Abstract
Over the past 25 years the broad field of epigenetics, and over the past decade in particular the emerging field of neuroepigenetics, have begun to have tremendous impact in the areas of learned behavior, neurotoxicology, CNS development, cognition, addiction, and psychopathology. However, epigenetics is such a new field that in most of these areas the impact is more in the category of fascinating implications as opposed to established facts. In this brief commentary I will attempt to address and delineate some of the open questions and areas of opportunity that discoveries in epigenetics are providing to the discipline of neuroscience.
Keywords: histone, methylation, active demethylation, nurture, development, learning, memory, transcription factor, gene expression, chromatin, DNA, histone, acetylation, epigenesis, DNA recombination, epigenetic, HDAC, drug addiction, imprinting, transgenerational effects, metaplasticity, homeostatic plasticity, neuroepigenetic
Introduction
Only infrequently do scientific discoveries force the recasting of a centuries-long philosophical debate. However, over the last 25 years and indeed largely over the last decade the emerging field of neuroepigenetics has necessitated the reformulation of the fundamental existential question of “nature versus nurture” (Sweatt, 2009). Based on recent discoveries in the broad field of epigenetics it no longer makes sense to debate nature versus nurture. There is no longer a mechanistic dichotomy between nature and nurture (or genes and environmental experience, as is the more modern phrasing). Rather, it is now clear that there is a dynamic interplay between genes and experience, a clearly delineated and biochemically driven mechanistic interface between nature and nurture. That mechanistic interface is epigenetics.
The term epigenetics derives from Waddington’s coining of the term “epigenesis” to capture his logical deduction that during organismal development a layer of mechanisms must surely exist that reside above (epi) the level of the genes, that control their output in order to specify cell fate determination. In terms of the underlying biochemistry, there are two main epigenetic mechanisms, DNA methylation and regulation of chromatin structure via histone modifications (although see Table 1). These mechanisms have been mostly explored in the context of organismal development. However, it is now clear that experience, be it environmental toxins, maternal behavior, psychological or physical stress, learning, drug exposure, or psychotrauma leads to active regulation of the chemical and three-dimensional structure of DNA in the nervous system, i.e. that experience regulates epigenetic mechanisms in the CNS (Borelli et al, 2008; Champagne and Curley, 2009; Day and Sweatt, 2010; Dulac, 2009; Renthal and Nestler, 2008). These epigenomic changes lead to alterations in gene readout (and who knows what else?) in cells in the nervous system that trigger lasting and in some cases perpetual changes in neural function.
Table 1.
Major Biochemical Mechanisms in Neuroepigenetics.
| Covalent Modification of DNA |
| DNA Cytosine Methylation |
| Active Cytosine DeMethylation |
| Hydroxymethyl-Cytosine Formation |
| Methyl-Cytosine Oxidation (5-formyl-Cytosine, 5-carboxyl-Cytosine) |
| Histone Post-translational Modifications |
| Lysine Acetylation |
| Lysine (mono/di/tri) and Arginine (mono/di) Methylation |
| Serine/Threonine Phosphorylation |
| Mono-Ubiquitination |
| Poly ADP-ribosylation |
| ATP-dependent Chromatin Remodeling (SWI-SNF) |
| Histone Subunit Exchange |
| H2A.Z |
| H3.3 |
| RE1-Silencing Transcription Factor (REST)/REST Co-repressor (Co-REST)/Sin 3A System |
| Non-Coding RNAs |
| piRNAs |
| microRNAs |
| small interfering RNAs (siRNAs) |
| small nuclear RNAs (snRNAs) |
| Line 1 Retrotransposition |
| Prion Protein-based Mechanisms |
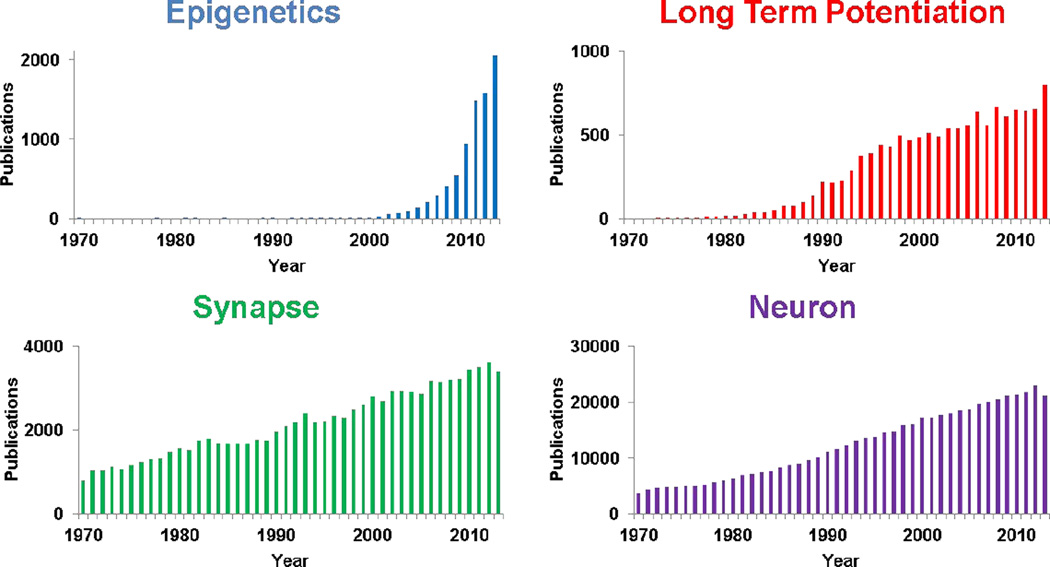
The field of epigenetics has undergone an exponential expansion of late. A quick check of the Pubmed publication database reveals that about 98% of all the research published in the broad area of epigenetics was published within the last 15 years. The search term epigenetics returns 1 publication in 1989, i.e. the year after Neuron was established. Last year (2012) over 1500 papers were published on epigenetics, an orders-of-magnitude increase over the 25-year time-span that is the focus of the this special Neuron anniversary issue. Interesting comparison searches for neuroscientists are memory, synapse, and long-term potentiation, to place these numbers in context (see Figure 1).
Figure 1. Yearly publications for the Pubmed search terms “epigenetics”, “long term potentiation”, “ synapse” and “ neuron” for the years 1970–2013.
Yearly data were generated using the Pubmed website and their metrics re-plotted for this figure. Please note that the numbers for the final year of these graphs (2013) are projections based on the numbers of publications in each category as of September 25, 2013.
The Biochemical Mechanisms of Epigenetics
I will not go into detail concerning the basic molecular and biochemical mechanisms that comprise the established epigenetic toolkit, because those mechanisms have been reviewed extensively in a number of other prior publications (Allis et al, 2007; Campos and Reinberg, 2009; Lee et al 2010; Levenson and Sweatt, 2006; Turner, 2007), and the topic is too broad to address in a short perspective article. However in Table 1 I have listed the major (known and emerging) players in the arena of neuroepigenetics, to introduce terms and provide some basic background. I also will briefly describe the major epigenetic molecular mechanisms listed in Table 1 in the following few paragraphs in order to help make the rest of this perspective piece comprehensible to those readers new to the epigenetics milieu. Thus, I will introduce a few terms that one needs to be familiar with before I launch into discussion of the “open questions in epigenetics” section that is the main thrust of this perspective piece. Please keep in mind that in only a few paragraphs I will be sparsely summarizing and broadly oversimplifying the results of literally thousands of recent publications – for a more comprehensive treatment please see the references by Allis et al (2008) and Sweatt et al (2013).
It is worth noting that all these modifications I will describe have the basic biochemical characteristics of both regulating gene function (transcription) without altering the DNA sequence directly, and of being (at least theoretically) capable of self-regeneration and self-perpetuation. In other words, of having the capacity to trigger a persisting change in gene function even in the face of subsequent cell division or even organismal procreation. The biochemical capacity of a specific chemical reaction to trigger self-perpetuation is the defining characteristic of a process involved in cellular information storage, as was initially commented upon in the neuroscience context by Francis Crick and John Lisman almost 30 years ago (Crick, 1984; Lisman, 1985). Epigenetic mechanisms also possess this defining characteristic (Holliday, 1999).
The Epigenetic Tookit
Covalent chemical modification of DNA, specifically cytosine 5’-methylation, has been referred to as the prima donna of epigenetics, because it is an extremely powerful regulator of gene transcription (Santos et al 2005). As a first approximation, DNA methylation is the proximal molecular mechanism that triggers, and perpetuates over the full lifespan, the complete gene silencing in cells that is part and parcel of cell fate determination and perpetuation (Bird, 2002). DNA cytosine methylation is a core mechanism for silencing all the non-neuronal genes in all the cells in the body that are not neurons, for example. DNA cytosine methylation is the core driver of the “epigenesis” mechanism that Waddington postulated to exist (Holliday, 2006). In the existing literature DNA cytosine methylation is described as occurring preferentially at cytosine-guanine dinucleotide sequences in DNA (so-called CpG sites), and is said to lead to attenuation of gene transcription. These generalizations are largely true, but based on recent discoveries it is clear that cytosine methylation also occurs at non-CpG sites, and that cytosine methylation can also be associated with transcriptional activation. This is the ambiguous nature of newly emerging fields.
Besides DNA cytosine methylation, other chemical modifications of cytosine in DNA have also been documented to exist, including 5-hydroxymethyl-Cytosine (hmC) formation and methyl-Cytosine oxidation to generate 5-formyl-Cytosine and 5-carboxyl-Cytosine. The functional role(s) of these novel modifications not fully established, and is a hot area of investigation in the field at present.
A central dogma of the epigenetics field has been that once DNA methylation patterns are established upon the genome in terminally differentiated cells, those modifications are permanent and essentially immutable. This view is aligned with the original conception of epigenesis by Waddington, wherein he reasoned that such mechanisms are necessary to perpetuate cellular phenotype over an entire lifespan (Holliday, 2006). However, of late it has become clear that so-called active cytosine de-methylation also occurs, wherein a previously-methylated cytosine can undergo a net re-conversion back to the un-methylated state. This mechanism (while likely rare in the overall context of the entire genome and epigenome) appears to be particularly prominent in two places: in the mature nervous system and in the fertilized zygote undergoing generation of totipotent embryonic stem cells. In other words, in the two most highly plastic tissues in the body. We will return to this idea later in the “open questions” section.
Histone post-translational modifications are the second major category of epigenetic biochemical mechanisms in cells, and this area has a broad and rich literature (Jenuwein and Allis, 2001). Histone post-translational modifications that have functional consequences on gene readout are multitudinous, including: lysine acetylation; lysine mono/di/tri-methylation; arginine mono/di-methylation; serine/threonine phosphorylation; histone mono-ubiquitination, and histone poly ADP-ribosylation. In the nucleus histone proteins exist largely as octameric complexes, which make up the core of the chromatin particle around which most DNA is wrapped, forming a three-dimensional histone/DNA complex that is itself a powerful regulator of transcriptional efficacy. Histone post-translational modifications regulate this structure in order to modulate transcriptional readout of the associated gene.
Individual isoforms of histone monomers can also be swapped in and out of the octamer, a regulatory mechanism referred to as histone subunit exchange. The histone H2A.Z and H3.3 isoforms, among others, are prominent participants in these subunit exchange mechanisms, and also regulate transcriptional efficacy in a manner reminiscent of histone post-translational modifications. Subunit exchange and post-translational modifications trigger either increases or decreases in transcription, depending upon the particular modification, the particular histone isoform involved, and even the context of other histone modifications in which the modification resides. This attribute of these mechanisms has given rise to the concept of a histone code, wherein histone modifications are interpreted in situ as a combinatorial code regulating gene transcription rates at specific loci across the genome (Jenuwein and Allis, 2001; Borelli et al, 2008, Lee et al 2010; Strahl and Allis, 2000; Wang et al 2010). The implications of this sort of molecular/cellular information processing within neurons is only beginning to be considered and addressed at present (Wood et al, 2006).
A variety of other epigenetic molecular mechanisms are also in play in neurons, however I will only be able to touch on these briefly due to space limitations.
ATP-dependent chromatin remodeling, which involves the SWI-SNF biochemical regulatory system, can regulate the affinity of the histone octamer core particle for its associated DNA, and in an energy-dependent fashion promote gene transcription through “loosening” chromatin 3-dimensional structure. This mechanism has specifically been implicated in human cognitive function based on genetic studies of intellectual disabilities (Ronan et al 2013).
The RE1-Silencing Transcription Factor (REST)/REST Co-repressor (Co-REST)/Sin 3A system is a well-established player in neuronal/non-neuronal cell fate determination, and indeed is likely the best-understood epigenetic mechanism in play related to neuronal function (Ballas and Mandel, 2005). This is a core mechanism that silences non-neuronal genes in non-neurons, and conversely that allows the broad segment of the genome that is specifically necessary for neuronal function to be selectively expressed in nerve cells.
A wide variety of non-coding RNAs have either been shown to be or hypothesized to be involved in regulating cell function in the nervous system, including: piRNAs, microRNAs, small interfering RNAs (siRNAs), and small nuclear RNAs (snRNAs) (Sun et al 2013; Tardito et al 2013). These mechanisms have in common the exquisite capacity for nucleotide sequence-specific effects, allowing them to affect the function of particular genes with high specificity. This is a burgeoning area for all of biology including most recently neurobiology.
Other relevant mechanisms include LINE 1 (Long Interspersed Nuclear Element 1, aka L1) retrotransposition, in which the L1 class of repeat sequences can recombine and re-insert themselves into the genome using a copy-and-paste mechanism. Through this mechanism L1 elements can dramatically affect gene transcription, and indeed the elements themselves are capable of self-regeneration – thus they qualify as “epigenetic” mechanisms based on these unifying criteria. However L1 element recombination does not fit the classical definition of an epigenetic mechanism by virtue of the fact they modify gene transcription through changing the genomic nucleotide sequence. Regardless of this, L1 retrotransposition in neurons has largely been semantically regarded as an epigenetic mechanism in the CNS due to its striking functional similarity to other epigenetic biochemical mechanisms. One compelling current model for L1 function in neurons is that the mechanism drives cellular heterogeneity at the genomic and functional level through insertional mutagenesis (Muotri and Gage, 2006). The broad context is that this allows individual neurons to achieve genomic diversity and distinction from their siblings, broadening the spectrum of cellular phenotypes driven by the single available genome in any particular neuronal subtype.
Finally, a provocative mechanism that has been proposed to be important in sustaining long-term function changes in neurons are prion protein-based mechanisms. Prion proteins function in yeast as non-genic heritable elements that potently regulate cellular function and phenotype. These prion protein elements are heritable, self-regenerating, and alter gene function, placing them clearly in the epigenetic realm of biochemical processes. Recent studies in the Aplysia model system for studying synaptic plasticity and memory have implicated a prion-protein-like mechanism as being a long-term controller of synaptic efficacy, specifically acting through the Aplysia cytoplasmic polyadenylation element binding protein (ApCPEB, Bailey et al, 2004; Si et al, 2004). This represents a particularly intriguing candidate for a novel epigenetic mechanism operating to regulate neuronal function.
The Emerging Sub-Discipline of Neuroepigenetics
Over the last decade there has been a great expansion of the number of research papers and reviews published concerning epigenetic mechanisms in the nervous system, especially as related to adult CNS function. These burgeoning neuroscience discoveries have necessitated a redefinition of epigenetics, at least in regard to epigenetic mechanisms in adult neurons (refs). As mentioned already, epigenetic mechanisms were originally defined as heritable either in a procreative organismal sense or at the cellular level across cell division. However, the discovery that those biochemical mechanisms listed in Table 1 are operating in adult neuronal function forces a re-assessment; because adult neurons are non-dividing cells obviously nothing happening in them is heritable in the traditional sense. An epigenetic molecular mark in an adult neuron can be long-lasting, permanent and self-regenerating, but cannot be inherited by a daughter cell since the neuron does not divide. This sets apart the roles of epigenetic mechanisms in adult neurons from their roles in developmental biology such as perpetuation of cell fate determination, heritability, genomic imprinting, etc. For this reason, along with other unique attributes of the role of epigenetic molecular mechanisms in adult CNS function, Jeremy Day and I have proposed adopting the term neuroepigenetic to help capture this distinction (Day and Sweatt, 2010). Regardless of that specific set of semantic conventions, it also seems clear that the term neuroepigenetic is emerging due to the discoveries of a wide variety of roles for epigenetic molecular mechanisms in the CNS regarding acquired behaviors, CNS disorders, neural plasticity, neurotoxicity, and drug addiction (Table 2). Thus, we have the emerging subdiscipline now being called neuroepigenetics.
Table 2.
Areas Where Epigenetic Mechanisms have been Implicated in Human Nervous System Function.
| Function or Disorder | Mechanism(s) Implicated |
|---|---|
| Learning and Memory | Histone Modifications, DNA Methylation piRNAs, miRNAs |
| Maternal Nurturing | Histone Modifications, DNA Methylation |
| Adult Neurogenesis | Histone Modifications, DNA Methylation |
| Stress Responses | Histone Modifications, DNA Methylation |
| Alzheimer’s Disease | Histone Modifications, DNA Methylation |
| Rett Syndrome | MeCP2 Methyl-Cytosine Binding |
| Fragile X Mental Retardation | DNA Methylation, miRNAs |
| Schizophrenia | DNA and Histone Methylation, miRNAs |
| Rubinstein-Taybi Syndrome | Histone Acetyl Transferase Deficiency |
| Angelman Syndrome | Genomic Imprinting (DNA Methylation) |
| Depression/Suicide | DNA Methylation |
| Bipolar Disorder | Histone Modifications, DNA Methylation miRNAs |
| Addiction and Reward Behavior | Histone Modifications, DNA Methylation miRNAs |
| PTSD | Histone Modifications, DNA Methylation |
| ATR-X Syndrome (α-Thalassemia MR) | SNF2 Chromatin Remodeling, H3.3 |
| Cognitive Aging | Histone Modifications, DNA Methylation |
| Coffin-Lowry Syndrome | Histone Phosphorylation |
| Kleefstra Syndrome | Histone Methylation |
| Epilepsy | Histone Modifications, DNA Methylation miRNAs |
| Autism | Histone and DNA Methylation? miRNAs? |
A Daunting Dozen for the Emerging Sub-Discipline of Neuroepigenetics
For the remainder of this commentary, I will present my perspective concerning several open questions in neuroepigenetics at present and for the next decade or so. I have tried to orient my thoughts towards capturing some of the most challenging, but vitally important, avenues of pursuit open to the field. I fully realize that this is an incomplete list and that others working in the area, such as Eric Nestler, Ted Abel, Li-Huei Tsai, Michael Meaney, and Schahram Akbarian, would come up with different lists (Sweatt et al, 2013). My hope in presenting the following brief thoughts is that they might catalyze debate, discussion, and further experimentation concerning the informational voids that are outlined.
With this in mind, here are my own personal “daunting dozen” hot questions in neuroepigenetics:
1. Are epigenomic marks a piece of the engram?
Epigenetic molecular mechanisms certainly are a component of developmental information storage, playing critical roles in cell fate determination and lifelong perpetuation of cellular phenotype in both dividing and non-dividing cells. This is the scientific context in which epigenetic mechanisms were originally proposed to exist, and in which they were discovered at the molecular level. A broader question is whether epigenetic mechanisms might be a more universal mechanism for cellular information storage, operating to subserve plastic change in the adult CNS and learned behavior at the organismal level.
The ability to form memories about both negative and positive biological and emotional events is critical for human adaptive behavior and decision making. Recent studies from a number of laboratories has demonstrated a role for active DNA methylation and demethylation in regulating learning and memory formation in the mammalian CNS (see Day and Sweatt, 2011 for a review). Our understanding of this basic process is beginning to have a far-reaching impact across disciplines, shedding new light on scientific research into learning, memory, addiction, stress disorders, and decision making. Thus, in recent years, epigenetic modifications of DNA and chromatin have been identified as essential mediators of memory formation through the regulation of gene expression (Sultan and Day, 2011), with methylation of cytosines at CpG dinucleotides playing a critical role in memory consolidation and stabilization over time (Feng et al., 2010a; Lubin et al., 2008; Miller et al., 2010; Miller and Sweatt, 2007; Monsey et al., 2011). However, a question in the field that has been only sparsely investigated is whether epigenetic mechanisms are necessary for ongoing storage of memory (ref Miller et al, 2010; Lesburgueres et al, 2011) – in other words, are epigenetic mechanisms a cog in the machinery of the engram? Answering this question will have important implications both regarding the long-standing question of the molecular biology of the engram and whether there are universally shared biochemical mechanisms for cellular information storage.
2. How do cell-wide epigenomic changes interface with synapse-specific mechanisms of neural plasticity?
One of the most intriguing aspects of epigenetic mechanisms is that they typically operate to drive cell-wide changes in gene expression. Given the emerging role of epigenetic mechanisms in learning and memory, this raises an apparent conundrum: how do cell-wide changes in the neuron that are driven by nuclear epigenomic marks fit into the well-established necessity for synapse-specific plasticity as a mediator of memory? One possibility is that they interdigitate with molecular species such as synaptic tags in order to participate in synapse-specific changes (Day and Sweatt, 2010). However, an intriguing alternative possibility is that epigenetic changes play to their strengths, and are purposely utilized for driving cell-wide functional changes. Thus, a speculative notion is that the neuronal epigenome may be preferentially involved in non-Hebbian plasticity. For example, epigenetic molecular mechanisms may be particularly relevant to various forms of metaplasticity, operating to establish a set-point for biasing the entire cell toward or against being susceptible to synapse-specific plasticity mechanisms such as long-term potentiation. Similarly, the neuronal or glial epigenome might be allocated to controlling intrinsic properties that are themselves cell-wide, such as excitability and activity-dependent synaptic scaling. Conceptually the epigenome, having the capacity to control the entire genomic output and sense pan-cellular signaling mechanism, might be the ideal control point for achieving coordinated orchestration of the readout of a plethora of ion channels, receptors, and trafficking mechanism in order to achieve homeostatic plasticity.
3. How is DNA methylation actively regulated in neurons?
While early studies identified 5- methyl-Cytosine as a stable transcriptional silencer based on its role in tissue-specific gene expression, X chromosome inactivation, and gene imprinting (Bonasio et al., 2010; Feng et al., 2010b), new evidence of rapid and reversible changes in DNA methylation at memory-associated genes implies the presence of both active DNA methylation and active DNA demethylation process in response to neuronal activity (see Miller and Sweatt, 2007; Lubin et al, 2008 for examples). The upstream signaling mechanisms that control both activity-dependent inducible increases in methylation and active cytosine demethylation in the nervous system are completely mysterious at present. Those signaling mechanisms regulating histone modifications are better understood, but our understanding of even those pathways may be best described as a working sketch (Bonasio et al, 2010). Thus, an important area for further research is investigating how things like action potential firing, membrane depolarization, and neurotransmitter/hormone receptor activation signal the epigenome to change. By extension, an open question in all of epigenetics is how histone modifications interact with the cytosine methylation apparatus in order to trigger and perpetuate changes in epigenomic structure.
The recent discovery of novel oxidative modifications of methyl-cytosine in the nervous system is quite exciting, and these findings further enrich the picture concerning how DNA methylation is regulated in the nervous system. Hydroxy-methyl-Cytosine is emerging as the active demethylation mark that targets a specific 5’-methyl group on cytosine for “net” removal by a complex base excision repair mechanism (Guo et al, 2011a, 2011b). Moreover, as was already mentioned, hmC is of highest abundance in the fertilized zygote and the adult CNS, tissues that might be thought of as hyper-plastic relative to other cell types in mammals, further implicating this chemical species as a potential plasticity mechanism for the epigenome.
Consistent with this idea hmC is involved as a specific mechanism for active cytosine demethylation, recent studies identified the Ten-eleven translocation (Tet) family of proteins in active DNA demethylation (Ito et al., 2010; Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Specifically, Tet1, Tet2 and Tet3 enzymes regulate the oxidation of 5mC to 5-hydroxymethyl cytosine (5hmC) (Ito et al., 2010; Kriaucionis and Heintz, 2009; Tahiliani et al., 2009), which is deaminated to 5-hydroxyuracil (5hmU) (Guo et al., 2011b; Popp et al., 2010; Zhu, 2009) to create a 5hmU:G mismatch that is recognized and removed by one of several glycosylases. This abasic site is then repaired by the base excision repair (BER) machinery, resulting in overall demethylation of a specific cytsosine. Further Tet-mediated oxidation of 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) can also occur prior to glycosylase excision and BER (Ito et al., 2011).
Recent studies specifically investigating the role of TET1 oxidase in the nervous system provided direct evidence for this model of TET Oxidase control of active DNA demethylation in the CNS, and indeed of a role for this pathway in memory formation and storage (Kaas et al, 2013; Rudenko et al, 2013). Overall these results mark a substantial advance and also reveal new information about how plasticity of neuroepigenetic marks regulates activity-dependent processes within the central nervous system.
4. How is one specific cytosine from a 3-billion-nucleotide cellular genome specified for chemical modification?
This is one of the biggest open questions in all of epigenetics, not just neuroepigenetics, and applies equally to both methylation and demethylation. It is clear on its face that mechanisms for identifying genomic sites for selective epigenetic modification must exist – the epigenome has specificity and structure, with specific individual genes, exons, promoter regions, gene bodies, alleles, and even specific cytosines being methylated or demethylated. Moreover, these modifications can occur at both CpG sites and non-CpG sites, so even the previously-held minimal methylation consensus sequence of a C-G dinucleotide no longer holds. But there is no current mechanistic explanation for how this specificity of cytosine methylation can happen. I and many others in the field speculate, based on first principles, that the mechanisms must be directed to specific loci in some fashion based on nucleotide sequence – it seems to be the only component of the system with adequate informational content. In this regard, non-coding RNAs serving as a targeting template is one appealing mechanism. Indeed the recent landmark finding from the Kandel lab regarding piRNAs directing activity-dependent site-specific DNA methylation in Aplysia sensory neurons may be key insight (Rajasethupathy et al, 2012; Landry et al, 2013). However, it is unclear if those mechanisms operate in the mammalian CNS at present and this is a hypothesis that warrants vigorous investigation.
5. What roles do epigenetic mechanisms play in complex human diseases of the nervous system?
As illustrated in the list in Table 2, this is an extremely broad question and obviously I cannot discuss it in detail in a brief commentary such as this. However, I will note that epigenetic mechanisms may be particularly relevant to multifactorial diseases with low genetic penetrance, such as schizophrenia and depression (Patronis, 2010). Thus, epigenomically based mechanisms for these disorders may help fill a void where historically genomic analyses have not led to clearly identifiable causes. In addition, disorders that are triggered by one single or only a few experiences, but that are henceforth enduring, also seem likely candidates to be epigenetically mediated. In this line of thinking, disorders such as drug addiction, PTSD, epilepsy, and schizophrenia might selectively involve the co-optation of epigenetic mechanisms used for development and learned behavior to subserve behaviorally disadvantageous but obdurate behavioral change.
6. Will epigenetically targeted drugs allow the development of novel neurotherapeutics?
This is the corollary to the preceding question – if epigenetic mechanisms are broadly involved in CNS disorders, might epigenetic targets as a category be broadly applicable to drug development? This is a very active area of investigation at present, with drug discovery efforts ongoing in the areas of: cognitive enhancers for learning disabilities, Alzheimer’s Disease, neurodegenerative disorders, schizophrenia, depression, addiction, generalized stress disorders, and PTSD (Kazantsez and Thompson, 2008; Fischer et al, 2007; Anier et al 2010; Kilgore et al, 2010; Peleg et al 2010; Renthal and Nestler, 2008; Szyf, 2009, Monsey et al, 2011; Oliviera et al, 2012).
7. Are acquired epigenetic marks transmitted across generations?
Some aspects of this question are among the most contentious areas in the epigenetics field at present. Broadly speaking, epigenetic “transgenerational” effects come in two flavors. The first type is not transgenerational in the heritable sense, but rather is experience-dependent. For example, Michael Meaney’s group, his collaborators and scientific descendents have demonstrated that maternal nurturing behavior regarding newborn pups triggers DNA methylation changes in CNS glucocorticoid receptors of offspring that persist into the adult and effect behavioral change (Champagne and Curley, 2009; Weaver et al 2004, 2005). Discovery of these experience-dependent changes in the epigenome is the prototype for the first category of transgenerational effects, and such experience-driven epigenomic changes in the CNS have been documented to occur with a number of both positive and negative environmental effects in offspring. Thus, several examples of the persisting CNS epigenomic effects on offspring of parental behavior and environmental insult have survived the rigors and skepticism of peer review (Champagne and Curley, 2009; Roth et al 2009). At the most basic level these studies demonstrate that early-life experience can trigger lifelong persisting epigenomic changes in brain of an individual, an observation which has clear implications for how epigenetic mechanisms might contribute to CNS health and pathogenesis over the lifespan.
The second flavor of transgenerational effect is the idea that experience-driven epigenetic changes in an animal might lead to heritable DNA methylation changes that are propagated through the germline, through many or all subsequent generations. This neo-Lamarckian scenario is a truly frightening possibility with interesting implications for topics such as free will, and is being hotly debated even as a possibility at present. A presumed evolutionary role for these types of mechanisms is “soft inheritance”, wherein environmental experience/exposure could trigger heritable epigenomic changes that improve survival over a few generations, but that are ultimately reversible because they are based upon epigenomic changes (epimutations) and not upon directly altering the offsprings’ DNA nucleotide sequence. There are several tantalizing and fascinating indications of experience-dependent heritable changes in the CNS epigenome in the literature at this point, involving maternal behavior, paternal behavior, diet, exposure to drugs of abuse, and endocrine disruption (Bohacek et al, 2013). Definitively determining whether experience-driven acquired epigenetic changes can propagate through the germline and effect behavioral change in subsequent generations is one of the most important areas of contemporary neuroepigenetics research in my opinion. Proof of the existence of such mechanisms has the potential to fundamentally change our outlook on evolutionary biology, psychobiology, and neurophilosophy.
8. What is the role of DNA recombination in CNS development and function?
In the background section above I included a brief description of LINE 1 retrotransposition in neurons, in which the L1 class of repeat sequences recombine and re-insert themselves into the genome. As I already alluded to, strictly speaking this is not an “epigenetic” mechanism because it involves a change in nucleotide sequence. However, this area has been “adopted” by neuroepigeneticists because the mechanistic and functional roles are so similar to epigenetic mechanisms, and this mechanism fits quite well into the current novelty and mysteriousness of epigenetic mechanisms in the nervous system.
The existence of this mechanism in neurons in the CNS implies the existence of a biochemical system that is capable of producing genomic diversity at the level of individual neurons, which in principle would be a potent force for generating idiosyncratic genotypes (presumably useful) for specific neurons or subgroups of neurons. The laboratory of Rusty Gage has led the way in establishing the existence of this mechanism in the CNS (Muotri and Gage, 2006). Testing the functional roles of this type of genomic diversity, both in the workings of the individual cell and in the context of organismal behavior may result in paradigm-shifting models for control of neuronal function. Regardless of the precise functional model for the effects of these transposable genetic elements in neurons, the existence of the requisite molecular machinery in the CNS is clear. Documentation of the phenomenon of genomic plasticity in the brain is iconoclastic in its own right, potentially akin to the discovery of DNA recombination as a driver for antibody diversity in the immune system.
9. What new roles for epigenetic mechanisms in neural development await discovery?
Epigenetic mechanisms were of course first hypothesized to exist, and then discovered to exist, in the field of developmental biology (Ng and Gurdon, 2008; Tate et al, 1996; Feng et al 2010b). Thus, our understanding of the developmental roles of epigenetic mechanisms is the most mature area of this relatively young field. Epigenetic mechanisms are a core process driving cell fate determination and especially cell fate perpetuation. However, by no means does this imply that novel developmental epigenetic regulators are not out there to be found, nor that distinct developmental uses of know mechanism cannot exist. This represents a rich field for additional research, especially in my opinion as relates to non-coding RNAs and their role in CNS development.
Moreover, the existing developmental models of epigenomic effects are largely based in the broad concept that epigenetic marks are essentially immutable once laid down, in order to perpetuate cellular phenotype over time. The new understanding of dynamic regulation of DNA methylation in the nervous system forces a re-thinking of the basic tenet of epigenetics. What new mechanisms are there to be found in terms of active regulation of the epigenome during neuronal, glial, and nervous system development, especially regarding the effects of neural activity and behavioral experience as it shapes the developing nervous system? Furthermore, the plastic nature of the neural epigenome has immense implications for “neurodevelopmental” disorders that were previously assumed to be irreversible, given that cells in the CNS might be subject to epigenetic reprogramming later in life (Ehninger et al 2008, Weeber and Sweatt, 2002, Jiang et al 1998).
Finally, a particularly intriguing area regarding this overall question is the phenomenon of genetic imprinting, wherein the paternal or maternal allele of a gene can be epigenetically tagged to modify its function. Allelic imprinting can go so far as to completely silence one allele of a given gene is a cell type or brain region. It has been proposed that imprinting mechanisms may bias one allele to be preferentially used at one developmental stage, essentially preserving an epigenetically fresh copy of the same gene for distinct epigenetic regulation somewhere down the time-line (Day and Sweatt, 2011; Gregg et al, 2010a, 2010b). Testing this idea awaits further investigation.
10. How can a single cell have both a stable developmental epigenome and a dynamically regulated one as well?
The discussion in point number nine leads us to the next question, because on its face it seems that there must be unique mechanistic challenges to utilizing the biochemical machinery that allows dynamic activity-dependent modification of the epigenome in a terminally differentiated and non-dividing cell such as a neuron (Feng et al 2010b). Thus, active regulation of epigenetic marks in a neuron must exist simultaneously alongside the stable epigenetic marks that perpetuate neruonal phenotype over the lifespan. How can a single genome be subject to both perpetual and immutable epigenetic marking at the same time it is subject to dynamic regulation in response to experience? This thought experiment tells us that some set of mechanisms must compartmentalize the “developmental” epigenome from the “dynamic” epigenome. These mechanisms are completely mysterious at present.
11. What is the extent of involvement of protein-based epigenetic mechanisms in nervous system regulation?
As described in the introductory section, epigenetic mechanisms are so powerful because they can self-perpetuate over time. Indeed this peculiar aspect of epigenetics is why Francis Crick first proposed DNA methylation as a component of memory storage in the nervous system, in a personal correspondence to the editor at Nature (Crick, 1984). However, as described above self-perpetuating epigenetic mechanisms are not limited to DNA modifications - prion-like mechanisms, histone subunit exchange, and histone methylation all have the demonstrated or at least hypothetical capacity for self-regeneration in the face of protein turnover. Presumably other self-reinforcing protein-based mechanisms await discovery, and their potential roles in neuronal information storage are tantalizing.
12. Can we really understand the CNS epigenome?
My final question for this perspective piece is whether as scientists we will ever be able to fully comprehend the mechanistic roles of neuroepigenetic mechanisms in any sort of compelling, understandable, and satisfying fashion. It might be a reality that the neuroepigenetic mechanisms operating in the CNS are so complex as to defy comprehensive explanation and understanding. I certainly hope this is not the case! But as a closing comment I would like to explain my fear in this regard.
Explaining how neuroepigenetic mechanisms serve as the interface between genes and experience, or nature and nurture as I mentioned to start this essay, is certainly going to be a “Big Data” endeavor. It is already clear that tracking epigenetic changes in the CNS over the lifespan is going to be a huge bio-informatics challenge (Lister et al, 2013). The biomedical poster-child for Big Data thus far has been sequencing the human genome, as well as the genomes of other species. This is the prototype for how we think about large-scale bio-informatics initiatives in biology – sequencing and annotating the 3 billion or so nucleotides comprising a mammalian genome. However, the genome in all its complexity is simply the basic first layer of infrastructure upon which epigenetic mechanisms operate. A single mammalian organism has a single genome, but that same organism has hundreds of cellular phenotypes, each of which has its own distinct epigenome. Each cellular epigenome itself comprises a hundred or so distinct epigenetic marks (Table 1), that can potentially be read out in a complex combinatorial fashion, that potentially can have functional consequences specifically localized to any one of the 3 billion or so individual nucleotides in the cell. So, conceptually one can multiply 3 billion nucleotides (the genome) times 100 potential epigenetic marks that may or may not be there at each nucleotide, each epigenetic mark of which exists in some background epigenome specifying cell type, and which may be read out in a combinatorial fashion depending upon nearby epigenetic modifications (Scharf and Imhof, 2010; D’Allessio and Szyf, 2006). The potential combinatorial complexity of this system is indeed daunting. Deciphering how the epigenome regulates the functional properties of neurons and glia in the brain is clearly going to be an immense bio-informatics challenge. The workings of the epigenomic code in the CNS will certainly be refractory to succinct and simple explanation. However, it already is clear that parsing that code will be required for any comprehensive model of how experience shapes function in the brain.
Acknowledgements
The author thanks Michael Meaney, Eric Nestler, Schahram Akbarian, and Li-Huei Tsai for many helpful discussions, and Felecia Hester for help in preparing the figure and manuscript. I apologize to the many authors whose primary work was not directly cited, owing to limitations of space. Research in the author’s laboratory is supported by funds from the NINDS, NIMH, NINR, NIDA, DARPA, the Pitt-Hopkins Syndrome Foundation, the Simons Foundation, the Ellison Medical Foundation, and the Evelyn F. McKnight Brain Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T, Reinberg D. Epigenetics. New York: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA Methylation Regulates Cocaine-Induced Behavioral Sensitization in Mice. Neuropsychopharmacology. 2010;35(12):2450–2461. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER, Di K. The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron. 2004;44:49–57. doi: 10.1016/j.neuron.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bohacek J, Gapp K, Saab BJ, Mansuy IM. Transgenerational epigenetic effects on brain functions. Biol Psychiatry. 2013;73:313–320. doi: 10.1016/j.biopsych.2012.08.019. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci Biobehav Rev. 2009;33:593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Crick F. Memory and molecular turnover. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- D'Alessio AC, Szyf M. Epigenetic tete-a-tete: the bilateral relationship between chromatin modifications and DNA methylation. Biochem Cell Biol. 2006;84:463–476. doi: 10.1139/o06-090. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Li W, Fox K, Stryker MP, Silva AJ. Reversing neurodevelopmental disorders in adults. Neuron. 2008;60:950–960. doi: 10.1016/j.neuron.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010a;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010b;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010a;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010b;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011a;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011b;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Is there an epigenetic component in long-term memory? J Theor Biol. 1999;200:339–341. doi: 10.1006/jtbi.1999.0995. [DOI] [PubMed] [Google Scholar]
- Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CD, Kandel ER, Rajasethupathy P. New mechanisms in memory storage: piRNAs and epigenetics. Trends Neurosci. 2013;36:535–542. doi: 10.1016/j.tins.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesburguères E, Gobbo OL, Alaux-Cantin S, Hambucken A, Trifilieff P, Bontempi B. Early tagging of cortical networks is required for the formation of enduring associative memory. Science. 2011;331:924–928. doi: 10.1126/science.1196164. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms: a common theme in vertebrate and invertebrate memory formation. Cell Mol Life Sci. 2006;63:1009–1016. doi: 10.1007/s00018-006-6026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman JE. A mechanism for memory storage insensitive to molecular turnover: a bistable autophosphorylating kinase. Proc Natl Acad Sci U S A. 1985;82:3055–3057. doi: 10.1073/pnas.82.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One. 2011;6:e19958. doi: 10.1371/journal.pone.0019958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Bading H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci. 2012;15:1111–1113. doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends Mol Med. 2008;14:341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai L-H. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos KF, Mazzola TN, Carvalho HF. The prima donna of epigenetics: the regulation of gene expression by DNA methylation. Braz J Med Biol Res. 2005;38:1531–1541. doi: 10.1590/s0100-879x2005001000010. [DOI] [PubMed] [Google Scholar]
- Scharf AN, Imhof A. Every methyl counts - Epigenetic calculus. FEBS Lett. 7. 2010;585(13):2001–2007. doi: 10.1016/j.febslet.2010.11.029. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel E. A possible epigenetic mechanism for the persistence of memory. Cold Spring Harb Symp Quant Biol. 2004;69:497–498. doi: 10.1101/sqb.2004.69.497. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sultan FA, Day JJ. Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011;3:157–181. doi: 10.2217/epi.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun AX, Crabtree GR, Yoo AS. MicroRNAs: regulators of neuronal fate. Curr Opin Cell Biol. 2013. 2013;25:215–221. doi: 10.1016/j.ceb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. Experience-dependent epigenetic modifications in the central nervous system. Biol Psychiatry. 2009;65:191–197. doi: 10.1016/j.biopsych.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD, Meaney MJ, Nestler EJ, Akbarian S. Epigenetic Regulation in the Nervous System. San Diego: Academic Press; 2013. [Google Scholar]
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardito D, Mallei A, Popoli M. Lost in translation. New unexplored avenues for neuropsychopharmacology: epigenetics and microRNAs. Expert Opin Investig Drugs. 2013;22:217–233. doi: 10.1517/13543784.2013.749237. [DOI] [PubMed] [Google Scholar]
- Tate P, Skarnes W, Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Sweatt JD. Molecular neurobiology of human cognition. Neuron. 2002;33:845–848. doi: 10.1016/s0896-6273(02)00634-7. [DOI] [PubMed] [Google Scholar]
- Wood MA, Hawk JD, Abel T. Combinatorial chromatin modifications and memory storage: a code for memory? Learn Mem. 2006b;13:241–244. doi: 10.1101/lm.278206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]