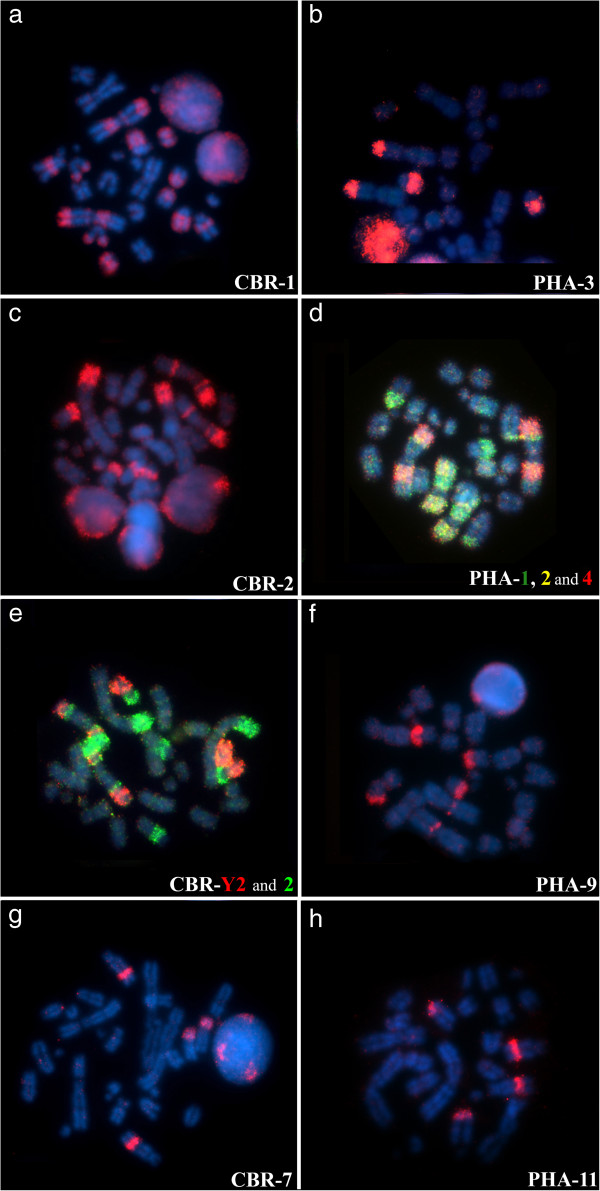
Hybridization of Phyllostomus hastatus (PHA) whole chromosome probes revealed 32 homologous segments on the M. hirsuta (MHI) genome (Figure 1). Three Phyllostomus probes (PHA-14, 15 and X) give just one fluorescent signal on the chromosomes of Micronycteris, corresponding to segments of MHI-3, MHI-5 and entirely on to X, respectively.
Eight paints of Phyllostomus yielded two hybridization signals, with each probe marking regions of two distinct chromosomes in Micronycteris: PHA-3 (MHI-1 and 4), PHA-5 (MHI-8 and 9), PHA-6 (MHI-1 and 7), PHA-7 (MHI-3 and 8), PHA-8 (MHI-5 and 6), PHA-10 (MHI-10 and 11), PHA-11 (MHI-3 and 4) and PHA-12 (MHI-1 and 6).
Two PHA paints showed two signals, but hybridized to parts of just one chromosome in Micronycteris: PHA-2 (MHI-2) and PHA-4 (MHI-1). Three paints of Phyllostomus each hybridized to three chromosomes of Micronycteris: PHA-1 (MHI-2, 3 and 5), PHA-9 (MHI-1, 6 and 7) and PHA-13 (MHI-2, 9 and 12) (Figure 1). The Figure 4(b, d, f, h) shows some hybridization with PHA probes on metaphases of MHI.
Figure 4.
Examples of chromosomal painting in Micronycteris hirsuta, using probes of CBR (left), and probes of PHA (right) onto: a) CBR-1 (pairs 1, 3, 4, 6, 8 and 9); b) PHA-3 (pairs 1 and 4); c) CBR-2 (pairs 1, 2, 5 and 7); d) PHA-1, 2 and 4 (pairs 1, 2, 3 and 5); e) CBR-Y2 and 2 (pairs 1, 2, 5 and 7); f) PHA-9 (pairs 1, 6 and 7); g) CBR-7 (pairs 3 and 4); h) PHA-11 (pairs 3 and 4).
Hybridization of Carollia brevicauda painting probes onto Micronycteris hirsuta
Comparative painting with Carollia brevicauda (CBR) probes revealed 35 homologous segments on the Micronycteris hirsuta (MHI) genome (Figure 1). Two paints of Carollia gave just one signal of hybridization on MHI metaphases: CBR-9 hybridized on the short arm of MHI-3 and CBR-X entirely hybridized on MHI-X.
Four paints of Carollia each hybridized to two Micronycteris chromosomes: CBR-4 (MHI-5 and 6), CBR-5 (MHI-1 and 12), CBR-7 (MHI-3 and 4) and CBR-8 (MHI-2 and 9). CBR-Y2 hybridized to two chromosomes, but showed three hybridization signals, two on MHI-2 and one on MHI-7.
CBR-2 hybridized to chromosomes MHI-1, 2, 5 and 7, showing three fluorescents signals on chromosome 2, and gave just one signal on other chromosomes. CBR-3 hybridized to five distinct segments on MHI chromosomes: 2, 3, 8, 10 and 11. CBR-1 hybridized to 8 regions over six different chromosomes: MHI-1 (three signals), 3, 4, 6, 8 and 9. The Figure 4(a, c, e, g) shows some hybridization with CBR probes on metaphases of MHI. The results of complete mapping of PHA and CBR whole chromosome probes on G-banded karyotype of MHI are on Figure 1.
Discussion
Karyotypic diversity in Micronycteris hirsuta
Here we describe for the first time cytogenetic data of Micronycteris hirsuta from the Amazon Region, Brazil. Specimens presents distinct karyotypes with 2n = 25 (7 M/SM + 2ST + 14A) and 26 (6 M/SM + 2ST + 16A) chromosomes, FN = 32 in both. The difference in diploid number is explained satisfactorily by a fusion involving chromosomes 4 and 10 (Figure 2). These karyotypes differ from those previously reported for this species [12,26,27,29], where the same type of rearrangement found here is believed to explain in part the karyotypic diversity observed in M. hirsuta.
The distribution pattern of constitutive heterochromatin in only a few chromosomes differs from that in Lampronycteris brachyotis (Micronycterinae) [28], where the entire chromosome complement presents conspicuous markings. This result found in Micronycteris hirsuta is in agreement with previous observations that bats have a strong tendency to reduction in genomic size (or C-value) and consequently a decrease of regions containing highly repetitive DNA [33,34].
The NOR was found interstitially in one chromosome pair in all specimens of Micronycteris hirsuta studied here. This same chromosome can be easily identified by the presence of a secondary constriction in specimens of M. hirsuta studied in Ecuador with 2n = 26 chromosomes [12]. The NORs labeled positive for fluorochrome CMA3, but negative for DAPI, indicating that the ribosomal DNA is interspersed with repetitive GC-rich DNA.
In addition to signals in telomeric regions, the telomeric probe produced unexpected signals in the centromeric regions of chromosomes in Micronycteris hirsuta. The intrachromosomal or interstitial telomeric sequences (ITSs) may correspond to sequences of repetitive DNA similar to telomeric sequences, but probably are not originating from the telomeres, as evidenced in other species of bats [35-37].
Considering that all species of the genus Micronycteris, except Micronycteris hirsuta, have a bi-armed X chromosome, it is likely that the acrocentric shape of the X is an autapomorphic character of this species, as previously suggested by Rodrigues et al.[38] and Noronha et al.[39]. In contrast, the primitive condition of X for phyllostomids would be a bi-armed form as found in Macrotus waterhousii, Phyllostomus discolor and P. hastatus[28,29,38,40].
Table 1 summarizes the karyotypic data of Micronycteris hirsuta analyzed to date. Comparing karyotypes studied here and those from Central and South America with 2n = 28 (Trinidad and Tobago) [26] and 30 chromosomes (Honduras, Nicaragua and Suriname) [26,27], the differences can be explained by two Robertsonian rearrangements (fusion/fission) between acrocentric chromosomes. Despite their similar diploid number (2n = 26) [12], populations from Ecuador and Brazil diverge by three rearrangements (2 fusion/fission and 1 pericentric inversion) resulting in morphological alterations of five chromosome pairs (1, 2, 5, 7 and 8 in Figure 1, present study). When compared with the 2n = 28 karyotype from Trinidad and Tobago, two rearrangements for the Ecuadorian population and a single for the Brazilian population are enough to explain their divergence. Therefore, we assume that karyotypes with 2n = 26 from Ecuador and Brazil may have evolved independently starting from an ancestral with 2n = 28.
The sharing of the NOR-bearing pair in the karyotype of specimens from Brazil and Ecuador (see Figure seven in reference [12]), reinforces the argument that karyotypic divergence occurred by centric fusion rearrangements that led to a reduction in the diploid numbers. The similarity between the specimens from Brazil and Ecuador is best explained by karyotypic homoplasy on the diploid number and it is highly likely that the karyotype 2n = 28 chromosomes represents the ancestral karyotype these cytotypes. Thus, karyotypic data suggest that the specimens of Trinidad and Tobago (2n = 28) are closer to the Brazilian (2n = 26, NF = 32) and equatorial (2n = 26, NF = 30) specimens.
Moreover the Andean Cordillera, a geographic barrier that crosses the center of Ecuador from the north to south, has probably contributed to isolating populations from Brazil and Ecuador. This hypothesis is plausible considering that the events which led to the diversification of the genus Micronycteris occurred during the Pliocene and Pleistocene epochs (5.0 and 0.6 million years ago) and the corresponding lineage of M. hirsuta that diversified about 1.0 million years ago (range 2.0 to 0.5 MYA) [4], while the uplift of the Andes was completed about 2.5 Million years ago during the Quaternary [41]. A tentative interpretation of karyotypic differentiation routes in M. hirsuta is shown in Figure 5, where examination of karyotypes seems to indicate that geographic variation of diploid numbers exhibits a decrease from North to South.
Figure 5.
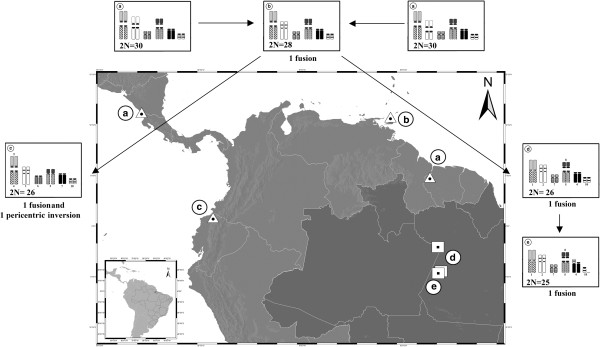
Map of compared samples of Micronycteris hirsuta. Triangles indicate the sites from which previous cytogenetic descriptions were performed. Squares represent the cytogenetic samples studied herein (see Table 1 for locality details). Idiograms plotted beside each collection site represent the unique or shared chromosomes among specimens. Diploid number (2n) of specimen and the rearrangements that differentiate the different cytotypes are indicated below each idiogram. Idiograms of MHI from a) MHI from Honduras, Nicaragua and Suriname; b) Trinidad and Tobago; c) Ecuador; d) and e) Present work. For specimens of Ecuador with 2n = 26 the numeration of chromosome arms is according Fonseca et al.[12]. For details see text and Table 1.
From Figure 5 we note that there are different diploid numbers in different locations but there is no information on the intermediary regions. Two possibilities can be suggested: 1) If intermediary karyotypes are found in the intermediary regions, this would be a typical case of chromosomal polymorphism. 2) If there are no intermediary karyotypes, the different populations with different diploid numbers are reproductively isolated, meaning that they are different species and Micronycteris hirsuta would not represent a monotypic taxon. Under this view, the karyotype with 2n = 25 (LR-275) can have three explanations: 1) this would be a single heterozygous specimen found as result of a balanced rearrangement Robertsonian (fusion) that occurred during gametogenesis of one of its parents. 2) An intermediary karyotype that is probably common, meaning that this is a chromosomal polymorphism. 3) An evidence of a hybrid zone where the heterozygote sample with 2n = 25 was a hybrid derived from a cross between two homozygous forms with 2n = 26 and 24 chromosomes (the latter remains to be found).
Analyses of mitochondrial (cytochrome b) and nuclear (intron 7 of the nuclear fibrinogen) genes demonstrate that populations of M. hirsuta are subdivided into three distinct clades [4,13]. Ecuadorian specimens with 2n = 26 are phylogenetic closer with populations of Panama (diploid number unknown). On the other hand, specimens of Trinidad and Tobago with 2n = 28 are more similar of phylogenetic standpoint with specimens collected in French Guiana (diploid number unknown) [4,12,13]. Although these data indicate a phylogenetic proximity between populations of Panama and Ecuador, the karyotypic data available are yet incipient to corroborate these phylogenetic relationships. In order to clarify this issue, obtaining cytogenetic data of intermediate populations across the geographic distribution of M. hirsuta that may reveal new karyotypic forms, and analysis of nuclear and mitochondrial genes are sorely needed.
The cytotypes described here, added to evidence suggesting some geographic differentiation of populations based in karyotypic, mitochondrial, nuclear, and morphological markers [26], strongly suggest that Micronycteris hirsuta do not represent a monotypic taxa.
Additionally, the record of Micronycteris hirsuta in two localities of eastern Amazonia described here is an increase in the distribution of this species in South America to about 700 km. Although rarely captured, species of big eared-bats represent a common component of Neotropical rain forests [11]. Therefore it is possible that the occurrence of M. hirsuta in Brazil and the Amazon region have been underestimated, since there are records of this species in the Atlantic Forest [42].
Micronycteris hirsuta and its correlation with Phyllostomidae
Recent data from chromosome painting, associated with G-banding, confirm that Robertsonian fusions with the complete conservation of chromosome or whole arms, and inversions are the main mechanisms of karyotypic differentiation in the Order Chiroptera [19,20,22,24,25,32]. Herein we found conservatism in many chromosomal segments shared between Micronycteris hirsuta, Phyllostomus hastatus and Carollia brevicauda, but homology of whole chromosomes was not detected in the autosomal set.
A comparison of Micronycteris hirsuta painted chromosomal map with other phyllostomids previously investigated [17,18,32], revealed that many rearrangements have occurred in the differentiation of the M. hirsuta karyotype from the phyllostomid common ancestor. Even the highly conserved syntenic group (PHA-11; CBR-7), shared by Phyllostomus, Carollia, Artibeus, Uroderma magnirostrum[17,18] and Desmodontinae bats [32], was broken in the MHI genome (MHI-3 and MHI-4) (Figure 1). In the other hand, although not broken, the chromosome PHA-14 is fusioned with PHA-11 in MHI, with PHA-9 in UBI and, in the Desmodontinae bats with two distinct chromosomal segments: 12 in DYO and 13 in DRO [32].
These chromosomes have been suggested to be present on the ancestral karyotype of the family Phyllostomidae, since it was present and conserved in the karyotype of such phylogenetically distant species of uncorrelated subfamilies mapping so far [17,18,32]. Although MHI represent a basal clade for Phyllostomidae, and has previously been suggested that basal taxa could contribute to confirm the chromosomal ancestrality [32], we argue that not necessarily basal taxa have primitive karyotypes. It is more probably that these chromosomes forms found in MHI correspond to derivative forms within Phyllostomidae and autoapomorphies in this species, without phylogenetic value.
Classical cytogenetic studies had already suggested a high rate of karyotype evolution within the big-eared bats lineage [28,43]. Arnold et al.[44] argue that, compared to their congeners, Micronycteris hirsuta has undergone extensive and independent karyotypic changes, which has been supported by other authors [3,12,13]. Our results corroborate the hypothesis of karyotypic megaevolution within Micronycteris, and strong evidence for this is that the entire chromosome complement of M. hirsuta proved to be derivative with respect to species compared in this study.
Conclusions
In conclusion, the karyotypes of Brazilian specimens of Micronycteris hirsuta (2n = 26 and 25, FN = 32), show variation in both diploid and fundamental number, in relation to specimens from Central and South America, and therefore are new karyotypic forms reported to this species. The entire autosomal chromosome complement of M. hirsuta seems to be highly derived in relation to that of other phyllostomid bats, and the finding of new cytotypes reinforce that Micronycteris hirsuta does not represent a monotypic taxon.
Methods
Specimens analyzed
Six specimens (four females and two males) of Micronycteris hirsuta were collected from natural populations during field expeditions to assess biodiversity in the Amazon Region, Brazil. The study sites were: Itaituba (S04°28′20, 5”/W56°17′03,7) and Faro (S02°04′43,4”/W 56°37′12,0”), municipalities of Pará, Brazil (Table 1). Voucher specimens (LR-194, LR-275, LR-1342, LR-2104, LR-2160 and LR-2428) were fixed in 10% formalin preserved in 70% ethanol and deposited in the mammal collection of the Museu Paraense Emilio Goeldi and Museu de Zoologia da Universidade Federal do Oeste do Pará.
Chromosomal preparation, chromosomal banding and staining with fluorochromes
Chromosomal preparations were obtained by direct extraction from bone marrow after Colchicine treatment following Baker et al.[45]. Conventional staining was used for diploid (2n) and fundamental numbers (FN) determination. G-banding followed two distinct methods: trypsin treatment [46], and saline solution (2xSSC) incubation [47]. In both methods the metaphases were stained with Wright’s solution. C-banding was carried out according to Sumner [48] and Ag-NOR staining followed Howell and Black [49]. For observation of GC/AT base pairs rich regions, metaphase chromosomes were stained with CMA3 following Schweizer [50] and DAPI according to Pieczarka et al.[51], both with modifications.
Fluorescence in situ hybridization (FISH)
FISH with digoxigenin labeled telomeric probes (All Human Telomere Probes, Oncor) were performed according to the manufacturer’s protolocol and 18S rDNA probes from Prochilodus argenteus[52], were labeled with biotin or digoxigenin by nick translation. Chromosome-specific painting probes from P. hastatus and C. brevicauda were generated from flow-sorted chromosomes and labeled by DOP-PCR (degenerate oligonucleotide-primed-polymerase chain reaction) amplification [53], and used to determine chromosomal homologies in M. hirsuta. The chromosome painting experiments were carried out following procedures previously described [17,54]. Briefly, the slides were incubated in pepsin solution, and dehydrated in an ethanol series (70, 90 and 100%), air-dried and aged in a 65°C incubator for two hours. Chromosomal DNA was denatured in 70% formamide/2xSSC for 40 seconds, and the slides immersed immediately in cold 70% ethanol for 4 minutes. For single-color detection, biotin-labeled probes were visualized with Cy3-avidin or FITC-avidin. For two-color detection, Cy3-labelled probes were used together with biotin-labeled probes in the same experiment. After hybridization and washing, the metaphases were stained with DAPI. Images were captured using the Axiovision 3.0 software with a CCD camera (Photometrics C250/A) coupled on a Zeiss-Axiophot 2 microscope or with a software Nis-Elements on a Nikon H550S microscope. For image processing Adobe Photoshop CS4 software was used.
The karyotypes of Desmodontinae bats [32], were used in the comparative analysis. The chromosome complement of Phyllostomus hastatus (2n = 32) was used as a reference for defining syntenic associations among the compared species, since previous G-banding results indicated that its karyotype retained most of the segments and the supposed ancestral chromosome arms of Phyllostomidae [28].
Abbreviations
CBR: Carollia brevicauda; CMA3: Chromomycin A3; DAPI: fluorochorme 4′-6′-diamidino-2-phenylindole; DRO: Desmodus rotundus; DOP-PCR: Degenerate oligonucleotide-primed-polymerase chain reaction; DYO: Diaemus youngi; FISH: Fluorescence In Situ Hybridization; FN: Fundamental number; PHA: Phyllostomus hastatus; MHI: Micronycteris hirsuta; SSC: Saline sodium citrate.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TFAR carried out classic cytogenetic and chromosome painting, organized undertook the bibliographic review, analyzed the data and wrote the paper. LRRR collected most the samples, helped in classic and molecular cytogenetic analysis and contributed to the discussion of data. CYN participated of the techniques development and contributed to the discussion of data. TCMB participated of the techniques development and carried out the FISH with telomeric and rDNA 18S probes. AJBG collected some samples, participated of the techniques development, helped in classic and molecular cytogenetic analysis and contributed to the discussion. PCMO carried out the whole chromosome probes from Carollia brevicauda and Phyllostomus hastatus. MFS revised the manuscript and contributed to the discussion of data. FY revised the manuscript and participated of the techniques development. JCP coordinated the study and contributed to the discussion. All authors read and approved the final version of the manuscript.
Contributor Information
Talita FA Ribas, Email: talitafernanda.ufpa@gmail.com.
Luis RR Rodrigues, Email: luisreginaldo.ufpa@hotmail.com.
Cleusa Y Nagamachi, Email: cleusanagamachi@gmail.com.
Anderson JB Gomes, Email: citand2004@yahoo.com.br.
Thayse CM Benathar, Email: thaysebenathar@yahoo.com.br.
Patricia CM O’Brien, Email: pco20@mole.bio.cam.ac.uk.
Fengtang Yang, Email: fy1@sanger.ac.uk.
Malcolm A Ferguson-Smith, Email: maf12@cam.ac.uk.
Julio C Pieczarka, Email: juliopieczarka@gmail.com.
Acknowledgements
The authors thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação de Amparo à Pesquisa do Estado do Pará (FAPESPA) for financial support (Edital BIONORTE - Proc 552032/2010-7) on a project coordinated by CY Nagamachi; the Universidade Federal do Oeste do Pará by scholarship IC-UFOPA to TFAR; the FAPESPA for financial support (Edital Vale – Proc 2010/110447) on a project coordinated by JC Pieczarka; the Wellcome Trust for a grant to MA Ferguson-Smith; the Biodinâmica-Rio and Aotus Consultoria Ambiental for financing the expeditions; the Sapopema and Conservação Internacional do Brasil for logistic support and financing the field trips; to Fábio Sarmento, Ramon Araújo and Eloisa Soares for help in the field work; to Maria da Conceição and Jorge Rissino for assistance in laboratory work. Sample collection was authorized by Instituto Brasileiro de Meio Ambiente e dos Recursos Nacionais Renováveis (IBAMA). TFAR is a recipient of FAPESPA Master Scholarship in Genetics and Molecular Biology; TCMB is a recipient of CNPq Master Scholarship in Genetics and Molecular Biology, AJB Gomes is a recipient of CAPES Doctor Scholarship in Genetics and Molecular Biology.
References
- Simmons NB. In: Mammal species of the world: a taxonomic and geographic reference: third edition. Wilson DE, Reeder DM, editor. Baltimore MD: Johns Hopkins Univ Press; 2005. Order chiroptera; pp. 312–529. [Google Scholar]
- Baker RJ, Hood CS, Honeycutt RL. Phylogenetic relationships and classification of the higher categories of the New World bat family Phyllostomidae. Syst Zool. 1989;14:228–238. doi: 10.2307/2992284. [DOI] [Google Scholar]
- Wetterer AL, Rockman MV, Simmons NB. Phylogeny of Phyllostomid bats (Mammalia: Chirpotera): data from diverse morphological systems, sex chromosomes and restriction sites. Bull Am Mus Nat Hist. 2000;14:1–200. [Google Scholar]
- Larsen AP, Siles L, Pedersenm SC, Kwiecinski GG. A new species of Micronycteris (Chiroptera: Phyllostomidae) from Saint Vincent, Lesser Antilles. Mammal Biol. 2011;14:687–700. [Google Scholar]
- Baker RJ, Bininda-Emonds ORP, Mantilla-Meluk H, Porter CA, Van Den Bussche RA. In: Evolutionary history of bats: fossils, molecules and morphology. Gunnell GF, Simmons NB, editor. Cambridge: Univ Press; 2012. Molecular timescale of diversification of feeding strategy and morphology in New world leaf-nosed bats (Phyllostomidae): a phylogenetic perspective; pp. 385–409. [Google Scholar]
- Sanbor CC. Bats of the genus micronycteris and its subgenera. Chicago Nat Hist Mus: EUA; 1949. [Google Scholar]
- Simmons NB. A new species of Micronycteris (Chiroptera:Phyllostomidae) from northeastern Brazil, with comments on phylogenic relationships. Am Mus Novit. 1996;14:1–34. [Google Scholar]
- Simmons NB, Voss RS. The mammals of paracou, French Guiana: a neotropical lowland rainforest fauna part 1: bats: New York. Bull Am Mus Nat Hist. 1998;14:1–219. [Google Scholar]
- Baker RJ, Porter CA, Patton JC, Van Den Bussche RA. Systematics of bats of the family Phyllostomidae based on RAG2 DNA sequences. Occas Pap Mus Texas Tech Univ. 2000;14:1–16. [Google Scholar]
- Baker RJ, Hoofer SR, Porter CA, Van Den Bussche RA. Diversification among new-world leaf-nosed bats: an evolutionary hypothesis and classification inferred from digenomic congruence of DNA sequence. Occasional Papers, Museum of Texas Tech University. 2003;14:1–32. [Google Scholar]
- Simmon NB, Voss RS, Fleck DW. A new Amazonian species of micronycteris (chiroptera: phyllostomidae) with notes on the roosting behavior of sympatric congeners. Am Mus Novit. 2002;14:1–14. [Google Scholar]
- Fonseca RM, Hoofer SR, Porter CA, Cline CA, Parish DA, Hoofman FG, Baker RJ. Morphological and molecular variation within little big-eared bats of genus Micronycteris (Phyllostomidae: micronycterinae) from San Lorenzo, Ecuador. Univ of California Publicat Zool. 2007;14:1–981. [Google Scholar]
- Porter CA, Hoofer SR, Cline AC, Hoofman FG, Baker RJ. Molecular Phyllogenetics of the Phyllostomid bat genus Micronycteris with descriptions of two new subgenera. J Mammal. 2007;14:1205–1215. doi: 10.1644/06-MAMM-A-292R.1. [DOI] [Google Scholar]
- Volleth M, Klett C, Kollak A, Dixkens C, Winter Y, Just W, Vogel W, Hameister H. ZOO-FISH analysis in a species of the order Chiroptera: Glossophaga soricina (Phyllostomidae) Chromosome Res. 1999;14:57–64. doi: 10.1023/A:1009227428727. [DOI] [PubMed] [Google Scholar]
- Volleth M, Heller KG, Pfeiffer RA, Hameister H. Comparative ZOO-FISH analysis in bats elucidates the phylogenetic relationships between Megachiroptera and five microchiropteran families. Chromosome Res. 2002;14:477–497. doi: 10.1023/A:1020992330679. [DOI] [PubMed] [Google Scholar]
- Volleth M, Yang F, Muller S. High resolution chromosome painting reveals the first signature for the chiropteran suborder Pteropodiformes (Mammalia: Chiroptera) Chromosome Res. 2011;14:507–519. doi: 10.1007/s10577-011-9196-5. [DOI] [PubMed] [Google Scholar]
- Pieczarka JC, Nagamachi CY, O’Brien PC, Yang F, Rens W, Barros RM, Noronha RC, Rissino J, de Oliveira EH, Ferguson-Smith MA. Reciprocal chromosome painting between two South American bats: Carollia brevicauda and Phyllostomus hastatus (Phyllostomidae, Chiroptera) Chromosome Res. 2005;14:339–347. doi: 10.1007/s10577-005-2886-0. [DOI] [PubMed] [Google Scholar]
- Pieczarka JC, Gomes AJ, Nagamachi CY, Rocha DC, Rissino JD, O’Brien PC, Yang F, Ferguson-Smith MA. A phylogenetic analysis using multidirectional chromosome painting of three species (Uroderma magnirostrum, U. bilobatum and Artibeus obscurus) of subfamily Stenodermatinae (Chiroptera-Phyllostomidae) Chromosome Res. 2013;14:383–392. doi: 10.1007/s10577-013-9365-9. [DOI] [PubMed] [Google Scholar]
- Ao L, Gu X, Feng Q, Wang J, O’Brien PCM, Fu B, Mao X, Su W, Wang Y, Volleth M, Yang F, Nie W. Karyotype relationships of six bat species (Chiroptera, Vespertilionidae) from China revealed by chromosome painting and G-banding comparison. Cytogenet Genome Res. 2006;14:145–153. doi: 10.1159/000095235. [DOI] [PubMed] [Google Scholar]
- Ao L, Mao X, Nie W, Gu X, Feng Q, Wang J, Su W, Wang Y, Volleth M, Yang F. Karyotypic evolution and phylogenetic relationships in the order Chiroptera as revealed by G-banding comparison and chromosome painting. Chromosome Res. 2007;14:257–268. doi: 10.1007/s10577-007-1120-7. [DOI] [PubMed] [Google Scholar]
- Mao X, Nie W, Wang J, Su W, Ao L, Feng Q, Wang Y, Volleth M, Yang F. Karyotype evolution in Rhinolophus bats (Rhinolophidae, Chiroptera) illuminated by cross-species chromosome painting and G-banded comparison. Chromosome Res. 2007;14:2–14. doi: 10.1007/s10577-007-1167-5. [DOI] [PubMed] [Google Scholar]
- Mao X, Nie W, Wang J, Weiting SU, Feng Q, Wang Y, Dobigny G, Yang F. Comparative cytogenetics of bats (Chiroptera): the prevalence of robertsonian translocation limits the power of chromosomal characters in resolving interfamily phylogenetic relationships. Chromosome Res. 2008;14:155–170. doi: 10.1007/s10577-007-1206-2. [DOI] [PubMed] [Google Scholar]
- Mao X, Wang J, Su W, Wang Y, Yang F, Nie W. Karyotypic evolution in family Hipposideridae (Chiroptera, Mammalia) revealed by comparative chromosome painting, G- and C-banding. Zoonoses Res. 2010;14:453–460. doi: 10.3724/SP.J.1141.2010.05453. [DOI] [PubMed] [Google Scholar]
- Kulemzina AI, Nie W, Trifonov VA, Staroselec Y, Vasenkov DA, Volleth M, Yang F, Graphodatsky AS. Comparative chromosome painting of four Siberian vespertilionidae species with Aselliscus stoliczkanus and human probes. Cytogenet Genome Res. 2011;14:200–205. doi: 10.1159/000328834. [DOI] [PubMed] [Google Scholar]
- Volleth M, Eick G. Chromosome evolution in bats as revealed by FISH: the ongoing search for the ancestral chiropteran karyotype. Cytogenet Genome Res. 2012;14:165–173. doi: 10.1159/000338929. [DOI] [PubMed] [Google Scholar]
- Baker RJ, Genoways HH, Bleier WJ, Warner JW. Cytotypes and a morphometrics of two Phyllostomatid bats, Micronycteris hirsuta and Vampyressa pussila. Mus Texas Tech Univ. 1973;14:1–11. [Google Scholar]
- Baker RJ, Genoways HH, Seyfarth PA. Results of the Alcoa foundation-Suriname expeditions: VI. additional chromosomal data for bats (Mammalia: Chiroptera) from Suriname. Ann Carnegie Mus of Nat Hist. 1981;14:333–344. [Google Scholar]
- Patton JC, Baker RJ. Chromosomal homology and evolution of phyllostomatoid bats. Sist Zool. 1978;14:449–462. doi: 10.2307/2412927. [DOI] [Google Scholar]
- Baker RJ. In: Biology of bats of the new world family Phyllostomatidae, part III. Baker RJ, Jones JKJr, Carter DC, editor. Lubbock, Texas, USA: Occas Pap Mus Texas Tech Univ; 1979. Karyology; pp. 107–155. [Google Scholar]
- Tucker PK, Bickham JW. Sex chromosome-autosome translocation in the leaf nosed bats: II. Meiotic analyses of the subfamilies Sternodermatinae and Phyllostominae. Cytogenet Cell Genet. 1986;14:27–28. doi: 10.1159/000132294. [DOI] [PubMed] [Google Scholar]
- Tucker PK. Sex chromosome-autosome translocations in the leaf-nosed bats, family Phyllostomidae. I. Mitotic analyses of the subfamilies Sternodermatinae and Phyllostominae. Cytogenet Cell Genet. 1986;14:19–27. doi: 10.1159/000132293. [DOI] [PubMed] [Google Scholar]
- Sotero-Caio CG, Pieczarka JC, Nagamachi CY, Gomes AJ, Lira TC, O’Brien PC, Ferguson-Smith MA, Souza MJ, Santos N. Chromosomal homologies among vampire bats revealed by chromosome painting (Phyllostomidae, Chiroptera) Cytogenet Genome Res. 2011;14:156–164. doi: 10.1159/000321574. [DOI] [PubMed] [Google Scholar]
- Baker RJ, Maltbie M, Owen JG, Hamilton MJ, Bradley RJ. Reduced number of ribosomal sites in bats: evidence for a mechanism to contain genome size. J Mamm. 1992;14:847–858. doi: 10.2307/1382206. [DOI] [Google Scholar]
- Van Den Bussche RA, Longmire JL, Baker RJ. How bats achieve a smal C-value: frequency of repetitive DNA in Macrotus. Mammal Genome. 1995;14:521–525. doi: 10.1007/BF00356168. [DOI] [PubMed] [Google Scholar]
- Meyne J, Baker RJ, Hobart HH, Hsu TC, Ryder OA, Ward OG, Wiley JE, Wurster-Hill DH, Yates TL, Moyzis RK. Distribution of non-telomeric sites of the (TTAGGG) n telomeric sequence in vertebrate chromosomes. Chromosoma. 1990;14:3–10. doi: 10.1007/BF01737283. [DOI] [PubMed] [Google Scholar]
- Finato AO, Varella-Garcia M, Tajara EH, Taddei VA, Morielle-Versute E. Intrachromosomal distribution of telomeric repeats in Eumops glaucinus and Eumops perotis (Molossidae, Chiroptera) Chromosome Res. 2000;14:563–569. doi: 10.1023/A:1009288121670. [DOI] [PubMed] [Google Scholar]
- Faria KC, Morielle-Versute E. In situ hybridization of bat chromosomes with human (TTAGGG) n probe, after previous digestion with Alu I. Genet Mol Biol. 2002;14:365–371. [Google Scholar]
- Rodrigues LRR, Barros RMS, Marques-Aguiar S, Assis MFL, Pieczarka JC, Nagamachi CY. Comparative cytogenetics of two phyllostomids bats: a new hypothesis to the origin of the rearranged X chromosome from artibeus lituratus. Caryologia (Firenze) 2003;14:413–419. doi: 10.1080/00087114.2003.10589352. [DOI] [Google Scholar]
- Noronha RCR, Nagamachi CY, O’Brien PCM, Ferguson-Smith MA, Pieczarka JC. Meiotic analysis of XX/XY and neo-XX/XY sex chromosomes in Phyllostomidae by cross-species chromosome painting revealing a common chromosome 15-XY rearrangement in Stenodermatinae. Chromosome Res. 2010;14:667–676. doi: 10.1007/s10577-010-9146-7. [DOI] [PubMed] [Google Scholar]
- Rodrigues LRR, Barros RMS, Assis MFL, Marques-Aguiar S, Pieczarka JC, Nagamachi CY. Chromosome comparison between two species of Phyllostomus (Chiroptera - Phyllostomidae) from Brazilian Amazonia, with some phylogenetic insights. Genet Mol Biol. 2000;14:595–599. doi: 10.1590/S1415-47572000000300016. [DOI] [Google Scholar]
- Hoorn C, Wesselingh FP, ter Steege H, Bermudez MA, Mora A, Sevink J, Sanmartín I, Sanchez-Meseguer A, Anderson CL, Figueiredo JP, Jaramillo C, Riff D, Negri FR, Hooghiemstra H, Lundberg J, Stadler T, Särkinen T, Antonelli A. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science. 2010;14:927–931. doi: 10.1126/science.1194585. [DOI] [PubMed] [Google Scholar]
- Gardner AL. Mammals of South America. Chicago: University of Chicago Press; 2007. [Google Scholar]
- Baker RJ, Bickham JW. Karyotypic evolution in bats: evidence of extensive and conservative chromosomal evolution in closely related taxa. Syst Zool. 1980;14:239–253. doi: 10.2307/2412660. [DOI] [Google Scholar]
- Arnold ML, Baker RJ, Honeycut RL. Genic differentiation and phylogenetic relationships within two new world Bat genera. Biochem Syst Ecol. 1983;14:295–303. doi: 10.1016/0305-1978(83)90067-4. [DOI] [Google Scholar]
- Baker RJ, Hamilton M, Parish DA. Preparations of Mammalian karyotypes under field conditions. Occ Papers, Mus Texas Tech Univ. 2003;14:1–7. [Google Scholar]
- Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971;14:971–972. doi: 10.1016/s0140-6736(71)90287-x. [DOI] [PubMed] [Google Scholar]
- Verma RS, Babu A. Human chromosomes: principles and techniques: second edition. New York: Health profession division, McGraw-Hill, EUA; 1995. [Google Scholar]
- Sumner AT. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972;14:304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- Howell WM, Black DA. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia. 1980;14:1014–1015. doi: 10.1007/BF01953855. [DOI] [PubMed] [Google Scholar]
- Schweizer D. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma. 1976;14:307–324. doi: 10.1007/BF00292840. [DOI] [PubMed] [Google Scholar]
- Pieczarka JC, Nagamachi CY, Souza ACP, Milhomem SSR, Castro RR, Nascimento AL. An adaptation to DAPI-banding to fishes chromosomes. Caryologia. 2006;14:43–46. [Google Scholar]
- Hatanaka T, Galetti PM Jr. Mapping of 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae) Genetica. 2004;14:239–244. doi: 10.1007/s10709-004-2039-y. [DOI] [PubMed] [Google Scholar]
- Telenius H, Ponder BAJ, Tunnacliffe A, Pelmear AH, Carter NP, Ferguson-Smith MA, Behmel A, Nordenskjöld M, Pfragner R. Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genomics. 1992;14:718–725. doi: 10.1016/0888-7543(92)90147-K. [DOI] [PubMed] [Google Scholar]
- Yang F, Carter NP, Shi L, Ferguson-Smith MA. A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma. 1995;14:642–652. doi: 10.1007/BF00357691. [DOI] [PubMed] [Google Scholar]