Abstract
Production of a polysaccharide matrix is a hallmark of bacterial biofilms, but the composition of matrix polysaccharides and their functions are not widely understood. Previous studies of the regulation of Escherichia coli biofilm formation suggested the involvement of an unknown adhesin. We now establish that the pgaABCD (formerly ycdSRQP) locus affects biofilm development by promoting abiotic surface binding and intercellular adhesion. All of the pga genes are required for optimal biofilm formation under a variety of growth conditions. A pga-dependent cell-bound polysaccharide was isolated and determined by nuclear magnetic resonance analyses to consist of unbranched β-1,6-N-acetyl-d-glucosamine, a polymer previously unknown from the gram-negative bacteria but involved in adhesion by staphylococci. The pga genes are predicted to encode envelope proteins involved in synthesis, translocation, and possibly surface docking of this polysaccharide. As predicted, if poly-β-1,6-GlcNAc (PGA) mediates cohesion, metaperiodate caused biofilm dispersal and the release of intact cells, whereas treatment with protease or other lytic enzymes had no effect. The pgaABCD operon exhibits features of a horizontally transferred locus and is present in a variety of eubacteria. Therefore, we propose that PGA serves as an adhesin that stabilizes biofilms of E. coli and other bacteria.
In many natural and artificial habitats, bacteria form sessile communities known as biofilms (9). Biofilms represent a distinct physiological state, designed in part to provide a protected environment for survival under hostile conditions. They are composed of cells embedded within a glycocalyx-like matrix, and their complex structures have been likened to primitive multicellular organisms (9, 10). Biofilms play important roles in interactions of both nonpathogenic and pathogenic bacteria with eucaryotic hosts. Nonpathogenic biofilms in the mammalian gut and on the roots of plants provide barriers to invading pathogens (47, 50). Biofilms protect pathogens from attack by the immune system, complicate chronic infections that are difficult to eliminate with antibiotic therapy, and are involved in prostatitis, biliary tract infection, and urinary catheter cystitis caused by Escherichia coli (10, 18).
Biofilm development is a complex process (10, 17, 37). In general, it is initiated by cell attachment to a surface and formation of “microcolonies” on that surface. A variety of surface factors facilitate attachment and microcolony formation by E. coli (13, 48, 61). Differentiating microcolonies produce a matrix that encloses the biofilm and typically contains polysaccharides as its major components (59). Ultimately, planktonic cells are released that can complete the development cycle and colonize elsewhere.
Despite intensive interest in biofilm development, the functions of the extracellular polysaccharides (EPS) that form the matrix remain largely undefined. Colanic acid of E. coli affects biofilm architecture but not the adhesion of bacteria to surfaces or to themselves (14, 32). Cellulose is needed for biofilm formation by certain morphotypes of Enterobacteriaceae (58, 69). Although some strains of E. coli synthesize cellulose and the biosynthetic genes are present in E. coli K-12, this strain does not synthesize cellulose under known conditions (70). The length of the O-antigenic side chain of lipopolysaccharide is inversely correlated with biofilm formation by Salmonella enterica (45) and E. coli (X. Wang and T. Romeo, unpublished data). Perhaps the most widely studied role of EPS in biofilm formation by a gram-negative bacterium is that of alginate, a uronic acid polymer responsible for the mucoid phenotype of Pseudomonas aeruginosa. Alginate is a virulence factor and affects biofilm architecture (see, for example, reference 29). Although it is often cited as a component important for biofilm formation, recent investigations have cast doubt on that idea (46, 67).
Staphylococcus epidermidis and S. aureus produce β-1,6-N-acetyl-d-glucosamine polymers (β-1,6-GlcNAc) that serve as adhesins and are required for biofilm formation. These polymers have been referred to as polysaccharide intercellular adhesin (PIA), PNAG (earlier PS/A), or SAE (28, 34, 39, 40, 42, 43). Although various staphylococcal β-1,6-GlcNAc fractions differ in size, degree of N acetylation, and substitution by phosphate, succinate, or other moieties, their synthesis depends upon the icaABCD locus, and the basis of the observed chemical differences in the isolated polysaccharides remains to be resolved (reviewed in reference 26).
Biofilm development is guided by several regulatory systems in E. coli (1, 8, 19, 32, 33, 49, 61). Perhaps the most dramatic effects are exhibited by the Csr (carbon storage regulatory) system. Csr is a global regulatory system that represses stationary phase processes (reviewed in reference 51), including glycogen synthesis and catabolism (52, 68), gluconeogenesis (54), and biofilm formation (32, 60, 64). Conversely, it activates glycolysis (54), motility (63), acetate metabolism (62), and biofilm dispersal (32). Its key component, CsrA, is an RNA-binding protein that binds to the untranslated leader sequences of target mRNAs and alters their translation and stability (5, 38, 63). Repression of biofilm formation by CsrA involves the synthesis and catabolism of intracellular glycogen (32). This finding highlights the importance of central carbon metabolism and its regulation in biofilm development. The effect of CsrA on biofilm formation did not require several known surface factors, suggesting that unknown adhesins or other factors are required (32).
We identify here a genetic locus of E. coli that promotes surface binding, intercellular adhesion, and biofilm formation. Biochemical and genetic experiments show that this involves the production of a β-1,6-GlcNAc polysaccharide. Loci that are homologous to pga are present in variety of bacterial pathogens, suggesting that they may synthesize related polysaccharide adhesins that contribute to biofilm-mediated diseases.
MATERIALS AND METHODS
Bacterial strains, phage, plasmids, and growth conditions.
All E. coli strains, phage, and plasmids used in the present study are listed in Table 1. Unless otherwise indicated, bacteria were routinely grown at 37°C in Luria-Bertani (LB) medium (44) containing 0.2% glucose. Biofilms were grown at 26°C in LB or colonization factor antigen (CFA) medium (1% Casamino Acids, 0.15% yeast extract, 0.005% MgSO4, and 0.0005% MnCl2; pH 7.4 [32]). Kornberg medium (1.1% K2HPO4, 0.85% KH2PO4, 0.6% yeast extract containing 0.5% glucose for liquid or 1% for solid medium) was used in the selection of transposon mutants. Semisolid CFA or tryptone medium (pH 7.4) containing 1% tryptone, 0.5% NaCl, and 0.35% agar was used to test motility (63). Minimal media (M9 and M63) were prepared as described by Miller (44). Media were supplemented with antibiotics, as needed, at the following final concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 100 μg/ml; and tetracycline, 10 μg/ml.
TABLE 1.
Strains, plasmids, and bacteriophage used in this study
| Strain, plasmid, or phagea | Description or genotype | Source or reference |
|---|---|---|
| E. coli K-12 strains | ||
| MG1655 | F− λ− | Michael Cashel |
| TRMG1655 | MG1655 csrA::kan | 52 |
| CF7789 | MG1655 ΔlacI-Z (MluI) | Michael Cashel |
| TRCF7789 | CF7789 csrA::kan | 60 |
| DJ4 | TRMG1655 cpsE::Tn10 | 32 |
| DJ6 | TRMG1655 ΔfimB-H | 32 |
| DJ2 | TRMG1655 csgA2::Tn105 | 32 |
| DJ24 | TRMG1655 ΔmotB uvrC-279::Tn10 | 32 |
| DJ25 | DJ24 ΔfimB-H | 32 |
| TRFMXWA* | DJ25 pgaA672::cam | This study |
| TRFMXWB* | DJ25 pgaB510::cam | This study |
| TRFMXWC* | DJ25 pgaC880::cam | This study |
| TRFMXWD* | DJ25 pgaD146::cam | This study |
| XWA672 | MG1655 pgaA672::cam | This study |
| XWB510 | MG1655 pgaB510::cam | This study |
| XWC880 | MG1655 pgaC880::cam | This study |
| XWD146 | MG1655 pgaD146::cam | This study |
| TRXWA672 | TRMG1655 pgaA672::cam | This study |
| TRXWB510 | TRMG1655 pgaB510::cam | This study |
| TRXWC880 | TRMG1655 pgaC880::cam | This study |
| TRXWD146 | TRMG1655 pgaD146::cam | This study |
| TRXWEC | DJ4 pgaC880::cam | This study |
| XWMGΔA | MG1655 ΔpgaA | This study |
| XWMGΔB | MG1655 ΔpgaB | This study |
| XWMGΔC | MG1655 ΔpgaC | This study |
| XWMGΔABCD | MG1655 ΔpgaABCD | This study |
| TRXWMGΔA | TRMG1655 ΔpgaA | This study |
| TRXWMGΔB | TRMG1655 ΔpgaB | This study |
| TRXWMGΔC | TRMG1655 ΔpgaC | This study |
| TRXWMGΔABCD | TRMG1655 ΔpgaABCD | This study |
| Plasmids | ||
| pCR-XL-TOPO | Cloning vector | Invitrogen |
| pCRPGA37 | E. coli pga locus in pCR-XL-TOPO | This study |
| pCR2.1-TOPO | Cloning vector | Invitrogen |
| pUC19 | Cloning vector | 55 |
| pPGA372 | pgaABCD in pUC19 | This study |
| pCRS5 | pgaA gene in pCR2.1-TOPO | This study |
| pCRR12 | pgaB gene in pCR2.1-TOPO | This study |
| pCRP93 | pgaD gene in pCR2.1-TOPO | This study |
| pETQwt | pgaC gene in pET-Blue | This study |
| pET-Blue | Expression vector | Novagen |
| pKD46 | For arabinose induction of λ Red system | 16 |
| pKD3 | Contains the cat gene | 16 |
| pCP20 | For FLP-recombinase production | 16 |
| Bacteriophage | ||
| P1vir | Strictly lytic P1 | Carol Gross |
| λNK1324 | Contains mini-Tn10cam transposon | 36 |
Original transposon insertion mutants, as displayed in Fig. 1A, used in the present study are indicated by an asterisk.
Isolation of biofilm mutants.
DJ25 strain (MG1655 ΔfimB-H ΔmotB csrA) was infected with λNK1324 containing mini-Tn10cam at a multiplicity of infection of 0.2, as described previously (36). Biofilm mutants were isolated as described previously (48), with minor modifications. Briefly, chloramphenicol-resistant colonies were selected on Kornberg agar plates containing 2.5 mM sodium pyrophosphate. Individual colonies (11,000) were inoculated into 96-well microtiter plates containing CFA medium and incubated at 26°C for 24 h. The cultures were then diluted 1:100 and subcultured into fresh microtiter plates. After 24 h of incubation at 26°C, cell growth and biofilms were measured as described below. Mutants with altered biofilm production and normal growth were isolated. To assess the genetic linkage between altered biofilm production and the transposon insertion, the mutations were transduced by P1vir into the wild-type E. coli K-12 strain MG1655 and/or its csrA mutant (TRMG1655). Transposon mutations that were genetically linked to the altered biofilm phenotypes (153 in all) were further analyzed by arbitrarily primed PCR and DNA sequencing (see below).
Quantitative biofilm assay.
Biofilms were assayed by crystal violet staining, as described previously (32). Overnight cultures were diluted 1:100 into fresh medium and grown in 96-well microtiter plates. Bacterial growth was determined by measuring the absorbance at 630 nm. At least six replicas were conducted for each sample, and each experiment was performed at least twice. The results were calculated as averages and standard errors of two or more experiments. Tukey multigroup analysis (StatView; SAS Institute, Inc., Cary, N.C.) was used for statistical analysis of data.
PCR amplification of insertion sites and DNA sequence analyses.
Chromosomal DNA flanking the transposon insertions was amplified by using arbitrarily primed PCR (25). Sequences of primers used in the first round (ARB1 and OUT1-L) and the second round (ARB2 and PRIMER1-L) of PCRs, as well as those of all other oligonucleotide primers, will be provided upon request. Arbitrarily primed PCR conditions were as described by Gibson and Silhavy (25). PCR products were purified by QIAquick gel extraction kit and sequenced by using PRIMER1-L. All DNA sequence analyses were conducted at the DNA Sequencing Facility at the University of Arizona. Disrupted genes were identified by basic local alignment search tool (BLAST) analysis (2) at the National Center for Biotechnology Information (NCBI) website.
Cloning of the pgaABCD locus and individual pga genes.
Molecular cloning of the pga genes involved PCR amplification of chromosomal or plasmid DNA by using Elongase enzyme (Invitrogen) under the reaction conditions described by the manufacturer, with annealing temperatures and extension times that were based on primer melting temperature (TM) and final product size, respectively. PCR fragments of 3,070, 2,167, 1,325, and 596 bp, corresponding to pgaA, pgaB, pgaC, and pgaD, respectively, were prepared. The pgaA, pgaB, and pgaD genes were cloned into the vector pCR2.1-TOPO, resulting in pCRS5, pCRR12, and pCRP93, respectively. The pgaC gene was cloned into pET-Blue (Novagen) to produce pETQwt. Clones of individual pga genes were sequenced and found to be free of mutations. To clone the intact pga locus with all noncoding flanking DNA, a 6,933-bp fragment was amplified by PCR with Elongase enzyme (Invitrogen) from chromosomal DNA of MG1655. The 6,933-bp fragment was purified by using the QIAquick gel extraction kit (Qiagen) and cloned into vector pCR-XL-TOPO by using DH5α as the host for transformation. The plasmid clone pCRPGA37 increased biofilm ∼6-fold when expressed in DH5α. This clone was subsequently treated with HindIII and XbaI, and the insert DNA was subcloned into pUC19 to yield pPGA372. The genomic DNA of this clone was completely sequenced and found to have a silent mutation in pgaB and two missense mutations in pgaA (Q130R and N195D). However, it fully complemented biofilm defects caused by mutations in each of the four individual pga genes, as well as a deletion of the entire pgaABCD operon.
Nonpolar deletion of pgaA, pgaB, and pgaC and deletion of the pgaABCD locus.
The chromosomal pgaABCD locus and pgaA, pgaB, and pgaC genes were deleted by targeted gene substitutions (16). The cat gene flanked by FLP recognition target (FRT) was amplified from pKD3 by PCR and introduced by electroporation into arabinose-treated MG1655(pKD46). Transformants were selected on chloramphenicol, and their insertion sites were confirmed by PCR. The camR marker of each strain was subsequently moved into TRMG1655 by P1vir transduction. The cat genes of these strains were eliminated as described previously (16). The resulting deletions were confirmed by PCR analyses. The single gene deletions were determined to be nonpolar by complementation of their biofilm phenotypes with plasmids containing the corresponding genes, pCRS5 (pgaA), pCRR12 (pgaB), and pETQwt (pgaC). pPGA372 complemented the pgaABCD deletion. The pgaD146 transposon insertion was complemented by pCRP93 (pgaD).
Detection of pga-dependent polysaccharide.
The pgaC880 mutant (TRXWEC) containing pPGA372 or pUC19 was grown for 24 h at 37°C with shaking at 250 rpm in CFA medium. Cells were harvested from 2 liters of each culture and resuspended in 20 ml of 50 mM Tris-Cl buffer (pH 8.0). Lysozyme (100 mg) and 4 ml of 0.1 M EDTA were added, and each suspension was incubated at room temperature for 30 min. Then, α-amylase (100 mg), DNase I (5 mg), and RNase A (20 mg) were added, and the suspension was incubated at room temperature for 1 h and then at 37°C for 2 h. Polysaccharide was separated from proteins, and cell debris by phenol extraction (65). The aqueous phase was extracted with an equal volume of chloroform, concentrated by ultrafiltration (Amicon YM-10 membrane; 10,000 molecular weight cutoff) and fractionated by fast protein liquid chromatography (FPLC) on Sephacryl S-200 (HiPrep 16/60; Amersham Pharmacia Biotech). The column was equilibrated with 0.1 M phosphate-buffered saline (PBS; pH 7.4) and eluted with the same buffer. Fractions (1.6 ml) were collected and assayed for hexosamine after acid hydrolysis with 3-methyl-2-benzothiazolone hydrazone hydrochloride (MBTH) (57). Neutral-sugar content was determined by using the phenol-sulfuric acid assay (20). N-Acetyl-d-glucosamine and d-glucose, respectively, were used as standards in these two assays. Proteins and nucleic acids were detected by UV absorbance and ethidium bromide staining after agarose gel electrophoresis, respectively. Spent medium was also collected from these two strains and treated with ethanol to precipitate polysaccharides, and the resulting precipitates were dissolved in 0.1 M PBS. Hexosamine content was determined by using the MBTH assay.
Nuclear magnetic resonance (NMR) analyses of pga-dependent polysaccharide.
A cell lysate from 24 liters of cell culture (135 g of cells) was prepared by using a pga wild-type strain (DJ4) carrying pPGA372. The polysaccharide was purified as described above, except that fractionation was performed on a semipreparative Sephacryl S-300 column (HiPrep 26/60; Amersham Pharmacia Biotech), and 5-ml fractions were collected. The hexosamine-containing high-molecular-weight fractions from three columns were combined, precipitated with 67% ethanol at 4°C, and collected by centrifugation. The resulting white precipitate (0.7 g containing ∼35-mg GlcNAc equivalents) was suspended in 10 ml of D2O (99.9%; Aldrich Chemical) and subjected to filtration and dialysis by using a Centriprep YM-10 (Amicon/Millipore). This process was repeated several times until the conductivity reached a level less than that of 0.02 M NaCl. The retentate was then lyophilized to provide 52 mg of a white residue. A portion (28 mg) was suspended in 2.0 ml of 5.0 M DCl, prepared from 35% DCl in D2O (99% D; Aldrich Chemical), resulting in a faintly turbid suspension after dissolution of most of the residue. The suspension was clarified by brief centrifugation (microfuge) and neutralized to pH 6 to 7 with 5.0 M NaOH dissolved in D2O. The faintly opalescent solution was further triturated with D2O by using the YM-10 Centriprep. The retentate solution, estimated to contain 4 mg of polymer/ml and 99.6% D2O, was analyzed by 1H- and 13C-NMR. NMR spectra were obtained by using a Bruker Avance 500 Console, a Magnex 11.75 T/54-mm magnet, and a 5-mm BBO probe. Acquisitions were obtained at 300°K without spinning. 1H spectra were obtained at 500 MHz; 13C spectra were obtained at 125 MHz. 1H shift assignments were established by COZY-45 and corroborated with 13C assignments by 500/125-MHz 1H/13C-HMQC spectral analyses.
Treatment of biofilms with HIO4 and hydrolytic enzymes.
Biofilms were grown for 24 or 48 h in 96-well microtiter plates containing CFA or LB media. The planktonic cells were aspirated, and the biofilms were incubated at 4°C for 23 h after adding 200 μl of the following agents, respectively: H2O, 40 mM NaIO4 (metaperiodate [pH 5.0]), 40 mM NaIO4 plus 40 mM glucose or lactose (quenched metaperiodate), 40 mM glucose or 40 mM lactose, 0.1 M PBS, 40 mM EDTA, and 40 mM NaCl. Dispersed cells were examined by microscopy, and biofilms were quantified with crystal violet staining. The quenched periodate was prepared by incubation with 40 mM glucose or lactose at 4°C for 23 h before it was added to preformed biofilms. For enzymatic treatment of biofilms, DNase I and RNase A were dissolved in 10 mM Tris-Cl (pH 8.0) containing 2 mM MgCl2 at final concentrations of 200 and 50 μg/ml, respectively. Proteinase K was dissolved in 100 mM Tris-Cl to a final concentration of 1 mg/ml (41). These enzyme solutions were added to biofilms (200 μl per well), and the reaction mixtures were incubated at 37°C for 3 or 6 h. For treatment with Trichoderma reesei cellulase, 200 μl of a mixture containing 70 U of enzyme/ml in 0.05 M sodium citrate buffer (pH 5.0) was added to the biofilm, and the reaction was incubated at 45°C for 72 h as previously described (58). Remaining biofilm was assayed by crystal violet staining.
Microscopy.
Sterile borosilicate coverslips were aseptically placed into 15-cm petri dishes containing 50 ml of a freshly inoculated (1:100) culture. The petri dishes were incubated at 26°C, and coverslips were removed at various times and rinsed gently with water. Adherent cells were viewed by transmitted light with an Olympus 1X71 microscope (×40 objective lens with a ×1.6 selector). The images were captured by using a charge-coupled device camera (COHU-4915) and Image Pro-Plus 4.1 software (MediaCybernetics) and then stored as separate digital files for subsequent analysis.
Molecular biology, genetics, and bioinformatics.
Standard procedures were used for plasmid isolation, restriction digests, ligations, transformation, and transduction of antibiotic markers (44, 55). Predictions of protein domains and envelope localization are available from the Entrez Protein database at the NCBI website.
RESULTS
Isolation of transposon insertions in pgaABCD.
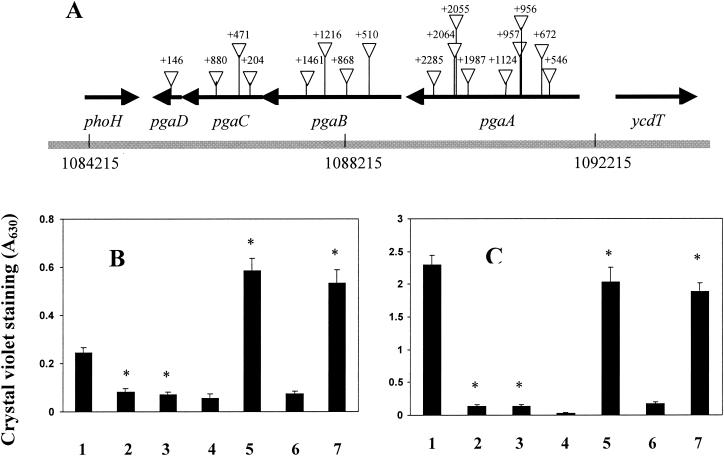
Random transposon mutagenesis to identify novel genes required for biofilm development was performed with a ΔmotB ΔfimB-H csrA mutant of MG1655 (DJ25) as the parent strain. This strain was designed to avoid the isolation of predominant mutants that affect type I pili and motility (48). Because the combined ΔmotB ΔfimB-H deletions severely disrupt biofilm development, a csrA mutation was introduced to increase biofilm formation and permit us to isolate biofilm-deficient mutants (Materials and Methods). Altogether, 17 such mutations were isolated in a predicted operon, ycdSRQP (6), renamed here as pgaABCD (Fig. 1A). These insertions were genetically linked to the biofilm-deficient phenotype and decreased biofilm ∼10-fold when transduced back into the parent strain (data not shown).
FIG. 1.
Transposon insertions in the pga locus of E. coli K-12 and their effects on biofilm formation. (A) Insertions are numbered relative to the first nucleotide (+1) of the coding region of the corresponding gene. The coordinates of this locus on the E. coli K-12 genome (6) are shown. (B) Biofilm formation in polystyrene microtiter plates. Isogenic strains are represented by bars numbered as follows: 1, MG1655; 2, XWC880 (pgaC880::cam); 3, XWA672 (pgaA672::cam); 4, XWC880(pUC19); 5, XWC880(pPGA372)(pgaABCD), 6, XWA672(pUC19); and 7, XWA672 (pPGA372). (C) Strains were as in panel B, except that they were csrA mutants. All biofilms were grown in LB medium at 26°C for 24 h. The asterisks denote significant differences relative to the corresponding parent strain (P < 0.001 [Tukey multigroup analysis]).
The intact pgaABCD locus is required for optimal biofilm formation.
Four insertions—pgaA672, pgaB510, pgaC880, and pgaD146—were tested and found to significantly decrease biofilm formation in both the MG1655 strain background (∼3-fold) and its isogenic csrA mutant (>10-fold) (Fig. 1B and C; compare bar 1 with bars 2 and 3; other data not shown). A plasmid containing the complete pga locus, pPGA372, complemented the biofilm defects caused by these mutations (Fig. 1B and C, compare bars 4 and 6 to bars 5 and 7, respectively; data not shown). In csrA wild-type strains, this plasmid complemented the pga defects and increased biofilm formation to a level ∼3-fold greater than that of the wild-type parent strain, MG1655 (Fig. 1B; compare bar 1 with bars 5 and 7). Similar effects of pga mutations were observed in all growth media that have been tested, including LB medium containing 0.2% glucose, CFA with or without 0.2% glucose, and minimal medium (M9 and M63) containing 0.2% glucose (data not shown). We also noticed that the parent strain (DJ25) and other csrA mutants formed distinct pellicles at the air-liquid interface in shaking flasks or borosilicate test tubes, whereas pga mutants of these strains did not (data not shown). The pga mutations did not affect the growth curves of the strains (data not shown), suggesting that they block one or more steps specifically needed for biofilm development.
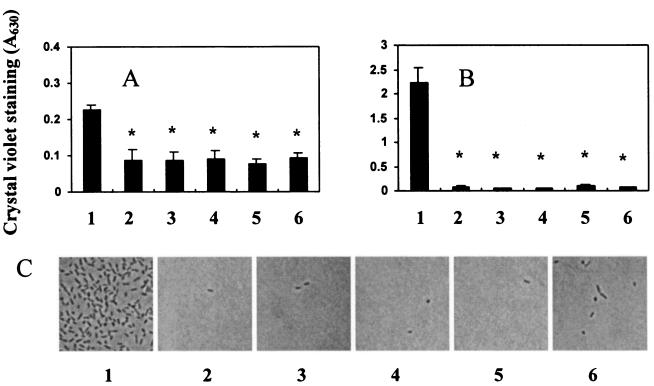
To investigate the roles of individual pga genes in biofilm formation, we constructed nonpolar pgaA, pgaB, and pgaC chromosomal deletions, as well as a deletion of the entire pgaABCD locus, in both MG1655 and its csrA mutant. These deletions and the pgaD146 insertion decreased biofilm formation in both strain backgrounds (Fig. 2A and B). Furthermore, they caused almost complete loss of the ability to adhere to borosilicate glass coverslips (Fig. 2C and data not shown). In similar experiments, the single-gene mutants were inoculated in all pairwise combinations, and biofilm formation was tested. No strain combination was able to restore biofilm development (data not shown), revealing that these mutations are incapable of intercellular complementation. Thus, all four pga genes must be functional within the cell to promote surface attachment and biofilm formation.
FIG. 2.
Effects of nonpolar pga gene disruptions on crystal violet binding of biofilms grown in polystyrene microtiter wells (A and B) and adherence of cells to borosilicate coverslips (C). Panels A and C depict results in the MG1655 strain background. Strains are represented by bars in panels A and B as follows: 1, MG1655; 2, XWMGΔABCD (ΔpgaABCD); 3, XWMGΔA (ΔpgaA); 4, XWMGΔB (ΔpgaB); 5, XWMGΔC (ΔpgaC); and 6, XWD146 (pgaD146::cam). (B) Strain identities were the same as in panel A, except that the strains were csrA mutants. Cultures were grown in LB medium at 26°C for 24 h. Asterisks denote significant differences relative to the corresponding parental strain (P < 0.001 [Tukey multigroup analysis]).
Predicted functions of pga genes.
A search for conserved protein domains (3) predicted that PgaC is a 441-amino-acid N-glycosyltransferase belonging to family 2 (GT-2; afmb.cnrs-mrs.fr/CAZY) (7). It is also predicted to be an inner membrane protein with two N-terminal and three C-terminal transmembrane domains. IcaA from S. epidermidis and NodC from Rhizobium loti are processive glycosyltransferases, which contain an N-terminal catalytic domain and a C-terminal domain that is needed both for catalysis and interaction with nascent polysaccharide chains (24, 56). Overall, IcaA and PgaC share 35% amino acid identity and 57% similarity. Five amino acids that appear to be essential for the catalytic activities of processive glycosyltransferases are found in PgaC (Asp163, Asp256, Gln292, Arg295, and Trp296) (24). These analyses suggested that PgaC is a polysaccharide polymerase that uses UDP-GlcNAc as a substrate.
PgaB is a predicted 672-amino-acid lipoprotein with a 20-amino-acid signal sequence and is homologous to the second gene product of the staphylococcal ica locus, IcaB. Both PgaB and IcaB contain putative polysaccharide N-deacetylase domains and belong to carbohydrate esterase family 4 (CE4) (11), suggesting that they may modify polysaccharides during synthesis.
PgaA is predicted to be a large (807-amino-acid) outer membrane protein, suggesting that it might mediate translocation and/or docking of PGA to the cell surface. PgaA has no functional homologues, including in the staphylococci, which lack outer membranes.
PgaD is predicted to be a small (137-amino-acid) inner membrane protein with two N-terminal membrane-spanning domains. IcaD of staphylococci is also a small cytoplasmic membrane protein, which enhances PIA synthesis by IcaA (24). Although PgaD and IcaD are not related in sequence, perhaps they may nevertheless function similarly.
Detection of a pga-dependent hexosamine-rich polysaccharide.
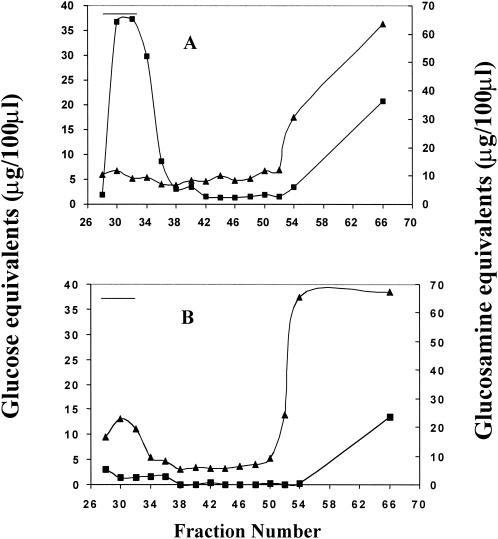
To test the hypothesis that the pgaABCD locus is required for the synthesis of a novel E. coli polysaccharide, two isogenic strains were constructed. One strain, TRXWEC (MG1655 csrA cpsE pgaC880) was defective for the pga locus in the chromosome and contained the plasmid vector pUC19, whereas the second strain contained a multicopy plasmid clone of the pgaABCD locus, pPGA372. The csrA mutation was introduced with the intention of enhancing central carbon flux into synthesis of the polysaccharide. Colanic acid production (cpsE) was eliminated to avoid contamination of the extracts with this polymer. The cpsE mutation does not affect the quantity of biofilm formed by these strains (32; data not shown). Cell extracts were prepared, fractionated by FPLC on Sephacryl S-200, and fractions were assayed for hexosamine, neutral sugars, proteins, and nucleic acids (Materials and Methods). The extract from the pga-overexpressing strain contained material rich in hexosamine, which eluted in the void volume of the column, and which was absent from extracts of the pga-defective strain (Fig. 3A and B, respectively). The separation range of Sephacryl S-200 for dextrans (1,000 to 80,000 Da) suggested that the hexosamine component is at least 80,000 Da. Chromatography on Sephacryl S-300 suggested a size of ≥400,000 Da (data not shown). The void fraction contained ≤10% neutral sugar with respect to hexosamine (Fig. 3A), and proteins and nucleic acids were not detectable (data not shown). The hexosamine-containing polymer was not detected in the spent medium from either strain (data not shown). These analyses revealed that E. coli synthesizes a cell-bound, hexosamine-rich polysaccharide whose production depends upon the pgaABCD locus.
FIG. 3.
Fractionation of polysaccharide extracts by gel filtration FPLC. Extracts from strain TRXWEC (MG1655 csrA cpsE pgaC880) containing either pPGA372 (pgaABCD) (A) or pUC19 (B) were fractionated by using a Sephacryl S-200 (HiPrep 16/60) column. Fractions (1.6 ml) were analyzed for neutral-sugar (▴) and, after hydrolysis, for hexosamine (▪). The column void volume, as determined with 2-MDa blue dextran, is indicated by a horizontal line.
NMR analysis of the purified polysaccharide.
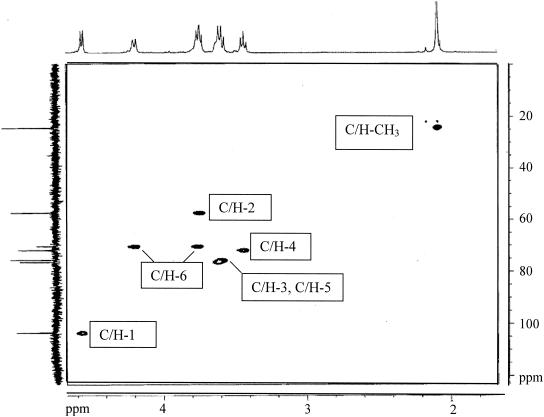
To determine the chemistry of the hexosamine-rich polysaccharide, ∼35-mg GlcNAc equivalents were prepared and then analyzed by NMR spectroscopy (see Materials and Methods). The 1H-NMR spectrum (Fig. 4, x axis) has three defined signals—d = 4.56 ppm, d = 3.45 ppm, and d = 2.08 ppm—that correspond to H-1, H-4, and N-acetyl protons, respectively, of β-1,6-linked GlcNAc residues, as originally reported for PIA of S. epidermidis (39). COZY spectra established that each of these signals represents 1H attached to a single carbon atom. The integration of these signals results in a 1.01:1.05:3.00 ratio that is consistent with these assignments and indicates that unacetylated glucosamine residues account for ≤3% of the residues of the polymer. A barely detectable broad signal at 2.875 ppm that could not be further defined by COZY analysis may be that of 1H of free amino groups of the unacetylated residues. Other signals have shift assignments that are essentially identical to those reported (39) and which are supported by COZY analysis. The spectrum is nearly identical to that of the polysaccharide adhesin (PS/A) from Staphylococcus aureus (42). The small signal at 2.17 ppm might represent 1H of the methylene carbons of succinyl groups, suggestive of a small extent of succinylation.
FIG. 4.
500/125 MHz 1H/13C-HMQC spectrum of E. coli pga-dependent polysaccharide with projected 1H (1.55 to 4.85 ppm) and 13C (0 to 124) spectra.
The 13C spectrum (Fig. 4, y axis) provided further definition to the structure, with chemical shifts of 102.5, 75.4, 74.8, 71.5, 68.5, 56.5, and 23 ppm corresponding to C-1, C-5, C-3, C-4, C-6, C-2, and CH3, respectively, values close to those reported for PIA (39). The 1H/13C-HMQC spectrum defined the exact relationships of 1H signals to the carbon atoms to which they are attached. This analysis further confirms the assignments made for β-1,6-GlcNAc polymers of S. aureus (42) and S. epidermidis (39). These data establish that the pga-dependent polysaccharide is a linear polymer of β-1,6-GlcNAc residues.
Time course of surface attachment by strains defective for PGA versus other components.
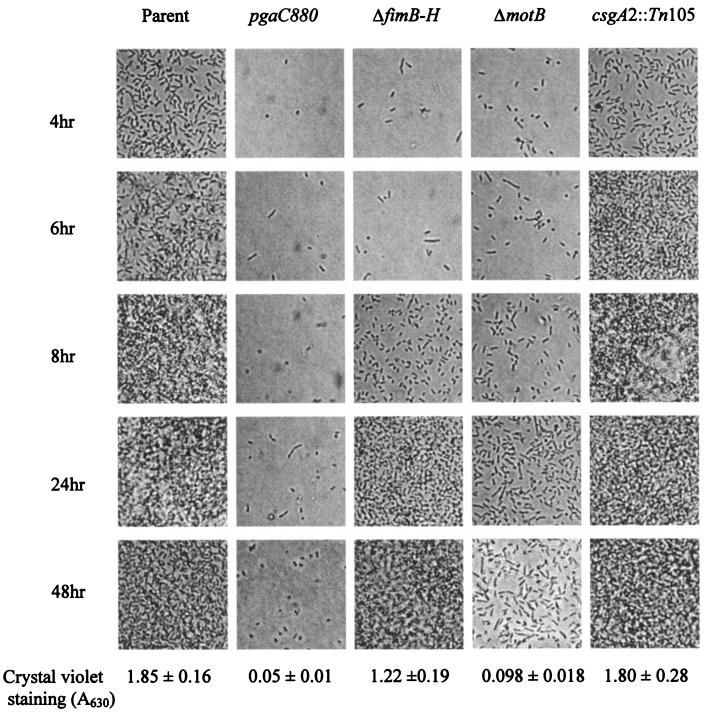
During biofilm development, initial attachment and subsequent microcolony formation utilize a variety of surface structures. To further investigate the role of PGA in biofilm development, isogenic strains that were defective for factors that promote biofilm formation, type I pili, motility, and curli were compared to a pgaC880::cam mutant for binding to microscope coverslips and biofilm formation on a polystyrene surface (Fig. 5). The parent strain (TRMG1655) began to form a dispersed monolayer on the glass coverslips by 4 h. Over the next several hours, a progressively more dense layer of cells was formed. By 8 h, multiple layers of cells were attached, which produce biofilm with a characteristic three-dimensional structure (32). The extent of biofilm formed by the mutant strains was closely correlated with the capacity to attach to coverslips. The pgaC mutant exhibited the most severe defect; few cells attached to coverslips during the entire 48 h of incubation. Mutations in motility (ΔmotB), type I pili, and curli caused defects of decreasing severity, as previously observed (32). Although the biofilm phenotypes of the pgaC and ΔmotB mutants were somewhat similar, the pgaC mutation did not affect the motility of these strains (data not shown). The addition of purified PGA (8 or 38 μg/ml) to the growth medium of a ΔpgaC nonpolar mutant (TRXWMGΔC) at 0, 16, or 20 h failed to restore biofilm development in this strain (data not shown). This suggested that PGA must be synthesized in situ to promote biofilm formation. These experiments revealed that disruption of the pga locus does not simply delay surface attachment and biofilm formation but causes severe, persistent defects in these processes.
FIG. 5.
Time course of adherence to coverslips by strains defective for PGA or other surface factors. The parent strain TRMG1655 and isogenic mutants defective in PGA production (pgaC880), type I pili (ΔfimB-H), motility (ΔmotB), or curli (csgA2::Tn105) were inoculated in parallel into petri dishes containing CFA medium and sterile borosilicate glass coverslips. Cultures were incubated at 26°C, and attached cells were analyzed at the indicated times. Representative fields are shown. The quantitative results of crystal violet staining of 24-h biofilms, grown in a polystyrene microtiter plate under similar conditions, are shown for comparison.
HIO4 treatment releases biofilm.
If PGA itself serves as an adhesin in E. coli, then its cleavage should disrupt a preformed biofilm. Although no enzyme is known to degrade PIA-like polysaccharides, metaperiodate (HIO4) is able to do so, by oxidizing the carbons (3 and 4) bearing vicinal hydroxyl groups and cleaving the C-C bonds (39, 42). Treatment of 24-h biofilms of TRMG1655 (csrA::kanR) at 4°C with metaperiodate (see Materials and Methods) led to near-complete release (>90%) of the biofilm, whereas similar incubation with 0.1 M PBS (pH 7.4), 40 mM EDTA (pH 8.0), or 40 mM NaCl had minimal or no effect (Table 2). To determine whether this requires metaperiodate to serve as an oxidant, i.e., to cleave C-C bonds, it was quenched by preincubation with glucose. In this case, metaperiodate-mediated biofilm release was inhibited (Table 2). Treatment of biofilm at 4°C with glucose alone had no effect (data not shown). Essentially the same results were observed when 48-h biofilms were examined (data not shown). Biofilm of a strain that was defective for colanic acid synthesis was released similarly by metaperiodate treatment (Table 2), revealing that colanic acid depolymerization does not account for the observed detachment. Similar results were obtained with an isogenic strain that cannot metabolize lactose and by quenching with lactose instead of glucose (Table 2). This control experiment confirmed that inhibition of biofilm release was not due to sugar metabolism. During these studies, we observed that metaperiodate-mediated dispersal of biofilm produced turbid suspensions comprised of intact cells (data not shown). This revealed, as expected, that metaperiodate does not release biofilm by causing cell lysis. Finally, an experiment was performed in which biofilm was grown on borosilicate test tubes and then removed by gentle scraping with a pipette tip. Under microscopic observation, the resulting cell aggregates were dissociated with metaperiodate treatment but not by treatment with the other agents that are shown in Table 2 (data not shown). These experiments suggest that polysaccharide(s) stabilizes the intercellular structure of biofilm.
TABLE 2.
Dispersal of biofilm by metaperiodatea
| Strain | Mean dispersal ± SE
|
|||||
|---|---|---|---|---|---|---|
| Control | HIO4 | HIO4 + polyolb | PBS | EDTA | NaCl | |
| Parent | 2.1 ± 0.3 | 0.08 ± 0.03 | 1.3 ± 0.3 | 2.1 ± 0.4 | 1.6 ± 0.2 | 1.8 ± 0.3 |
| cpsE | 2.0 ± 0.3 | 0.07 ± 0.03 | 2.0 ± 0.3 | 1.8 ± 0.4 | 1.5 ± 0.2 | 1.6 ± 0.3 |
| ΔlacIZ | 2.3 ± 0.2 | 0.09 ± 0.02 | 1.5 ± 0.1 | 2.7 ± 0.2 | 2.7 ± 0.1 | 2.7 ± 0.2 |
Biofilms formed after 24 h by TRMG1655 and isogenic mutants defective for colanic acid production (cpsE) or lactose metabolism (ΔlacIZ) were treated at 4°C with the indicated reagents, and the remaining biofilm was quantified by crystal violet staining as described in Materials and Methods. Values represent the mean ± the standard error of two experiments with six replicates per sample.
The polyol used for quenching metaperiodate was glucose for the parental strain and cpsE mutant or lactose for the ΔlacIZ strain.
Macromolecules that have been reported to stabilize biofilm structure include cellulose, nucleic acids, and proteins (12, 48, 58, 59, 66). However, treatment of E. coli biofilms that were grown as described above with DNase I, RNase A, proteinase K, or cellulase (see Materials and Methods) failed to disrupt biofilm or cause the release of cells (data not shown).
Phylogenetic distribution of pga homologues.
BLAST analysis (2) at the NCBI website with the pga gene products as query sequences revealed homologous loci in eubacteria but not in archaea or eukaryotes. Species with complete pgaABCD loci include E. coli O157:H7 and uropathogenic E. coli strain CFT073, Yersinia pestis, Y. entercolitica, Xanthomonas axonopodis, and Pseudomonas fluorescens. Species with a locus containing pgaABC but lacking an apparent pgaD homologue include Actinobacillus actinomycetemcomitans, A. pleuropneumoniae, Ralstonia solanacearum megaplasmid, Bordetella pertussis, B. parapertussis and B. bronchiseptica. A pga locus was not apparent in some closely related species, e.g., Salmonella and Shigella species, P. aeruginosa, or Vibrio cholerae. The gram-positive species Lactococcus lactis contains colocalized homologues of the E. coli pgaC and pgaB genes (staphylococcal icaA and icaB). This locus of L. lactis also contains a homologue of staphylococcal icaC, a third gene involved in PIA production. Streptomyces coelicolor and Streptomyces avermitilis have pgaC homologues that are predicted to encode proteins with N-deacetylase activity, a characteristic suggestive of bifunctional PgaC-PgaB enzymes.
Many species contain genes with limited sequence similarity to pgaC, which may encode glycosyltransferases, but without colocalization of the other pga genes. For example, Streptococcus pyogenes hasA encodes an enzyme that synthesizes hyaluronic acid, another GlcNAc-containing polysaccharide. Thus, the presence of a pgaC homologue alone is not a reliable indicator of PGA synthesis.
The observation that diverse species contain loci homologous to pga even though these loci are not typical of all members of a given family or genus suggests that these genes are horizontally transferred (21). This view is further supported by the low G+C content of the pgaABCD locus of E. coli (44 versus 51% for the genome), as recognized previously (23). The homologous locus of Y. pestis is present on an unstable region of the genome (22), a finding consistent with this possibility.
DISCUSSION
A crucial biological function that could be provided by EPS, either solely or in complex with ions, proteins such as lectins, or other matrix components, is that of an adhesin. A biofilm adhesin should permit bacterial cells to bind to a surface and/or to each other and thereby stabilize the structure of the biofilm. The present study provides evidence that the pgaABCD locus of E. coli is needed for the biosynthesis and function of a polysaccharide that is a biofilm adhesin, i.e., PGA. This polysaccharide affects biofilm in either the presence or absence of other factors, curli fimbriae, type I pili, or motility. This class of polysaccharides was previously unknown from E. coli and other gram-negative bacteria but serves an adhesive role in staphylococcal biofilms. Although these are not the only polysaccharides that may serve as biofilm adhesins (59), we expect them to function as adhesins in species that produce them.
What evidence supports the role of PGA as a biofilm adhesin? First, the pga locus is needed for attachment to abiotic surfaces, intercellular adherence, biofilm formation, and the accumulation of PGA (Fig. 1, 2, and 5). Second, PGA is structurally related to the staphylococcal polysaccharide adhesins. Third, PGA is cell bound and was not detected in the spent medium. The four pga gene products are predicted to be cell envelope proteins (discussed below), further suggesting that PGA is surface associated. Fourth, the dissociation of biofilm by metaperiodate suggests that polysaccharide(s) maintains its structure (Table 2). Neither colanic acid (Table 2) nor cellulose (data not shown) performs this function. Furthermore, enzymatic digestion of RNA, DNA, or protein failed to cause release (data not shown), suggesting that under our experimental conditions these macromolecules are not responsible for maintenance of biofilm structure. Viewed together, these findings offer a compelling case that PGA serves as a biofilm adhesin in E. coli.
The predicted localization and biological functions of the Pga gene products suggest a model in which PGA is synthesized at the cytoplasmic side of the inner membrane, where UDP-GlcNAc is present, and is transported through the cell surface by an envelope complex of the Pga proteins. Biofilm formation by nonpolar pga mutants was not restored by intercellular complementation or by providing external PGA to growing cultures (data not shown). These observations are fully consistent with this model.
Although the picture is far from complete, some information is available concerning the metabolic processes involved in PGA production. A large number of transposon insertions that disrupt biofilm formation were isolated in the pgaABCD locus (Fig. 1 and 2). Because UDP-GlcNAc is an essential sugar nucleotide, no mutations that disrupt the synthesis of this apparent precursor of PGA were isolated. CsrA represses biofilm formation in E. coli and its relatives, as well as the synthesis and catabolism of intracellular glycogen, which are required in this process (32). Thus, we previously proposed that redirection of central carbon flux through a reserve polymer might be a general principle of biofilm formation, designed to provide precursor(s) for adhesins or other factors (32). The main product of glycogen catabolism, glucose-1-phosphate, is a precursor via fructose-6-phosphate of the apparent substrate for PGA synthesis, UDP-GlcNAc. In view of the present findings on the role of PGA, we hypothesize that the main influence of glycogen synthesis and catabolism in biofilm formation is to overcome a limitation for UDP-GlcNAc in PGA synthesis, which must compete with peptidoglycan and lipopolysaccharide for this precursor. The finding that pga mutations cause more severe defects in biofilm formation in csrA mutant strains (Fig. 1), which exhibit elevated glycogen synthesis and subsequent catabolism (52, 68), supports this notion. It is notable that although Salmonella enterica lacks a pga locus, glycogen synthesis nevertheless is positively correlated with biofilm formation and CsrA represses biofilm development in this bacterium (discussed in reference 32). This example suggests that relatives of E. coli that do not synthesize PGA may use similar strategies for regulating central carbon flux into other polysaccharide adhesins. Unlike bacterial cellulose (58), synthesis of staphylococcal PIA does not require a lipid carrier (24), and we failed to isolate any mutants that were consistent with this requirement for PGA biosynthesis.
Chromatographic and NMR analyses revealed that PGA consists of high-molecular-mass β-1,6-GlcNAc (≥400,000 Da) containing <3% deacetylated residues and no other major substituents (Fig. 3 and 4). The PGA was prepared from a strain containing multiple copies of the pga locus, and even in this strain there was only ∼1 mg of PGA per liter of culture. Although we recognize that the recombinant genotype of the strain may have affected the structure of the polysaccharide, overexpression of the pga operon greatly enhanced biofilm formation, indicating that the resulting polysaccharide is functional. Furthermore, the staphylococcal β-1,6-GlcNAc polymers appear to vary in molecular weight, N acetylation, phosphorylation, and succinylation, and yet all apparently function as adhesins. It has been suggested that structural variations in the staphylococcal polysaccharides may reflect different growth conditions or purification conditions (34). Whether such structural variations have subtle effects on adhesion or other biological functions remains to be determined.
The production of β-1,6-GlcNAc may affect a variety of disease processes that involve biofilm development. In the most definitive example, PIA has been shown to be a virulence factor in model catheter and foreign body infections caused by S. epidermidis (see reference 53 and references therein). The hmsHFRS locus of Y. pestis, the plague bacillus, is homologous to pgaABCD and is needed for biofilm formation in vitro (27) and in invertebrates (15). Part of the life cycle of this bacterium involves blockage of the flea gut by the accumulation of an extracellular material that depends upon the hmsHFRS locus and appears to protect the bacteria from expulsion from the gut (30). In conjunction with this information, the present study raises the possibility that β-1,6-GlcNAc may be important in the transmission of this deadly disease. Although definitive evidence is lacking, other diseases might also be affected by this polysaccharide, based on the presence of pga loci in the etiological agents. These include bladder infections by uropathogenic strains of E. coli, which produce substantial amounts of EPS (4), and food-borne illness by enterohemorrhagic E. coli, which may be transmitted through biofilm formation on food or food processing equipment (31). β-1,6-GlcNAc might also participate in microbial interactions with higher plants, e.g., formation of a protective biofilm on plant roots by P. fluorescens (discussed in reference 47) or diseases caused by Xanthomonas or Ralstonia species (35).
Acknowledgments
We thank Konstantin Agladze for help with the microscopy, Rebecca Desplas for help with the sequencing of transposon insertions and preparation of pETQwt, and June Scott and Rod Donlan for reviewing the manuscript. Acquisition of NMR data was supported through the National High Magnetic Field Laboratory and obtained at the Advanced Magnetic Resonance Imaging and Spectroscopy (AMRIS) facility in the McKnight Brain Institute of the University of Florida. We thank James R. Rocca for these analyses.
These studies were funded in part by the National Science Foundation (MCB-9726197), the National Institutes of Health (GM066794), and Kane Biotech, Inc. Kane is developing applications related to the findings herein. T. Romeo serves as Chief Scientific Advisor for, owns equity in, and may receive royalties from this company. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict of interest policies. NMR studies were supported in part by CRIS Project R-01049 of the University of Florida Institute of Food and Agricultural Sciences and are acknowledged as journal series no. R-09696.
REFERENCES
- 1.Adams, J. L., and R. J. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. [DOI] [PubMed] [Google Scholar]
- 5.Baker, C. S., I. Morozov, K. Suzuki, T. Romeo, and P. Babitzke. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 44:1599-1610. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326(Pt. 3):929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corona-Izquierdo, F. P., and J. Membrillo-Hernandez. 2002. A mutation in rpoS enhances biofilm formation in Escherichia coli during exponential phase of growth. FEMS Microbiol. Lett. 211:105-110. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 11.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 12.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 14.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 20.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for the determination of sugars and related substances. Anal. Chem. 28:350-356. [DOI] [PubMed] [Google Scholar]
- 21.Dutta, C., and A. Pan. 2002. Horizontal gene transfer and bacterial diversity. J. Biosci. 27:27-33. [DOI] [PubMed] [Google Scholar]
- 22.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Vallve, S., E. Guzman, M. A. Montero, and A. Romeu. 2003. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 31:187-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Gotz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 25.Gibson, K. E., and T. J. Silhavy. 1999. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator sprE. J. Bacteriol. 181:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 27.Hare, J. M., and K. A. McDonough. 1999. High-frequency RecA-dependent and -independent mechanisms of Congo red binding mutations in Yersinia pestis. J. Bacteriol. 181:4896-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 29.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinnebusch, B. J., E. R. Fischer, and T. G. Schwan. 1998. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J. Infect. Dis. 178:1406-1415. [DOI] [PubMed] [Google Scholar]
- 31.Hood, S. K., and E. A. Zottola. 1997. Adherence to stainless steel by food-borne microorganisms during growth in model food systems. Int. J. Food Microbiol. 37:145-153. [DOI] [PubMed] [Google Scholar]
- 32.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson, D. W., J. W. Simecka, and T. Romeo. 2002. Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 184:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joyce, J. G., C. Abeygunawardana, Q. Xu, J. C. Cook, R. Hepler, C. T. Przysiecki, K. M. Grimm, K. Roper, C. C. Ip, L. Cope, D. Montgomery, M. Chang, S. Campie, M. Brown, T. B. McNeely, J. Zorman, T. Maira-Litran, G. B. Pier, P. M. Keller, K. U. Jansen, and G. E. Mark. 2003. Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr. Res. 338:903-922. [DOI] [PubMed] [Google Scholar]
- 35.Kang, Y., H. Liu, S. Genin, M. A. Schell, and T. P. Denny. 2002. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46:427-437. [DOI] [PubMed] [Google Scholar]
- 36.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 37.Kolter, R., and R. Losick. 1998. One for all and all for one. Science 280:226-227. [DOI] [PubMed] [Google Scholar]
- 38.Liu, M. Y., H. Yang, and T. Romeo. 1995. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177:2663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwarzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62:3244-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Mireles, J. R., II, A. Toguchi, and R. M. Harshey. 2001. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183:5848-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O-acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 48.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 49.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reid, G., J. Howard, and B. S. Gan. 2001. Can bacterial interference prevent infection? Trends Microbiol. 9:424-428. [DOI] [PubMed] [Google Scholar]
- 51.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 52.Romeo, T., M. Gong, M. Y. Liu, and A. M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175:4744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rupp, M. E., P. D. Fey, C. Heilmann, and F. Gotz. 2001. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 183:1038-1042. [DOI] [PubMed] [Google Scholar]
- 54.Sabnis, N. A., H. Yang, and T. Romeo. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 270:29096-29104. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Saxena, I. M., R. M. Brown, Jr., M. Fevre, R. A. Geremia, and B. Henrissat. 1995. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J. Bacteriol. 177:1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, R. L., and E. Gilkerson. 1979. Quantitation of glycosaminoglycan hexosamine using 3-methyl-2-benzothiazolone hydrazone hydrochloride. Anal. Biochem. 98:478-480.496014 [Google Scholar]
- 58.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 59.Sutherland, I. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3-9. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki, K., X. Wang, T. Weilbacher, A. K. Pernestig, O. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei, B., S. Shin, D. LaPorte, A. J. Wolfe, and T. Romeo. 2000. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J. Bacteriol. 182:1632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245-256. [DOI] [PubMed] [Google Scholar]
- 64.Weilbacher, T., K. Suzuki, A. K. Dubey, X. Wang, S. Gudapaty, I. Morozov, C. S. Baker, D. Georgellis, P. Babitzke, and T. Romeo. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48:657-670. [DOI] [PubMed] [Google Scholar]
- 65.Westphal, O., and K. Jann. 1965. In R. L. Whistler (ed.), Methods in carbohydrate chemistry, vol. V, p. 83-91. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 66.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 67.Wozniak, D. J., T. J. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, H., M. Y. Liu, and T. Romeo. 1996. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J. Bacteriol. 178:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zogaj, X., W. Bokranz, M. Nimtz, and U. Romling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]