Abstract
More than 2 billion people suffer from iron (Fe) deficiency, and developing crop cultivars with an increased concentration of micronutrients (biofortification) can address this problem. In this review, we describe seven transgenic approaches, and combinations thereof, that can be used to increase the concentration of Fe in rice seeds. The first approach is to enhance the Fe storage capacity of grains through expression of the Fe storage protein ferritin under the control of endosperm-specific promoters. Using this approach, the concentration of Fe in the seeds of transformants was increased by approximately 2-fold in polished seeds. The second approach is to enhance Fe translocation by overproducing the natural metal chelator nicotianamine; using this approach, the Fe concentration was increased by up to 3-fold in polished seeds. The third approach is to enhance Fe influx to the endosperm by expressing the Fe(II)-nicotianamine transporter gene OsYSL2 under the control of an endosperm-specific promoter and sucrose transporter promoter, which increased the Fe concentration by up to 4-fold in polished seeds. The fourth approach is introduction of the barley mugineic acid synthesis gene IDS3 to enhance Fe uptake and translocation within plants, which resulted in a 1.4-fold increase in the Fe concentration in polished seeds during field cultivation. In addition to the above approaches, Fe-biofortified rice was produced using a combination of the first, second, and third approaches. The Fe concentration in greenhouse-grown T2 polished seeds was 6-fold higher and that in paddy field-grown T3 polished seeds was 4.4-fold higher than in non-transgenic seeds without any reduction in yield. When the first and fourth approaches were combined, the Fe concentration was greater than that achieved by introducing only the ferritin gene, and Fe-deficiency tolerance was observed. With respect to Fe biofortification, the introduction of multiple Fe homeostasis genes is more effective than the introduction of individual genes. Moreover, three additional approaches, i.e., overexpression of the Fe transporter gene OsIRT1 or OsYSL15, overexpression of the Fe deficiency-inducible bHLH transcription factor OsIRO2, and knockdown of the vacuolar Fe transporter gene OsVIT1 or OsVIT2, may be useful to further increase the Fe concentration of seeds.
Keywords: Biofortification, Iron, Zinc, Transgenic rice, Nicotianamine, YSL, Ferritin, IDS3, Mugineic acids, Anemia
Introduction
Iron (Fe) is an essential micronutrient for most organisms, including all plants and animals. Fe deficiency is one of the most prevalent micronutrient deficiencies globally, affecting an estimated two billion people (Stoltzfus et al. 1998) and causing 0.8 million deaths annually worldwide (WHO 2002). Fe deficiency is ranked sixth among the risk factors for death and disability in developing countries with high mortality rates (WHO 2002).
There are three basic approaches to alleviate micronutrient deficiencies: micronutrient supplementation, food fortification, and biofortification. Traditional public health interventions, including nutritional supplementation and industrial food fortification programs, have reduced micronutrient deficiencies worldwide. However, these interventions require infrastructure to produce micronutrient supplements or food fortifications, as well as purchasing power or access to markets and health care systems for their success, which are often not available to individuals living in remote rural areas (Mayer et al. 2008). These projects required continuous costs and are difficult to perform in developing countries. In contrast, biofortification (i.e., increasing the bioavailable concentration of essential elements in edible portions of crop plants through conventional breeding or genetic engineering) does not require specific processing after harvesting or a dedicated infrastructure (Grusak and DellaPenna 1999; Mayer et al. 2008). Therefore, biofortification is advantageous for both individuals and governments, as it is inexpensive and sustainable (Mayer et al. 2008). This can complement current efforts of nutritional supplementation and food fortification to address micronutrient deficiencies. Moreover, biofortification is beneficial for individuals who find it difficult to change their dietary habits owing to financial, cultural, regional or religious restrictions.
Rice is a particularly suitable target for biofortification because Fe-deficiency anemia is a serious problem in developing countries where rice is a major staple crop (WHO 2002; Juliano 1993). Brown rice is rich in mineral value. However, rice is mostly consumed in the polished form (the endosperm tissue), which contains low mineral levels (Grusak and Cakmak 2005). Biofortification can be applied to increase the Fe concentrations in polished seeds and achieve the target Fe demand for human nutrition.
Among the methods available for Fe biofortification of rice, transgenic methods can most efficiently increase Fe concentration in rice seeds. In this report, we describe seven recently reported transgenic approaches used to increase the Fe concentration of rice seeds (Figure 1, Table 1), and we propose some additional prospective target genes for the Fe biofortification of rice.
Figure 1.
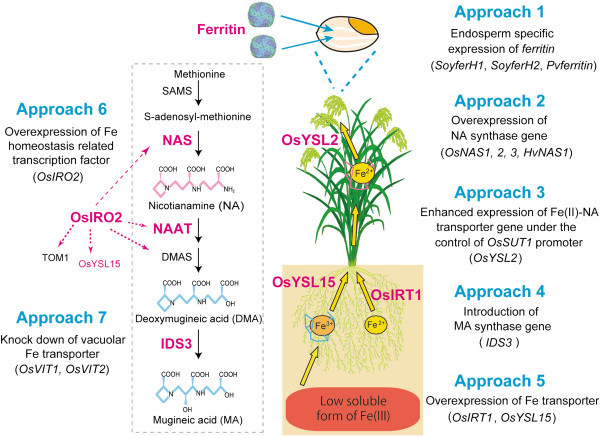
Seven transgenic approaches to Fe biofortification of rice. The pathway inside the gray dashed-line rectangle shows the biosynthetic pathway for mugineic acid family phytosiderophores (MAs) in graminaceous plants. SAMS, S-Adenosyl-methionine synthase; NAS, NA synthase; NAAT, NA aminotransferase; DMA, 2′-deoxymugineic acid; DMAS, DMA synthase; IDS3, MA synthase (dioxygenase that catalyzes the hydroxylation of DMA and epiHDMA at the 2′ position); Ferritin, iron storage protein; OsYSL2, Fe(II)-NA and Mn(II)-NA transporter; OsIRO2, Fe deficiency-inducible bHLH transcription factor related to Fe homeostasis in rice; OsIRT1, ferric transporter; OsYSL15, Fe(III)-DMA transporter; TOM1, MA transporter. Rice lacks the two dioxygenase genes (IDS2 and IDS3) and secretes only DMA. Approach 1: Enhancing Fe accumulation in seeds by introducing the Fe storage protein, ferritin gene, SoyferH1, SoyferH2 or Pvferritin, under the control of endosperm-specific promoters. Approach 2: Enhancing Fe transport within the plant body by the overexpression of NAS. Approach 3: Enhancing Fe influx to seeds by expression of the Fe(II)-NA transporter gene OsYSL2 under the control of the OsSUT1 promoter. Approach 4: Enhancing Fe uptake and translocation by introduction of the phytosiderophore synthase gene IDS3. Approach 5: Enhanced Fe uptake from soil by overexpression of the Fe transporter gene OsIRT1 or OsYSL15. Approach 6: Enhanced Fe uptake and translocation by overexpression of the OsIRO2 gene. Approach 7: Enhanced Fe translocation from flag leaves to seeds by knockdown of the vacuolar Fe transporter gene OsVIT1 or OsVIT2. The ferritin image was kindly provided by Dr. David S. Goodsell (Scripps Research Institute, La Jolla, CA, USA) and the RCSB PDB.
Table 1.
Approaches of Fe biofortification of rice: single transgenic approaches
| Approach | Introduced genes | Rice cultivar | Cultivation condition | Fold increase in Fe concentration compared to non-transgenic rice a | References |
|---|---|---|---|---|---|
| Approach 1 | OsGluB1 pro-SoyferH1 | Japonica cv. Kitaake | Soil cultivation in greenhouse | 2 fold (polished seeds) | Goto et al. 1999 |
| Enhancement of Fe storage in rice seeds by ferritin | |||||
| 3 fold (brown seeds) | |||||
| OsGluB1 pro- SoyferH1b | Japonica cv. Kitaake | Soil cultivation in greenhouse | 1.5 fold (brown seeds) | Qu et al. 2005 | |
| OsGlb1 pro-SoyferH1b | |||||
| OsGluB1 pro-SoyferH1 | Japonica cv. Taipei 309 | Soil cultivation in greenhouse | 2.2 fold (brown seeds) | Lucca et al. 2002 | |
| OsGluB1 pro-SoyferH1 | Indica cv. IR68144 | Soil cultivation in greenhouse | 3.7 fold (polished seeds) | Vasconcelos et al. 2003 | |
| (High Fe breeder line) | |||||
| OsGluA2 pro-OsFer2 | Indica cv. Pusa-Sugandh II (Aromatic rice) | Soil cultivation in greenhouse | 2.1 fold (polished seeds) | Paul et al. 2012 | |
| Approach 2 | OsActin1 pro-HvNAS1c | Japonica cv. Tsukinohikari | Soil cultivation in greenhouse | 2 fold (polished seeds) | Masuda et al. 2009 |
| Enhancement of Fe translocation by overexpression of NAS | |||||
| 35S pro-HvNAS1c | |||||
| Activation tag line of OsNAS3 | Japonica cv. Dongjin | Soil culture in greenhouse | 3 fold (polished seeds) | Lee et al. 2009c | |
| 35S pro- OsNAS1, 2, 3 | Japonica cv. Nipponbare | Soil cultivation in greenhouse | 4 fold (polished seeds) | Johnson et al. 2011 | |
| Approach 3 | OsSUT1 pro-OsYSL2 | Japonica cv. Tsukinohikari | Soil cultivation in greenhouse | 4 fold (polished seeds) | Ishimaru et al. 2010 |
| Enhancement of Fe transportation by Fe transporter | |||||
| Approach 4 | Barley IDS3 genome fragment | Japonica cv. Tsukinohikari | Andosol soil in paddy field | 1.4 fold (polished seeds) 1.3 fold (brown seeds) |
Masuda et al. 2008 |
| Enhancement of Fe uptake and translocation by IDS3 gene | |||||
| Calcareous soil in paddy field | 1.3 fold (brown seeds) | Suzuki et al. 2008 | |||
| Approach 5 | Ubiquitin pro-OsIRT1 | Japonica cv. Dongjin | Paddy field | 1.7 fold (leaves) | Lee et al.2009a |
| Overexpression of Fe transporter | 1.1 fold (brown seeds) | ||||
| OsActin1 pro-OsYSL15 | Japonica cv. Dongjin | Paddy field | 1.3 fold (brown seeds) | Lee et al. 2009b | |
| Approach 6 | 35S pro-OsIRO2 | Japonica cv. Tsukinohikari | Calcareous soil in greenhouse | 3 fold (brown seeds) | Ogo et al. 2011 |
| Overexpression of transcription factor | |||||
| Approach 7 |
OsVIT1 or OsVIT2 T-DNA insertion mutant lines |
Japonica cv. Zhonghua11 Japonica cv. Dongjin | Hydroponic culture | 1.4 fold (brown seeds) | Zhang et al. 2012 |
| Knockdown of OsVITs genes | |||||
| Paddy field | 1.4 fold (brown seeds) | ||||
|
OsVIT2 T-DNA insertion mutant line |
Japonica cv. Dongjin | Soil cultivation in greenhouse | 1.3 fold (brown seeds) | Bashir et al. 2013 | |
| 1.8 fold (polished seeds) |
aThe tissue name written in Parentheses is the rice tissue where Fe concentration was increased. bThey introduced these two genes into same transgenic lines. c These two genes were introduced separately into rice and they analyzed these two types of transgenic lines.
Review
Approach 1: Enhancing Fe accumulation in seeds by expression of the Fe storage protein, ferritin gene, SoyferH1 and SoyferH2, under the control of endosperm-specific promoters
The first approach to increasing Fe concentration in rice seeds involves enhancing Fe accumulation in rice seeds by expressing the ferritin gene under the control of endosperm-specific promoters (Figure 1; Approach 1). Ferritin is a ubiquitous Fe storage protein that sequesters as many as 4,000 Fe atoms in a complex (Theil 2003). The Fe stored in soybean ferritin is readily absorbed by the human gastrointestinal tract (Lonnerdal 2009). The molecular mechanism underlying the uptake of Fe stored in food ferritin to the human body has been revealed (Theil 2011). Thus, Fe stored in ferritin is an important source for humans to use to avoid Fe deficiency (Theil et al. 2012).
Goto et al. (1999) generated transgenic rice plants expressing the soybean ferritin gene SoyferH1 in endosperms using the rice endosperm-specific, 1.3-kb OsGluB1 promoter. Transformants showed 3-fold higher levels of Fe accumulation in brown seeds (Table 1). Additionally, the concentration of Fe in the endosperm was increased 2-fold. This endosperm-specific expression of ferritin has been used to generate the following increases in Fe concentration in rice plants: a 2.2-fold increase in brown seeds of transgenic japonica cv. Taipei 309 (Lucca et al. 2002), a 3.7-fold increase in polished seeds of indica cv. IR68144 (Vasconcelos et al. 2003), and a 2.1-fold increase in polished seeds of indica cv. Pusa Sugandhi II (Paul et al. 2012) (Table 1).
Furthermore, Qu et al. (2005) expressed the SoyferH1 gene under the control of two endosperm specific promoters, OsGlb1 promoter and 1.3-kb OsGluB1 promoter, to increase further the concentration of Fe in the seed. However, using multiple promoters to increase the level of ferritin expression in rice seeds did not significantly increase the Fe concentration compared to expression driven by a single endosperm-specific promoter (Table 1; Qu et al. 2005).
In our previous study, the Fe concentration in brown seeds was not increased in transgenic rice introduced only ferritin compared to non transgenic line (Masuda et al. 2013). Moreover, the introduction of ferritin gene alone produced symptoms of Fe deficiency in the leaves of transgenic plants (Qu et al. 2005; Masuda et al. 2013). Thus, improving ferritin expression may not be sufficient to increase Fe concentration in rice grains.
Generally, Fe translocation ability to endosperm is low in rice. In fact, the Fe content of endosperm is significantly lower than in other rice tissue, including leaves, stems, roots, husk, or the aleurone layer. 0.5% of total Fe content in aerial parts of plant body was found in polished seeds (endosperm) of Tsukinohikari rice variety, despite 3.6% in brown seeds, 3.4% in husk, 2.4% in rachis and 90.6% in straw, respectively (Masuda et al. 2008). Rice plants uptake certain amount of Fe, but the plant may not translocate it to endosperm positively. A strict regulation system may exist to control Fe translocation into endosperm in rice plants. This may explain why this approach does not significantly increase the Fe concentration in polished seeds.
Therefore, in addition to increased Fe storage in seeds, enhanced uptake of Fe from the soil and enhanced translocation of Fe within the plant body are required to improve further the Fe biofortification of rice seeds. Thus, the following approaches (from approach 2 to 7) are considered as ways to increase the Fe concentration of the rice endosperm (Figure 1).
Approach 2: Enhancing Fe transport within the plant body by overexpression of the nicotianamine synthase gene NAS
The uptake, translocation, and homeostasis of Fe in rice are beginning to be understood at the molecular level, and many of the associated genes have been identified (Bashir et al. 2010; Kobayashi et al. 2012). As a second approach to increase Fe concentrations in rice, we overexpressed the nicotianamine (NA) synthase gene (NAS) (Figure 1; Approach 2).
NA, which is a chelator of metal cations such as Fe(II) and zinc (Zn)(II), is biosynthesized from methionine via S-adenosyl methionine synthase (SAMS) and NAS (Figure 1; Higuchi et al. 1994). All higher plants synthesize and utilize NA for the internal transport of Fe and other metals (Hell and Stephan 2003; Takahashi et al. 2003). Indeed, the NA-defective tomato mutant chloronerva (Rudolph et al. 1985) has a phenotype that is indicative of Fe deficiency (Pich and Scholz 1996;Stephan et al. 1996). Takahashi et al. (2003) produced NA-deficient transgenic tobacco plants by constitutively expressing the barley NA aminotransferase (NAAT) gene, HvNAAT-A. The obtained transformants showed interveinal chlorosis in the young leaves, and the Fe and Zn concentrations in the leaves and flowers decreased as a result of disrupted internal metal transport. Conversely, overexpression of the barley NAS gene HvNAS1 increased the concentrations of Fe and Zn in the leaves, flowers, and seeds of tobacco plants (Takahashi et al. 2003).
Rice has three NAS genes; OsNAS1, OsNAS2, and OsNAS3. Although these genes are differentially regulated by Fe, all of them are expressed in cells involved in the long-distance transport of Fe (Inoue et al. 2003). These results suggest that NAS and NA play important roles in the long-distance transport of Fe in rice, in addition to their roles in phytosiderophore synthesis.
Higuchi et al. (2001a) produced transgenic rice expressing HvNAS1 under the control of the cauliflower mosaic virus constitutive CaMV35S promoter. NA concentration in plant shoot increased 3-fold in this transgenic rice. Based on these results, we hypothesized that enhancing the expression of NAS could increase the NA concentration within the plant body and, as a consequence, increase the Fe concentration in seeds. Therefore, we produced transgenic rice overexpressing HvNAS1 under the control of the OsActin1 promoter or 35S promoter (Masuda et al. 2009). HvNAS1-overexpressing transgenic rice showed increased HvNAS1 expression and increased endogenous NA levels in the shoots and seeds by 5 to 10-fold (Masuda et al. 2009). The Fe and Zn concentrations in the T1 polished seeds from the transgenic plants increased more than 3-fold and 2-fold, respectively. The Fe concentrations also increased 2-fold in the T2 polished seeds (Table 1). Lee et al. (2009c) and Johnson et al. (2011) showed that NAS overexpression increased the Fe concentration in polished rice seeds by 3–4-fold (Table 1). Thus, Fe transport within the plant body, including the phloem, may be improved by NAS overexpression (Masuda et al. 2009; Lee et al. 2009c; Johnson et al. 2011). Moreover, Lee et al. (2009c) reported that Fe-biofortified rice overexpressing OsNAS3 by virtue of an enhancer tag mitigated Fe-deficiency anemia in mice to a greater degree than non-transgenic rice seeds.
Importantly, not only the NA content but also the deoxymugineic acid (DMA) content was increased by overexpressing HvNAS1 in rice (Masuda et al. 2009, Lee et al. 2009c). In graminaceous plants, including rice, DMA is synthesized from NA by NAAT and DMA synthase (DMAS) (Figure 1) (Takahashi et al. 1999; Bashir et al. 2006; Inoue et al. 2008). Overexpression of AtNAS1 together with endosperm expression of Pvferritin in rice enhanced the expression of OsSAMS2, OsNAS1, OsNAS3 and OsDMAS1 in roots (Wang et al. 2013). Enhanced expression of endogenous NAS and DMAS might contribute to DMA production in rice. In addition, DMA concentration in rice xylem sap was increased in Fe deficient rice (Kakei et al. 2009). This suggested that DMA contributes to the transport of Fe from root to shoot through xylem.
Moreover, OsYSL15, a Fe(III)-DMA transporter, is expressed in the root epidermis and stele, and contributes to the internal translocation of Fe (Inoue et al. 2009). Aoyama et al. (2009) showed that OsYSL18, a Fe(III)-DMA transporter, was expressed in reproductive organs and phloem of lamina joints. Kakei et al. (2012) also showed that OsYSL16, another Fe(III)-DMA transporter, was expressed in both root epidermis and vascular bundles of whole plants. Furthermore, Nishiyama et al. (2012) described that Fe(III)-DMA complexes were detected as low-molecular-weight chemical forms of Fe in rice phloem sap, and DMA may play a role in long-distance Fe transport in phloem sap. These results suggest that, similar to NA, DMA is involved in Fe distribution from roots to seeds in rice plant. Thus, the overproduction of NA and subsequent increase in DMA content may enhance the translocation of Fe and Zn into rice grains (Masuda et al. 2009). Moreover, an increase in the DMA content of rice may increase DMA secretion from the roots and contribute to enhancing the uptake of Fe from the soil. Therefore, increasing the NA and DMA concentrations by enhancing NAS expression may increase the levels of Fe and Zn in rice grains.
Approach 3: Enhancing Fe influx to seeds by expressing the Fe (II)-NA transporter gene OsYSL2 under the control of the OsSUT1 promoter
Manipulating the expression of membrane transporters can increase Fe concentrations in crops (Schroeder et al. 2013). Thus, the third approach to increase Fe concentrations in rice was to enhance Fe influx to seeds by expressing the Fe(II)-NA transporter gene OsYSL2 (Figure 1; Approach 3). Koike et al. (2004) identified the rice OsYSL2 gene, which is preferentially expressed in leaf phloem cells, the vascular bundles of flowers and developing seeds, suggesting a role in internal Fe transport. The Fe concentration in OsYSL2 knockdown rice was 18% lower in brown seeds and 39% lower in polished seeds compared with non-transgenic rice (Ishimaru et al. 2010). Thus, OsYSL2 plays an important role in the transport of Fe to rice seeds.
Therefore, we hypothesized that enhanced expression of OsYSL2 could increase the Fe concentration in rice seeds. However, overexpression of OsYSL2 under the control of the CaMV35S promoter did not increase the concentration of Fe in the seeds, although it did increase the Fe concentration in the roots (Ishimaru et al. 2010). Constitutive expression of OsYSL2 may disrupt Fe translocation in rice.
A rice sucrose transporter, OsSUT1, was expressed in companion cells of the phloem in flag leaves and the rachis (Scofield et al. 2007). OsSUT1 was also strongly expressed in the panicle and immature seeds during seed maturation (Aoki et al. 2003). Moreover, OsSUT1 antisense rice showed significant reductions in sucrose uptake and filling rates in seeds (Ishimaru et al. 2001). OsSUT1 is a key transporter of sucrose from the phloem to seeds. Therefore, the OsSUT1 promoter is a suitable promoter to control seed metal concentrations in rice. Takahashi et al. (2012) also expressed the Zn and Cd transporter OsHMA2 under the control of the OsSUT1 promoter, and successfully increased Zn concentration by 20% and decreased Cd concentration by 50% in brown seeds compared to non-transgenic rice.
Therefore, to effectively target OsYSL2 expression, we engineered a construct in which OsYSL2 expression was controlled by OsSUT1 promoter, and introduced the construct into rice to enhance Fe(II)-NA translocation into seeds (Ishimaru et al. 2010). We found that the enhanced OsYSL2 expression under the control of the OsSUT1 promoter increased the Fe concentration up to 4-fold in polished rice seeds (Table 1) (Ishimaru et al. 2010). Thus, expression of the Fe(II)-NA transporter gene OsYSL2 under the control of the OsSUT1 promoter is one of the remarkable approaches to increase the Fe concentration in rice seeds.
Combination of Approaches 1–3
Wirth et al. (2009) combined the first and second approaches by introducing the CaMV35S promoter-AtNAS1 and Globulin promoter-Pvferritin genes into the Japonica cv. Taipei 309 rice variety (Table 2). They also introduced the Globulin promoter-Afphytase gene cassette to reduce the phytate content and improve Fe bioavailability in rice seeds. This rice, named NFP rice, showed increased Fe concentrations by up to 6-fold in hydroponic cultures under various Fe nutrient conditions (Table 2).
Table 2.
Approaches of Fe biofortification of rice: multi transgenic approaches
| Approach | Introduced genes a | Rice cultivar | Cultivation | Fold increase in Fe concentration compared to non-transgenic rice b . | References |
|---|---|---|---|---|---|
| condition | |||||
| Combination of approaches 1 and 2 | OsGlb pro-Pvferritin | Japonica cv. Taipei 309 | Hydroponic culture | 6 fold (polished seeds) | Wirth et al. 2009 |
| 35S pro-AtNAS1 | |||||
| OsGlb pro-Afphytase | |||||
| Combination of approaches 1, 2 and 3 | OsGluB1 pro-SoyferH2 | Japonica cv. Tsukinohikari | Soil cultivation in greenhouse | 6 fold (polished seeds) | Masuda et al. 2012 |
| OsGlb1 pro-SoyferH2 | |||||
| OsActin1 pro-HvNAS1 | Paddy field | 4.4 fold (polished seeds) | |||
| OsSUT1 pro-OsYSL2 | |||||
| OsGlb1 pro-OsYSL2 | |||||
| OsGluB1 pro-SoyferH2 | Tropical Japonica cv. Paw San Yin (Myanmar High Quality Rice) | Soil cultivation in greenhouse | 3.4 fold (polished seeds) | Aung et al. 2013 | |
| OsGlb1 pro-SoyferH2 | |||||
| OsActin1 pro-HvNAS1 | |||||
| OsSUT1 pro-OsYSL2 | |||||
| OsGlb1 pro-OsYSL2 | |||||
| Combination of approaches 1 and 4 | OsGluB1 pro-SoyferH2 | Japonica cv. Tsukinohikari | Normal soil in greenhouse | 4 fold (polished seeds) | Masuda et al. 2013 |
| OsGlb1 pro-SoyferH2 | |||||
| HvNAS1, HvNAAT-A,-B and IDS3 genome fragments | Calcareous soil in greenhouse | 2.5 fold (polished seeds) |
aThese gene expression cassettes were introduced concomitantly. b.The tissue name written in parentheses is the rice tissue where Fe concentration was increased.
We generated Fe-biofortified rice using a combination of the first, second, and third approaches (Figure 1). We produced 45 independent transgenic rice lines (Japonica cv. Tsukinohikari; Fer-NAS-YSL2 lines), which included the OsGluB1 promoter–SoyferH2, OsGlb1 promoter–SoyferH2, OsActin1 promoter–HvNAS1, OsSUT1 promoter–OsYSL2, and the OsGlb1 promoter–OsYSL2 gene cassettes (Table 2) (Masuda et al. 2012). The Fe concentrations in the T2 seeds were increased up to 6-fold in the Fer-NAS-YSL2 lines and 3-fold in the OsActin1 promoter-HvNAS1 lines compared with the non-transgenic line under soil cultivation in a greenhouse (Figure 2a). For practical applications, we explored whether the Fer-NAS-YSL2 lines set seeds with increased Fe concentrations under actual paddy field conditions. To accomplish this, selected T2 lines were cultivated in an isolated paddy field. We found that the Fe concentration in the paddy field-grown T3 polished seeds was 4.4-fold higher than in non-transgenic seeds (Figure 2b), without any loss of yield. Moreover, the transgenic seeds accumulated Zn at levels up to 1.6-fold higher in the field. Our results demonstrated that the introduction of multiple Fe homeostasis genes is more effective at achieving Fe biofortification than the introduction of individual genes under soil cultivation in greenhouse and paddy field conditions.
Figure 2.
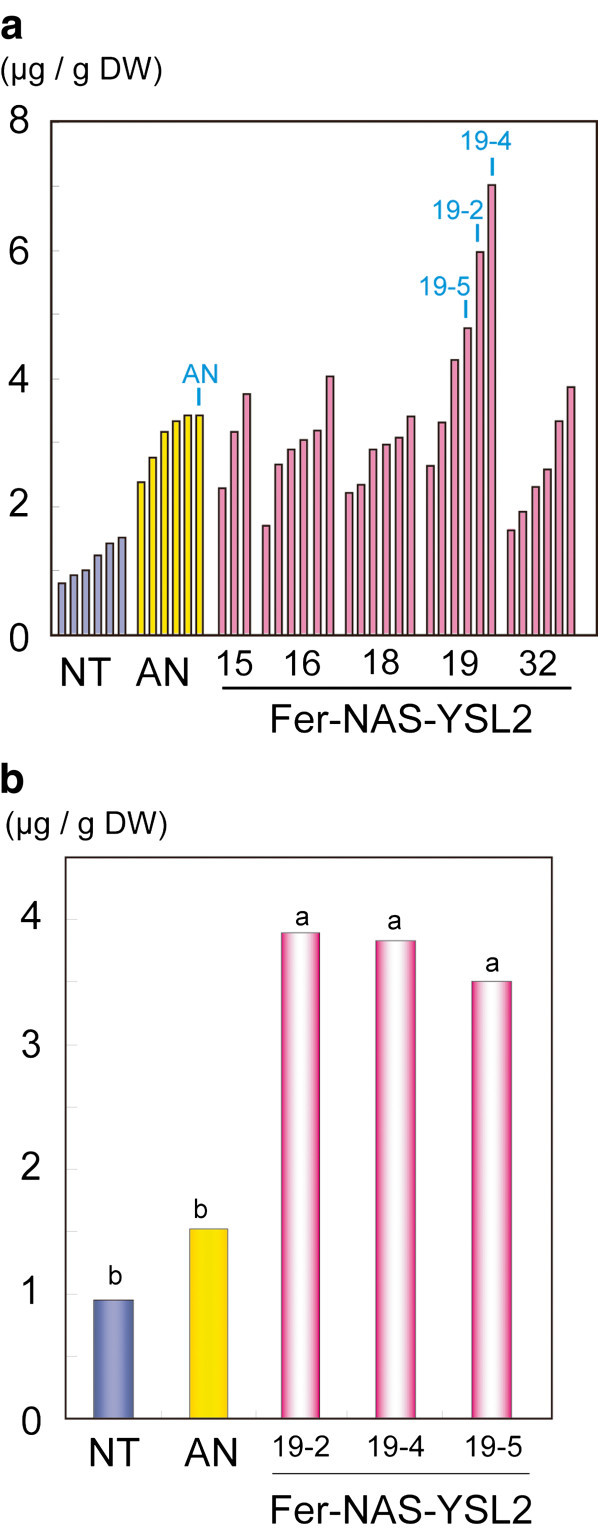
Fe concentration in the Fer-NAS-YSL2 lines (Masuda et al.2012). a; The Fe concentrations in T2 polished rice seeds (Oryza sativa cv. Tsukinohikari). Bars represent the Fe concentrations in polished seeds obtained from individual transgenic or non-transgenic plants. The numbers indicate the line numbers of the independent T1 lines. The arrows and numbers above the bars show the lines that contained high levels of Fe, which are shown in b for the subsequent generation. b; Fe concentrations in the T3 polished rice seeds (Oryza sativa cv. Tsukinohikari) harvested from the paddy field. ANOVA with the Tukey–Kramer HSD test was used for each four-block dataset (n = 4). The letters above the bars indicate significant differences (P < 0.05). NT, Non-transgenic rice; AN, OsActin1 promoter–HvNAS1 transgenic rice line No. 8 (Masuda et al. 2009); Fer-NAS-YSL2, transgenic rice lines that carry the OsGlb1 promoter–SoyferH2, OsGluB1 promoter–SoyferH2, OsSUT1 promoter–OsYSL2, OsGlb1 promoter–OsYSL2, and OsActin1 promoter–HvNAS1 (Masuda et al. 2012).
Fe concentration was increased significantly in Fer-NAS-YSL2 rice (Masuda et al. 2012) because the introduced genes, ferritin, HvNAS1 and OsYSL2, functioned synergistically as follows: Overexpression of the NAS gene increased the NA and DMA concentrations in the plant body. Abundant NA and DMA facilitated the formation of Fe(II)–NA or Fe(III)–DMA. Additionally, Fe(II) or Fe(III) transport in the plant body, including in the phloem, was improved by NAS overexpression. For effective translocation of enhanced Fe(II)–NA in phloem sap, we engineered the Fe(II)–NA transporter OsYSL2 to be under the control of the OsSUT1 promoter. Additionally, OsYSL2 expressed in endosperm cells under the control of the OsGlb1 promoter may enhance transport of Fe(II)–NA into endosperm cells. Moreover, ferritin expressed under the control of the OsGlb1 promoter and the OsGluB1 promoter accumulated Fe in seed endosperm cells. As a result, Fe concentration in the polished seeds of Fer-NAS-YSL2 rice was markedly elevated than Fe concentration achieved by a single-transgenic approach (Masuda et al. 2012).
Fe deficiency anemia is prevalent in many populations in Myanmar (MOH 2003). Myanmar has one of the highest per capita rice consumption rates (average, 578 g per day) globally (Maclean et al. 2002). Thus, Myanmar rice is a suitable target for Fe biofortification. Therefore, we introduced this Fer-NAS-YSL2 gene into the Paw San Yin variety, which is a popular high-quality rice variety in Myanmar (Aung et al. 2013). Paw San Yin transgenic rice was produced successfully, and the concentration of Fe in the Paw San Yin Fer-NAS-YSL2 line increased 3.4-fold in polished seeds, which was close to the dietary target level for people in Myanmar (Table 2) (Aung et al. 2013).
Approach 4: Enhancing Fe uptake and translocation by introducing the phytosiderophore synthase gene IDS3
Graminaceous plants secrete mugineic acid family phytosiderophores (MAs), which are natural Fe(III) chelators crucial to take up Fe from the rhizosphere (Takagi 1976; Mihashi and Mori 1989). Methionine has been identified as the precursor of MAs (Figure 1) (Mori and Nishizawa 1987). In graminaceous plants including rice, DMA is synthesized from NA by NAAT and DMAS (Figure 1), and in barley and some graminaceous species, other types of MAs are synthesized from DMA by Fe deficiency-specific clones no. 2 (IDS2) and no. 3 (IDS3, mugineic acid synthase) (Nakanishi et al. 2000; Kobayashi et al. 2001). Graminaceous plant roots secrets MAs, which chelate insoluble Fe(III) in the rhizosphere. The resulting Fe(III)-MAs complexes are absorbed into the roots via the transporter YS1 or YSL (Curie et al. 2001). In rice, DMA is secreted from roots by the transporter of mugineic acid family phytosiderophores 1 (TOM1) in rice (Nozoye et al. 2011), and Fe(III)-DMA complexes are thought to be absorbed via the OsYSL15 transporter (Inoue et al. 2009; Lee et al. 2009b).
Among graminaceous plants, barley is highly tolerant to Fe deficiency and possesses a series of biosynthetic genes for MAs, including HvNAS1, HvNAAT-A, HvNAAT-B, HvDMAS1, IDS2, and IDS3, the expression levels of which are up-regulated in Fe-deficient barley roots (Higuchi et al. 1999; Takahashi et al. 1999; Nakanishi et al. 2000; Bashir et al. 2006). In contrast, rice lacks IDS2 and IDS3 and secretes only DMA. This is thought to be one of the reasons why barley has a higher tolerance than rice to Fe deficiency (Kobayashi et al. 2001).
Thus, a fourth approach to Fe biofortification involves enhancing Fe uptake and translocation by introducing genes responsible for biosynthesis of MAs (Figure 1; Approach 4). Three transgenic rice lines, which were transformed with the barley genome fragments involving mugineic acid synthase genes, HvNAS1 or HvNAS1 and HvNAAT-A,-B or IDS3, were produced (Higuchi et al. 2001b; Kobayashi et al. 2001; Masuda et al. 2008; Suzuki et al. 2008). The Fe concentrations in the seeds of transgenic lines were analyzed after cultivation in the paddy field in Fe-sufficient (andosol) or low Fe-available (calcareous) soil (Masuda et al. 2008; Suzuki et al. 2008). The IDS3 rice lines produced Fe concentrations that were 1.4-fold and 1.3-fold higher in the polished and brown seeds than in non-transgenic rice respectively after cultivation in Fe-sufficient soil (Table 1) (Masuda et al. 2008). Fe concentrations of the IDS3 rice lines were also 1.3-fold higher in the brown seeds in low Fe-available soil (Table 1) (Suzuki et al. 2008).
In addition to DMA, the introduction of IDS3 conferred upon the rice the ability to secrete MA (Kobayashi et al. 2001). As MA showed greater Fe(III)-complex stability than DMA at a slightly acidic pH (von Wirén et al. 2000), the production of MA via IDS3 may be beneficial for Fe translocation in rice. Furthermore, since these transformants contained introduced barley genome fragments, expression of the genes responsible for the biosynthesis of MAs was regulated by their own promoters. In rice, these promoters induce expression in response to Fe deficiency in roots and leaves (Higuchi et al. 2001b; Kobayashi et al. 2001; Takahashi et al. 2001). Thus, these genes are likely expressed where and when the requirement for Fe is elevated.
Combination of Approaches 1 and 4
We attempted to increase the Fe concentrations in rice seeds using a combination of the first and fourth approaches (Figure 1). We introduced the SoyferH2 gene driven by two endosperm-specific promoters (the OsGluB1 promoter and OsGlb1 promoter), together with three fragments of the barley genome (5.9 kb of HvNAS1, 11 kb of HvNAAT-A,-B, and 6 kb of IDS3) to enhance MA production in rice plants (Table 2) (Masuda et al. 2013). Representative lines were selected and grown in commercially supplied soil (Fe-sufficient condition) or calcareous soil (low Fe-available condition). Lines expressing both ferritin and the MA biosynthetic genes showed signs of tolerance to Fe deficiency in the calcareous soil. The concentrations of Fe in the T3 polished seeds were increased by 4- and 2.5-fold compared with the levels in non-transgenic lines grown in normal soil and calcareous soil, respectively (Table 2). During calcareous soil cultivation, the Fe concentration in polished seeds was increased in Fer-NAS-NAAT-IDS3 lines, but not in the Ferritin-introduced line compared to non-transgenic lines. These results indicated that the concomitant introduction of the ferritin and MA biosynthetic genes effectively increased Fe levels in seeds without inducing Fe sensitivity under Fe-limited conditions.
Approach 5: Enhanced Fe uptake from soil by overexpression of the Fe transporter gene OsIRT1 or OsYSL15
Fe concentrations in rice seeds are generally low (Grusak and Cakmak 2005). Especially, rice is mainly consumed as polished seeds, and Fe levels are diminished by milling and polishing; thus, the remaining endosperm contains low Fe levels. In addition, within a rice-consuming country, rice consumption is lower in urban areas than in rural areas (Juliano, 1993). Therefore, a higher target Fe concentration in polished rice grain is desirable.
Rice consumption per capita per day ranges from 300 to 600 g in South East Asia and China (Maclean et al. 2002). The Fe requirement for adult females is 15–18 mg per day (Food and Nutrition Board 2001). Thus, the target Fe concentration in rice ranges from 7 to 14 ppm in the polished grain for individuals that consume 600 g or 300 g per day, respectively. This would account for ~25% of the daily Fe requirement. Pfeiffer and McClafferty (2007) also proposed similar target level for Fe biofortification.
Fe concentrations in various indica rice varieties harvested in the paddy field mostly ranged from 1 to 2 ppm (Aung et al., unpublished data). Therefore, the Fe concentration should be increased by 4–10-fold to reach the target levels. This target Fe requirement has not been achieved using approaches 1-4 alone, which resulted in a maximum 4-fold increase (Table 1). A combination of approaches 1-3 achieved a 6-fold increase (Table 2). However, this may not be sufficient in some cases. Thus, we proposed three additional approaches (approaches 5, 6 and 7) to further Fe biofortification in seeds. Approach 5 involves enhancing Fe uptake by overexpressing the Fe transporter (Figure 1; Approach 5).
Previous studies have reported increased concentrations of Fe in seeds after overexpressing the Fe transporter. Lee et al. (2009a) produced transgenic rice that expressed the rice ferric ion transporter gene OsIRT1 under the control of the Ubiquitin promoter. This rice showed a 13% increase in Fe concentration in the brown seeds (Table 1), while the Fe concentration in the leaves increased 1.7-fold. The authors suggested that OsIRT1 could be used to enhance Fe levels in rice grains. Next, Lee et al. (2009b) reported that OsYSL15 overexpression using the OsActin1 promoter increased the concentration of Fe in brown seeds by approximately 1.3-fold compared with non-transgenic rice (Table 1). In addition, Gómez-Galera et al. (2012) produced transgenic rice that overexpressed the barley Fe(III)-MA transporter gene HvYS1 under the control of the CaMV35S promoter. The concentration of Fe in the transgenic leaves was 1.5-fold higher than in the non-transgenic leaves. In this previous study, it was suggested that Fe uptake from the rhizosphere could be enhanced by expressing HvYS1. Although overexpression of OsIRT1, OsYSL15 or HvYS1 increased the Fe concentrations in the leaves, it did not significantly increase Fe concentrations in the seeds (Table 1) (Lee et al. 2009a; Lee et al. 2009b; Gómez-Galera et al. 2012). Lee et al. (2009a) reported that plants overexpressing OsIRT1 were shorter and had fewer tillers. As constitutive expression of OsIRT1 may disturb metal homeostasis in rice, they suggested that targeted expression of OsIRT1 using specific promoters might solve this problem.
Application of this approach 5 alone did not remarkably increase the Fe concentrations in seeds. However, it is possible that this approach in combination with other approaches (i.e., Approaches 1–3) could increase Fe levels.
Approach 6: Enhanced Fe uptake and translocation by overexpression of the Fe homeostasis-related transcription factor OsIRO2
Ogo et al. (2006) identified a Fe-deficiency-inducible basic helix–loop–helix (bHLH) transcription factor, OsIRO2, in rice. OsIRO2 is responsible for regulation of the key genes involved in MAs-related Fe uptake; e.g., OsNAS1, OsNAS2, OsNAAT1, OsDMAS1, TOM1, and OsYSL15 (Figure 1) (Ogo et al. 2007; Ogo et al. 2011). Ogo et al. (2007) introduced OsIRO2 under the control of the CaMV35S promoter into rice plants. Rice that overexpressed OsIRO2 secreted more DMA than non-transgenic rice, and exhibited enhanced Fe-deficiency tolerance in calcareous soils (Ogo et al. 2007; Ogo et al. 2011). Moreover, the concentration of Fe in the transgenic brown seeds was increased 3-fold when the transgenic rice was cultivated in calcareous soil (Table 1) (Ogo et al. 2011). Therefore, this approach can be used to increase the Fe concentrations in seeds in soils with low Fe availability.
Approach 7: Enhanced Fe translocation from flag leaves to seeds by knockdown of the vacuolar Fe transporter gene OsVIT1 or OsVIT2
Kim et al. (2006) have reported that the Arabidopsis vacuolar Fe transporter, VIT1, is highly expressed in developing seeds and transports Fe and manganese into the vacuole. Zhang et al. (2012) reported that disruption of the rice VIT orthologues (OsVIT1 and OsVIT2) increased the Fe concentrations by 1.4-fold in brown seeds (Table 1) and decreased the Fe concentrations by 0.8-fold in the source organ flag leaves. A possible explanation for these results is that the VIT genes are highly expressed in rice flag leaves. Bashir et al. (2013) also reported that an OsVIT2-knockdown mutant showed 1.3-fold and 1.8-fold increases in Fe concentrations in the brown and polished rice seeds, respectively (Table 1). They suggested that disruption of OsVIT1 or OsVIT2 enhanced Fe translocation between the source and sink organs, and proposed this as a novel strategy for producing Fe-biofortified rice (Figure 1).
Zhang et al. (2012) and Bashir et al. (2013) reported that the Cd concentration was also increased in VIT knockdown rice. Therefore, this approach should be avoided in Cd-contaminated soils. Although further studies are required for this approach, likewise approach 5 and 6, approach 7 may also be applied in combination with other approaches to further increase Fe concentrations in polished seeds.
Alternative strategies to increase Fe transport within rice plants, with the goal of increasing the Fe concentration in seeds
An alternative approach to achieving Fe biofortification involves enhancing Fe chelate export by the overexpression of genes for mugineic acid or protocatechuic acid exporter. The MA transporter gene, TOM1, is expressed in restricted regions of the exodermis of roots under Fe-sufficient conditions and throughout the roots under Fe-deficient conditions (Nozoye et al. 2011). TOM1 is also expressed in the vascular bundles of the leaf sheaths and leaf phloem, the pollen, and the dorsal vascular bundle in developing seeds. Nozoye et al. (2011) produced rice that overexpressed TOM1; in this rice, DMA secretion was enhanced, the Fe concentration was increased 1.2-fold, the Zn concentration was increased 1.6-fold in the rice seeds, and tolerance to Fe deficiency was increased.
Plants secrete phenolics to absorb apoplasmic precipitated Fe, such as protocatechuic acid (PCA) (Cesco, et al. 2010). Ishimaru et al. (2011) identified a phenolics efflux transporter, PEZ1, in rice. PEZ1 localized to the plasma membrane and transported PCA when expressed in Xenopus laevis oocytes. PEZ1 is responsible for increasing the PCA concentration in the xylem sap, and is essential for the utilization of apoplasmic precipitated Fe in the stele. PEZ1 localized mainly to the stele of the roots. The concentration of Fe in the leaves of transgenic rice lines containing the CaMV35S promoter-PEZ1 gene cassette was increased 3-fold. The concentration of Fe in the roots of this transgenic rice was also increased 2-fold due to the high solubilization level of apoplasmic precipitated Fe in the stele (Ishimaru et al. 2011). Consequently, the concentration of Fe in seeds may increase by overexpression of PEZ1.
Therefore, the seed Fe concentration may be increased through chelate export using the TOM1 or PEZ1 gene and a combination of the other approaches; e.g., endosperm-specific expression of the ferritin gene (Approach 1; Figure 1), overexpression of the NAS gene (Approach 2; Figure 1), or OsYSL2 expression under the control of the OsSUT1 promoter (Approach 3; Figure 1).
Mining of high-Fe rice varieties or other target genes for Fe biofortification of rice
Increases in Fe concentrations using transgenic approaches are dependent on the cultivar. The target Fe concentration using transgenic approaches should be considered based on the Fe concentration level of the host rice variety. Based on this information, the required increase in Fe concentration can be calculated. Therefore, to produce rice lines with higher Fe concentrations, it is worthwhile to apply transgenic methods to original high-Fe rice varieties obtained through mining among extant rice varieties, or high-Fe rice lines produced by conventional breeding, or high-Fe mutant rice lines.
The mining of high-Fe rice varieties or the identification of novel target genes is important to Fe biofortification of rice. Anuradha et al. (2012) found seven quantitative trait loci (QTL) and selection markers related to the concentrations of Fe in rice seeds. They performed mapping of QTL using Madhukar × Swarna indica rice varieties and identified genes related to Fe homeostasis, such as OsYSLs, OsNASs, OsNRAMP1, OsIRT1, OsZIPs and APRT, as candidate genes that effect the concentration of Fe in seeds.
Sperotto et al. (2010) analyzed the gene expression profiles of 25 metal-related genes, including rice homologues of YSL2, NRAMPs, ZIPs, IRT1, VIT1, NASs, FROs and NAC5, in eight rice varieties with different Fe and Zn concentrations in the seeds. They also identified putative target genes that contribute to increasing the Fe and Zn concentrations in rice grains.
Jeng et al. (2012) discovered mutant lines that have higher Fe or Zn concentrations in polished seeds by searching among NaN3-induced mutant lines (Oryza sativa cv. IR64). Ruengphayak et al. (2012) screened 12,000 fast neutron-irradiated M4 mutant lines (Oryza sativa cv. Jao Hom Nin) and identified 76 mutant lines that contained higher Fe densities in the grains. Using these high-Fe mutant rice lines, it is possible to identify novel candidate genes to improve Fe biofortification of rice.
Some studies have been conducted to improve mineral nutrition in rice seeds through traditional breeding or marker-assisted breeding. IR68144 rice derived from conventional breeding method was shown to have 2-fold higher Fe concentrations in its seeds (Gregorio et al. 2000). This IR68144 rice has been shown to be superior to normal rice in improving the Fe status of women (Haas et al. 2005).
Conclusions
We generated transgenic rice by introducing multiple genes, including ferritin under the control of endosperm-specific promoters, NAS overexpression, OsSUT1 promoter-driven OsYSL2 expression, and the barley IDS3 genome fragment, and showed increased concentrations of bioavailable Fe. This technique could be applied to mitigate the global problem of Fe-deficiency anemia. However, further efforts in Fe biofortification of rice are required to increase further Fe concentrations in polished seeds and reach the recommended levels. Increasing the expression of OsIRT1, OsYSL15, and OsIRO2, or knockdown of OsVIT1 or OsVIT2 are the candidate approaches to improve Fe biofortification of rice seeds. Further attempts are required to evaluate high-Fe rice varieties or other target genes. A combination of these approaches will be beneficial to future Fe biofortification work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
The manuscript was written by HM and improved by MSA and NKN. All the authors read and approved the final manuscript.
Contributor Information
Hiroshi Masuda, Email: masuda@ishikawa-pu.ac.jp.
May Sann Aung, Email: mmgster@gmail.com.
Naoko K Nishizawa, Email: annaoko@mail.ecc.u-tokyo.ac.jp.
Acknowledgments
We thank Dr. Khurram Bashir (RIKEN, Yokohama, Japan) for his kind reading of the manuscript and valuable suggestions. This work was supported by a grant from the HarvestPlus project of CGIAR and the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
References
- Anuradha K, Agarwal S, Rao YV, Rao KV, Viraktamath BC, Sarla N. Mapping QTLs and candidate genes for iron and zinc concentrations in unpolished rice of Madhukar×Swarna RILs. Gene. 2012;508:233–240. doi: 10.1016/j.gene.2012.07.054. [DOI] [PubMed] [Google Scholar]
- Aoki N, Hirose T, Scofield NG, Whitfeld RP, Furbank TR. The sucrose transporter gene family in rice. Plant Cell Physiol. 2003;44:223–232. doi: 10.1093/pcp/pcg030. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Kobayashi T, Takahashi M, Nagasaka S, Usuda K, Kakei Y, Ishimaru Y, Nakanishi H, Mori S, Nishizawa NK. OsYSL18 is a rice iron(III)-deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Mol Biol. 2009;70:681–692. doi: 10.1007/s11103-009-9500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung MS, Masuda H, Kobayashi T, Nakanishi H, Yamakawa T, Nishizawa NK. Iron biofortification of Myanmar rice. Front Plant Sci. 2013;4:158. doi: 10.3389/fpls.2013.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir K, Inoue H, Nagasaka S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J Biol Chem. 2006;281:32395–32402. doi: 10.1074/jbc.M604133200. [DOI] [PubMed] [Google Scholar]
- Bashir K, Ishimaru Y, Nishizawa NK. Iron uptake and loading into rice grains. Rice. 2010;3:122–130. [Google Scholar]
- Bashir K, Takahashi R, Akhtar S, Ishimaru Y, Hiromi N, Nishizawa NK. The knockdown of OsVIT2 and MIT2 affects iron localization in rice seed. Rice. 2013;6:31. doi: 10.1186/1939-8433-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesco S, Neumann G, Tomasi N, Pinton R, Weisskopf L. Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant Soil. 2010;329:1–25. [Google Scholar]
- Curie C, Panavience Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL. Maize yellow stripe1 encodes a membrane protein directly involved in Fe (III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- Food and Nutrition Board. Dietary reference intakes from vitamin A, vitamin K, arsenic, boron, chromium, copper, iodine, iron, manganese, molybdenum, nickel, silicon, vanadium, and zinc. Institute of Medicine. Washington, DC: National Academic Press; 2001. [Google Scholar]
- Gómez-Galera S, Sudhakar D, Pelacho AM, Capell T, Christou P. Constitutive expression of a barley Fe phytosiderophore transporter increases alkaline soil tolerance and results in iron partitioning between vegetative and storage tissues under stress. Plant Physiol Biochem. 2012;53:46–53. doi: 10.1016/j.plaphy.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F. Iron fortification of rice seed by the soybean ferritin gene. Nat Biotechnol. 1999;17:282–286. doi: 10.1038/7029. [DOI] [PubMed] [Google Scholar]
- Gregorio GB, Senadhira D, Htut H, Graham RD. Breeding for trace mineral density in rice. Food Nutr Bull. 2000;21:382–386. [Google Scholar]
- Grusak MA, Cakmak I. In: Plant Nutritional Genomics. Broadley MR, White PJ, editor. Oxford: Blackwell; 2005. Methods to improve the crop delivery of minerals to humans and livestock; pp. 265–286. [Google Scholar]
- Grusak MA, DellaPenna D. Improving the nutrient composition of plants to enhance human nutrition and health. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:133–161. doi: 10.1146/annurev.arplant.50.1.133. [DOI] [PubMed] [Google Scholar]
- Haas JD, Beard JL, Murray-Kolb LE, del Mundo AM, Felix A, Gregorio GB. Iron-biofortified rice improves the iron stores of nonanemic Filipino women. J Nutr. 2005;135:2823–2830. doi: 10.1093/jn/135.12.2823. [DOI] [PubMed] [Google Scholar]
- Hell R, Stephan UW. Iron uptake, trafficking and homeostasis in plants. Planta. 2003;216:541–551. doi: 10.1007/s00425-002-0920-4. [DOI] [PubMed] [Google Scholar]
- Higuchi K, Kanazawa K, Nishizawa NK, Chino M, Mori S. Purification and characterization of nicotianamine synthase from Fe- deficient barley roots. Plant Soil. 1994;165:173–179. [Google Scholar]
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S. Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 1999;119:471–479. doi: 10.1104/pp.119.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi K, Takahashi M, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S. Analysis of transgenic rice containing barley nicotianamine synthase gene. Soil Sci Plant Nutr. 2001a;47:315–322. [Google Scholar]
- Higuchi K, Watanabe S, Takahashi M, Kawasaki S, Nakanishi H, Nishizawa NK, Mori S. Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. Plant J. 2001;25:159–167. doi: 10.1046/j.1365-313x.2001.00951.x. [DOI] [PubMed] [Google Scholar]
- Inoue H, Higuchi K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. Three rice Nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. The Plant J. 2003;36:366–381. doi: 10.1046/j.1365-313x.2003.01878.x. [DOI] [PubMed] [Google Scholar]
- Inoue H, Takahashi M, Kobayashi T, Suzuki M, Nakanishi H, Mori S, Nishizawa NK. Identification and localization of the rice nicotianamine aminotransferase gene OsNAAT1 expression suggests the site of phytosiderophore synthesis in rice. Plant Mol Biol. 2008;66:193–203. doi: 10.1007/s11103-007-9262-8. [DOI] [PubMed] [Google Scholar]
- Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, Nakazono M, Nakanishi H, Mori S, Nishizawa NK. Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem. 2009;284:3470–3479. doi: 10.1074/jbc.M806042200. [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirose T, Aoki N, Takahashi S, Ono K, Yamamoto S, Wu J, Saji S, Baba T, Ugaki M, Matsumoto T, Ohsugi R. Antisense expression of a rice sucrose transporter OsSUT1 in rice (Oryza sativa L.) Plant Cell Physiol. 2001;42:1181–1185. doi: 10.1093/pcp/pce148. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Masuda H, Bashir K, Inoue H, Tsukamoto T, Takahashi M, Nakanishi H, Aoki N, Hirose T, Ohsugi R, Nishizawa NK. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010;62:379–390. doi: 10.1111/j.1365-313X.2010.04158.x. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Kakei Y, Shimo H, Bashir K, Sato Y, Sato Y, Uozumi N, Nakanishi H, Nishizawa NK. A rice phenolic efflux transporter is essential for solubilizing precipitated apoplasmic iron in the plant stele. J Biol Chem. 2011;286:24649–24655. doi: 10.1074/jbc.M111.221168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng TL, Lin YW, Wang CS, Sung JM. Comparisons and selection of rice mutants with high iron and zinc contents in their polished grains that were mutated from the indica type cultivar IR64. J Food Compos Anal. 2012;28:149–154. [Google Scholar]
- Johnson AAT, Kyriacou B, Callahan DL, Carruthers L, Stangoulis J, Lombi E, Tester M. Constitutive overexpression of the OsNAS gene family reveals single gene strategies for effective iron- and zinc-biofortification of rice endosperm. PLoS ONE. 2011;6:e24476. doi: 10.1371/journal.pone.0024476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano BS. Rice In Human Nutrition. Los Banos: IRRI; 1993. [Google Scholar]
- Kakei Y, Yamaguchi I, Kobayashi T, Takahashi M, Nakanishi H, Yamakawa T, Nishizawa NK. A highly sensitive, quick and simple quantification method for nicotianamine and 2'-deoxymugineic acid from minimum samples using LC/ESI-TOF-MS achieves functional analysis of these components in plants. Plant Cell Physiol. 2009;50:1988–1993. doi: 10.1093/pcp/pcp141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakei Y, Ishimaru Y, Kobayashi T, Yamakawa T, Nakanishi H, Nishizawa NK. OsYSL16 plays a role in the allocation of iron. Plant Mol Biol. 2012;79:583–594. doi: 10.1007/s11103-012-9930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science. 2006;314:1295–1298. doi: 10.1126/science.1132563. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. Iron Uptake Translocation and Regulation in Higher Plants. Annu Rev Plant Biol. 2012;63:131–52. doi: 10.1146/annurev-arplant-042811-105522. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nakanishi H, Takahashi M, Kawasaki S, Nishizawa NK, Mori S. In vivo evidence that IDS3 from Hordeum vulgare encodes a dioxygenase that converts 2′-deoxymugineic acid to mugineic acid in transgenic rice. Planta. 2001;212:864–871. doi: 10.1007/s004250000453. [DOI] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. OsYSL2 is a rice metal-Nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004;39:415–424. doi: 10.1111/j.1365-313X.2004.02146.x. [DOI] [PubMed] [Google Scholar]
- Lee S, An G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009a;32:408–416. doi: 10.1111/j.1365-3040.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Chiecko JC, Kim SA, Walker EL, Lee Y, Guerinot ML, An G. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 2009b;150:786–800. doi: 10.1104/pp.109.135418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Jeon US, Lee SJ, Kim YK, Persson DP, Husted S, Schjørring JK, Kakei Y, Masuda H, Nishizawa NK, An G. Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc Natl Acad Sci USA. 2009c;106:22014–22019. doi: 10.1073/pnas.0910950106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonnerdal B. Soybean ferritin: implications for iron status of vegetarians. Am J Clin Nut. 2009;89:1680S–1685S. doi: 10.3945/ajcn.2009.26736W. [DOI] [PubMed] [Google Scholar]
- Lucca P, Hurrell R, Potrykus I. Fighting iron deficiency anemia with iron-rich rice. J Am Coll Nutr. 2002;21:184–190. doi: 10.1080/07315724.2002.10719264. [DOI] [PubMed] [Google Scholar]
- Maclean JL, Dawe DC, Hardy B, Hettel GP, editor. Rice almanac, third edition, source book for the most important economic activity on earth. Oxfordshire: CABI Publishing; 2002. [Google Scholar]
- Masuda H, Suzuki M, Morikawa KC, Kobayashi T, Nakanishi H, Takahashi M, Saigusa M, Mori S, Nishizawa NK. Increase in iron and zinc concentrations in rice grains via the introduction of barley genes involved in phytosiderophore synthesis. Rice. 2008;1:100–108. [Google Scholar]
- Masuda H, Usuda K, Kobayashi T, Ishimaru Y, Kakei Y, Takahashi M, Higuchi K, Nakanishi H, Mori S, Nishizawa NK. Overexpression of the barley nicotianamine synthase gene HvNAS1 increase iron and zinc concentrations in rice grains. Rice. 2009;2:155–166. [Google Scholar]
- Masuda H, Ishimaru Y, Aung MS, Kobayashi T, Kakei Y, Takahashi M, Higuchi K, Nakanishi H, Nishizawa NK. Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci Rep. 2012;2:534. doi: 10.1038/srep00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Kobayashi T, Ishimaru Y, Takahashi M, Aung MS, Nakanishi H, Mori S, Nishizawa NK. Iron-biofortification in rice by the introduction of three barley genes participated in mugineic acid biosynthesis with soybean ferritin gene. Front Plant Sci. 2013;4:132. doi: 10.3389/fpls.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer JE, Pfeiffer WH, Beyer P. Biofortified crops to alleviate micronutrient malnutrition. Curr Opin Plant Biol. 2008;11:166–170. doi: 10.1016/j.pbi.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Mihashi S, Mori S. Characterization of mugineic acid-Fe transporter in Fe-deficient barley roots using the multicompartment transport box method. Biol Met. 1989;2:164–154. [Google Scholar]
- MOH. National Haemoglobin Survey. National plan of action for food and nutrition 2006-2010. Yangon: Nutrition Section, Department of Health, Ministry of Health (MOH); 2003. [Google Scholar]
- Mori S, Nishizawa N. Methionine as a dominant precursor of phytosiderophore in graminaceae plant. Plant Cell Physiol. 1987;28:1081–1092. [Google Scholar]
- Nakanishi H, Yamaguchi H, Sasakuma T, Nishizawa NK, Mori S. Two dioxygenase genes, Ids3 and Ids2, from Hordeum vulgare are involved in the biosynthesis of mugineic acid family phytosiderophores. Plant Mol Biol. 2000;44:199–207. doi: 10.1023/a:1006491521586. [DOI] [PubMed] [Google Scholar]
- Nishiyama R, Kato M, Nagata S, Yanagisawa S, Yoneyama T. Identification of Zn-nicotianamine and Fe-2′-Deoxymugineic acid in the phloem sap from rice plants (Oryza sativa L.) Plant Cell Physiol. 2012;53:381–390. doi: 10.1093/pcp/pcr188. [DOI] [PubMed] [Google Scholar]
- Nozoye T, Nagasaka S, Kobayashi T, Takahashi M, Sato Y, Sato Y, Uozumi N, Nakanishi H, Nishizawa NK. Phytosiderophore Efflux Transporters Are Crucial for Iron Acquisition in Graminaceous Plants. J Bio Chem. 2011;286:5446–5454. doi: 10.1074/jbc.M110.180026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, Takahashi M, Mori S, Nishizawa NK. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J Exp Bot. 2006;57:2867–2878. doi: 10.1093/jxb/erl054. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, Kobayashi T, Takahashi M, Mori S, Nishizawa NK. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe deficient conditions. Plant J. 2007;51:366–377. doi: 10.1111/j.1365-313X.2007.03149.x. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Kobayashi T, Aung MS, Nakanishi H, Nishizawa NK. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol Biol. 2011;75:593–605. doi: 10.1007/s11103-011-9752-6. [DOI] [PubMed] [Google Scholar]
- Paul S, Ali N, Gayen D, Datta SK, Datta K. Molecular breeding of Osfer2 gene to increase iron nutrition in rice grain. GM Crops Food. 2012;3:310–316. doi: 10.4161/gmcr.22104. [DOI] [PubMed] [Google Scholar]
- Pfeiffer WH, Mcclafferty B. HarvestPlus: Breeding crops for better nutrition. Crop Sci. 2007;47:S88–S105. [Google Scholar]
- Pich A, Scholz G. Translocation of copper and other micronutrients in tomato plants (Lycopersicon esculentum Mill.): nicotianamine-stimulated copper transport in the xylem. J Exp Bot. 1996;294:41–47. [Google Scholar]
- Qu LQ, Yoshihara T, Ooyama A, Goto F, Takaiwa F. Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta. 2005;222:225–233. doi: 10.1007/s00425-005-1530-8. [DOI] [PubMed] [Google Scholar]
- Rudolph A, Becker R, Scholz G, Procházka Z, Toman J, Macek T, Herout V. The occurrence of the amino acid nicotianamine in plants and microorganisms: a reinvestigation. Biochemie und Physiologie der Pflanzen. 1985;180:557–563. [Google Scholar]
- Ruengphayak S, Ruanjaichon V, Phromphan S, Chaichumpoo E, Saensuk C, Toojinda T, Tragoonrung S, Kongkachuichai R, Vanavichit A. Reverse and forward genetics approaches to identify mutants conferring high grain Fe density and tolerance to Fe toxicity. Chiang Mai: Paper presented at the 10th International Symposium on Rice Functional Genomics, Lotus Hotel Pang Suan Kaew; 2012. [Google Scholar]
- Schroeder JI, Delhaize E, Frommer WB, Guerinot ML, Harrison MJ, Herrera-Estrella L, Horie T, Kochian LV, Munns R, Nishizawa NK, Tsay YF, Sanders D. Using membrane transporters to improve crops for sustainable food production. Nature. 2013;497:60–66. doi: 10.1038/nature11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield GN, Hirose T, Aoki N, Furbank TR. Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J Exp Bot. 2007;58:3155–3169. doi: 10.1093/jxb/erm153. [DOI] [PubMed] [Google Scholar]
- Sperotto RA, Boff T, Duarte GL, Santos LS, Grusak MA, Fett JP. Identification of putative target genes to manipulate Fe and Zn concentrations in rice grains. J Plant Physiol. 2010;167:1500–1506. doi: 10.1016/j.jplph.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Stephan UW, Schmidke I, Stephan VW, Scholz G. The nicotianamine molecule is made-to-measure for complexation of metal micronutrients in plants. Biometals. 1996;9:84–90. [Google Scholar]
- Stoltzfus RJ, Dreyfuss ML. Guidelines for the use of iron supplements to prevent and treat iron deficiency anemia. Washington DC: ILSI Press; 1998. [Google Scholar]
- Suzuki M, Morikawa KC, Nakanishi H, Takahashi M, Saigusa M, Mori S, Nishizawa NK. Transgenic rice lines that include barley genes have increased tolerance to low iron availability in a calcareous paddy soil. Soil Sci Plant Nutr. 2008;54:77–85. [Google Scholar]
- Takagi S. Naturally occurring iron-chelating compounds in oat-and rice-root washings. Soil Sci Plant Nutr. 1976;22:423–433. [Google Scholar]
- Takahashi M, Yamaguchi H, Nakanishi H, Shioiri T, Nishizawa NK, Mori S. Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (Strategy II) in graminaceous plants. Plant Physiol. 1999;121:947–956. doi: 10.1104/pp.121.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S. Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes. Nat Biotech. 2001;19:466–469. doi: 10.1038/88143. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK. Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell. 2003;15:1263–1280. doi: 10.1105/tpc.010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T, Nishizawa NK, Nakanishi H. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012;35:1948–1957. doi: 10.1111/j.1365-3040.2012.02527.x. [DOI] [PubMed] [Google Scholar]
- Theil EC. Ferritin: at the crossroads of iron and oxygen metabolism. J Nutr. 2003;133:1549–1553. doi: 10.1093/jn/133.5.1549S. [DOI] [PubMed] [Google Scholar]
- Theil EC. Iron Homeostasis and Nutritional Iron Deficiency. J Nutr. 2011;141:724S–728S. doi: 10.3945/jn.110.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil EC, Chen H, Miranda C, Janser H, Elsenhans B, Núñez MT, Pizarro F, Schümann K. Absorption of Iron from Ferritin Is Independent of Heme Iron and Ferrous Salts in Women and Rat Intestinal Segments. J Nutr. 2012;142:478–483. doi: 10.3945/jn.111.145854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos M, Datta K, Oliva N, Khalekuzzaman M, Torrizo L, Krishnan S, Oliveira M, Goto F, Datta SK. Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci. 2003;164:371–378. [Google Scholar]
- von Wirén N, Khodr H, Hider RC. Hydroxylated phytosiderophore species from rye and barley possess an enhanced chelating efficiency and affinity for iron(III) Plant Physiol. 2000;124:1149–1157. doi: 10.1104/pp.124.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Gruissem W, Bhullar NK. Nicotianamine synthase overexpression positively modulates iron homeostasis-related genes in high iron rice. Front Plant Sci. 2013;4:156. doi: 10.3389/fpls.2013.00156. doi:10.3389/fpls.2013.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. The World Health Report. Reducing Risks, Promoting Healthy Life. Geneva: Word Health Organization (WHO); 2002. [Google Scholar]
- Wirth J, Poletti S, Aeschlimann B, Yakandawala N, Drosse B, Osorio S, Tohge T, Fernie AR, Günther D, Gruissem W, Sautter C. Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotech J. 2009;7:1–14. doi: 10.1111/j.1467-7652.2009.00430.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu YH, Yi HY, Gong JM. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012;72:400–410. doi: 10.1111/j.1365-313X.2012.05088.x. [DOI] [PubMed] [Google Scholar]


