Abstract
Pseudomonas aeruginosa is an opportunistic bacterial pathogen which poses a major threat to long-term-hospitalized patients and individuals with cystic fibrosis. The capacity of P. aeruginosa to form biofilms is an important requirement for chronic colonization of human tissues and for persistence in implanted medical devices. Various stages of biofilm formation by this organism are mediated by extracellular appendages, such as type IV pili and flagella. Recently, we identified three P. aeruginosa gene clusters that were termed cup (chaperone-usher pathway) based on their sequence relatedness to the chaperone-usher fimbrial assembly pathway in other bacteria. The cupA gene cluster, but not the cupB or cupC cluster, is required for biofilm formation on abiotic surfaces. In this study, we identified a gene (mvaT) encoding a negative regulator of cupA expression. Such regulatory control was confirmed by several approaches, including lacZ transcriptional fusions, Northern blotting, and transcriptional profiling using DNA microarrays. MvaT also represses the expression of the cupB and cupC genes, although the extent of the regulatory effect is not as pronounced as with cupA. Consistent with this finding, mvaT mutants exhibit enhanced biofilm formation. Although the P. aeruginosa genome contains a highly homologous gene, mvaU, the repression of cupA genes is MvaT specific. Thus, MvaT appears to be an important regulatory component within a complex network that controls biofilm formation and maturation in P. aeruginosa.
In their natural environment, bacteria grow predominantly in organized communities termed biofilms. The ability to attach to solid surfaces and the subsequent formation of an organized bacterial biofilm community are also important steps in the establishment of chronic bacterial infections and persistence in host tissues (4). It was recently demonstrated that Pseudomonas aeruginosa grows as a biofilm in the lungs of cystic fibrosis patients (13, 31). These microbial communities then develop unique characteristics and properties that protect them from external influences, most significant of which is the enhanced resistance of bacteria within the biofilm to antibiotics and host defenses (18). In P. aeruginosa, the adherence phenotype is mostly driven by extracellular appendages, such as flagella and type IV pili (22). Flagellar motility appears to be required for approaching surfaces and counteracting repulsive forces. Twitching motility, a mechanism that is mediated by type IV pili, is required for subsequent spreading over the surface. Following formation of microcolonies, the biofilm structure undergoes maturation, where the bacterial community assumes a characteristic architecture. The production of the extracellular polysaccharide alginate and the rhamnolipid surfactant allow the maintenance of biofilm architecture and provide the biofilm with a degree of physical resistance (6, 11, 21). The maturation process is accompanied by the engulfment of microcolonies within a glycocalyx matrix containing aqueous channels allowing the flow of nutrients and waste products but also the transport of oxygen and various signaling molecules. Biofilm maturation in Pseudomonas (7), Burkholderia (14), and Aeromonas (17) has been reported to depend on N-acylhomoserine lactone-dependent quorum sensing, although not under all conditions (12). Although the role of quorum sensing as an important mechanism for regulating biofilm maturation cannot be doubted, the overall process is much more complex and involves the integration of many diverse signals from the environment (26), including the availability of nutrients. It is very likely that successful cellular adaptations within the community necessitate the coordination of activities of many genes through complex regulatory networks (16).
Recently, we identified three P. aeruginosa gene clusters, termed cup for chaperone-usher pathways, which may be involved in the assembly of novel P. aeruginosa fimbrial structures that are radically different from the type IV pili (5, 38). Such structures and the pathways involved in their assembly in P. aeruginosa have not previously been described. The chaperone-usher pathway takes its name from the components required for assembling the pilin subunits into pili (fimbriae) on the surface of many gram-negative bacteria. The assembly pathway has been most extensively studied in Escherichia coli. A so-called chaperone protein binds fimbrial subunits that are secreted into the periplasm through the general export pathway (25, 36). The chaperone-pilin complex is subsequently delivered to the usher protein, which forms a pore in the outer membrane. This usher protein allows the pilin subunits to be finally released from the chaperone and assembled into fibrils while crossing the outer membrane (36). Among the genes encoding putative chaperones and usher proteins, those in the cupA gene cluster were found to be required for adhesion to inert surfaces, and thus their products are likely to be important in the initiation of biofilm formation (38). Such a requirement was shown to be independent of type IV pili under the conditions tested. The two additional clusters that were identified, cupB and cupC, had no obvious influence on surface attachment in these conditions (38).
To advance our understanding of the sequential involvement of these new fimbriae during the establishment of a P. aeruginosa biofilm, we sought to determine how the cup gene clusters are regulated. In addition, we would like to understand how their regulation is coordinated with the expression of other adhesins and with the general transcriptional response during biofilm formation. Indeed, the identification of a substantial number of genes that are under the control of common biofilm regulators could lead to the development of new therapeutic strategies directed towards the prevention of biofilm formation during infections. Here we describe the identification of a negative regulator of cupA gene expression, MvaT. Interestingly, an mvaT mutant also exhibits enhanced expression of the cupB and cupC gene clusters and displays increased surface coverage and greater biofilm depth.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Strains were grown at 30 or 37°C in L broth (LB), in M63 minimal medium, or on LB agar plates. E. coli TG1 and TOP10F′ were used to propagate plasmids. Plasmids were introduced into P. aeruginosa strains by electroporation (32) or triparental mating by using plasmid pRK2013 as a helper (9). The transformants were selected on Pseudomonas isolation agar containing antibiotics. Plasmids were maintained by adding ampicillin and tetracycline (50 and 15 μg/ml, respectively) for E. coli and carbenicillin, tetracycline, and gentamicin (500, 200, and 50 μg/ml, respectively) for P. aeruginosa.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | supE Δ(lac-proAB) thi hsdRΔ5 (F′traD36 proA+B+lacIqZΔM15) | Laboratory collection |
| TOP10F′ | F′ [lacIq Tn10(Tetr)] mrcA Δ(mrr-hsdRMS-mcrBC)φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG | Invitrogen |
| S17λpir | thi pro hsdR hsdM+recA RP4-2-Tc::Mu-Km::Tn7 λpir | 30 |
| P. aeruginosa | ||
| PAO1 | Wild type | Laboratory collection |
| PAO1 lecA::lux | lecA::luxABCDE genomic reporter fusion in PAO1 | 42 |
| PAO-P10 | Tn5-B21 mvaT mutant derived from PAO1 lecA::lux | 8 |
| PAO-P48 | mvaU chromosomal deletion mutant derived from PAO1 lecA::lux | This work |
| PAO-P47 | mvaT chromosomal deletion mutant derived from PAO1 | 8 |
| PAO-P46 | mvaU chromosomal deletion mutant derived from PAO1 | This work |
| PAO-MC | cupA3 mutation in PAO-P47 | This work |
| Tn5G library | PAKΔpilA::Tn5G collection | 38 |
| Plasmids | ||
| pCR2.1 | Apr ColE1 f1 ori | Invitrogen |
| pDM4 | Cmr, suicide vector carrying the sacBR genes for sucrose selection | 19 |
| pMP220 | Tcr IncP, vector for lacZ transcriptional fusions | 33 |
| pMMB190 | Apr, pMMB66EH, tac promoter, lacZα | 20 |
| pRK2013 | Kmr ColE1 with Tra+ Mob+ | 9 |
| pUC18 | Apr ColE1 lacI φ80 lacZα | Boehringer |
| pUCmvaU | 1.4-kb BamHI-PstI PAO1 chromosomal DNA fragment containing mvaU in pUC18 | This work |
| pUCΔmvaU | 300-bp mvaU deletion in pUCmvaU | This work |
| pDM4ΔmvaU | pUCΔmvaU DNA insert cloned in pDM4 | This work |
| pMPFCAS | cupA1S-lacZ transcriptional fusion in pMP220 | This work |
| pMPFCAL | cupA1L-lacZ transcriptional fusion in pMP220 | This work |
| pMMBmvaT | 513-bp PAO1 chromosomal DNA fragment containing mvaT in pMMB190 | This work |
| pUCP18 | Same as pUC18, 1.8-kb stabilizing fragment for maintenance in Pseudomonas spp. | 29 |
| pUCPmvaT | 1.6-kb PAO1 DNA fragment containg mvaT cloned in pUCP18 | 8 |
Plasmid constructions and DNA manipulation.
The cupA1S- and cupA1L-lacZ transcriptional fusions were constructed by PCR amplification of upstream DNA regions from the cupA1 gene (370 and 1,017 bp, respectively) by using the oligonucleotide couples OFN65-OFN2 and OFN1-OFN2, respectively. PCR amplification products were directly cloned into the pCR2.1 vector by using the TA cloning kit (Invitrogen), nucleotide sequences were verified (Genome Express, Grenoble, France), and EcoRI DNA fragments were subcloned into the low-copy-number vector pMP220 (33), yielding pMPFCAS and pMPFCAL, respectively. The 513-bp fragment containing the mvaT gene was PCR amplified using the oligonucleotide pair M1/M2, cloned into pCR2.1, and subsequently subcloned into the pMMB190 vector (20), yielding pMMBmvaT. The mvaT gene was also cloned in pUCP18 (29), yielding pUCPmvaT, as described previously (8). Chromosomal DNA was prepared by using the Nucleospin C+T kit (Macherey-Nagel), and PCR amplifications were carried out using HiFi polymerase (Roche). All the oligonucleotides used in this study are listed in Table 2.
TABLE 2.
Oligonucleotide sequences
| Oligonucleotide | Nucleotide sequence | Gene or DNA region |
|---|---|---|
| OFN1 | 5′-GGAATACCAGTCGGAGGCGGGAT-3′ | cupAS-lacZ |
| OFN65 | 5′-CCATAGTCGGAAATACAAGCGTTG-3′ | cupAL-lacZ |
| OFN2 | 5′-GCTGAATGTGATAGTGTTTGCCG-3′ | cupAS and L-lacZ |
| M1 | 5′-CCACCGCCACTCAGCACAGACAAGG-3′ | mvaT |
| M2 | 5′-CGCTATTCGCTGGAGACTTGAGGGC-3′ | mvaT |
| OTn1 | 5′-GCGCGGATCCTGGAAAACGGGAAAG-3′ | Tn5G |
| OTn2 | 5′-CCATCTCATCAGAGGGTAGT-3′ | Tn5G |
| MA1O1 | 5′-GGCAAACACTATCACATTCAGCGG-3′ | cupA1 (A1 probe) |
| MA1O2 | 5′-GGCGGCGGTGTTCACCAGGTTCCC-3′ | cupA1 (A1 probe) |
| NB1O1 | 5′-TCGTCAACTTCTCGGGCAACATCAC-3′ | cupB1 (B1 probe) |
| NB1O2 | 5′-CAGCGGGATCTTCGTGGTGTTGGTC-3′ | cupB1 (B1 probe) |
| NC1O1 | 5′-CAGACGGCACCATCAATTTCAAAGG-3′ | cupC1 (C1 probe) |
| NC1O2 | 5′-ATAGATGCCAATACCGACGCCAGTG-3′ | cupC1 (C1 probe) |
| NA2O1 | 5′-CGGGCAAGCGTGGTGGTGACCG-3′ | cupA2 (A2 probe) |
| NA2O2 | 5′-TTGTCCACCCGCACCGCCCAGC-3′ | cupA2 (A2 probe) |
| NA3O1 | 5′-TCCAACTACACCTATTCCCGCTAC-3′ | cupA3 (A3 probe) |
| NA3O2 | 5′-CCGTCGTAGAAATCGCTGGAGGAG-3′ | cupA3 (A3 probe) |
| NA4O1 | 5′-GGGACGCCGTCTACGAGATGTTTTC-3′ | cupA4 (A4 probe) |
| NA4O2 | 5′-TGTAGGAATAGGGCTGVGAGAGGAT-3′ | cupA4 (A4 probe) |
| NA5O1 | 5′-CTGCCGCCCGTATTCCGCCTGG-3′ | cupA5 (A5 probe) |
| NA5O2 | 5′-AGTACGGCGTCGGGTTGTCCAC-3′ | cupA5 (A5 probe) |
| NB3O1 | 5′-CCTGTCTGCTGGCACTGTTTC-3′ | cupB3 (B3 probe) |
| NB3O2 | 5′-AATAGCTGGGCACCGAGACATA-3′ | cupB3 (B3 probe) |
| NC3O1 | 5′-AGGTGTCCGTCTATTCCAGGT-3′ | cupC3 (C3 probe) |
| NC3O2 | 5′-GGTACGGTTGCTACTGAACTTG-3′ | cupC3 (C3 probe) |
| 4648O1 | 5′-GCAGCCATTCCGACCTCACCAACG-3′ | PA4648 probe |
| 4648O2 | 5′-CGTCGGCGTGGGTGTGCTTTGTCC-3′ | PA4648 probe |
| 4651O1 | 5′-ACCTACCTCTCCGCCCCAGCCGCT-3′ | PA4651 probe |
| 4651O2 | 5′-CCTGTTCCTCGACCAGCCGCCAGG-3′ | PA4651 probe |
| Pil1 | 5′-ACCCGCTGAAGACCACTGTTGAAGA-3′ | pilA probe |
| Pil2 | 5′-ATTTACAAGCCCAGACCCCATCCGC-3′ | pilA probe |
| mvaUUF | 5′-CGGCTGGATCCGCGACTTCGCGGAAAT-3′ | Upstream mvaU |
| mvaUDR | 5′-GACGCCTGCAGTTCGGCTCCCCGGCC-3′ | Downstream mvaU |
| mvaUUR | 5′-TGCCTCGCGGAACTGTGCACGTTTGGACAT-3′ | Upstream mvaU |
| mvaUDF | 5′-ACCGTCGAATCCTGGGTGCACCGCTAAGCCG-3′ | Downstream mvaU |
Screening of the P. aeruginosa Tn5G library.
Construction of the Tn5G library was described previously (38). The plasmids carrying lacZ transcriptional fusions were introduced into the Tn5 library by using triparental mating. Mutants containing the plasmid were selected on LB plates supplemented with 200 μg of tetracycline/ml and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (125 μg/ml). After overnight growth, blue colonies were identified and isolated on the same medium.
Inverse PCR.
Inverse PCR was performed as described previously (38). Briefly, chromosomal DNA from each mutant was prepared by using the Nucleospin C+T kit. DNA was digested with AluI and religated. The circular DNA was used as the matrix for PCR amplification using oligonucleotides OTn1 and OTn2. The DNA fragments thus amplified were cloned into the pCR2.1 vector by using the TA cloning kit and transformed into E. coli TOP10F' (Invitrogen). DNA sequencing was performed by Genome Express.
Computer analysis.
The DNA sequences obtained following inverse PCR were analyzed with the BLASTN program and compared with the PAO1 genome at http://www.pseudomonas.com.
β-Galactosidase assay.
Overnight cultures, grown in LB, were diluted in LB to an optical density at 600 nm (OD600) of 0.1. Growth and β-galactosidase activity were monitored by harvesting samples at different time intervals. β-Galactosidase activity was measured according to the Miller method, based on o-nitrophenyl-β-d-galactopyranoside hydrolysis (28). β-Galactosidase activities were expressed in Miller units. β-Galactosidase activity was also observed by looking at blue colonies on plates containing 125 μg of X-Gal per ml of LB agar.
Northern blot hybridization.
Total RNA was isolated from 50-ml cultures of strains PAO1, PAO-P46, and PAO-P47 grown in LB. Following centrifugation, bacterial cells pellets were resuspended in 30 ml of Trizol reagent (Gibco BRL). After chloroform extraction and isopropanol precipitation, samples were treated with 20 U of RQ1 DNase (Promega) and cleaned using the RNeasy kit (Qiagen). One percent agarose gels for size fractionating RNA were cast and run using NorthernMax-Gly Gel prep/running buffer (Ambion). Glyoxal Load Dye (Ambion) was used for denaturation of sample RNA, with Millenium Marker (Ambion) included as a molecular weight marker. Twenty micrograms of total RNA was used for analysis.
Transfer of RNA to nylon membranes (Hybond N+ membranes; Amersham Pharmacia Biotech) was performed using transfer buffer (Ambion). Every step was performed following the manufacturer's instructions. Digoxigenin (DIG)-labeled DNA probes were generated using the PCR DIG Probe synthesis kit (Roche). The oligonucleotide pairs MA101/MA102, NB101/NB102, and NC101/NC102 were used to generate probes for cupA1 (A1, 260 bp), cupB1 (B1, 330 bp), and cupC1 (C1, 339 bp), respectively. DNA probes for cupA2 to cupA5, cupB3, and cupC3 were also generated by using the oligonucleotide pairs NA2O1/NA2O2, NA3O1/NA3O2, NA4O1/NA4O2, NA5O1/NA5O2, NB3O1/NB3O2, and NC3O1/NC3O2, respectively. Finally, the DNA probes for PA4648 and PA4651 were generated by using the oligonucleotide pairs 4648O1/4648O2 and 4651O1/4651O2, respectively. Hybridization was performed overnight at 50°C using at least 5 ng of labeled probe per ml of DIG Easy Hyb buffer (Roche). Washing and blocking reagents came from the DIG Wash and Block buffer set. Blots were developed using Fab fragments from an anti DIG-alkaline phosphatase conjugate (Roche), with detection with CDP-star (Roche). Autoradiography was performed with X-Omat films (Kodak). Every step was performed according to the manufacturer's instructions.
Microarray analysis.
Five-hundred-microliter portions of bacterial cultures were treated with 100 μl of lysozyme (1 mg/ml), the samples were left for 15 min at room temperature, and total RNA was isolated by using the Qiagen RNeasy kit. RQ1 DNase (Promega) was used for DNase treatment, and RNAs were checked on 1% agarose gels (RNase free). Targets for microarrays were subsequently prepared according to the Affymetrix Expression Analysis Protocol Guide with few modifications, and RNA spike controls provided by the Cystic Fibrosis Foundation were included. Briefly, 12 μg of RNA was used and treated with 1 U of RNase inhibitor (Ambion) per μl during cDNA synthesis using Superscript II (Invitrogen). cDNA was purified by using a PCR purification kit (Qiagen), and cDNA fragmentation was performed by using 0.06 U of DNase I (Roche) per microgram of cDNA at 37°C for 10 min followed by 10 min at 100°C. The extent of cDNA fragmentation was verified on a 2% agarose gel, and then cDNA was end labeled with biotin ddUTP by using the Enzo BioArray Terminal Labeling kit. Target hybridization to GeneChip P. aeruginosa genome arrays (Affymetrix), washing, and scanning were performed using a hybridization oven, a Fluidics station, and a scanner obtained from Affymetrix. Hybridization intensity data were extracted from the scanned array images, and intrachip normalizations were performed using Affymetrix Microarray Suite 5.0 software. The data were subsequently analyzed and experimental comparisons were made using GeneSpring version 4.2.1 (Silicon Genetics).
Construction of P. aeruginosa mutants.
The cupA3 mutation was engineered in the mvaT mutant (PAO-P47) as described previously (38), yielding a mvaT cupA3 mutant termed PAO-MC. An mvaU chromosomal deletion mutant of PAO1 lacking 300 internal nucleotides was constructed as follows. Using PAO1 DNA as a template, the intact mvaU gene (PA2667) (354 bp) plus 528 bp of upstream flanking DNA and 526 bp of downstream flanking DNA was amplified by using the primer pair mvaUUF, containing a BamHI restriction site, and mvaUDR, containing a PstI restriction site. The resulting PCR product was digested with BamHI and PstI and cloned into similarly digested pUC18, resulting in plasmid pUCmvaU. To introduce a deletion of the recombinant mvaU gene, the primer pair mvaUUR and mvaUDF were used in conjunction with inverse PCR, using pUCmvaU DNA as a template. The resulting blunt-ended PCR product containing a 300-bp deletion in mvaU was self-ligated, resulting in plasmid pUCΔmvaU. The PCR product was excised from the vector by using BamHI and SphI and cloned into the vector pDM4 (19), digested with BglII and SphI, resulting in plasmid pDM4ΔmvaU. Allelic exchange using pDM4ΔmvaU contained in E. coli S17-1 λpir with PAO1 resulted in a P. aeruginosa strain (PAO-P46) containing an in-frame deletion of the mvaU gene. This deletion was confirmed by both PCR and Southern blot analysis (data not shown). The mvaU deletion mutant of the PAO1 lecA::lux strain was constructed by allelic exchange using pDM4ΔmvaU contained in E. coli S17-1 λpir with PAO1 lecA::lux, resulting in strain PAO-P48. The deletion was confirmed by PCR (data not shown).
Biofilm formation assay.
Biofilm formation on stainless steel was assayed as follows. Prior to inoculation, 10 ml of LB was seeded with fresh bacterial colonies and incubated overnight at 37°C. The turbidity of the cultures (600 nm) was adjusted to 1.0 (±0.1), and 100-μl aliquots were used to inoculate petri dishes containing three stainless steel coupons in 10 ml of 0.1× LB. Plates were incubated at 37°C on a rotary shaker (60 rpm) for 72 h.
Visualization and analysis of biofilm structure.
Stainless steel coupons were rinsed twice in sterile phosphate-buffered saline and air dried. They were then heat fixed, stained in 0.1% acridine orange for 2.5 min, and rinsed in phosphate-buffered saline. The coupons were then examined for bacterial attachment by using an inverted fluorescence microscope. Surface coverage was estimated as described previously (17) using the LUCIA image analysis program (version 4.11).
Time- and cell-density-dependent measurement of bioluminescence.
Bioluminescence was determined as a function of cell density using a combined automated luminometer-spectrometer (Anthos Labtech LUCYI) as described previously (42). Overnight cultures of P. aeruginosa were diluted 1:1,000 in fresh LB medium, and 0.2 ml was inoculated into microtiter plates. Luminescence and turbidity of the cultures at 495 nm (OD495) were automatically determined every 30 min (as relative light units per unit of OD495).
RESULTS
Expression of the cupA gene fusion in P. aeruginosa PAO1.
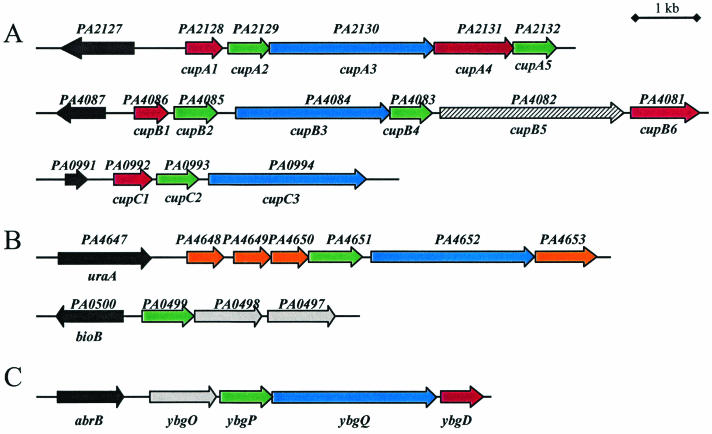
To investigate the activity of the cupA promoter region, two transcriptional fusions were constructed using various lengths of DNA located upstream from the cupA1 start codon. The cupA1 gene is the first gene of a cluster encoding components involved in fimbrial biogenesis via the chaperone-usher pathway (cupA1 to cupA5) (Fig. 1A). An open reading frame (ORF) (PA2127 in the PAO1 genome annotation) encodes a protein which shares 40% identity with the hypothetical E. coli ybdN gene product. It is located 850 bp upstream of cupA1 and transcribed in the reverse orientation. Fragments of 370 and 1,017 bp, the last one containing the whole PA2127-cupA1 intergenic region, were PCR amplified using the oligonucleotide pairs OFN65/OFN2 and OFN1/OFN2, respectively. The DNA fragments were cloned into the pMP220 vector, yielding the transcriptional fusions cupA1S-lacZ (S for short) and cupA1L-lacZ (L for large) carried on plasmids pMPFCAS and pMPFCAL, respectively. The fidelity of each cloned fragment was verified by DNA sequencing.
FIG. 1.
Organization of fimbrial gene clusters. Genes encoding putative fimbrial subunits are indicated in red, chaperones are indicated in green, and ushers are indicated in blue. Genes located upstream of the putative promoter region of the gene clusters are represented in black. (A) cupA, cupB, and cupC gene clusters from P. aeruginosa. The hatched cupB5 gene encodes a protein homologous to filamentous hemagglutinin. (B) Additional P. aeruginosa gene clusters containing fimbrial assembly genes. Genes represented in orange encode proteins containing the COG5430 domain. In grey are ybgO homologues that are found in an E. coli K-12 fimbrial gene cluster. (C) Fimbrial ybg gene cluster from E. coli K-12.
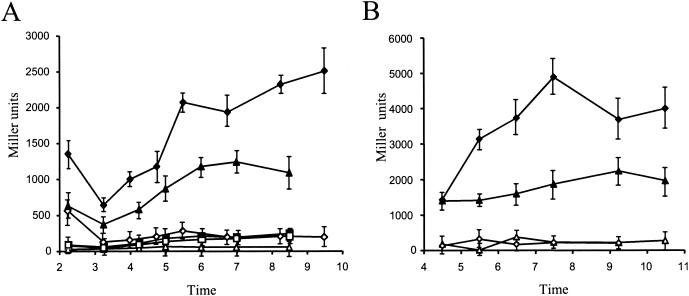
The recombinant plasmids were introduced into the P. aeruginosa wild-type strain PAO1. The recombinant strains were subsequently grown in LB medium, and β-galactosidase activity was measured at different time points over the growth period (Fig. 2A). Both transcriptional fusions exhibited similar β-galactosidase activity levels compared with that observed for the pMP220 control. The same comparative results were obtained when these strains were grown in minimal medium (M63) supplemented with glucose and Casamino Acids at different temperatures (30 or 37°C) in flask cultures or in microtiter plate wells (data not shown).
FIG. 2.
Expression of the cupA gene cluster. (A) The expression level of the cupA1S-lacZ (pMPFCAS) (diamonds) and cupA1L-lacZ (pMPFCAL) (triangles) transcriptional fusions from the wild-type PAO1 strain (open symbols) or the isogenic mvaT mutant (filled symbols) were compared to those obtained with the pMP220 cloning vector control (squares). (B) Expression of the cupA1S-lacZ (pMPFCAS) (diamonds) and cupA1L-lacZ (pMPFCAL) (triangles) transcriptional fusions from the mvaT mutant containing a plasmid encoding the mvaT gene (pMMBmvaT) (open symbols) or the cloning vector (pMMB190) (filled symbols). Each experiment was independently repeated three times, and error bars indicate standard deviations.
Isolation of P. aeruginosa mutants exhibiting detectable cupA gene expression.
Because of the low β-galactosidase activity originating from the cupA1 promoter-lacZ constructs in P. aeruginosa, we reasoned that cupA gene expression in the P. aeruginosa wild-type strain PAO1 was repressed in the conditions of our assay. The pMPFCAL plasmid, carrying the cupA1L-lacZ fusion, was thus introduced into a P. aeruginosa mutant collection generated by random insertion of a Tn5 modified transposon, Tn5G (38). The recombinant clones were plated on selective media containing X-Gal. Three clones that exhibited an intense blue color, revealing an increased level of β-galactosidase activity, were selected and further analyzed. The genomic DNA from these clones was isolated and subjected to inverse PCR using divergent oligonucleotides that match the 3′ end of the transposon, as previously described (38). Determination of the nucleotide sequence of the PCR products obtained revealed that, in all three clones, the transposon had inserted into a gene that was annotated as PA4315 on the P. aeruginosa genome website (www.pseudomonas.com) (34). This ORF encodes a protein that is 82% similar to the P16 subunit of the Pseudomonas mevalonii heteromeric transcriptional regulator MvaT (27). This gene has also recently been found to encode a novel regulator of virulence gene expression in P. aeruginosa, which is termed MvaT (8).
Examination of the P. aeruginosa PAO1 genome database revealed the presence of a second MvaT P16 subunit homologue (PA2667). It is 64% similar to that of P. mevalonii (it is also 51% identical and 68% similar to PA4315). We have therefore designated PA4315 as mvaT and PA2667 as mvaU. Further analysis of the genome databases indicates that MvaT P16 subunit homologues, with sequence conservation spread evenly throughout the proteins, are highly conserved in the Pseudomonas genus genomes, including P. putida, P. syringae, P. fluorescens, P. aeruginosa, P. mevalonii, and the closely related Azotobacter vinelandii. Recently, these MvaT proteins have been described as a novel class of H-NS-like proteins. Indeed, even if they show no significant homology with any known H-NS protein at the sequence level, MvaT proteins are predicted to share the same three-dimensional structure as the E. coli H-NS and seem to be functionally related (35).
MvaT specifically down-regulates the cupA1 promoter.
The plasmid pMPFCAL was introduced into a PAO1 mvaT deletion mutant (PAO-P47), and the β-galactosidase activity was determined to confirm the influence of MvaT on gene expression from the cupA1 promoter. In these conditions, the level of β-galactosidase was increased up to sixfold when compared with the expression level observed in the wild-type strain PAO1 (Fig. 2A). Furthermore, there is a reproducible difference in the activation of gene expression when the two cupA transcriptional fusions are considered. That containing the shorter region (cupA1S-lacZ) is twofold up-regulated when compared with the larger fusion (cupA1L-lacZ), which encompasses an additional 550-bp DNA region (Fig. 2A). This suggests that an additional, as yet uncharacterized, cis-controlling element might exist within the upstream DNA fragment of the larger fusion. To verify that the increase in β-galactosidase activity from the cupA-lacZ transcriptional fusion was a direct consequence of the absence of MvaT, the mvaT gene was cloned into the broad-host-range pMMB190, yielding pMMBmvaT. This recombinant plasmid was introduced into PAO-P47 (mvaT) containing pMPFCAL or pMPFCAS to complement the mvaT mutation. Interestingly, the level of β-galactosidase was strongly reduced in the mvaT mutant carrying pMMBmvaT whereas it remained high when the plasmid control pMMB190 was used (Fig. 2B). This observation confirmed that expression from the cupA1 promoter is repressed when MvaT is present.
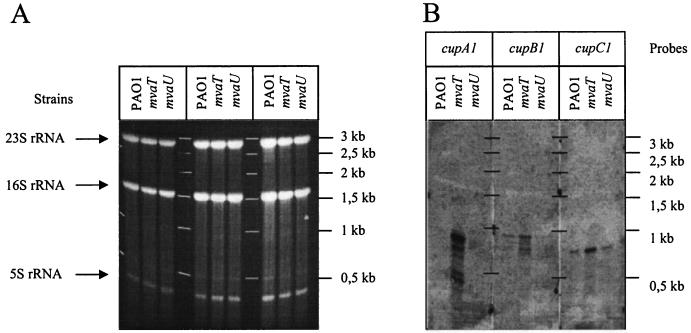
The influence of MvaT on cupA gene cluster expression was also analyzed by Northern blot hybridization in order to evaluate which of the cupA genes could be transcribed from the cupA1 promoter. RNAs were extracted from PAO1 and PAO-P47 (mvaT) as described in Materials and Methods. RNA samples were run on a 1% agarose gel and transferred onto a nylon membrane. DNA probes, which match with each of the cupA genes (Fig. 1A and Table 2), were engineered using the PCR DIG Probe synthesis kit (Roche) and used for hybridizing the membrane. The position of the transcripts was visualized by using anti-DIG antibodies and autoradiography (Fig. 3). Using the cupA1 probe, we clearly detected a transcript of approximately 780 nucleotides, slightly longer than the full-length cupA1 gene (551 bp), in the mvaT mutant but not in the wild-type strain. This observation confirmed that transcription from the cupA1 promoter could not clearly be detected in the wild-type background but that a deletion of the gene encoding the MvaT transcriptional regulator considerably enhanced transcription of the cupA1 gene. Moreover, the size of the mRNA indicates that the cupA1 gene is monocistronic or that its transcript is rapidly processed from a larger precursor. Transcripts corresponding to the cupA2 to cupA5 cluster region could not be detected (data not shown). This might be due to a much lower level of transcription of these genes or mRNA instability compared to cupA1. Finally, using a pilA control probe, we could detect no variation in the transcript level of this gene between the PAO1 wild-type strain and the mvaT mutant (data not shown).
FIG. 3.
Northern blot analysis. (A) Total RNA was extracted from various P. aeruginosa strains, including PAO1 and the mvaT (PAO-P47) and mvaU (PAO-P46) isogenic mutants, and loaded on a 1% agarose gel. The positions of the major bands corresponding to the 23S, 16S, and 5S rRNA are indicated on the left, whereas the molecular size standards are indicated on the right. (B) After electrophoresis the RNA was transferred onto nitrocellulose and hybridized with one of various probes, including cupA1, cupB, and cupC1 probes. Each experiment was independently repeated up to three times with no significant variation observed.
mvaT mutants exhibit enhanced biofilm formation.
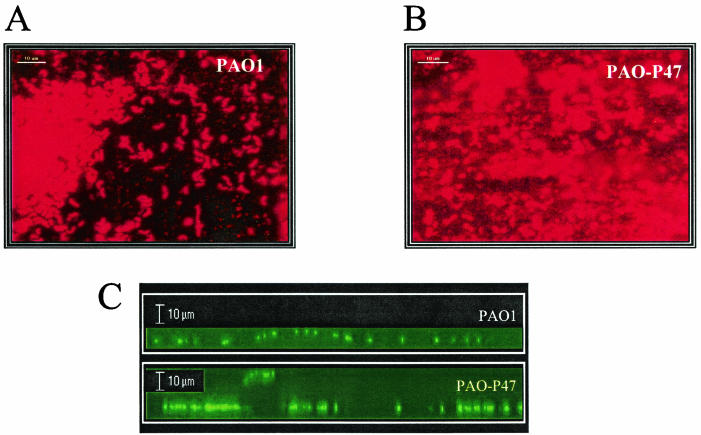
Since the cupA gene cluster is required for biofilm formation on abiotic surfaces and as MvaT negatively regulates cupA gene cluster expression, we examined the biofilm phenotype of the mvaT mutant on stainless steel surfaces. The surface coverage of the biofilms formed by each P. aeruginosa strain on stainless steel coupons in static biofilm experiments after 72 h at 37°C was analyzed (see Materials and Methods). The data were obtained from 60 independent measurements of biofilms stained with 0.1% (wt/vol) acridine orange and revealed a mean twofold increase in surface coverage for the mvaT mutant PAO-47 compared with the wild type (data not shown). Figure 4 illustrates the enhanced biofilm depth and surface coverage of the mvaT mutant as compared with the wild-type strain. We also used another technique to quantify bacterial attachment to polystyrene surfaces, a crystal violet-based assay (23), and similarly PAO-P47 showed enhanced biofilm formation (data not shown). These data are consistent with a role for mvaT as a negative regulator of cupA gene expression since these genes have previously been shown to be involved in initial attachment (38). Moreover, on stainless steel, we observed a significant increase in biofilm depth for PAO-P47 as compared with the wild-type strain PAO1. This increase is revealed by the large augmentation of microcolony size, as shown in Fig. 4C.
FIG. 4.
Biofilm formation by P. aeruginosa PAO1 and PAO-P47 (mvaT mutant) on stainless steel. (A and B) Characteristics of the biofilms formed by PAO1 and PAO-P47, respectively. (C) Depth of biofilms formed by PAO1 and PAO-P47 in a static model as determined using confocal laser scanning microscopy.
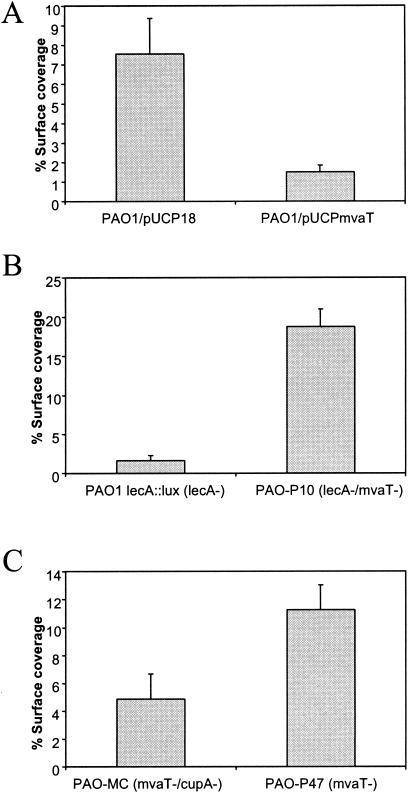
To supplement these data, we tested the influence of mvaT with respect to surface coverage on stainless steel in different genetic backgrounds. For example, overexpression of mvaT from pUCPmvaT in a wild-type strain of P. aeruginosa resulted in reduced surface coverage (Fig. 5A), which could be a result of down-regulation of a gene such as cupA1 or lecA (8) (Fig. 2). However, surface coverage of a mvaT lecA double mutant, PAO-P10 (8), was increased as compared to the lecA single mutant (PAO1 lecA::lux) (Fig. 5B). This indicates that adhesive structures encoded by genes different from lecA, but also overexpressed in the mvaT background, may be responsible for the increased biofilm phenotype. Finally, surface coverage of a mvaT cupA3 double mutant (PAO-MC) was decreased as compared to the mvaT single mutant (PAO-P47) (Fig. 5C), suggesting that in the mvaT background, the CupA system strongly contributes to the enhanced biofilm phenotype.
FIG. 5.
Quantitation of biofilm formation by P. aeruginosa on stainless steel. (A) Influence of overexpression of mvaT from pUCPmvaT in the wild-type PAO1 strain. (B) Influence of the mvaT mutation within a lecA genetic background (PAO1-P10). PAO lecA::lux is the parental lecA mutant. (C) Influence of the cupA3 mutation within a mvaT-negative background (PAO-MC). PAO-P47 is the parental mvaT mutant.
Influence of MvaT on cupB and cupC gene expression.
Two additional gene clusters encoding components of the chaperone-usher pathway have previously been identified in P. aeruginosa (5, 38). These gene clusters, termed cupB1 to cupB6 (PA4086 to PA4081) and cupC1 to cupC3 (PA0992 to PA0994) (Fig. 1A), were examined with respect to their expression in a P. aeruginosa wild-type or mvaT-negative genetic background.
We analyzed the transcription of cupB and cupC gene clusters using Northern blot hybridization. DNA probes were designed for two of the cupB and cupC genes (cupB1 and -3 and cupC1 and -3), encoding, respectively, the predicted major pilin subunit and the usher for each system (Fig. 1). In contrast to what we observed with the cupA gene cluster, transcripts could be detected in the wild-type PAO1 strain with B1 and C1 probes (Fig. 3). Interestingly, the RNA levels appeared to be increased by two- to threefold in both cases in PAO-P47 (mvaT) (Fig. 3). These results suggest that cupB1 and cupC1 gene expression might also be controlled in an MvaT-dependent manner. The sizes of the transcripts detected with B1 and C1 are approximately 885 and 730 nucleotides, respectively (Fig. 3). For information, the length of the cupB1 and cupC1 genes is 569 and 617 bp, respectively. Similarly as with the cupA gene cluster, it seems that only transcripts corresponding to the first gene of the cluster could be detected, since no RNA could be detected using either B3 or C3 probes in an mvaT or wild-type genetic background (data not shown).
Analysis of cup gene expression in an mvaT genetic background using DNA microarrays.
We examined the range of genes controlled by MvaT, because there was a reasonable probability that in addition to a set of gene clusters (cup genes) it may exert a global influence on other genes involved in bacterial attachment and biofilm formation. The microarray used (Affymetrix GeneChip P. aeruginosa genome array), contains probes representing the whole PAO1 genome. We were therefore able to compare global gene expression of the parental PAO1 strain and its isogenic mvaT mutant. Bacterial strains were grown at 37°C to OD600 of 3 in LB medium. We chose these conditions for the analysis since they were those in which we observed a large change in cupA1 gene expression with lacZ transcriptional fusions as well as Northern blot studies. Two independent microarray experiments were performed, and the data obtained were merged and quantified by using the Microarray Suite (Affymetrix) and GeneSpring (Silicon Genetics) softwares. Strikingly, cupA1 gene (PA2128) expression was increased 15-fold in the mvaT mutant, which is the most drastic change observed in gene expression in this analysis, whereas other cupA genes presented a 4.12-fold (cupA2) or 2.96-fold (cupA3) up-regulation (Table 3). These values are in agreement with the previous results obtained by Northern blot analysis indicating that only cupA1 transcripts are clearly detectable in an mvaT background and thus could be considered as good internal controls to validate the variation in expression of other genes. The up-regulation of cupB and cupC gene expression was also confirmed by the microarray analysis. Specifically, cupB1 and cupC1 transcripts had a 2.43- and 2.9-fold increase, respectively, in the mvaT mutant (Table 3). Based on the microarray analysis, 156 genes showed at least a twofold change in their mRNA levels in the mvaT mutant compared to the wild-type (see Tables S4 and S5 in the supplemental material). Among these, 104 genes showed a twofold or greater increase in mRNA levels in the mvaT mutant (see Table S4 in supplemental material). Interestingly, one of the genes (annotated PA4651) that shows the highest level of differential expression (5.1-fold) encodes a probable pilus assembly chaperone (Table 3). This gene belongs to a larger cluster in which a gene encoding an usher homologue could be found (PA4652) but no genes encoding fimbrial subunits were identified (Fig. 1B). However, most of the genes in the cluster (PA4648, -4649, -4650, and -4653) are at least threefold up-regulated in the mvaT mutant (Table 3). Using Northern blot analysis and DNA probes directed against PA4648 and PA4651, we observed higher transcript levels corresponding to these genes in the mvaT mutant than in the wild-type strain (data not shown), thus confirming the microarray data. Four of the genes (PA4648, PA4649, PA4650, and PA4653) encode proteins with similarities to protein U, which is found at the surface of the Myxococcus xanthus spore (10). Moreover, another gene, PA0498, was identified with a 3.13-fold up-regulation (Table 3). This ORF is found clustered with PA0499 and PA0497, with PA0499 encoding a probable pilus assembly chaperone (Fig. 1B). PA0497 and PA0498 encode highly homologous proteins that are 38% similar to YbgO from E. coli (Fig. 1C). Strikingly, the ybgO gene belongs to a cluster containing genes coding for a chaperone (YbgP), an usher (YbgQ), and a fimbrial subunit (YbgD) (3). The fold change in expression of all cup-related genes in the mvaT mutant is reported in Table 3. Another interesting genetic locus up-regulated in mvaT (see Table S4 in the supplemental material) contains the genes PA4306 to PA4297, which encode components reported in Actinobacillus actinomycetemcomitans as being important for tight adherence (Tad) (15). Finally, the lecA gene, encoding the PA-IL lectin, has previously been described as an MvaT target (8). We found a 4.02-fold up-regulation of lecA in the mvaT mutant (see Table S4 in the supplemental material), confirming our previous results (8) and again validating the overall data obtained with the microarray.
TABLE 3.
cup-related genes that are up-regulated in the mvaT mutanta
| PA No. | Gene | Gene product characteristic | Fold change |
|---|---|---|---|
| PA2128 | cupA1 | Fimbrial subunit | 15.920 |
| PA2129 | cupA2 | Pillus assembly chaperone | 4.120 |
| PA2130 | cupA3 | Usher protein | 2.960 |
| PA2131 | cupA4 | Putative adhesin subunit | 1.830 |
| PA2132 | cupA5 | Pilus assembly chaperone | 1.485 |
| PA4086 | cupB1 | Fimbrial subunit | 2.430 |
| PA4085 | cupB2 | Pilus assembly chaperone | 2.400 |
| PA4084 | cupB3 | Usher protein | 0.739 |
| PA4083 | cupB4 | Pilus assembly chaperone | 1.519 |
| PA4081 | cupB6 | Putative adhesin subunit | 1.728 |
| PA0992 | cupC1 | Fimbrial subunit | 2.900 |
| PA0993 | cupC2 | Pilus assembly chaperone | 1.950 |
| PA0994 | cupC3 | Usher protein | 1.595 |
| PA4648 | Hypothetical protein (COG5430) | 5.360 | |
| PA4649 | Hypothetical protein (COG5430) | 3.700 | |
| PA4650 | Hypothetical protein (COG5430) | 6.580 | |
| PA4651 | Pilus assembly chaperone | 5.100 | |
| PA4652 | Usher protein | 2.560 | |
| PA4653 | Hypothetical protein (COG5430) | 3.480 | |
| PA0498 | Hypothetical protein-YbgO homolog | 3.130 |
Genes with a change above twofold are indicated in bold.
MvaU does not influence cup gene expression or biofilm surface coverage.
The gene annotated PA2667 encodes a second P. aeruginosa regulator belonging to the MvaT family (MvaU). To determine whether mvaU is also involved in the regulation of cupA gene expression and biofilm formation, a P. aeruginosa mvaU deletion mutant was constructed (PAO-P46). Interestingly, when the cupAL-lacZ transcriptional fusion was introduced into this strain, no increase in β-galactosidase activity was observed (data not shown). Similar results were obtained with the cupAS-lacZ transcriptional fusion (data not shown). In addition, Northern blot analysis revealed that no cupA1 transcript could be detected in the mvaU mutant, in contrast to that observed for the mvaT mutant (Fig. 3). The two- to threefold increase in cupB1 and cupC1 mRNA levels observed in the mvaT mutant in Northern blot analysis was also absent in the mvaU mutant. These results indicate that the repression exerted on the cupA1 promoter, and to a lesser extent on the cupB1 and cupC1 promoters, is MvaT dependent and does not appear to involve any cross-talk with the MvaU protein. Furthermore, analysis of surface coverage on stainless steel coupons indicates that there is no significant difference between the mvaU mutant (PAO-P46) biofilm and that of the PAO1 wild type (data not shown). However, MvaT and MvaU may share a number of targets, as is the case for lecA, the gene encoding the PA-IL lectin. Indeed, we showed that in both the mvaT and mvaU mutants, the expression of lecA is enhanced (Fig. 6).
FIG. 6.
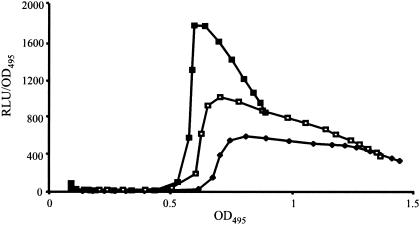
Mutation in mvaU derepresses lecA. Influence of mvaT and mvaU on the expression of a P. aeruginosa lecA::lux chromosomal fusion (PAO1 lecA::lux). Lux-related bioluminescence in PAO1 lecA::lux (closed diamonds), a PAO1 lecA::lux-containing mvaT mutant (PAO-P10) (closed squares), and a PAO1 lecA::lux-containing mvaU mutant (PAO-P48) (open squares) was measured as a function of light output per cell. Relative light units and OD495 were determined as described in the Materials and Methods.
DISCUSSION
Biofilm formation in a variety of model systems has been used to understand many intriguing aspects of microbial life in communities within a particular niche such as the environment or an infected host. In recent years, genetic approaches have facilitated characterization of the molecular mechanisms involved in this process (21). We have also used genetic approaches to isolate mutants of P. aeruginosa affected in biofilm formation, and this led to the identification of cup genes, encoding components of a so-called chaperone-usher pathway, that play an important role at certain stages of biofilm formation (38). In this study, we (i) investigated the expression of the P. aeruginosa cup gene clusters; (ii) uncovered the role of MvaT, a transcriptional regulator belonging to a family of proteins which so far have been found only in the pseudomonads, but which could be functionally related to the H-NS proteins (8, 35); and (iii) established the contribution of MvaT to biofilm formation.
When compared with the parental wild-type strain, a P. aeruginosa cupA mutant was unable to attach to the wells of polystyrene microtiter plates when grown at 30 or 37°C in minimal medium supplemented with Casamino Acids (38). This observation suggested that under these growth conditions, in which a distinct CupA-dependent phenotype is apparent, the cupA genes must be expressed at a significant level. However, using different lacZ transcriptional fusions to the cupA1 promoter region, we were unable to detect high β-galactosidase activity during the growth of P. aeruginosa in microtiter plate wells or in flasks in either minimal or rich medium at 30 or 37°C. One obvious explanation is that the cupA genes are only poorly expressed. Nevertheless, it is a possibility that certain adhesins, such as those related to the cupA gene cluster, are only transiently expressed, i.e. not before bacterial cells reach, or contact, a surface and not after the biofilm reaches a certain level of maturation. Involvement of regulatory circuits, such as the CpxRA pathway, in biofilm formation following surface contact has already been proposed (24). Moreover, it is noteworthy that the cupA1 gene (PA2128) is down-regulated once the biofilm established (41). It is thus a possibility that only those cells which reach the wall of the microtiter plate wells will support strong induction of cup genes, whereas most of the planktonic bacterial culture, or the cells embedded within the biofilm structure, will keep a low level of cup gene expression. To determine whether cupA expression is repressed at some stages via a regulatory element, we introduced a cupA1 promoter fusion into a library of random P. aeruginosa Tn5 mutants and screened for strains in which cupA1 expression was up-regulated. Three distinct mutants were obtained, each of which contained a Tn5 insertion in the same gene, mvaT. This gene encodes a homologue of the P16 subunit of the P. mevalonii transcriptional regulator MvaT (27).
MvaT was originally identified as a regulator of the P. mevalonii operon mvaAB, which encodes two enzymes required for mevalonate catabolism, (27). MvaT was shown to bind to a region located at positions −48 to −84 upstream from the mvaA translational start codon (39). Analysis of the cupA1 promoter regions, as well as those of cupB1, cupC1, and lecA, analyzed since they also appeared to be down-regulated by MvaT, failed to identify any significant regions of homology with the mvaA cis-acting element. Moreover, no obviously identifiable repeats or conserved DNA domains could be detected when these promoter regions were compared. Finally, and because of the lack of consensus sequences, it is possible that the control exerted by MvaT on its various targets, including cupA1, might be direct or indirect.
Interestingly, the cup gene clusters are similar to the fimbrial gene clusters of other organisms in that they contain genes encoding an usher, a chaperone, and one or more fimbrial subunits. Strikingly, Northern blot analysis showed that in all three cases, only a transcript with a size corresponding to that of the first gene of each cluster, cupA1, cupB1, and cupC1, could clearly be identified. It was previously shown that with the E. coli gene clusters encoding the Pap or F1845 fimbriae, a polycistronic precursor mRNA is degraded to a relatively stable monocistronic transcript. Such a transcript exclusively contains the gene encoding the major fimbrial subunit, papA or daaE, respectively (1, 2). These observations strongly suggest that CupA1, CupB1, and CupC1 are major subunits of fimbrial structures assembled by a chaperone-usher pathway. Remarkably, in E. coli, the H-NS protein is known to be involved in regulating the expression of genes responsible for the assembly of the Pap pili and the type 1 pili, the best-studied of the chaperone-usher pathways. The assertion by Tendeng and collaborators (35) that MvaT proteins and H-NS proteins are functionally linked is thus reinforced by our observations, which indicate that P. aeruginosa MvaT and E. coli H-NS are both involved in fimbrial gene expression control.
In P. aeruginosa, biofilm formation is strongly dependent on the expression of extracellular appendages such as flagella, type IV pili, or the putative fimbrial structures encoded by the cupA genes (22, 38). Given the inability of cupA mutants to attach to an abiotic surface and the negative effect exerted through MvaT on cupA expression, mvaT mutants would be predicted to exhibit enhanced adhesion properties. Consistent with this prediction, we have shown that the biofilm produced on stainless steel coupons by the P. aeruginosa mvaT mutant shows greater surface coverage and depth than that produced by the wild type. However, MvaT may control the expression of other yet uncharacterized genes and some may also contribute to the enhanced biofilm-forming capacity of the mvaT mutant. Transcriptional profiling of the mvaT mutant clearly showed that the absence of this regulatory factor can indeed cause both an increase and a decrease in the levels of mRNA for a substantial number of genes (see Tables S4 and S5 in the supplemental material). In addition to the already known cup genes, we identified novel, but incomplete, clusters containing genes encoding a chaperone and an usher protein homologue (Fig. 1B and Table 3), which were also dependent on MvaT. A gene cluster containing PA4651 and PA4652, the chaperone- and usher-encoding genes, respectively, was found up-regulated in the mvaT background. Intriguingly, no gene encoding a protein with significant similarities to fimbrial subunits could be identified in this cluster. However, the four other ORFs (PA4648, PA4649, PA4650, and PA4653) encode proteins that contain a characteristic domain found in the protein U precursor of M. xanthus (31). This protein is produced as a precursor with a canonical signal peptide and is assembled on the spore surface after crossing the membrane. The protein U domain, called SCPU (pfam05229), is included in a larger conserved domain identified as COG5430. The COG5430 domain could represent a novel type of putative pilin domain, and the PA4648-PA4653 gene cluster could be involved in the assembly of fimbrial structures according to the chaperone-usher model. A similar gene cluster has recently been identified in Acinetobacter baumanii and has been shown to be involved in biofilm formation by this organism (37). A second but incomplete fimbrial assembly system was found to be up-regulated in the mvaT mutant. In this cluster no usher-encoding gene could be identified but only an ORF encoding a putative chaperone, PA0499. The two other ORFs of the cluster, PA0497 and PA0498, encode signal peptide-containing proteins similar to the secretory protein YbgO (Fig. 1B). In E. coli, the ybgO gene is found clustered with genes encoding chaperone-usher components (3) (Fig. 1C). Whether the putative structures assembled by these novel systems are important for adhesion under certain conditions, as is the case for the CupB and CupC systems, remains to be determined.
We also report in this study the existence of a second MvaT-like regulator, MvaU. We could show that in common with MvaT, MvaU is a negative regulator of lecA gene expression, encoding the PA-IL lectin, which has been reported to function as an adhesin (40), but in contrast has no influence on cup gene expression. This indicates that the MvaT-dependent expression of the cupA, cupB, and cupC genes is strictly dependent on MvaT. Intriguingly, microarray data revealed that mvaU is up-regulated in an mvaT background (see Table S4 in the supplemental material). Yet, in this context, increased MvaU levels do not seem sufficient to prevent up-regulation of lecA in response to the lack of a functional MvaT protein. This observation may confirm that both regulators, even though homologous, are not exchangeable and the relationship, if any, might be complex. To help with understanding these data, it should be noted that the repression effect on lecA due to MvaT is nearly twice that due to MvaU (Fig. 6). Finally, because the mvaU mutant biofilm exhibited no clear increased surface coverage, we concluded that in the P. aeruginosa mvaT mutant, the up-regulation of lecA is not sufficient to unmask the enhanced biofilm phenotype of the mvaT strain. In addition, the up-regulation of the cupA genes in the mvaT mutant might thus strongly contribute to the enhancement of biofilm formation in this strain. To support this suggestion, we introduced a cupA mutation in the mvaT mutant, which resulted in decreased bacterial surface coverage (Fig. 5C). In contrast, introduction of the mvaT mutation in a lecA mutant still increased bacterial surface coverage (Fig. 5B). In light of these observations, it is possible that the products of the cupA genes play an important role in the initial colonization of surfaces, whereas the lecA- encoded protein contributes to cell-cell interaction or biofilm maturation at a later stage.
In conclusion, MvaT in P. aeruginosa is likely to be an important component of a complex regulatory network involved in biofilm formation. In future work, we will try to determine which conditions affect the repression of the cup genes and how this relates to the biofilm developmental cycle. Furthermore, it will also be necessary to define in detail the nature of the MvaT regulon and identify its target DNA binding sites in P. aeruginosa. For this purpose, we will purify MvaT in order to perform DNA band shift assays of the putative target genes. Finally, it is challenging to believe that understanding the function of MvaT and its contribution to the lifestyle of P. aeruginosa may uncover new opportunities for controlling biofilm-associated infections.
Supplementary Material
Acknowledgments
We thank the P. aeruginosa sequencing consortium for providing updated annotations at http://www.pseudomonas.com. We are grateful to Meghan Gilmore and Andy Goodman for their assistance in the use of microarrays and in data analysis.
We thank Cystic Fibrosis Foundation Therapeutics, Inc., for subsidizing the GeneChip P. aeruginosa genome arrays. Research in the Marseilles laboratory was supported by EU grant QLK2 CT2001 01339 and a grant from the French Cystic Fibrosis Foundation (Vaincre La Mucoviscidose). Work in the Nottingham laboratory was supported by a research studentship from the Biotechnology and Biological Sciences Research Council, United Kingdom (S.P.D.), and by a Medical Research Council Co-operative Group core grant (G9800920), which is gratefully acknowledged. Research in the laboratory of Stephen Lory is supported by Public Health Service grant AI21451 from the National Institutes of Health and by several grants from the U.S. Cystic Fibrosis Foundation.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baga, M., M. Göransson, S. Normark, and B. E. Uhlin. 1988. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. EMBO J. 52:197-206. [DOI] [PubMed] [Google Scholar]
- 2.Bilge, S. S., J. M. Apostol, Jr., M. A. Aldape, and S. L. Moseley. 1993. mRNA processing independent of RNase III and RNase E in the expression of the F1845 fimbrial adhesin of Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 5.Croft, L., S. A. Beatson, C. B. Whitchurch, B. Huang, R. L. Blakeley, and J. S. Mattick. 2000. An interactive web-based Pseudomonas aeruginosa genome database: discovery of new genes, pathways and structures. Microbiology 146:2351-2364. [DOI] [PubMed] [Google Scholar]
- 6.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 8.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Camara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gollop, R., M. Inouye, and S. Inouye. 1991. Protein U, a late developmental spore coat protein of Myxococcus xanthus, is a secretory protein. J. Bacteriol. 173:3567-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heydorn, A., B. Ersboll, J. Kato, M. Hentzer, M. R. Parsek, T. Tolker-Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoiby, N., H. Krogh Johansen, C. Moser, Z. Song, O. Ciofu, and A. Kharazmi. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 3:23-35. [DOI] [PubMed] [Google Scholar]
- 14.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 15.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, and D. H. Figurski. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9:429-437. [DOI] [PubMed] [Google Scholar]
- 16.Kjelleberg, S., and S. Molin. 2002. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol. 5:254-258. [DOI] [PubMed] [Google Scholar]
- 17.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 18.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 19.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 21.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 22.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 23.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 24.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigue, A., Y. Quentin, A. Lazdunski, V. Mejean, and M. Foglino. 2000. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 8:498-504. [DOI] [PubMed] [Google Scholar]
- 27.Rosenthal, R. S., and V. W. Rodwell. 1998. Purification and characterization of the heteromeric transcriptional activator MvaT of the Pseudomonas mevalonii mvaAB operon. Protein Sci. 7:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Schweizer, H. P. 1991. Improved broad host range lac-based plasmid vectors for the isolation and characterization of protein fusions in Pseudomonas aeruginosa. Gene 103:87-92. [DOI] [PubMed] [Google Scholar]
- 30.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 31.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 32.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spaink, P. H., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of Rhizobium leguminosarum Sym plasmid pRJL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 34.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 35.Tendeng, C., E. Krin, O. A. Soutourina, A. Marin, A. Danchin, and P. N. Bertin. 2003. A Novel H-NS-like protein from an antarctic psychrophilic bacterium reveals a crucial role for the N-terminal domain in thermal stability. J. Biol. Chem. 278:18754-18760. [DOI] [PubMed] [Google Scholar]
- 36.Thanassi, D. G., E. T. Saulino, and S. J. Hultgren. 1998. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1:223-231. [DOI] [PubMed] [Google Scholar]
- 37.Tomaras, A. P., C. W. Dorsey, R. E. Edelmann, and L. A. Actis. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473-3484. [DOI] [PubMed] [Google Scholar]
- 38.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Y. L., and V. W. Rodwell. 1991. The cis-acting regulatory element of the mvaAB operon of Pseudomonas mevalonii. J. Bacteriol. 173:3803-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wentworth, J. S., F. E. Austin, N. C. Garber, N. Gilboa-Garber, C. A. Paterson, and R. J. Doyle. 1991. Cytoplasmic lectins contribute to the adhesion of Pseudomonas aeruginosa. Biofouling 4:99-104. [Google Scholar]
- 41.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 42.Winzer, K., C. Falconer, N. C. Garber, S. P. Diggle, M. Camara, and P. Williams. 2000. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 182:6401-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.