Abstract
What is the best way to identify regulatory DNA sequences, such as enhancers, promoters, insulators and silencers? A recent study shows that specific binding by a co-activator protein identifies enhancers that are invisible to common methods based on evolutionary constraint.
Much effort has been devoted to accurately predicting the regulatory DNA sequences required for correct gene expression. Unfortunately, molecular biologists and biochemists are struggling to develop even a rudimentary set of rules that can identify regulatory modules using solely DNA sequence. Comparisons of homologous DNA between related species has provided a valuable approach for identifying regulatory sequences. Thousands of human noncoding DNA sequences have sustained very few alterations over vertebrate evolution, showing that they are under stringent evolutionary constraint 1. Many of these are enhancers of expression in some tissues, such as brain (Figure 1) 2. However, enhancers active in other tissues, such as heart, are rarely found by this approach. A new study on p. XXXX of this issue by Len Pennacchio and colleagues 3 uses the presence of a transcriptional co-activator bound to DNA to identify heart enhancers (Figure 1). They show that many regulatory sequences are preserved over a limited phylogenetic distance, and suggest that the pattern of evolutionary constraint may vary by tissue type.
Figure 1.
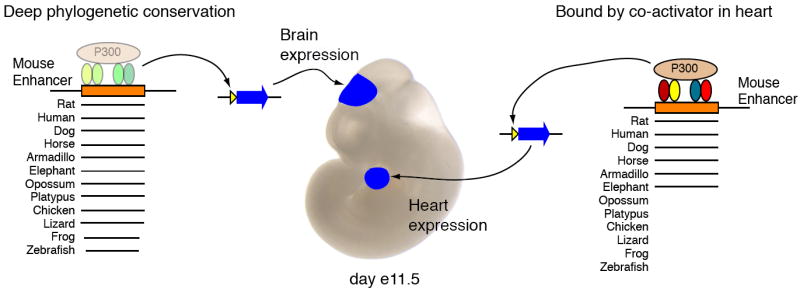
Binding by the transcriptional co-activator p300 identifies enhancers that are not conserved in all vertebrates. (Left) Deep phylogenetic conservation of noncoding DNA is a good method for finding tissue-specific developmental enhancers, as assayed in transient transgenic mice. Such evolutionarily-constrained enhancers are evident in some tissues, such as brain, but not in others, such as heart. (Right) Direct binding by the co-activator p300 identifies tissue-specific enhancers that are not necessarily deeply conserved (i.e. conserved only in placental mammals). Lines after each species indicate that sequences homologous to the mouse enhancer are present in that species.
Variability in evolutionary constraint
Blow et al. 3 identified DNA segments bound by the common transcriptional co-activator protein, p300, in mouse embryonic heart tissue by chromatin immunoprecipitation. After deep sequencing of the DNA associated with p300, approximately 3600 DNA segments that were specifically bound by this co-activator were identified and nominated as candidate heart-specific enhancers.
Blow et al. find two important features of the candidate heart enhancers. First, 75% of candidate sequences were validated as enhancers in vivo, and of these validated enhancers, the vast majority (84%) were active in the developing heart. Second, these candidate heart enhancers differ dramatically from other tissue-specific enhancers in their evolutionary signatures. Most of the heart enhancers are not subject to the stringent evolutionary constraint that is characteristic of forebrain enhancers active at the same developmental timepoint [AU: ok?] [OK – Ross]. While 65% of heart enhancers are detectably conserved in placental mammals, over half (56%) of the forebrain enhancers are more deeply conserved in birds, with some even conserved in amphibians and fish (Figure 1).
Clearly, stringent constraint is not a feature shared by all regulatory regions, so other approaches are required to identify these sequences in the genome. In fact, genome-wide mapping studies of biochemical features associated with gene regulatory regions 4,5 are becoming the method of choice for finding regulatory sequences.
The results from Blow et al.3 and related work point to a complicated pattern of evolutionary constraint. The profile of evolutionary constraint, at least at the developmental timepoint investigated, appears to be distinctive to enhancers from different tissues, with heart enhancers being under weak constraint, forebrain enhancers under strong constraint, and enhancers in other tissues showing an intermediate pattern. Previous work has suggested that the evolutionary patterns of regulatory regions can provide predictive power about other aspects of their functions, e.g. rapidly evolving sequences are implicated in adaptive responses, and sequences conserved in one clade (e.g. placental mammals) appear to regulate classes of genes that differ from those regulated by sequences conserved in other clades (e.g. vertebrates)6. Thus, even as biochemical features take the forefront in predicting regulatory regions, detailed studies of their evolutionary signatures, as revealed by alignments of DNA from multiple species across a wide phylogenetic spectrum, will continue to illuminate aspects of DNA function.
Value of interspecies alignments
One may wonder if there is any role left for comparative sequence analysis in studying regulatory regions. For a given DNA sequence that acts as a regulatory module in human, how frequently do homologs in other species serve a similar function? Recent investigations of specific binding by liver transcription factors in five vertebrate species show that occupancy of a small minority (10%-22%) of the bound sites are shared between two different mammals 7. This indicates that much of the binding of transcription factors is species-specific, suggesting that some of the bound sites may be evolving adaptively. Perhaps many of the bound sites are playing a passive role (e.g. for storage), and may be evolving close to neutrally.
Indeed, previous studies of tissue-specific enhancers found many sequences that were not subject to strong evolutionary constraint across the entire tissue-specific regulatory module, with some enhancers being species-specific and others showing highly localized constraint (i.e. in certain binding sites for particular transcription factors) 6,8. Could it be that the level of constraint on enhancers is a characteristic of particular biological properties of regulatory modules? Perhaps enhancers active in some tissues may be subject to strong constraint, while those active in other tissues may be evolving more rapidly. Certainly, DNA sequences regulating genes whose products are subject to positive selection, providing an adaptive advantage, would be expected to change as well, and not be strongly constrained. If evolutionary profiles in regulatory regions prove to be truly distinctive for functional categories of target genes or patterns of tissue-specific expression, then comparative genomics of regulatory regions will be most useful for deducing aspects of their function.
Biological implications
Interpreting this apparent tissue-specificity in the levels of constraint on regulatory DNA sequences is a formidable challenge. For instance, it is not at all clear why enhancers leading to heart-specific gene expression would be evolving more rapidly than those leading to forebrain expression. The heart is an ancient organ, one that is obviously critical to organisms that have it. One may have expected that the mechanisms of gene regulation leading to heart formation would be preserved over a phylogenetic distance comparable to that of species having hearts. However, at least for the enhancers described to date, that is not the case. Is the pattern of heart gene regulation actually well-preserved in vertebrates, or does it vary? If it is well-preserved, are the heart-specific enhancers re-invented independently in different lineages? What evolutionary mechanisms provide the sequence changes that could fuel that re-invention? What evolutionary profiles dominate in enhancers for other tissues? These are but a few of the questions raised by these recent results. Happily, many of them should be answerable with the growing genomic analyses of gene regulation. These answers should deepen our understanding of gene regulation in complex organisms, and improve our ability to interpret genetic variation affecting gene regulation in humans.
References
- 1.Dermitzakis ET, Reymond A, Antonarakis SE. Conserved non-genic sequences - an unexpected feature of mammalian genomes. Nat Rev Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 2.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 3.Blow MJ, et al. ChIP-seq Identification of Weakly Conserved Heart Enhancers. Nature Genetics. 2010 doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 5.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King DC, et al. Finding cis-regulatory elements using comparative genomics: some lessons from ENCODE data. Genome Res. 2007;17:775–786. doi: 10.1101/gr.5592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margulies EH, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]


