Abstract
Haloviruses HF1 and HF2 were isolated from the same saltern pond and are adapted to hypersaline conditions, where they infect a broad range of haloarchaeal species. The HF2 genome has previously been reported. The complete sequence of the HF1 genome has now been determined, mainly by PCR and primer walking. It was 75,898 bp in length and was 94.4% identical to the HF2 genome but about 1.8 kb shorter. A total of 117 open reading frames and five tRNA-like genes were predicted, and their database matches and characteristics were similar to those found in HF2. A comparison of the predicted restriction digest patterns based on nucleotide sequence with the observed restriction digest patterns of viral DNA showed that, unlike the case for HF2, some packaged HF1 DNA had cohesive termini. Except for a single base change, HF1 and HF2 were identical in sequence over the first 48 kb, a region that includes the early and middle genes. The remaining 28 kb of HF1 showed many differences from HF2, and the similarity of the two genomes over this late gene region was 87%. The abrupt shift in sequence similarity around 48 kb suggests a recent recombination event between either HF1 or HF2 and another HF-like halovirus that has swapped most of the right-end 28 kb. This example indicates there is a high level of recombination among viruses that live in this extreme environment.
Our understanding of viral and prokaryotic evolution has been radically changed over the last 10 years as a result of the ability to sequence whole genomes of organisms, including a large number of viruses (3). One of the most striking discoveries is that viral genomes are often mosaics (22), consisting of many small genetic fragments from different sources yet making up a functional whole (17, 38). Despite this mosaicism, the arrangement and order of genes (synteny), particularly within functional modules such as DNA packaging and virus structural genes, are often well conserved (2, 4, 17, 18, 21).
About 204 prokaryotic viruses have been fully sequenced (National Center for Biotechnology Information [NCBI] database, December 2003), and many are related (for example, the dairy bacteriophages and the lambdoid phages). Comparison of related viruses is important as it can shed light on their evolution and structure-function relationships, but to adequately assess the breadth of virus diversity and the degree of genetic exchange between viruses, novel viruses from across the two prokaryotic domains need to be isolated and sequenced (13). While thousands of bacteriophages are known, it is unfortunate that only a small number of archaeal viruses have been isolated and few of these have had their genomes completely sequenced (20, 24, 33-35, 38). Most of these genome sequences are very different from each other (and from those of other viruses), but some are sufficiently close that comparisons can be made (e.g., viruses of Sulfolobus [36] and Methanobacterium [24]), and these have given some indication about how archaeal viruses have evolved.
The first haloarchaeal virus to be discovered, Hs1, was described in 1974 (41). Thirty years later, only about 15 haloviruses have been described, with most having head-tail morphologies and linear double-stranded DNA (dsDNA) genomes and being specific for Halobacterium salinarum (reviewed in references 12 and 48). Of these, the best studied at the molecular level include ΦH (see reference 37 and references therein), ΦCh1 (20), and HF2 (38). There have also been some elegant studies of halovirus ecology (7, 43, 44), the presence of high concentrations of virus-like particles in natural hypersaline waters (14, 31), and restriction systems (8).
Haloviruses HF1 and HF2 were isolated from the same pond at the same time, and although they were initially thought to be distinct, they have subsequently been found to be closely related (30). They have identical head-tail morphologies, contractile tails, and similar protein profiles by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and their genomes showed a cross-hybridization of at least 80%. However, they differed in their sensitivity to inactivation by chloroform and have significantly different and nonoverlapping host ranges (29, 30). Unlike other haloviruses, HF1 has a very broad host range, including genetically tractable species such as Halobacterium salinarum and Haloferax volcanii. The genomes of both of these species are either complete (28) or near completion (C. Daniels, personal communication), and the combination of a fully sequenced halovirus with experimentally convenient hosts would provide a valuable system for the study of archaeal viruses and their genes. Here, we report the complete HF1 DNA sequence, an analysis of its predicted open reading frames (ORFs), a reappraisal of the previously published restriction map, and a comparison between the HF1 and HF2 genomes. This represents the first comparison of two related haloviruses for which both genome sequences are complete.
MATERIALS AND METHODS
Halovirus HF1, haloarchaeal strains, and media.
Halovirus HF1 was isolated from a solar saltern in southern Australia (30). Haloterrigena trapanicum DS66, a clonal derivative of NCIMB 784 (42), and Haloferax lucentensis (wild-type strain, Phenon K, strain Aa2.2 [previously “Haloferax alicantei”]) (15, 39) were used for virus growth and plaque assay. Modified growth medium (18%, wt/vol) was used to culture these host strains, as described previously (30).
Halovirus HF1 DNA isolation.
A 50-μl sample of filtered (0.22-μm-pore-size filter) virus lysate (108 PFU/ml) was diluted 1/10 with water, and proteinase K and sodium dodecyl sulfate were added to final concentrations of 50 μg/ml and 0.1%, respectively. The sample was mixed, incubated at 37°C for 1 h, and then extracted once with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol) and a further one or two times with chloroform-isoamyl alcohol (24:1, vol/vol). After centrifugation to separate the phases, the aqueous layer was removed to a clean plastic tube. DNA was precipitated by adding sodium acetate (to 0.3 M) and ethanol (2 volumes) and incubating on ice for 15 min. The precipitate was collected by centrifugation (16,060 × g, 15 min, room temperature), washed with 70% ethanol (∼1 ml), dried under vacuum, and redissolved in 50 μl of pure water.
Sequencing of HF1 genomic DNA.
Short genomic fragments were amplified from HF1 DNA preparations (described above) by PCR (Hot-Star Taq kit; Qiagen, Hilden Germany) under high-stringency annealing conditions. Many of the primers originally designed for HF2 sequencing could be used to amplify and sequence HF1 DNA fragments. Where this was not possible, primer walking was performed with newly designed primers. The primer sequences are available upon request. The quality of each PCR fragment was first examined by agarose gel electrophoresis and, if it was satisfactory, sequencing reactions were performed with the purified DNA fragments as templates with specific primers. Automated dsDNA sequencing was carried out on an Applied Biosystems model 373A DNA sequencing system, using a PRISM terminator cycle sequencing kit, according to the manufacturer's instructions. The raw sequences were trimmed and assembled, and errors were corrected manually, using the program Sequencher version 3.01 (Gene Codes Corp.). More than 99% of the sequence was determined twice, independently. Only a few bases with clear-cut sequence chromatography were determined once.
Bioinformatics analysis.
Sequence similarity comparisons (BLASTN, BLASTP, and BLASTX) were carried out against the NCBI nonredundant protein and nucleotide databases during December 2003 (http://www.ncbi.nlm.nih.gov/). Searching of the Clusters of Orthologous Groups databases of protein sequence families was performed with COGnitor, which is available at the NCBI website (http://www.ncbi.nlm.nih.gov/COG/). Analysis, ORF determination, and annotation of the HF1 genome sequence were with the software Sequin (NCBI website) and Glimmer2 (http://www.tigr.org/software/) (9). ORFs with fewer than 50 codons were included if they (i) overlapped with or were closely adjacent to flanking ORFs, (ii) showed a pI of less than 7, or (iii) showed predicted amino acid similarity to an HF2 or GenBank database sequence. HF1 ORFs were numbered from left to right, using the same genome orientation as described for HF2 (38).
Global and dot-plot alignments between HF1 ORFs and HF2 ORFs were performed with Lasergene software (DNAstar Inc.). The methods used included the Wilbur-Lipman method, the Martinez-NW method (26), and the Lipman-Pearson method (46). For tRNA searching, tRNAscan (23) (http://www.genetics.wustl.edu/eddy/tRNAscan-SE/) was used to scan sequences for tRNA sequences in the viral genome, and the Mfold program (27) (http://bioinfo.math.rpi.edu/∼mfold/rna) was used to predict tRNA folding.
Protein secondary structure predictions, protein molecular weight determinations, amino acid composition determinations, and similar analyses were performed with DNA Strider (25), Lasergene, GeneticLab, or proteomic programs available at the ExPASy mirror site (http://au.expasy.org) of the Swiss Institute of Bioinformatics. Protein domain searches used the InterProScan program available at the ExPASy site.
Electron microscopy.
Purified virus preparations were applied to Formvar-coated copper grids and allowed to adsorb for 15 min, and then the grids were blotted dry and treated with 0.1% glutaraldehyde for 5 min at room temperature. Negative staining was with 2% (wt/vol) uranyl acetate. Grids were examined on a Hitachi H300 electron microscope at machine magnifications of ×20,300. Catalase crystals, negatively stained with 2% uranyl acetate, were used as an internal calibration standard (47).
Nucleotide sequence accession number.
The complete HF1 genome sequence has been deposited in the GenBank database (accession number AY190604).
RESULTS
Sequence of the HF1 genome and overview of its analysis.
The HF1 genome was fully sequenced by using custom primers to directly sequence PCR-amplified fragments of the genome. It was 75,898 bp long, which is about 1.8 kb shorter than the genome of HF2 (77,670 bp) (38). The overall G+C content was 55.8 mol%, the same as for HF2, and putative ORFs and noncoding regions had average values of 56.7% and 55.3%, respectively. ORFs were predicted by using the same criteria as used previously for HF2 (38). HF1 contained 117 ORFs, mostly closely spaced or overlapping by a few nucleotides. As the first 48 kb of HF1 was found to be nearly identical to the previously described HF2 genome sequence (38) (see also below), only the ORFs in the remaining 28 kb (ORFs 83 to 117) are presented in Table 1. Haloarchaeal proteins are usually highly acidic, and the average pI value of the proteins predicted from all HF1 ORFs was less than 5. Sequence similarity searches (BLASTP) using the predicted protein sequences against the GenBank database showed almost the same information as found previously for HF2; i.e., 12 ORFs had clear database homologues (e.g., DNA polymerase, methyl transferase, ribonucleotide reductase, terminase, etc.) BLASTN and tRNA-SE scans of the nucleotide sequence revealed the same five tRNA-like sequences as found in HF2, i.e., Arg-tRNA (UCU), Asx-tRNA (GUU), Tyr-tRNA (GUA), Pro-tRNA (UGG), and Thr-tRNA (GGU, truncated) (Table 1). Long repeat sequences found throughout the HF2 genome were also present in HF1. The HF1 ORFs that differed completely from those of HF2 showed no matches to other organisms in the sequence databases. In total, 87% of the predicted HF1 ORFs have no homologues of known function.
TABLE 1.
Comparison of HF1 ORFs 83 to 117 with those of HF2
| HF1 ORF | Right end/left end (bp)a | kDa (aa) | pI | Related ORFs in HF2
|
Similarity and commentsb | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ORF | Right end/ left end (bp)a | kDa (aa) | pI | % aa similarityc | % DNA similarityc | |||||
| 83 | 48119/49003 | 33.3 (294) | 4.5 | 83 | 48119/49003 | 33.3 (294) | 4.44 | 75.5 | 78.0 | Similar (E = 10−40) to hypothetical proteins from Sinorhizobium (AAK64736) and Mesorhizobium (NP_108298); 55 sc, 40 mc, 28 snc |
| 84 | 49008/49502c | 18.8 (164) | 4.7 | 84 | 49008/49502c | 18.8 (164) | 4.76 | 98.2 | 95.4 | 16 sc, 2 mc, 1, snc |
| 85 | 49524/49910c | 14.0 (128) | 3.9 | 85 | 49523/49933c | 14.5 (136) | 3.62 | 47.5 | 58.4 | 23 sc, 47 mc, 17 snc, 8 indel |
| 86 | 49927/50265 | 11.3 (112) | 4.45 | 0.0 | 0.0 | Deletion-insertion | ||||
| 86 | 49913/50950c | 33.1 (345) | 4.0 | 87 | 49931/50861c | 29.0 (309) | 4.49 | 31.6 | 29.4 | 3 nt to ORF 85; 68 sc, 190 mc, 52 indel |
| 87 | 50971/52380c | 52.9 (468) | 4.4 | 88 | 50976/52673c | 61.8 (565) | 4.06 | 52.1 | 49.6 | First 70 codons almost identical, after which there are 78 sc, 85 snc, 137 mc, and 128 indel, including a long 97-codon indel |
| 88 | 52384/53034c | 24.3 (216) | 4.1 | 89 | 52677/53327c | 24.2 (216) | 4.05 | 96.3 | 96.6 | 7 sc, 5 snc, 3 mc |
| 89 | 53039/54274c | 43.7 (411) | 4.1 | 90 | 53332/54567c | 43.7 (411) | 4.19 | 95.6 | 93.5 | COG3299 family of conserved phage proteins; 45 sc, 8 snc, 10 mc |
| 90 | 54271/54597c | 12.2 (108) | 4.2 | 91 | 54564/54890c | 12.2 (108) | 4.43 | 95.4 | 95.7 | Overlaps ORF 89; 5 sc, 2 snc, 3 mc |
| 91 | 54662/55027c | 12.3 (121) | 4.4 | 92 | 54955/55209c | 8.7 (84) | 4.16 | 86.9 | 88.2 | 11 sc, 4 snc, 7 mc, 37 indel |
| — | 93 | 55314/56249c | 30.1 (311) | 4.10 | 0.0 | 0.0 | Deletion-insertion | |||
| 92 | 55024/55461c | 15.4 (145) | 4.2 | 94 | 56252/57232c | 34.3 (326) | 3.90 | 67.1 | 43.5 | Very similar N-terminal sequences, then codon 157 of gene 92 (HF1) has changed to a nonsense codon; within the 156 codons, 21 sc, 10 snc, 36 mc, 1 indel, and 89 same; overlaps ORF 91. |
| 93 | 55463/56905c | 55.1 (480) | 4.6 | 95 | 57234/58676c | 55.0 (480) | 4.63 | 99.2 | 98.3 | 2 nt to ORF 92; 16 sc, 3 snc, 1 mc |
| 94 | 56902/57435c | 19.9 (177) | 4.3 | 96 | 58673/59206c | 19.9 (177) | 4.26 | 100 | 99.8 | Overlaps ORF 93; 1 sc |
| 95 | 57436/59253c | 63.6 (605) | 4.4 | 97 | 59207/61024c | 63.5 (605) | 4.41 | 99.8 | 99.4 | 1 nt to ORF 94; 8 sc, 1 mc |
| 96 | 59256/59492c | 8.6 (78) | 5.3 | 98 | 61027/61263c | 8.6 (78) | 5.35 | 98.7 | 99.6 | 3 nt to ORF 95; 1 mc |
| 97 | 59569/60039c | 17.6 (156) | 4.0 | 99 | 61340/61618c | 10.2 (92) | 3.95 | 98.8 | 98.2 | 3 sc, 1 snc, 64-codon indel (at the beginning of the ORF) |
| 98 | 60092/60634c | 19.4 (180) | 3.8 | 100 | 61863/62405c | 19.4 (180) | 3.81 | 97.2 | 96.1 | 8 sc, 2 snc, 3 mc |
| 99 | 60641/61954c | 46.3 (437) | 4.3 | 101 | 62434/63726c | 44.9 (430) | 4.07 | 89.2 | 89.8 | 47 sc, 14 snc, 32 mc, 7 indel (between codons 100 to 250 and the last 20) |
| 100 | 61963/62472c | 18.8 (169) | 4.3 | 102 | 63735/64244c | 18.8 (169) | 4.33 | 100 | 99.8 | 2 sc |
| 101 | 62509/62730c | 8.3 (73) | 6.1 | 103 | 64281/64502c | 8.3 (73) | 6.60 | 98.6 | 99.5 | 1 mc |
| 102 | 62732/65845c | 116.5 (1,037) | 4.1 | 104 | 64504/67617c | 116.5 (1,037) | 4.12 | 98.7 | 98.9 | 2 nt to ORF 101; 17 sc, 6 snc, 7 mc |
| 103 | 65848/66627c | 29.4 (259) | 5.1 | 105 | 67620/67958c | 12.9 (112) | 6.76 | 99.1 | 99.7 | ORF 103 appears to be a fusion of HF2 ORFs 105 (1 snc) and 106 (3 sc, 2 snc, 6 mc, 1 indel), frameshift; 3 nt to ORF 101 |
| 106 | 67958/68401c | 16.3 (147) | 4.49 | 98.6 | 94.2 | |||||
| 104 | 66671/67627c | 36.3 (318) | 4.5 | 107 | 68445/69062c | 23.1 (205) | 4.67 | 98.5 | 99.3 | ORF 104 (HF1) appears to be a fusion of HF2 ORF 107 (1 sc, 1 snc, 12 mc) and 108 (3 sc, 2 snc, 3 mc); ORF 108 (HF2) starts at codon 104 of ORF 104 (HF1) |
| 108 | 69064/69399c | 12.8 (111) | 4.43 | 91.3 | 97.2 | |||||
| 105 | 67669/68184c | 18.5 (171) | 3.8 | 109 | 69441/69956c | 18.4 (171) | 3.85 | 99.4 | 99.6 | 1 sc, 1 mc |
| 106 | 68208/68639c | 16.2 (143) | 3.5 | 110 | 69980/70411c | 16.2 (143) | 3.46 | 100.0 | 100.0 | Highly acidic central region. |
| 107 | 68646/69410c | 27.9 (254) | 4.0 | 111 | 70418/71182c | 27.9 (254) | 4.00 | 99.2 | 99.7 | 1 sc, 1 snc |
| 108 | 69407/69691c | 10.3 (94) | 6.6 | 112 | 71179/71463c | 10.3 (94) | 6.62 | 98.9 | 98.9 | Overlaps ORF 107; 1 sc, 1 snc |
| 109 | 69694/71133c | 53.1 (479) | 4.4 | 113 | 71466/72905c | 53.1 (479) | 4.37 | 99.8 | 99.1 | 3 nt from ORF 108; 12 sc, 1 snc |
| 110 (ter) | 71148/72839c | 63.6 (563) | 5.2 | 114 | 72920/74611c | 63.5 (563) | 5.23 | 100.0 | 99.6 | Terminase (large subunit); 17 nt to next RH gene (tRNA), 5 sc |
| 111 | 72857/72927 | NAd | NA | 115 | 74629/74699 | NA | NA | 100.0 | 100.0 | Pro-tRNA |
| 112 | 72932/73128c | 7.7 (68) | 4.1 | 116 | 74704/74910c | 7.7 (68) | 4.11 | 100.0 | 100.0 | 4 nt to PRO-tRNA |
| 113 | 73174/73368c | 6.9 (64) | 3.4 | 117 | 74946/75140c | 6.9 (64) | 3.44 | 100.0 | 100.0 | |
| 114 | 73372/73479c | 4.1 (35) | 3.7 | 118 | 75144/75251c | 4.1 (35) | 3.74 | 100.0 | 100.0 | |
| 115 | 73590/73895c | 11.4 (101) | 5.7 | 119 | 75362/75668c | 11.4 (101) | 5.72 | 100.0 | 100.0 | |
| 116 | 73846/74622c | 28.9 (258) | 4.3 | 120 | 75618/76394c | 28.9 (258) | 4.27 | 100.0 | 99.9 | Overlaps ORF 115; 1 sc |
| 117 | 75031/75501 | 17.5 (156) | 5.6 | 121 | 76803/77273 | 17.5 (156) | 5.60 | 100 | 100 | |
c, complementary or lower DNA strand (according to orientation in Fig. 1).
RH, right hand; sc, silent nucleotide change; mc, missense change; snc, synonymous change (codon change specifies a different but functionally equivalent amino acid, e.g., Lys → Arg) noc, nonsense change; indel, insertion-deletion.
Similarity formulas used in the alignment were Wilbur-Lipman for DNA (ktuple, 3; gap penalty, 3; window, 20) and Lipman-Pearson for protein (ktuple, 2; gap penalty, 4; gap length, 12). 0.0, zero similarity shown for predicted proteins of HF2 that have no HF1 homologue.
NA, not applicable.
Comparison of HF1 and HF2 genomes by using dot-plot and global alignment analyses.
The genomes of HF1 and HF2 were compared by using DNA and predicted-protein alignment methods, and a summary of the more significant similarity values is given in Table 2. While the HF1 sequence was 94.4% identical to that of HF2, the differences between them were not uniformly spread. The first 48 kb were identical except for a single (silent) base change (Gly [GGG] to Gly [GGT] at nucleotide [nt 23318), whereas the last third was only 87% identical and contained many base changes, insertions, and deletions. An ORF diagram of HF1 and HF2, from kb 41 to their right ends, is shown in Fig. 1, along with a plot of predicted protein similarity. Two regions of particularly low similarity (<86% nucleotide identity), termed major different regions (MDRs), stand out starkly as peaks in the inverse similarity plot in Fig. 1. The MDRs contain significant insertions and deletions as well as many base changes. The genomes become identical again (except for one nucleotide change) over the last 3 kb, i.e., after the Pro-tRNA gene (trn5) (Table 1). By inference from the HF2 transcription data (38) and the database matches shown by ORFs, the region from kb 48 to the right terminus is part of the late gene region, a region that contains all of the significant sequence variation between HF1 and HF2.
TABLE 2.
Alignment statistics for HF1 and HF2 DNAs and predicted proteins
| Alignment of HF1-HF2a | % Similarity |
|---|---|
| DNA alignment | |
| Of entire genomes | 94.4 |
| Before kb 48 | 99.99 |
| After kb 48 | 85.0 |
| After kb 48 within ORFs | 87.0 |
| From kb 48 to 71.2 | 80.7 |
| Protein alignment | |
| Before kb 48 | 100 |
| After kb 48 | 87.5 |
DNA alignment was by the Martinez-Needleman-Wunsch method (minimum match, 9; gap penalty, 1.10, gap length penalty, 0.33). Protein alignment was by the nonexhaustive global Lipman-Pearson method (ktuple, 2; gap penalty, 4; gap length penalty, 12).
FIG. 1.
ORF alignment and similarity plot from kb 41 to the right ends of the HF1 and HF2 genomes. The upper section shows the ORFs of each virus as shaded bars (with ORF numbers underneath), drawn to scale, and a graduated scale bar (in kilobases) between them. The intensity of shading of ORF bars indicates the level of similarity between corresponding predicted proteins, from white (0% similarity) to black (100% similarity). Regions of high similarity are indicated by shading between corresponding regions of the two genomes. The lower section shows a plot of predicted protein similarity for each ORF pair (y axis) plotted along the length of the HF2 ORF map. The y-axis scale is drawn so that similarity decreases in the upward direction, with peaks in the plot corresponding to unique HF2 ORFs (0% similarity). Numbers along the x axis show the corresponding ORF numbers (with HF1 above HF2).
Considering only the alignment after kb 48, the DNA sequences of HF1 and HF2 within predicted ORFs had a significantly higher similarity (2% higher) than the entire DNA sequences in this region, indicating a higher conservation within coding regions (Table 2). The alignments of predicted protein sequences showed a slightly higher similarity (about 0.5%) than the DNA sequences of the same ORFs, indicating that there were a number of silent (or synonymous) base changes. On a closer inspection, these average values were found to result from a widely uneven spread of differences across this late gene region.
Detailed genomic comparison between HF1 and HF2 ORFs after kb 48.
The significant differences between HF1 and HF2 occur between kb 48 and 73 (of HF1) and are detailed in Table 1. The predicted ORFs in this region vary in similarity from 30 to 100% (at both the nucleotide and amino acid sequence levels). The major differences were tightly clustered within the two MDRs, while ORFs adjacent to MDRs were highly conserved (Fig. 1). Only ORFs that differ significantly between the two viruses are described below.
(i) Unique ORFs.
Homologues of HF2 ORFs 86 and 93 were not present in HF1. Both ORFs had no database matches, were relatively long (112 and 311 codons), and had predicted pIs of less that 4.5. No transcript corresponding to HF2 ORF 86 was detected by Northern blot hybridization (38), and the transcriptional direction of this ORF would be rightward, unlike that of most other late genes (Table 1). It is embedded in a larger ORF (HF2 ORF 87) on the complementary strand, and this larger ORF differs significantly in its nucleotide and predicted protein sequences from its HF1 homologue, ORF 86 (see below). The second unique ORF, HF2 ORF 93, was shown to be transcribed (38), and it is oriented in the same direction as other late genes.
(ii) HF1 ORFs with less than 70% amino acid identity to their HF2 homologues.
The HF1 ORFs with less than 70% amino acid identity to their HF2 homologues included HF1 ORFs 85, 86, 87, and 92. They are all clustered within the two MDRs. Although these ORFs share significant protein sequence similarity (31.6 to 67%) to their corresponding HF2 ORFs and were certainly homologues, the number, even distribution, and range of mutations (Table 1) indicate that they have undergone extensive evolutionary change.
HF1 ORF 86 (33.1 kDa) showed only 29.4% amino acid similarity to its HF2 homologue, ORF 87 (29 kDa). There were 69 synonymous and 190 nonsynonymous nucleotide changes, as well as several insertions and deletions. Changes occurred evenly throughout the two sequences. The predicted proteins of these ORFs are rich in glycine residues, which occur in strings of four or five. They also differ significantly in size.
HF1 ORF 87 (52.9 kDa) is a large protein and corresponds to HF2 ORF 88 (61.8 kDa). They show 49.6% amino acid identity and also differ in size, but the first 70 amino acids (aa) are almost identical. After this, they rapidly diverge. The size difference is largely due to the HF1 ORF terminating earlier than the corresponding HF2 ORF. In addition, the nucleotide sequence shows numerous small repeat sequences within the HF1 ORF 87 that have a similar core but vary in length, i.e., CCX(A or G)CC, CCX(A or G)CCX(A or G)CC, or CCX(A or G)CCX(A or G)CCX(A or G)CC. The repetitions are in various reading frames, and the predicted protein does not contain highly repeated amino acids.
HF1 ORF 91 is larger than its corresponding HF2 ORF 92 because the latter ORF does not have the first 37 aa. The remaining sequence is almost the same as that of HF2 ORF 92. The nucleotide sequence upstream of HF2 ORF 92 was about 87% similar to HF1 over the region of the missing 37 aa. A close scrutiny of the nucleotide sequence showed that there was a frameshift (relative to the HF1 ORF) caused by an insertion of a few bases (at nt 55231 in HF2), producing a nonsense codon (TGA, nt 55246 to 55248) in the upstream sequence of HF2 ORF 92. No transcripts in this region were detected in HF2 (38).
HF1 ORF 92 (15.4 kDa) corresponds to part of HF2 ORF94 (34.3 kDa). A nonsense mutation terminates HF1 ORF 92 at about half the length of the HF2 ORF. However, a comparison of the surrounding nucleotide sequences indicates that the termination of HF1 ORF 92 was not due to a base change leading to a nonsense mutation but was due to a deletion (relative to HF2) that removed all of the rest of HF1 ORF 92 coding sequence (Table 1).
HF1 ORF 85 (14.0 kDa) corresponded to HF2 ORF 85 (14.5 kDa). They are similar in size but have an amino acid sequence similarity of only 47.5%, with the C-terminal half displaying a higher similarity than the N-terminal half. HF1 ORF 85 terminates eight codons earlier than HF2 ORF 85.
(iii) HF1 ORFs that are split in HF2.
Two ORFs found in HF1 have corresponding HF2 ORFs that are split into two smaller ORFs (Table 1). The sequencing data for these regions of both viruses have been carefully checked. In the first case, one half of HF1 ORF 103 is homologous to HF2 ORF106 (98.6% amino acid identity), and the other half is homologous to the adjacent HF2 ORF 105 (99.1% amino acid identity). The other case is HF1 ORF 104, in which the first third is similar to HF2 ORF 108 (91.1%) and the remainder corresponds to HF2 ORF 107 (98.5%). In both cases, the two smaller ORFs in HF2 (corresponding to the larger HF1 ORF) are within 2 nt of each other and are likely to be transcribed together. The region of the HF2 genome corresponding to these HF1 ORFs was shown to be transcribed (38).
(iv) Predicted and observed restriction fragments of the HF1 genome.
Restriction digest patterns of HF1 DNA have been published previously (30), and these were compared to restriction fragments predicted from the sequence, assuming a linear genome like that of HF2. Certain restriction digests clearly contained one additional fragment not predicted by the sequence (Fig. 2), and the terminal fragments were underrepresented. For example, DraI digestion produced all of the predicted fragments as well as a 6.2-kb band that was not predicted, and a HindIII digest was predicted to give 11 fragments but was observed to give 12, with the extra fragment being about 14 kb. The extra bands were discrete (not smeared), and their ethidium bromide staining intensities indicated a relative molarity that was about half of that of the other fragments in each digest (Fig. 2), excluding the terminal fragments. To check whether the stock of virus used in the present study had changed from that used earlier, we obtained a stock of HF1 that had been stored at 4°C since 1993 and sequenced the regions encompassing all 10 predicted HindIII sites. No changes were observed.
FIG. 2.
Agarose gel electrophoresis of restriction digests of HF1 DNA. Lane 1, DNA size standard (HindIII-cut lambda DNA), with fragment sizes indicated at the left. Lane 2, undigested HF1 DNA. Lanes 3 to 6, HF1 DNA cut with DraI, AseI, HpaI, and HindIII, respectively. Arrows point to extra fragments not predicted by the linear genome sequence. R, right terminal restriction fragment; L, left terminal restriction fragment. Some fragments in the lower part of the gel are indicated by white dots. These fragments were also expected from the linear sequence but were faint on the gel, as the ethidium staining intensity decreases with fragment size. Not all terminal fragments were large enough to show on this gel.
In light of the genome sequence data, we reappraised the HF1 restriction digestion data and found that the sizes of the additional fragments that were seen in the restriction digests of HF1 DNA, and that were unexpected if the genome was linear, could be explained if a proportion of genomic molecules in the preparation were joined head to tail, as either circles or concatemers. For example, the linear sequence predicts terminal DraI fragments of 4.8 and 1.7 kb, and the length of the unexpected fragment observed in DraI digests was about 6.4 kb, which is very close to the sum of the lengths of the terminal fragments (observed and predicted). The relative reduction in staining intensity of the two terminal fragments in these digests would fit with them being incorporated into joined fragments. Restriction digests of HF2 DNA gave only fragments of the sizes expected for a linear genome, and no additional fragments were ever observed.
In sequencing the HF1 termini, terminal restriction fragments were first treated with T4 DNA polymerase (to blunt end any overhangs) before cloning, a process that can remove 3′ overhangs and fill in 5′ overhangs. Primer walking near the termini of HF1 DNA (using DNA polymerase) was also used, but this can check the length of only the 5′ end and not that of the 3′ end. We believe that the genome sequence of HF1 is complete, since (i) the 5′ ends of (free) terminal repeats (TRs) have been sequenced, (ii) the sequence across TR borders (with internal genomic sequence) has been determined, and (iii) concatemer junctions derived from HF1-infected cells have been PCR amplified and sequenced (unpublished data). All give the same sequence, which is identical to that of the TR of HF2. However, our observations would be consistent with the presence of single-stranded cohesive termini, possibly 5′ overhangs.
(v) Head size of virus particles.
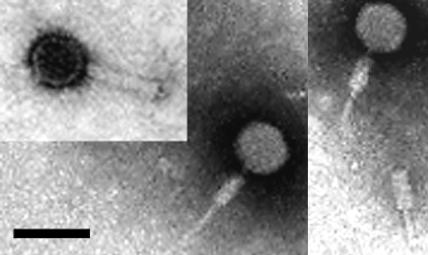
Our original description of HF1 and HF2 (30) reported head diameters of 58 nm, but the genomes of these viruses are unusually large for isometric heads of this size and would give DNA densities far higher than those of structurally well-studied bacteriophages such as T7 and lambda (e.g., 0.45 g/cm3 for T7 [5]). The head size of HF1 was reexamined. Preparations of HF1 were studied by negative-stain electron microscopy (Fig. 3), and particle dimensions were measured from digitized photographic negatives. Catalase crystals were used as an internal calibration standard (47) (major lattice spacing of 8.75 nm). The head diameter was found to be 67.8 ± 3 nm, which is significantly larger than previously determined, and would give an estimated DNA density (0.51 g/cm3) close to those of bacteriophages such as lambda, Mu, and T7 and to that of halovirus ΦH. The tail length of HF1 (excluding the connector) was found to be 90 ± 2 nm.
FIG. 3.
Electron micrographs of purified HF1 virus, stained with 2% uranyl acetate. Bar, 100 nm. This is a composite picture of three different particles, one with an uncontracted tail and two with contracted tails. A contracted tail without a head is also visible near the lower right. Magnification, ×20,300.
DISCUSSION
HF1 and HF2 show evidence of a recent recombination.
While a high level of sequence similarity between HF1 and HF2 was anticipated from previous cross-hybridization data (30), a comparison of their genome sequences has now revealed that the distribution of sequence conservation between them is extraordinarily uneven. Their nearly identical sequences over the first 48 kb would most simply be explained by a very recent divergence; they have not had time to show an appreciable number of neutral or selected changes. Even severe selection pressure would not be expected to preserve near perfect sequence identity over 48,000 bases while at the same time the right end has diverged so much. For example, the rate of spontaneous mutation in a diverse range of dsDNA coliphages, including T4, is about 100 times higher than that in their host (11). In addition, genomic data from relatives of the same phage species (e.g., dairy phages [2], T4-like phages, the lambda supergroup [18], etc.) invariably show considerable change over most of their genomes.
Assuming a recent, common origin of the left ends of HF1 and HF2, then the extensive differences seen in the region from 48 kb to the right end speaks of a much longer history of evolutionary change. The most likely scenario for this lopsided pattern of divergence is that a recent recombination event has swapped most or all of the right end of one of the two viruses for that of a third, HF-like virus. More than one recombinatorial cross (within the right end) may have occurred before they were isolated in the laboratory, but the near identity of the left 48 kb favors a very recent divergence. Recombination events between viruses are common, both in nature and in the laboratory, and genetic crosses between lytic coliphages such as T4 were an important genetic tool in early molecular biology. Recent comparative sequence studies of natural T4-like viruses show high levels of recombination, sometimes of large genomic segments (10), although a natural recombination event of the same apparent magnitude as observed in this study is difficult to find in the literature. While homologous recombination in haloarchaea is well documented (6), experimental crosses between archaeal viruses have not yet been demonstrated.
In a recent study very relevant to this one, Pajunen et al. (32) proposed that phage T3 was probably the result of a natural recombinatorial cross between a T7-like phage and a yersiniophage. Like HF1 and HF2, these viruses have different hosts (and their genomes have direct TRs). The authors showed that such crosses were possible by crossing T7 and φYeO3-12, after first constructing an Escherichia coli strain that expressed Yersinia O antigens in order to infect it with both viruses simultaneously. Recombinants were mainly T7 derived and were all the result of double-crossover events (so maintaining the T7 TRs).
The two MDRs found in HF1 and HF2 appear to be recombinational hot spots that have undergone considerable evolutionary change. The high conservation of genes surrounding the MDRs may reflect functional constraints; i.e., these genes cannot easily be replaced by genes with equivalent function but different sequence. This would be consistent with the nature of the late region, as it probably encodes all of the structural proteins and these must assemble into multimeric components that fit together precisely and function as a whole virus particle. One structural protein that is, by contrast, highly variable in sequence is the tail fiber, which carries the cell adhesin domain enabling the virus to bind to specific host cells (16). HF1 and HF2 have different host ranges and show small differences in their major structural proteins (30) but the identity of the tail fiber adhesin has not been experimentally determined.
The only other pair of haloarchaeal viruses that have substantial similarity and for which sequence data are available are the temperate viruses ΦH and ΦCh1 (20). The ΦCh1 genome is complete, but only 60% of the ΦH genome has been determined. They share 50 to 95% nucleotide sequence identity over the regions for which both sequences are available, and their gene arrangements are very similar. Klein and colleagues (20) detected one inversion and a likely gene deletion. In summary, the two viruses have diverged substantially, yet they share extensive homology over much of their genomes. In addition, a 1982 study comparing the restriction fragments of ΦH and halovirus Hs1 DNAs indicated they share significant sequence similarity (40), indicating that this virus group may be common.
HF1 genome termini.
Previous studies on halovirus HF2 showed conclusively that its dsDNA genome was packaged into virions as linear molecules with blunt-ended termini (29). These termini occurred at the left or right end of the 306-nt direct TR found at each end of the linear genome. From the type of termini, their presence in single copy between genomes in concatemeric DNA in infected cells, and sequence characteristics in and around the TR, it was speculated that the replication and packaging strategy may resemble that of the T7 group of bacteriophages (19, 29, 32). However, in at least a portion of HF1 DNA, the termini were bound to each other even though the TR and flanking sequences are identical to those of HF2. How the termini bind to each other has not been determined, but the join must be head to tail only, as restriction fragments arising from head-head or tail-tail fusions were not seen.
Although HF1 DNA may readily be able to circularize once injected into a host cell, the evidence we have so far indicates that HF1 is strictly lytic. No prophage state has been found, despite repeated attempts in this laboratory. Instead, both HF1 and HF2 can form unstable carrier states, with low levels of continuous virus production, but cells are not immune from superinfection and these laboratory cultures eventually collapse with complete lysis (unpublished data). HF1 and HF2 show a notable difference in the state of their DNA in infected cells. For HF1-infected cultures, Southern blot studies showed that TRs were not detected on free ends but were detected only as fragments where the TR was joined (at both ends) to viral genomic DNA (unpublished data), suggesting that the viral DNA is largely circular (or perhaps in very long concatemers). However, in HF2-infected cells, TRs at the ends of the linear viral genome (or concatemers) are readily detected, in addition to joined (concatemeric or circular) TRs (see Fig. 6 of reference 29). Concatemers have been directly observed in HF2-infected cells (29). Together, these results point to some difference in the processing of the termini of HF1 and HF2, a difference that could be either virus or cell specific.
Late gene organization and host range.
The late regions of bacteriophages include genes for structural proteins, packaging DNA, and cell lysis. Commonly, the genes are closely spaced, sometimes overlapping, and are transcribed from the same strand. The gene order (synteny) is remarkably conserved across a wide variety of examples (2), so that once the location of one or more highly conserved genes within a late gene region of a newly sequenced genome is discovered, the functions of neighboring ORFs can be tentatively predicted. In the case of HF1, two highly conserved genes in the late region include the terminase (ORF 110) and COG3299 (ORF 89; Mu gp47-like family), and these are usually situated at the beginning (terminase) and about two-thirds (COG3299) of the length of the late region. The positions and orientations of these two “landmark” ORFs, and the close spacing of the ORFs between them, provide a tentative framework. For example, the portal protein ORF is usually large, and just downstream (in the transcriptional sense) of the terminase ORF, and HF1 ORF 109 is of an appropriate size and position for a portal protein. Major tail and head protein genes are usually next (and some of these structural proteins are known to vary between HF1 and HF2). A tail fiber gene(s) is usually not far downstream of the COG3299 gene, and the tape measure gene is usually just upstream of it.
The distinct host ranges of HF1 and HF2 probably reflect different cell adhesins carried on virus particles. By analogy to bacteriophages with head-tail morphology and contractile tails, their adhesins are likely to be tail fiber proteins. Well-preserved HF1 and HF2 particles show a compact baseplate and small (approximately 20- to 25-nm) filamentous structures extending from the edges of the baseplate (30). If these are the tail fibers, then calculations based on a straight fiber structure that is largely alpha-helical would indicate a protein size of at least 150 aa. The tail tape measure protein forms a helical fiber that determines the tail length of tailed phages. If an alpha-helical protein spanned the 90 nm, it would be at least 600 aa long (assuming 3.6 aa per turn and a pitch of 0.54 nm). There are only two proteins in the late region with sizes of 600 aa or more; one is 605 aa (ORF 95) and the other is 1,037 aa (ORF 102). Future studies to map all of the virus structural proteins to their corresponding genes will require the isolation and analysis of individual proteins from purified virus particles.
Relationship of HF1 to other haloviruses.
HF1 and HF2 are clearly related to each other to the extent that they should be classified as members of the same virus group. At a morphological level they are remarkably similar to head-tail bacteriophages, and this is supported by the general features of their genomes, including the makeups and organizations of their genes. However, they share only a very distant relationship to other sequenced viruses and should be classified as a novel genus within the Myoviridae. Among other haloviruses being studied in this laboratory, they represent one of at least three broad morphological groups, the others being spindle-shaped haloviruses, such as His1 (1), and round viruses with an internal lipid layer, such as SH1 (12). The known diversity of haloviruses can be expected to increase with improvements in the ability to culture a wider variety of haloarchaea, particularly those species that are dominant members of their natural environment but have not yet been cultured, e.g., members of the SHOW group (square haloarchaea of Walsby) (12, 45).
Acknowledgments
S.-L. Tang was the recipient of a University of Melbourne International Postgraduate scholarship and an Australian Government International Postgraduate Research scholarship. This research was funded by a grant from the Australian Research Council.
We thank Helen Camakaris for critical reading of the manuscript and C. Bath for technical assistance. Students of the 2001 environmental project practical class assisted with analyzing the early-passage HF1 culture.
REFERENCES
- 1.Bath, C., and M. L. Dyall-Smith. 1998. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J. Virol. 72:9392-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brussow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 3.Brussow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 4.Casjens, S., G. Hatfull, and R. Hendrix. 1992. Evolution of dsDNA tailed-bacteriophage genomes. Semin. Virol. 3:383-397. [Google Scholar]
- 5.Cerritelli, M. E., N. Cheng, A. H. Rosenberg, C. E. McPherson, F. P. Booy, and A. C. Steven. 1997. Encapsidated conformation of bacteriophage T7 DNA. Cell 91:271-280. [DOI] [PubMed] [Google Scholar]
- 6.Cline, S. W., L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J. Bacteriol. 171:4987-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniels, L. L., and A. C. Wais. 1990. Ecophysiology of bacteriophage S5100 infecting Halobacterium cutirubrum. Appl. Environ. Microbiol. 56:3605-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels, L. L., and A. C. Wais. 1984. Restriction and modification of halophage S45 in Halobacterium. Curr. Microbiol. 10:133-136. [Google Scholar]
- 9.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with Glimmer. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desplats, C., and H. M. Krisch. 2003. The diversity and evolution of the T4-type bacteriophages. Res. Microbiol. 154:259-267. [DOI] [PubMed] [Google Scholar]
- 11.Drake, J. W. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyall-Smith, M., S. L. Tang, and C. Bath. 2003. Haloarchaeal viruses: how diverse are they? Res. Microbiol. 154:309-313. [DOI] [PubMed] [Google Scholar]
- 13.Forterre, P. 1999. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 33:457-465. [DOI] [PubMed] [Google Scholar]
- 14.Guixa-Boixereu, N., J. I. Calderon-Paz, M. Heldal, G. Bratbak, and C. Pedros-Alio. 1996. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat. Microb. Ecol. 11:215-227. [Google Scholar]
- 15.Gutierrez, M. C., M. Kamekura, M. L. Holmes, M. L. Dyall-Smith, and A. Ventosa. 2002. Taxonomic characterization of Haloferax sp. (“H. alicantei”) strain Aa 2.2: description of Haloferax lucentensis sp. nov. Extremophiles 6:479-483. [DOI] [PubMed] [Google Scholar]
- 16.Haggard-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 18.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J. S., S. H. Kim, and Y. B. Chung. 1997. Defects in concatemer processing of bacteriophage T7 DNA deleted in the M-hairpin region. Virology 236:37-46. [DOI] [PubMed] [Google Scholar]
- 20.Klein, R., U. Baranyi, N. Rossler, B. Greineder, H. Scholz, and A. Witte. 2002. Natrialba magadii virus ΦCh1: first complete nucleotide sequence and functional organization of a virus infecting a haloalkaliphilic archaeon. Mol. Microbiol. 45:851-863. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V., L. Aravind, and A. S. Kondrashov. 2000. The impact of comparative genomics on our understanding of evolution. Cell 101:573-576. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, J. G. 1997. Selfish operons and speciation by gene transfer. Trends Microbiol. 5:355-359. [DOI] [PubMed] [Google Scholar]
- 23.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo, Y., P. Pfister, T. Leisinger, and A. Wasserfallen. 2001. The genome of archaeal prophage ΨM100 encodes the lytic enzyme responsible for autolysis of Methanothermobacter wolfeii. J. Bacteriol. 183:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marck, C. 1988. ′DNA Strider': a ′C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 16:1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez, H. M. 1983. An efficient method for finding repeats in molecular sequences. Nucleic Acids Res. 11:4629-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 28.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, S. DasSarma, et al. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nuttall, S. D., and M. L. Dyall-Smith. 1995. Halophage HF2: genome organization and replication strategy. J. Virol. 69:2322-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuttall, S. D., and M. L. Dyall-Smith. 1993. HF1 and HF2: novel bacteriophages of halophilic archaea. Virology 197:678-684. [DOI] [PubMed] [Google Scholar]
- 31.Oren, A., G. Bratbak, and M. Heldal. 1997. Occurrence of virus-like particles in the Dead Sea. Extremophiles 1:143-149. [DOI] [PubMed] [Google Scholar]
- 32.Pajunen, M. I., M. R. Elizondo, M. Skurnik, J. Kieleczawa, and I. J. Molineux. 2002. Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J. Mol. Biol. 319:1115-1132. [DOI] [PubMed] [Google Scholar]
- 33.Palm, P., C. Schleper, B. Grampp, S. Yeats, P. McWilliam, W. D. Reiter, and W. Zillig. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242-250. [DOI] [PubMed] [Google Scholar]
- 34.Peng, X., H. Blum, Q. She, S. Mallok, K. Brugger, R. A. Garrett, W. Zillig, and D. Prangishvili. 2001. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291:226-234. [DOI] [PubMed] [Google Scholar]
- 35.Pfister, P., A. Wasserfallen, R. Stettler, and T. Leisinger. 1998. Molecular analysis of Methanobacterium phage ΨM2. Mol. Microbiol. 30:233-244. [DOI] [PubMed] [Google Scholar]
- 36.Prangishvili, D., K. Stedman, and W. Zillig. 2001. Viruses of the extremely thermophilic archaeon Sulfolobus. Trends Microbiol. 9:39-43. [DOI] [PubMed] [Google Scholar]
- 37.Stolt, P., and W. Zillig. 1994. Gene regulation in halophage ΦH—more than promoters. Syst. Appl. Microbiol. 16:591-596. [Google Scholar]
- 38.Tang, S. L., S. Nuttall, K. Ngui, C. Fisher, P. Lopez, and M. Dyall-Smith. 2002. HF2: a double-stranded DNA tailed haloarchaeal virus with a mosaic genome. Mol. Microbiol. 44:283-296. [DOI] [PubMed] [Google Scholar]
- 39.Torreblanca, M., F. Rodriguez-Valera, G. Juez, A. Ventosa, M. Kamekura, and M. Kates. 1986. Classification of non-alkaliphilic halobacteria based on numerical taxonomy and polar lipid composition, and description of Haloarcula gen. nov. and Haloferax gen. nov. Syst. Appl. Microbiol. 8:89-99. [Google Scholar]
- 40.Torsvik, T. 1982. Characterization of four bacteriophages for Halobacterium, with special emphasis on phage Hs1, p. 351. In O. Kandler (ed.), Archaebacteria. Gustav Fischer, Stuttgart, Germany.
- 41.Torsvik, T., and I. D. Dundas. 1974. Bacteriophage of Halobacterium salinarium. Nature 248:680-681. [DOI] [PubMed] [Google Scholar]
- 42.Ventosa, A., M. C. Gutierrez, M. Kamekura, and M. L. Dyall-Smith. 1999. Proposal to transfer Halococcus turkmenicus, Halobacterium trapanicum JCM 9743 and strain GSL-11 to Haloterrigena turkmenica gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:131-136. [DOI] [PubMed] [Google Scholar]
- 43.Wais, A. C., and L. L. Daniels. 1985. Populations of bacteriophage infecting Halobacterium in a transient brine pool. FEMS Microbiol. Ecol. 31:323-326. [Google Scholar]
- 44.Wais, A. C., M. Kon, R. E. MacDonald, and B. D. Stollar. 1975. Salt-dependent bacteriophage infecting Halobacterium cutirubrum and H. halobium. Nature 256:314-315. [DOI] [PubMed] [Google Scholar]
- 45.Walsby, A. E. 1980. A square bacterium. Nature (London) 283:69. [Google Scholar]
- 46.Wilbur, W. J., and D. J. Lipman. 1983. Rapid similarity searches of nucleic acid and protein data banks. Proc. Natl. Acad. Sci. USA 80:726-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wrigley, N. G. 1968. The lattice spacing of crystalline catalase as an internal standard of length in electron microscopy. J. Ultrastruct. Res. 24:454-464. [DOI] [PubMed] [Google Scholar]
- 48.Zillig, W., W.-D. Reiter, P. Palm, F. Gropp, H. Neumann, and M. Rettenberger. 1988. Viruses of archaebacteria, p. 517-558. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Publishing Corp., New York, N.Y.