Abstract
Background
Current diagnostic methods of renal allograft rejection are neither sensitive nor specific. Needle biopsies are invasive and associated with patient morbidity. Thus, it is desirable to develop noninvasive tests to predict and diagnose rejection.
Methods
Using a case-control approach, surface-enhanced laser desorption/ionization time-of-flight mass spectrometry was used to identify plasma proteins associated with renal allograft rejection. From each rejection patient (n=16), two plasma samples (one near the biopsy date and the other at a time postbiopsy) were compared. Biopsy-confirmed nonrejection patients (n=48) were further analyzed as controls. Antibody-based quantitative enzyme-linked immunosorbent assay was performed to validate candidate biomarker apolipoprotein A1 (Apo A1) in a subset of the original and a second cohort of biopsy-confirmed rejection (n=40) and nonrejection (n=70) patients.
Results
Twenty-two proteins/peptides showed significant differences between rejection and postrejection samples. Peptides 5191 Da and 4467 Da detected rejection with 100% sensitivity and 94% specificity. The 4467 Da peptide was identified as the C-terminal fragment of α-1 antichymotrypsin and a 28 kDa protein was determined as Apo A1. Both protein levels were significantly lower at rejection compared with postrejection. Protein levels of nonrejection patients were similar to the postrejection samples. Apo A1 enzyme-linked immunosorbent assay results showed significantly lower Apo A1 levels (P=0.001 for the original and P=4.14E-11 for the second cohort) at the time of rejection compared with nonrejection which coincides with the SELDI findings.
Conclusions
Together α-1 antichymotrypsin, Apo A1, and the unidentified 5191 Da peptide provide a plasma molecular profile, and this is associated with acute cellular renal allograft rejection.
Keywords: Acute cellular rejection, Transplantation, Proteomics, Biomarker, Renal
The diagnosis of acute rejection is suspected by clinical presentation and confirmed by biopsy. Serum creatinine levels increase during allograft dysfunction, but this measure is neither sensitive nor specific for acute rejection (1, 2). The gold standard for diagnosing rejection is needle biopsy, which is invasive, painful, and as sociated with patient morbidity (1–4). It would be advantageous to develop a noninvasive test that could detect rejection, improve transplant outcomes, and reduce the cost of care. The use of human plasma for biomarker discovery provides a means to monitor disease states in a relatively noninvasive manner (5, 6). Plasma comes in contact with tissues which are apt to release protein components that reflect the disease-altered state of the tissue (6).
The SELDI ProteinChip System (Bio-Rad, Hercules, CA) uses solid phase extraction of proteins and peptides from biological mixtures followed by detection using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. ProteinChip arrays use a variety of chromatographic and biological surfaces to bind subsets of proteins from complex biological samples (7). The bound proteins are used to generate protein profiles used for biomarker discovery (7, 8). Urine protein profiles from renal allograft rejection patients have identified potential biomarker candidates and generated valuable insight into the biology of graft injury (9–15). The utilization of plasma for finding candidate bio-markers of renal allograft rejection holds promise as a biological fluid that can provide informative proteomic signatures of renal allograft rejection (16).
This study analyzes the plasma proteomes of renal transplant patients by SELDI. Twenty-two proteins/peptides had significant differences when comparing plasma during rejection with postrejection. The combination of two candidate proteins had a high discriminatory value for detecting rejection. Two of the 22 candidates were identified and one, apolipoprotein A1 (Apo A1), was validated by enzyme-linked immunosorbent assay (ELISA) in the original and a larger independent cohort.
Results
Variations in Plasma Protein/Peptide Levels Between Rejection and Postrejection Samples
The peak intensity differences across spectra between the rejection and postrejection groups were analyzed. In total, 653 peak clusters were detected, including 235 peaks in the pH 4 fraction, 186 peaks in the pH 6 fraction, and 232 peaks in the pH 9 fraction.
The peak intensities of rejection and postrejection samples from the same patient were analyzed using a binomial model. Eighty-one peak intensities (see Table S1A, Supplemental Digital Content 1, http://links.lww.com/TP/A465) were significantly increased, whereas 63 peaks (see Table S1B, Supplemental Digital Content 1, http://links.lww.com/TP/A465) were significantly decreased in the rejection samples. After Bonferroni correction for the total number of peaks in each condition, 24 peaks (see Table S2, Supplemental Digital Content 1, http://links.lww.com/TP/A465) were significantly different between rejection and postrejection samples by the linear mixed model. Twenty-two significant protein peaks were detected by both the binomial model and the linear mixed model (Table 1).
Table 1. Significant peaks found by both the linear mixed model and binomial model.
| Conditiona | Protein peak m/z (Da) | P for mixed model | P for binomial model | Protein intensity ratioa (>1.5) | |
|---|---|---|---|---|---|
| 1 | 4CL | 4467 | 3.06E-04 | 0.004 | B/A |
| 2 | 4125 | 1.35E-04 | 0.011 | A/B | |
| 3 | 5191 | 1.15E-11 | 0.000 | A/B | |
| 4 | 4SH | 50,835 | 5.04E-06 | 0.003 | A/B |
| 5 | 143,718 | 3.62E-05 | 0.001 | B/A | |
| 6 | 6CL | 4564 | 4.41E-05 | 0.030 | A/B |
| 7 | 4933 | 1.80E-04 | 0.030 | A/B | |
| 8 | 5191 | 1.10E-06 | 0.002 | A/B | |
| 9 | 6SH | 59,256 | 2.24E-04 | 0.025 | B/A |
| 10 | 60,334 | 3.72E-04 | 0.025 | B/A | |
| 11 | 63,615 | 5.74E-04 | 0.000 | B/A | |
| 12 | 123,329 | 2.10E-04 | 0.025 | B/A | |
| 13 | 9CL | 5051 | 3.29E-05 | 0.001 | A/B |
| 14 | 5083 | 6.40E-05 | 0.020 | A/B | |
| 15 | 5190 | 8.58E-07 | 0.020 | A/B | |
| 16 | 5265 | 1.21E-05 | 0.001 | A/B | |
| 17 | 9SH | 28,186 | 2.18E-04 | 0.002 | B/A |
| 18 | 28,336 | 1.42E-04 | 0.007 | B/A | |
| 19 | 28,574 | 3.00E-04 | 0.007 | B/A | |
| 20 | 29,134 | 1.40E-04 | 0.026 | B/A | |
| 21 | 109,796 | 1.05E-04 | 0.007 | B/A | |
| 22 | 188,758 | 2.59E-04 | 0.026 | B/A |
Under “Condition,” the number refers to the pH fraction. The “Ratio” indicates whether the protein intensity is higher in rejection (A) or postrejection (B) patients.
C, alpha-cyano-4-hydroxy cinnamic acid; S, sinapinic acid; L, low laser energy; H, high laser energy; m/z, mass to charge ratio.
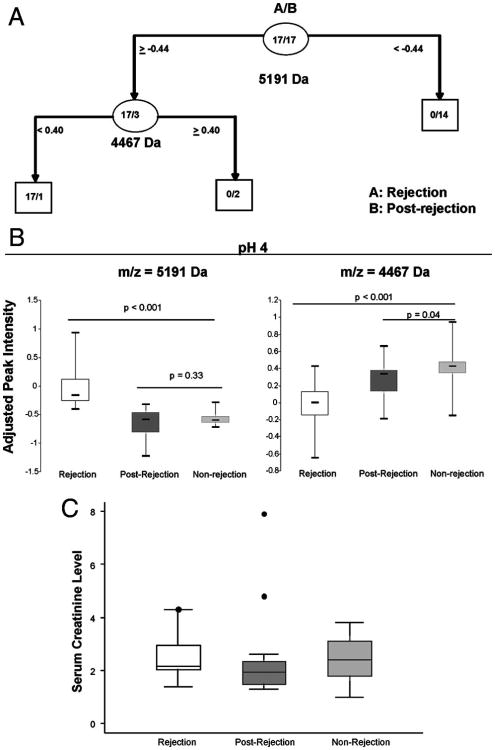
The 17 independent rejection events were applied to a Classification and Regression Tree (CART) analysis and produced a combination of two peaks (4467 Da and 5191 Da) (Fig. 1A), which best differentiate rejection from postrejection samples. All the rejection samples were correctly detected, whereas only one postrejection sample was incorrectly detected; showing 100% sensitivity and 94% specificity. The intensity of the 5191 Da peak was significantly increased during rejection (P<0.001), whereas the 4467 Da peak was significantly decreased during rejection (P<0.004) (Fig. 1B). The ROC plots produced cut points, which were similar to those found by CART analysis (see Figure S1, Supplemental Digital Content 1, http://links.lww.com/TP/A465).
Figure 1.
Candidate plasma proteins which detect renal allograft rejection. (A) Classification and Regression Tree (CART) analysis assessing the biomarker candidates for detecting rejection vs. postrejection samples. (B) Box plots of the adjusted intensities of the 4467 Da and 5191 Da biomarker candidates in rejection (group A, n=17), postrejection (group B, n= 17), and nonrejection (group C, n=48) plasma samples. (C) Box plots of the serum creatinine levels measured at the time of biopsy for rejection patients at the time of rejection and postrejection (n=16) compared with biopsy-proven nonrejection patients (n=19).
The peak intensities of these two proteins were compared with the nonrejection group (group C) (Fig. 1B). There was no significant difference in 5191 Da peak intensity between groups B and C (P=0.33). However, in rejection (group A), the 5191 Da peak was elevated over nonrejection (group C) (P<0.001). The 4467 Da peak intensity was lower during rejection (group A) (P<0.001) compared with nonrejection (group C), and nonrejection (group C) was only slightly higher than the postrejection samples (group B, P=0.04).
Although the levels of the 5191 DA and 4467 DA protein peaks were significantly associated with diagnosis of rejection, their intensities did not appear to discriminate among Banff grades of rejection. Because of the limited numbers, rejections were classified as either mild (≤Banff 1A, n=10) or severe (≥Banff 1B, n=7). The two groups individually showed the same intensity patterns (versus nonrejection) as listed earlier for the combined cases of rejection (data not shown).
Of the plasma samples used for protein profiling, blood sample serum creatinine levels were available in 16 of 17 rejection samples and 19 of 48 controls. There was no significant difference in serum creatinine levels between postrejection and samples collected during rejection. Although the creatinine levels in the postrejection samples (median=1.9) were in fact lower in all patients compared with the time of rejection (median=2.2), this difference was not statistically significant. Creatinine recovery can be slower due to a number of factors including type of rejection, the rejection therapy, and the donor condition.
Identification of Biomarker Candidates
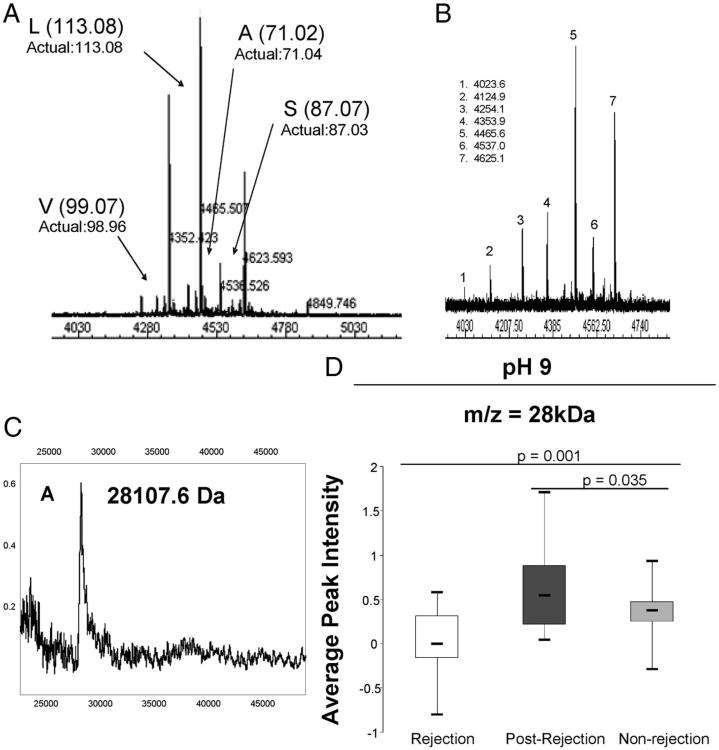
The 4467 Da peptide was detected in the pH 4 fraction, and further purification revealed a cluster of peaks (Fig. 2A) with mass differences matching the monoisotopic masses of valine, leucine, alanine, and serine giving a terminal sequence of SALV (S, serine; A, alanine; L, leucine; and V, valine), which corresponds to the C-terminal fragment of human α-1-antichymotrypsin (AACT). High-resolution MALDI analysis of immunoprecipitated plasma peptides confirmed the identity as AACT (Fig. 2B).
Figure 2.
Identification of the C-terminal fragment of α-1 antichymotrypsin and apolipoprotein A1 (Apo A1). (A) High-resolution MALDI-TOF mass spectrum of C8 high-performance liquid chromatography (HPLC)-purified fraction containing the 4467 Da biomarker candidate peptide (average mass as measured by SELDI-TOF MS). Monoisotopic mass differences indicate a sequence ladder of SALV (S, serine; A, alanine; L, leucine; and V, valine). (B) Immunoprecipitation of plasma samples with monoclonal antibody against α-1-antichymotrypsin. Numbered peptides with average masses (Da) are presented. (C) Immunoprecipitation of plasma samples with a monoclonal antibody against human Apo A1. (D) Box plots of the SELDI adjusted peak intensities of Apo A1 in rejection (group A, n=17), postrejection (group B, n=17) and nonrejection (group C, n=48) plasma samples.
To further develop a profile associated with renal allograft rejection, a cluster of peaks at ∼28 kDa found among the list of 22 significant proteins was analyzed (Table 1). Separation of the 28 kDa peaks by sodium dodecyl sulfate-PAGE followed by in-gel tryptic digestion produced peptides that were analyzed by MALDI-TOF-MS and searched on MASCOT to identify the protein as Apo A1. Immunoprecipitation using a monoclonal antibody confirmed the identity (Fig. 2C). The Apo A1 peak intensity was significantly higher in postrejection compared with the rejection and nonrejection patients and were more similar to the postrejection (P=0.035) than the rejection samples (P=0.001) (Fig. 2D). Although Apo A1 protein levels were significantly associated with diagnosis of acute rejection, they were not associated with an individual Banff grade of rejection. Logistic regression and CART analysis of the 4467 Da and the 28 kDa peaks showed that they differentiate rejection from postrejection with 59% sensitivity and 100% specificity (see Figure S2, Supplemental Digital Content 1, http://links.lww.com/TP/A465).
Validation of Apo A1
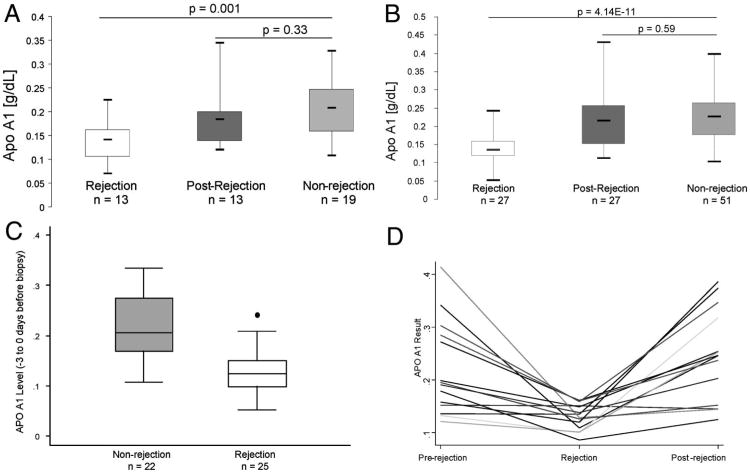
To validate the SELDI results, a quantitative Apo A1 ELISA was performed on a subset of the original rejection/postrejection samples (n=13) and the nonrejection samples (n=19). The pattern of Apo A1 levels measured by ELISA (Fig. 3A) is consistent with the SELDI intensity peak levels (Fig. 2D). To further substantiate these findings, the Apo A1 ELISA was performed on an independent cohort consisting of 27 plasma samples from patients with biopsy-confirmed acute cellular rejection (ACR) with a corresponding postrejection sample and 51 patients with biopsy-proven nonrejection. In the second cohort, Apo A1 levels by ELISA are significantly lower during rejection (mean=0.14±0.04), compared with postrejection (mean=0.22±0.09) and nonrejection samples (mean=0.23± 0.07, P=4.14E-11) (Fig. 3B), a finding that is consistent with the original cohort.
Figure 3.
Validation of apolipoprotein A1 (Apo A1) by enzyme-linked immunosorbent assay (ELISA) for association with acute cellular rejection. (A) Box plots of the ELISA results of plasma samples of a subset of the original cohort. (B) Box plots of the ELISA results of the plasma samples of the second independent cohort. (C) Box plots of Apo A1 levels in recipients that had a sample collected within – 3 to 0 days before the renal biopsy. Twenty-five rejection and 22 nonrejection samples were compared. The mean Apo A1 levels in nonrejection was 0.22 SD±0.06 and 0.13 SD±0.04 in rejection. The levels of Apo A1 were significantly lower at the time of rejection (P value <0.0001) compared with recipients with biopsy-proven nonrejection. (D) Line graph indicating the individual Apo A1 results for 14 patients who had plasma samples from all three time periods (prerejection, rejection, and postrejection). (E) ROC analysis comparing the rejection samples to the nonrejection samples from both the original cohort and the second cohort combined showing a cut point of 0.167 mg/dL and 76% sensitivity and 86% specificity.
Among both cohorts, samples which were collected within –3 to 0 days before the renal biopsy were examined. Twenty-five rejection and 22 nonrejection samples were compared, and Apo A1 levels were significantly lower at the time of rejection (P value<0.0001) compared with recipients with biopsy-proven nonrejection (Fig. 3C).
A subset of the new cohort (n= 14) also had available a plasma sample collected at a time point before the rejection episode (median = −50 days, range = − 180 to −11 days). The prerejection samples demonstrated Apo A1 levels nearly identical to the average levels of the postrejection samples (Fig. 3D).
An ROC analysis was performed using Apo A1 levels of the rejection and nonrejection samples from both the original and second cohort of patients (Fig. 3E). The results show a cut point of 0.167 g/dL provides a sensitivity of 76% and a specificity of 86% correctly detecting 80% of the patients. Twenty-six of the 27 rejection patients had Banff classification data as follows: borderline changes=2, Banff 1A=13, Banff 2A=6, Banff 3 vascular=1, and Banff antibody-mediated rejection (AMR)=4. The patients were grouped into categories based on mild ACR (borderline and Banff 1A=15), severe ACR (Banff 2A and Banff 3 vascular=7), or AMR with no ACR (n=4). The Apo A1 medial level was 0.14 for all three groups indicating that the severity of rejection was not distinguished by the Apo A1 level.
Discussion
Twenty-two peptides/proteins discovered by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry were significantly associated with the diagnosis of acute allograft rejection. The identity of two of these proteins, AACT and Apo A1, was elucidated. The C-terminal fragment of AACT is significantly lower during rejection compared with postrejection and nonrejection. A different variation of the AACT fragment (4354 Da) is found at higher levels in urine of patients with acute rejection compared with those with stable transplants (14). Pimenta et al. (17) examined the hydrolysis of AACT by cathepsin D using high-performance liquid chromatography isolation and MADLI and found three peptides: 4354 Da, 4468 Da, and 4625 Da. These cleavage products are identical to the 4467 Da peak identified in plasma and the 4354 Da found in urine based on the sequence data, suggesting that both may be produced by cathepsin D.
O'Riordan et al. (14) examined the levels of total AACT in the kidney by immunohistochemistry and found no difference between rejection and controls and suggested that finding a peptide AACT whose level varied was the result of cleavage after target protease interaction, and therefore, the peptide level variation between patients is a reflection of the activity of the AACT target proteases. We hypothesize that variations in the peptide level may be a result of total AACT being blocked or bound to other substrates for cathepsin D such as cystatin C or kininogen (18). Alternatively, during rejection, total AACT may instead inhibit its primary target, cathepsin G (19), an inflammation-related neutrophil protease, which has been linked to allograft function (20, 21).
The 28 kDa peak was identified as Apo A1. Apo A1 and synthetic peptides, which mimic Apo A1 activity, have strong antiinflammatory and antioxidant properties (22, 23). The Apo A1 mimetic peptide, D-4F, reduces intimal lesions caused by chronic rejection in a mouse model. The mechanism of this effect involves induction of the antioxidant gene heme oxygenase-1 in the graft and/or a direct effect on T-lymphocyte proliferation and effector cytokine production (24).
Apo A1 was validated by ELISA in both the original and an independent cohort. Comparisons of the rejection, postrejection, and nonrejection samples in both cohorts were consistent with the SELDI findings indicating that lower levels of Apo A1 are associated with rejection. The prerejection time point in the independent cohort could not predict the onset of the rejection episode, indicating that the Apo A1 drop is closely associated with the time of rejection. In addition, the Apo A1 ROC analysis of all patients suggests that Apo A1 could potentially be part of a signature used to describe rejection. Apo A1 was also identified as a candidate biomarker in pulmonary arterial hypertension, colorectal cancer, and ovarian cancer (25–27) and more recently was associated with Chagas disease, polycystic ovarian syndrome, and chronic obstructive pulmonary disease (28–30). In all these studies, Apo A1 levels were lower in the disease state compared with nondisease.
Infections can lead to changes in serum lipid profiles, and decreased Apo A1 has been postulated to link infection with chronic inflammation (31). The use of long-term immunosuppressive therapy leaves transplant patients susceptible to infection (32). We examined the 51 nonrejection patients from the second cohort and found five patients with evidence of bacterial infection at the time the plasma sample used for this study was collected. Apo A1 levels were not significantly different in nonrejection patients with infection compared with those without for this small subset of patients. This contributes to the hypothesis that for renal transplant patients, low levels of Apo A1 are associated with the rejection episode, but more studies are necessary to confirm this.
Isolation and identification of the 5191 Da peptide have been hindered due to its low abundance in plasma, its copurification with other more abundant peptides, and the inability to directly fragment it. For unknown reasons, some peptides fragment poorly and yield MS/MS spectra that cannot be deciphered (33, 34). A combination of the 5191 and 4467 Da peaks robustly assessed rejection verses nonrejection, whereas the combination of the 4467 and 28 kDa peaks was not as valuable, thus identification of the 5191 Da peak is important. We are currently exploring alternative methods to better isolate and fragment this peptide for identification.
A limitation of our study design is that the plasma was collected at routine patient visits, which was not always on the same day that the renal biopsy was performed. Also, the clinical parameters of our study population were not uniform with respect to immunosuppression. Furthermore, only two patients in the original cohort had AMR, and therefore, a signature for AMR could not be determined. A majority of patients rejected within the first 100 days, and thus, we were limited to only explore early rejection. In the future, it will be also valuable to explore these markers in patients undergoing late renal allograft rejection. Additionally, we did not find an association between severity of rejection based on Banff classification and Apo A1 levels. It would be of interest to attempt this comparison again in a larger cohort of subjects. Finally, we recognize that to better assess these markers, a prospective study is necessary to determine their true value in the clinical setting. To show the potential of a prospective study, we examined both cohorts of patients and found 25 rejection and 22 nonrejection samples collected –3 to 0 days before the renal biopsy. When we compared the Apo A1 levels between these groups, the rejection levels were significantly lower (P<0.0001), suggesting that a prospective study would likely have differentiated these patients at the time of biopsy.
This study demonstrates that searching for candidate biomarkers of renal allograft rejection in plasma has a promising clinical benefit. At the time point these plasma samples were collected, the candidate biomarkers showed variations that were more informative than serum creatinine level changes. Therefore, future studies should not only interrogate Apo A1 but also other markers for their ability to accurately detect renal allograft rejection. Finally, the two candidate markers identified are antiinflammatory proteins, and their functional characterization should be explored.
Materials and Methods
Study Design and Patient Population
Renal transplant patients were enrolled in the University of California Los Angeles Institutional Review Board-approved immune monitoring study by written consent. Sixteen renal transplant recipients contributed 34 blood specimens associated with 17 “independent” episodes of biopsy-confirmed ACR. One recipient had two rejection episodes far enough apart (4 months), and the intermediate serum creatinine was normal, so we considered the second rejection an independent event. Patients with biopsy-proven nonrejection were also selected (n=48). All biopsies were performed for cause and taken after a serum creatinine rise or delayed graft function. Rejection biopsies occurred within the first 100 days posttransplant with the exception of one which occurred at 656 days (median=27 days posttransplant). Banff classification was used to score acute rejection (35, 36). Two biopsies with ACR also showed evidence of AMR and were C4d positive. The rejection patients were studied at two time points: rejection specimen (group A) were selected at a time point closest to the positive biopsy date (median= −1 day, range: −16 to +8 days); and postrejection samples (group B) were selected closest to 14 days after biopsy date (median=28 days, range: 14–180 days). Twelve of 17 rejection samples (group A) were collected within 8 days before biopsy, and 5 of 17 were obtained within 8 days after the biopsy. For the postrejection samples (group B), 16 of 17 were collected within 60 days after the biopsy, and 1 of 17 was collected at 180 days after the biopsy. The nonrejection plasma samples (defined as controls, group C) were selected at one time point closest to the negative biopsy (median=1 day, range: −35 to +44 days). Table 2 illustrates the demographics of the study population and shows no significant differences with respect to age, gender, race, transplant number, or degree of human leukocyte antigen mismatch. The percentage of deceased and living donors was similar to U.S. Organ Procurement Transplant Network national data.
Table 2. Demographic characteristics of patients.
| Parameters | Rejection patients (n=16) | Nonrejectors (n=48) | Pa |
|---|---|---|---|
| Age (yr: mean±SD) | 48.7± 12.2 | 49.5±13.9 | 0.84 |
| Gender (male/female) | 14/2 | 32/16 | 0.20 |
| Race | 0.74b | ||
| Asian | 19% | 5% | |
| Black | 25% | 20% | |
| White | 44% | 54% | |
| Other | 12% | 21% | |
| Regraft | 0.74 | ||
| First allograft | 81% | 76% | |
| Regraft | 19% | 24% | |
| Multiorgan transplants | 0.16 | ||
| + Pancreas | 11% | 2% | |
| + Liver | 0% | 2% | |
| HLA mismatch (mean±SD) | 2.7±0.7 | 2.2± 1.3 | 0.15 |
| Type of donor | 0.21 | ||
| Deceased | 56% | 74% | |
| Living | 44% | 26% | |
| Induction | 0.74c | ||
| Anti-IL2Rα | 74% | 72% | |
| ATG | 22% | 21% | |
| Combined | 4% | 7% | |
| Time of blood draw relative to biopsy (days: median/range) | 0.37 | ||
| Rejection sample | −1/−16 to 8 | ||
| Postrejection sample | 28/14 to 147 | ||
| Nonrejection sample | 1/−35 to 44 |
P values for categorical variables from Fisher exact test, and P values for continuous variables from t test.
Race (black vs. other).
Induction (ATG vs. any anti-IL2Rα).
ATG, antithymoglobulin; IL2Rα, interleukin-2 receptor alpha chain; SD, standard deviation; HLA, human leukocyte antigen.
A second cohort of recipients with biopsy-proven renal allograft rejection (n=27) were selected, and again two plasma samples were studied (rejection and postrejection time points). All biopsies were performed for cause and taken after a serum creatinine rise or delayed graft function. Two of the ACR positive biopsies also had evidence of AMR, whereas four of the biopsies had evidence of onlyAMR. Rejection specimens (n=27) were selected at a time point within −7 days of the biopsy (median= −1 day, range=− 7 to 0 days). The average rejection episode was 122 days after transplantation. Twenty of the 27 biopsies were within the first 3 months, 5 of 27 were within the first 9 months, and 2 of 27 were greater than 1 year. Postrejection samples were selected more than 8 days after the biopsy (median=32 days, range=8–573 days). For the postrejection samples, 20 of 27 were collected within 3 months after the biopsy, 3 of 27 were within 6 months, 1 of 27 was collected at 9 months, and 3 of 27 were collected 1 year after the biopsy. In addition, a cohort of 51 biopsy-proven nonrejection samples were selected at a time closest to the negative biopsy (median=8 days, range=−22 to 581 days). For the biopsy-proven nonrejection samples, 38 of 51 were collected within 3 months after the biopsy, 5 of 51 were within 6 months, 4 of 51 were collected within 9 months after the biopsy, and 4 of 51 were collected after 1 year. All biopsies from this group of nonrejectors were performed for cause and taken after a serum creatinine rise or delayed graft function.
For both cohorts, maintenance immunosuppression included a calcineurin inhibitor (tacrolimus or cyclosporine), an antiproliferative agent (mycophenolate mofetil or sirolimus), and prednisone. In some cases without evidence of pretransplant sensitization, steroids were withdrawn.
Plasma Albumin and Immunoglobulin-G Depletion and Anion Exchange Fractionation
Blood samples were collected in BD Vacutainer ACD solution A tubes (Becton, Dickinson and Company, Franklin Lakes, NJ). Plasma was separated and stored. Antibody spin columns (Qproteome, Qiagen, Valencia, CA) were used to deplete human serum albumin and immunoglobulin-G. Complete Mini (ethylenediaminetetraacetic acid) Protease Inhibitor (Roche, Palo Alto, CA) was added. AcroPrep Mustang Q (hydrophilic polyethersulfone memebrane) Anion Exchange Resin 96-well plates (Pall, East Hills, NY) were used to separate the depleted plasma into the pH 9, pH 6, and pH 4 fractions.
ProteinChip Array preparation
ProteinChip arrays (CM10 and Q10) were preequilibrated for binding. pH fractions were bound to the arrays and analyzed in triplicate. Six pooled human serum (Sigma, St. Louis, MO) samples were used as internal controls on each array set. After sample array processing, matrix was applied: either saturated sinapinic acid (SPA, LaserBio Labs, Sophia-Antipolis, France) or 20% saturated alpha-cyano-4-hydroxy cinnamic acid (CHCA, LaserBio Labs). A Biomek 2000 liquid handling robot (Beckman, Fullerton, CA) was used for matrix spotting.
Mass Spectrum Data Analysis
A ProteinChip Biology System (PBS IIc) mass spectrometer using Ciphergen ProteinChip Software 3.2 was calibrated in the high mass range before reading the SPA-spotted arrays (laser energies of 150 and 170). A peptide mixture was used for calibration of the CHCA-spotted arrays (laser energies of 120 and 135). Mass spectrum from each spot was an average of 240 laser shots. All spectra were preliminarily analyzed in CiphergenExpress Client Software 3.0 to create clusters by Expression Difference Mapping analysis. A cluster is a group of peaks with the same mass/charge (m/z) ratio and treated as the same protein or peptide across multiple spectra. Peak intensities from each spectrum were used to measure relative protein/peptide amounts. Total ion current was used to normalize the signals. The reproducibility of peak heights was similar to what was previously published (37), showing a CV of 15% to 30%.
Protein Identification
MALDI-TOF-MS was performed with a prOTOF 2000 orthoganol-TOF mass spectrometer (Perkin Elmer, San Jose, CA). The high-performance liquid chromatography system was Prominence 2000 (Shimadzu Scientific Instruments, Columbia, MD). All protocol and reagents were in accordance with manufacturer's suggestions.
4467 Da Peptide
Q fractions with this peptide were loaded on a reverse phase C8 column (Axxiom, Moorpark, CA). The C8 fractions were subjected to collision-induced dissociation MS/MS fragmentation in a Thermo-Finnigan LTQ-FT electrospray mass spectrometer (Waltham, MA). The MS/MS data were searched by NCBI Blast. Immunoprecipitation was performed using a polyclonal antibody against α-1-antichymotrypsin (Lab Vision Corporation, Fremont, CA) followed by MALDI analysis.
28 kDa Protein
Pooled samples fractions with high levels of the protein were separated by sodium dodecyl sulfate-PAGE. The gel was stained using the ProteoSilver Plus kit (Sigma). Bands near 28 kDa were excised, and in-gel tryptic digestion was performed. Peptides were analyzed by MALDI. Intact tryptic masses were searched in MASCOT with online protein fragment database searching. Immunoprecipitation was done using a monoclonal antibody against human Apo A1 (R&D Systems, Minneapolis, MN) followed by MALDI analysis.
Statistical Analyses
Nonparametric (binomial) and parametric (mixed effects and linear regression) models were used to identify significant peaks. Two-sided P values less than 0.05 were considered significant, and Bonferroni correction was used as needed. CART analysis was conducted to select a subset of candidate biomarkers and determine their ability to correctly classify rejection. Adjusted intensities of the selected protein/peptide peaks were used as the predictor variables in CART training. The R software package (version 3.1, http://www.r-project.org/) was used for statistical computations. Traditional logistic regression analysis was performed, and ROC plots were generated using STATA data analysis and statistical software. Cut points were selected by the maximum correctly classified. Details of the statistical analyses are presented in the Supplemental material (see Supplemental Digital Content 1, http://links.lww.com/TP/A465).
ELISA for Quantitation of Plasma Apo A1 Levels
Total Human Apolipoprotein A1 ELISA Assay (Alerchek, Portland, ME) was used according to the manufacturer's protocol. Samples were run in duplicate along with a control plasma sample. Analysis was performed on the SpectraMax M2 plate reader with the SoftMax Pro 5.4 software (Molecular Devices, Sunnyvale, CA).
Supplementary Material
Acknowledgments
The authors thank Giovanni Lopez and Tiffany Smith for their technical assistance.
This work was supported by the NSF Graduate Research Fellowship (M.E.Z.), the NSF DMS-0707160 (K.C.L.), and the NIH U01AI077821, NIH AI042819, and NHLBI HL090995 (E.F.R.).
Footnotes
The authors declare no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal's Web site (www.transplantjournal.com).
References
- 1.Li B, Hartono C, Ding R, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344:947. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 2.Clarke W. Proteomic research in renal transplantation. Ther Drug Monit. 2006;28:19. doi: 10.1097/01.ftd.0000194500.40021.37. [DOI] [PubMed] [Google Scholar]
- 3.Beckingham IJ, Nicholson ML, Bell PR. Analysis of factors associated with complications following renal transplant needle core biopsy. Br J Urol. 1994;73:13. doi: 10.1111/j.1464-410x.1994.tb07449.x. [DOI] [PubMed] [Google Scholar]
- 4.Rush DN, Henry SF, Jeffery JR, et al. Histological findings in early routine biopsies of stable renal allograft recipients. Transplantation. 1994;57:208. doi: 10.1097/00007890-199401001-00009. [DOI] [PubMed] [Google Scholar]
- 5.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6:6326. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lescuyer P, Hochstrasser D, Rabilloud T. How shall we use the proteomics toolbox for biomarker discovery? J Proteome Res. 2007;6:3371. doi: 10.1021/pr0702060. [DOI] [PubMed] [Google Scholar]
- 7.Reddy G, Dalmasso EA. SELDI ProteinChip(R) array technology: Protein-based predictive medicine and drug discovery applications. J Biomed Biotechnol. 2003;2003:237. doi: 10.1155/S1110724303210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albitar M, Potts SJ, Giles FJ, et al. Proteomic-based prediction of clinical behavior in adult acute lymphoblastic leukemia. Cancer. 2006;106:1587. doi: 10.1002/cncr.21770. [DOI] [PubMed] [Google Scholar]
- 9.Clarke W, Silverman BC, Zhang Z, et al. Characterization of renal allograft rejection by urinary proteomic analysis. Ann Surg. 2003;237:660–4. doi: 10.1097/01.SLA.0000064293.57770.42. discussion 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaub S, Wilkins J, Weiler T, et al. Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int. 2004;65:323. doi: 10.1111/j.1523-1755.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- 11.O'Riordan E, Orlova TN, Mei JJ, et al. Bioinformatic analysis of the urine proteome of acute allograft rejection. J Am Soc Nephrol. 2004;15:3240. doi: 10.1097/01.ASN.0000145241.83482.68. [DOI] [PubMed] [Google Scholar]
- 12.Voshol H, Brendlen N, Muller D, et al. Evaluation of biomarker discovery approaches to detect protein biomarkers of acute renal allograft rejection. J Proteome Res. 2005;4:1192. doi: 10.1021/pr050060+. [DOI] [PubMed] [Google Scholar]
- 13.Jahnukainen T, Malehorn D, Sun M, et al. Proteomic analysis of urine in kidney transplant patients with BK virus nephropathy. J Am Soc Nephrol. 2006;17:3248. doi: 10.1681/ASN.2006050437. [DOI] [PubMed] [Google Scholar]
- 14.O'Riordan E, Orlova TN, Podust VN, et al. Characterization of urinary peptide biomarkers of acute rejection in renal allografts. Am J Transplant. 2007;7:930. doi: 10.1111/j.1600-6143.2007.01733.x. [DOI] [PubMed] [Google Scholar]
- 15.Ling XB, Sigdel TK, Lau K, et al. Integrative urinary peptidomics in renal transplantation identifies biomarkers for acute rejection. J Am Soc Nephrol. 2010;21:646. doi: 10.1681/ASN.2009080876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freue GV, Sasaki M, Meredith A, et al. Proteomic signatures in plasma during early acute renal allograft rejection. Mol Cell Proteomics. 2010;9:1954. doi: 10.1074/mcp.M110.000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pimenta DC, Chen VC, Chao J, et al. Alpha1-antichymotrypsin and kallistatin hydrolysis by human cathepsin D. J Protein Chem. 2000;19:411. doi: 10.1023/a:1026432402259. [DOI] [PubMed] [Google Scholar]
- 18.Lenarcic B, Krasovec M, Ritonja A, et al. Inactivation of human cystatin C and kininogen by human cathepsin D. FEBS Lett. 1991;280:211. doi: 10.1016/0014-5793(91)80295-e. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Janciauskiene S. Multi-functional capability of proteins: Alpha1-antichymotrypsin and the correlation with Alzheimer's disease. J Alzheimers Dis. 2002;4:115. doi: 10.3233/jad-2002-4206. [DOI] [PubMed] [Google Scholar]
- 20.Gibson TL, Cohen P. Inflammation-related neutrophil proteases, cathepsin G and elastase, function as insulin-like growth factor binding protein proteases. Growth Horm IGF Res. 1999;9:241. doi: 10.1054/ghir.1999.0115. [DOI] [PubMed] [Google Scholar]
- 21.Koo DD, Welsh KI, Roake JA, et al. Ischemia/reperfusion injury in human kidney transplantation: An immunohistochemical analysis of changes after reperfusion. Am J Pathol. 1998;153:557. doi: 10.1016/S0002-9440(10)65598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navab M, Anantharamaiah GM, Hama S, et al. Oral administration of an Apo A-I mimetic Peptide synthesized from D-amino acids dramatically reduces atherosclerosis in mice independent of plasma cholesterol. Circulation. 2002;105:290. doi: 10.1161/hc0302.103711. [DOI] [PubMed] [Google Scholar]
- 23.Navab M, Anantharamaiah GM, Reddy ST, et al. Apolipoprotein A-I mimetic peptides. Arterioscler Thromb Vasc Biol. 2005;25:1325. doi: 10.1161/01.ATV.0000165694.39518.95. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh GR, Schnickel GT, Garcia C, Shefizadeh A, Fishbein MC, Ardehali A. Inflammation/oxidation in chronic rejection: Apolipoprotein a-i mimetic peptide reduces chronic rejection of transplanted hearts. Transplantation. 2007;84:238. doi: 10.1097/01.tp.0000268509.60200.ea. [DOI] [PubMed] [Google Scholar]
- 25.Yuditskaya S, Tumblin A, Hoehn GT, et al. Proteomic identification of altered apolipoprotein patterns in pulmonary hypertension and vasculopathy of sickle cell disease. Blood. 2008;113:1122. doi: 10.1182/blood-2008-03-142604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engwegen JY, Helgason HH, Cats A, et al. Identification of serum proteins discriminating colorectal cancer patients and healthy controls using surface-enhanced laser desorption ionisation-time of flight mass spectrometry. World J Gastroenterol. 2006;12:1536. doi: 10.3748/wjg.v12.i10.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak KR, Su F, Whitelegge JP, et al. Characterization of serum biomarkers for detection of early stage ovarian cancer. Proteomics. 2005;5:4589. doi: 10.1002/pmic.200500093. [DOI] [PubMed] [Google Scholar]
- 28.Ndao M, Spithill TW, Caffrey R, et al. Identification of novel diagnostic serum biomarkers for Chagas' disease in asymptomatic subjects by mass spectrometric profiling. J Clin Microbiol. 2010;48:1139. doi: 10.1128/JCM.02207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi DH, Lee WS, Won M, et al. The apolipoprotein A-I level is down-regulated in the granulosa cells of patients with polycystic ovary syndrome and affects steroidogenesis. J Proteome Res. 2010;9:4329. doi: 10.1021/pr100008e. [DOI] [PubMed] [Google Scholar]
- 30.Nicholas BL, Skipp P, Barton S, et al. Identification of lipocalin and apolipoprotein A1 as biomarkers of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;181:1049. doi: 10.1164/rccm.200906-0857OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burger D, Dayer JM. High-density lipoprotein-associated apolipoprotein A-I: The missing link between infection and chronic inflammation? Autoimmun Rev. 2002;1:111. doi: 10.1016/s1568-9972(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 32.Fishman JA, Rubin RH. Infection in organ-transplant recipients. N Engl J Med. 1998;338:1741. doi: 10.1056/NEJM199806113382407. [DOI] [PubMed] [Google Scholar]
- 33.Shevchenko A, Chernushevich I, Wilm M, et al. De Novo peptide sequencing by nanoelectrospray tandem mass spectrometry using triple quadrupole and quadrupole/time-of-flight instruments. Methods Mol Biol. 2000;146:1. doi: 10.1385/1-59259-045-4:1. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko A, Chernushevich I, Ens W, et al. Rapid ‘de novo’ peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 1997;11:1015. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1015::AID-RCM958>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 35.Racusen LC, Colvin RB, Solez K, et al. Antibody-mediated rejection criteria—An addition to the Banff 97 classification of renal allograft rejection. Am J Transplant. 2003;3:708. doi: 10.1034/j.1600-6143.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 36.Racusen LC, Solez K, Colvin RB, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55:713. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 37.Albrethsen J. Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin Chem. 2007;53:852. doi: 10.1373/clinchem.2006.082644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.