Abstract
In 1995, The Institute for Genomic Research completed the genomic sequence of a rough derivative of Haemophilus influenzae serotype d, strain KW20. This sequence, though extremely useful in understanding the basic biology of H. influenzae, has yet to provide significant insight into our understanding of disease caused by nontypeable H. influenzae (NTHI), because serotype d strains are not generally pathogens. In contrast, NTHI strains are frequently mucosal pathogens and are the primary pathogens of chronic otitis media as well as a significant cause of acute otitis media in children. Thus, it is of great importance to further understand their biology. We used a DNA-based microarray approach to identify genes present in a clinical isolate of NTHI that were absent from strain Rd. We also sequenced the genome of a second NTHI isolate from a child with chronic otitis media to threefold coverage and then used an array of bioinformatics tools to identify genes present in this NTHI strain but absent from strain Rd. These methods were complementary in approach and results. We identified, in both strains, homologues of H. influenzae lav, an autotransported protein of unknown function; tnaA, which encodes tryptophanase; as well as a homologue of Pasteurella multocida tsaA, which encodes an alkyl peroxidase that may play a role in protection against reactive oxygen species. We also identified a number of putative restriction-modification systems, bacteriophage genes and transposon-related genes. These data provide new insight that complements and extends our ongoing analysis of NTHI virulence determinants.
Otitis media (OM) is a highly prevalent pediatric disease worldwide (9, 26, 31). The most recently available statistics indicate that 24.5 million physician office visits were made for OM in 1990, representing a >200% increase over those reported in the 1980s (41). OM is the most frequently diagnosed illness in children (<15 years) and is the primary cause for emergency room visits (9). While OM is only very rarely associated with mortality, the morbidity associated with OM is significant. Hearing loss is the most common complication of OM (4, 25, 31) with behavioral, educational, and language development delays being additional consequences of early-onset OM with effusion (26, 31, 49). The socioeconomic impact of OM is also great, with direct and indirect costs of diagnosing and managing OM exceeding $5 billion annually in the United States alone (1, 8, 27, 31).
Clearly, there is a tremendous need to develop effective and accepted approaches to the management and, preferably, the prevention of OM. Vaccine development holds the greatest promise and would be the most cost-effective method to accomplish this goal (22, 28). Progress in this area is most advanced for Streptococcus pneumoniae, a causative agent of acute OM, and in fact, capsular-conjugate vaccines are currently licensed for use. Less progress has been made for nontypeable Haemophilus influenzae (NTHI), the gram-negative pathogen that predominates in chronic OM with effusion (13, 32, 46) and is also associated with approximately one-third of all cases of acute OM (30). Hampering development of effective vaccines against NTHI is the as-yet-incomplete understanding of the pathogenesis of NTHI-induced middle ear disease. There are multiple gaps in our understanding of the dynamic interplay between microbe-expressed virulence factors and the host's immune response as the disease progresses from one of host immunological tolerance of a benign nasopharyngeal commensal to that of an active defensive reaction to an opportunistic invader of the normally sterile middle ear space.
In 1995, a group from The Institute for Genomic Research sequenced the genome from H. influenzae strain Rd KW20 (20). Strain Rd is a nonencapsulated derivative of a serotype d organism. Although strain Rd has some virulence properties, serotype d strains are generally considered to be commensals; they do not frequently cause disease (11). It is therefore important to determine the differences between disease-causing strains of H. influenzae and strain Rd. Since we lack complete genome sequence for pathogenic strains, which would allow an exhaustive comparison, this question may be approached by analyses that indicate which genes are present in disease-causing strains of H. influenzae that are absent from strain Rd. Suppressive subtractive hybridization has been employed to identify genomic differences between isolates of a species. In Haemophilus, this technique was used to identify genomic differences between H. influenzae biogroup aegyptius strains of the Brazilian purpuric fever clonal group and conjunctivitis isolates (33, 45). Bergman and Akerley employed a position-based PCR scanning technique to compare an H. influenzae serotype b genome with the strain Rd genome (6). With respect to NTHI, most studies have focused on differences in the sequence of single genes or small gene clusters or the presence or absence of specific genes, although Williams et al. identified and characterized a bacteriophage that was present in an invasive nontypeable isolate (50). From an epidemiologic perspective, Pettigrew et al. employed pulse field gels, ribotyping and enterobacterial repetitive intergenic consensus typing to examine strain differences among NTHI (42). Similarly, Meats et al. have employed multilocus sequence typing for epidemiologic characterization of isolates (36).
In an effort to more broadly approach the identification of virulence determinants in NTHI, we have employed two methods to identify genes in NTHI that are absent from H. influenzae strain Rd. First, we employed a DNA-based microarray approach to identify genes present in NTHI strain 1885MEE that were absent from strain Rd. We also sequenced the genome of a pathogenic NTHI strain, designated 86-028NP, to threefold coverage. Assuming a Poisson distribution, the probability of any given base being sampled at threefold coverage is 95%, although practical limitations, due to the nonrandom nature of biological sequences, cause the actual percentage of sequence identified to be somewhat lower than the theoretical model predicts. With these partial sequence data, a bioinformatics approach was used to identify genes that were not present in strain Rd. Several genes of interest present in both stains were further characterized.
MATERIALS AND METHODS
Choice of strains for the projects.
Epidemiologic studies of NTHI have indicated that the strains are heterogeneous with respect to outer membrane protein profiles (5), enzyme allotypes (38), and readouts generated via other commonly used epidemiologic tools. There have been several attempts to subtype NTHI, but the methodologies have not been useful with respect to identification of an isolate that is most suitable for both genomic sequencing and genomic comparison with strain Rd. We therefore chose the low-passage-number isolates, strains 1885MEE and 86-028NP, that were independently recovered from children with chronic OM. Both have subsequently been well characterized in vitro (3, 24) as well as in chinchilla models of OM, with strain 86-028NP being the better understood (2, 14, 48).
Library constructions.
Chromosomal DNA was prepared from strains 86-028NP and 1885MEE using Puregene reagents (Gentra Systems, Minneapolis, Minn.). DNA from strain 1885MEE was partially digested with Sau3A, and fragments in the range 0.5 to 1.5 kb were isolated by preparative agarose gel electrophoresis and then ligated into the BamHI site of a pUC18 derivative, modified by substitution of the bla gene with the kanamycin resistance gene of Tn903. The transformation mixture was transformed into Escherichia coli TOP10 cells and clones containing inserts were identified by blue/white selection as outlined above. Six thousand white clones were saved.
Genomic DNA from strain 86-028NP was sheared to 2 to 4 kb with a nebulizer (10). The sheared DNA was ethanol precipitated, its ends were repaired using the End-It Repair Kit (Epicentre), and it was subjected to size selection by agarose gel electrophoresis to obtain 2- to 4-kb fragments (10, 44). These fragments were cloned into the vector pUC18 that had been cleaved with SmaI and was then phosphatase-treated. The ligation mixture was transformed into E. coli XL-1 Blue MRF′ and clones that contained inserts were identified by blue-white screening on 2× yeast tryptone (YT)-ampicillin plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Insert-containing clones were transferred into 96-deep-well plates containing 1.5 ml of 2 × YT-ampicillin broth. The deep-well-plate cultures were grown overnight (18 to 22 h) at 37°C, with shaking.
Plasmid preparation and DNA microarray analysis.
Plasmid DNA from the strain 1885MEE library was purified by alkaline lysis in a 96-well format on a Beckman Biomek 2000 automated robotics workstation, using a typical sodium dodecyl sulfate (SDS)-NaOH lysis protocol (10). The final ethanol-precipitated templates (in a 96-well plate) were each dissolved in 100 μl of double-distilled H2O. DNA from random clones was analyzed by gel electrophoresis to determine the size of the insert. After demonstrating that the library was comprised of clones with inserts of the appropriate size, plasmids were denatured by boiling in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-400 mM NaOH-10 mM EDTA (pH 8.0) and spotted onto SuperChip I, aminopropylsilane coated slides (Perkin-Elmer Life Sciences Inc., Boston, Mass.) using an Affymetrix 417 four-pin arrayer (MWG Biotech Inc., High Point, N.C.). To generate fluorescent targets, genomic DNA was directly labeled with either Cy5-dUTP or Cy3-dUTP using a BioPrime DNA labeling kit that utilized a random primed Klenow method (Amersham Biosciences [Little Chalfont, Buckinghamshire, United Kingdom] and Invitrogen Corporation [Carlsbad, Calif.]). For hybridization, each slide was blocked at 42°C with 5× SSC-0.5% SDS-1% bovine serum albumin, and then a LifterSlip (Erie Scientific Co., Portsmouth, N.H.) was placed over the arrayed DNA, the denatured probe was pipetted under it, and the slide was incubated at 50°C for 16 h. Posthybridization, the slide was washed with 0.5× SSC-0.01% SDS and then 0.06× SSC-0.01% SDS followed by 0.06× SSC alone, and then the slide was dried and scanned using an Affymetrix 428 Array Scanner (MWG Biotech Inc.). Using a GenePix Pro3 analysis package (Axon Instruments, Union City, Calif.), each wavelength image was assigned a color and a ratio image of the two was built. Inserts from positive clones were sequenced using ABI Big-Dye terminators and M13 universal primers and then analyzed on an Applied Biosystems (Foster City, Calif.) 3100 capillary electrophoresis DNA sequencer.
Genomic sequencing and assembly.
Template from the 86-028NP library was prepared as above using the alkaline lysis procedure. Cycle-sequencing reactions were run in 96-well format using PE Big-Dye terminators and universal primers (M13 forward and reverse). Excess dye terminators were removed with Sephadex G-50 columns and analyzed on an ABI 3700 capillary electrophoresis DNA sequencer. Phred/Phrap was used for data assembly, employing the default assembly parameters (17, 18, 23).
Data analysis.
Using the BLAST algorithms, the sequences from clones identified as strain 1885MEE-specific, were searched against the NCBI nucleotide and protein databases and the previously published strain Rd genome.
In order to determine whether there was a high level of synteny between the Rd and 86-028NP genomes, the consensus sequences (contigs) from our threefold assembly was cross-compared to the strain Rd genome sequence, using a two-dimensional version of tricross (43). The tricross ordering for the 86-028NP contigs was based on the Rd chromosome coordinates, with each contig anchored at the start coordinate of the earliest Rd gene hit.
To generate the list of genes observed in strain 86-028NP which are not found in strain Rd, the following procedure was used. The strain 86-028NP threefold-coverage contigs were analyzed by BLAST (BLASTX) against the NCBI's nonredundant (NR) database (data current as of 8 May 2003). This was accomplished using a Perl wrapper for NCBI's blastcl3 interface. The results were accumulated in a relational database (MySQL). The top 100 hits to NR (10−4 or better) were layered over the contig sequence in a best-score first fashion. For each BLAST hit, contig bases that were not covered by a better-scoring hit were marked as belonging to this hit in the relational database. For each hit that claimed more than 10 unique bases to itself, the translation of the entire region that generated the hit was analyzed by BLAST (TBLASTX, using NCBI's blastall software, to a locally generated strain Rd database) against the H. influenzae strain Rd gene set (built from NC_000907.ffn). Regions that found hits to strain Rd sequence at an E score of 0.01 or better were marked as strain Rd genes and excluded. All hits to NCBI's NR database that claimed more than 10 unique bases and that found no strain Rd hits, are reported as genes potentially unique to strain 86-028NP compared to strain Rd. The BLAST parameters for analysis of the contig set versus the NR database were chosen to accommodate the partial sequence and low-quality sequencing regions, and the presence of false positives in the analysis is expected. Likewise, the BLAST parameters for excluding genes present in Rd were chosen to eliminate sequences with even cursory similarity to Rd and are expected to limit the result set to only those sequences for which we have a high confidence in their uniqueness.
The strain 1885MEE data set was compared by BLAST with the threefold-contig data from strain 86-028NP using the BLAST program implemented at http://www.microbial-pathogenesis.org.
RESULTS AND DISCUSSION
Genomic differences between NTHI strain 1885MEE and H. influenzae strain Rd.
In order to develop a methodology that could be used to compare the genomes of a panel of NTHI isolates, a DNA microarray approach was adopted as this approach has provided useful data in other systems (16). Chromosomal DNA from strain 1885MEE was labeled with Cy5, and chromosomal DNA from H. influenzae strain Rd was labeled with Cy3, using a Klenow DNA polymerase-based direct incorporation method. Equal amounts of labeled genomic DNA were then pooled and used to hybridize to the strain 1885MEE genomic DNA array. The fluorescence signals from Cy5 and Cy3 were measured independently for each spot, using an Affymetrix 428 Array Scanner. Using GenePix Pro 3.0, the two channels were combined and a ratio image was generated. By comparing the relative fluorescence values for the Cy5 channel and the Cy3 channel, it was then possible to determine which clones contained DNA that was specific for strain 1885MEE, relative to strain Rd.
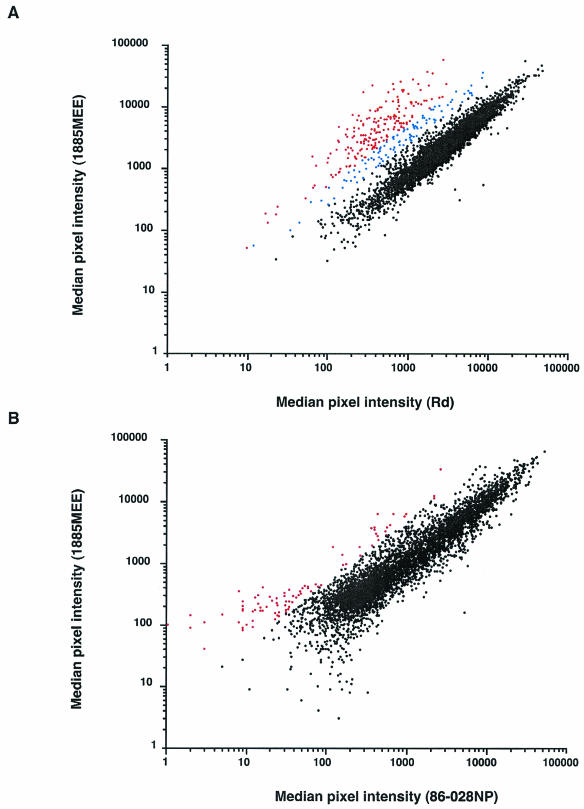
There is little precedent for the analysis of data derived from genomic DNA arrays, although work with Campylobacter jejuni indicates that a ratio of signal intensities between the two channels of twofold or higher, could be indicative of a clone containing a strain-specific insert (16). Thus, using the signal generated by labeled strain 1885MEE genomic DNA hybridizing with strain 1885MEE arrayed DNA as the control signal, a minimum fluorescence ratio of 2.5 (1885MEE/Rd) was used to select clones for further analysis (Fig. 1A). First, as the library was spotted in duplicate, only clones in which both replicates met this threshold were further characterized. Of 268 clones that met this criterion, viable sequence was obtained from 263 clones, vector sequence was trimmed and batch-wise analysis was conducted using the BLAST programs (http://www.microbial-pathogenesis.org). Thirty-two percent of the clones had homology to genes in both strains Rd and 1885MEE. Reanalysis of the data set, choosing clones having a fluorescence ratio of 5.0 (1885MEE/Rd), reduced the number of false positives from 32 to 8%, with the loss of only five strain 1885MEE-specific clones (Fig. 1A).
FIG. 1.
Genome differences between strains 1885MEE and Rd and between strains 1885MEE and 86-028NP as determined by microarray analysis. (A) The median pixel intensities, with background subtracted, of signals derived from the strain 1885MEE array analyzed with a mixture of labeled genomic DNA from strains 1885MEE and Rd. Spots with a signal intensity ratio greater than 5.0 (1885MEE/Rd) are plotted in red. Spots with a signal intensity ratio between 2.5 and 5.0 (1885MEE/Rd) are plotted in blue. Spots with a signal intensity ratio of less than 2.5 (1885MEE/Rd) are plotted in black. (B) The median pixel intensities, with background subtracted, of signals derived from the strain 1885MEE array analyzed with a mixture of labeled genomic DNA from strains 1885MEE and 86-028NP. Spots with a ratio greater than 5.0 (1885MEE/86-028NP) are plotted in red. Spots with a ratio of less than 5.0 (1885MEE/86-028NP) are plotted in black.
A total of 55 loci were identified as being unique to strain 1885MEE. A large number of these were conserved hypothetical genes. Some of the identified genes were of immediate interest due to their putative functions (Table 1). The translated sequence from clone 32-G11 had homology to hypothetical proteins from Pasteurella multocida and Neisseria meningitidis, both known pathogens. Similarly, the translated sequence from clone 62-G04 had homology to a hypothetical protein from Pseudomonas aeruginosa and a hypothetical membrane-associated transport protein from Helicobacter pylori, organisms both associated with human disease. Translated sequence from clone 17-H05 was 100% identical to the sequence of HicA from strain INT1, an invasive NTHI isolate from a meningitis patient. The translated amino acid sequence from clone 66-E09 was 80% identical to a portion of the amino acid sequence of Lav from H. influenzae strain INT1. Translated sequence from clone 17-H02 had 95% amino acid identity with a portion of the amino acid sequence of TNase (TnaA) from H. influenzae serotype b strain Eagan (34). Translated sequence from clone 14-A12 was 85% identical to the amino acid sequence of TsaA from P. multocida. TsaA is an alkyl peroxidase, which has a role in protection against reactive oxygen species and is potentially important in survival during phagocytosis (21, 35). These data, and the database reference of their respective homologue, are shown in Table 1.
TABLE 1.
Protein similarities identified by sequence analysis of clones identified by microarray as unique to strain 1885MEEa
| Clone | E value | Hit | Database reference | Organism |
|---|---|---|---|---|
| 14-A12 | 2.00E-66 | TsaA | ref|NP_245732.1| | Pasteurella multocida |
| 12-H10 | 4.00E-59 | Lavb | gb|AAK76425.1| | Haemophilus influenzae strain Int1/R2866 |
| 17-H02 | 1.00E-154 | TnaAb | gb|AAB96579.1| | Haemophilus influenzae strain b (Eagan) |
| 13-G04 | 4.00E-25 | Transcriptional factor MdcH | gb|AAF20290.1| | Acinetobacter calcoaceticus |
| 17-H05 | 9.00E-40 | HicAb | gb|AAC35809.1| | Haemophilus influenzae strain R3001 |
| 02-E06 | 5.00E-57 | Modification methylase HaeII-cytosine-specific methyltransferase HaeII | sp|O30868| | Haemophilus aegyptius |
| 59-F03 | 7.00E-07 | Modification methylase HphlA | sp|P50192| | Haemophilus parahaemolyticus |
| 63-E02 | 8.00E-95 | Type I restriction enzyme HsdR, putativeb | ref|NP_231400.1| | Vibrio cholerae |
Sequences identified in this study with homology to transposons, bacteriophage and hypothetical genes are not shown in the print version but are available on our Web site. (http://www.microbial-pathogenesis.org/hinf/files/20030917_Table1a.html). The complete alignment of each database hit as well as a link to the homologue in the NCBI database is also available on our Web site. In the online table, database hits to sequences also identified in the strain 86-028NP dataset are highlighted.
Database hits to sequences also identified in strain 86-028NP (see Table 2).
Sequences in 29 clones had homology to phage genes, many of which were similar to genes from Haemophilus phages HP1 and HP2. Of more interest, was the identification of strain 1885MEE-specific homologues of genes from the Gifsy 1 and 2 and Fels-2 prophages. Gifsy 1 and 2 were originally identified in S. enterica serovar Typhimurium and have been implicated in Salmonella virulence (19). Fels-2 has been identified in S. enterica (7). Sequences with homology to a number of other hypothetical genes were observed. The database reference for each hit as well as each alignment is available on our Web site.
Genomic differences between two NTHI clinical isolates analyzed by microarray.
To determine further the genetic content of strain 1885MEE compared to strain 86-028NP, genomic DNA was labeled with either Cy5 (1885MEE) or Cy3 (86-028NP), and equal amounts of labeled genomic DNA were pooled and used to hybridize to the strain 1885MEE genomic DNA array. The relative fluorescence values for the Cy3 channel and the Cy5 channel were then compared to determine which clones contained DNA that was specific for strain 1885MEE, relative to strain 86-028NP. Again, a fluorescence ratio of 5.0 (1885MEE/86-028NP) was used to choose putative strain 1885MEE-specific clones, relative to strain 86-028NP (Fig. 1B). Plasmid DNA was purified and valid sequence obtained from 72 of the 83 clones thought to contain sequence unique to strain 1885MEE. Sixty-eight of the clones had homology to genes in strain 86-028NP. The remaining four clones had homology to two hypothetical genes from H. influenzae strain Rd (HI1029 and HI1317), one from Vibrio vulnificus and one from P. multocida, respectively.
Comparison of the partial genome sequence of strain 86-028NP with the strain Rd genome sequence.
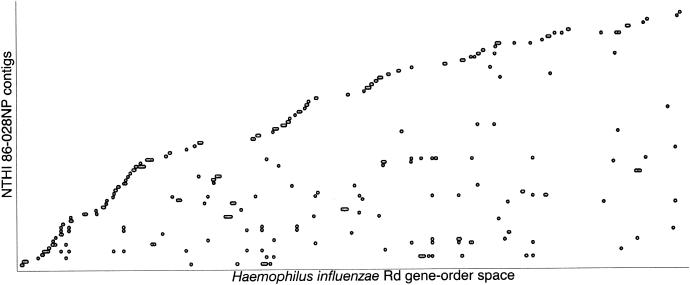
The genome of strain 86-028NP was sequenced to threefold coverage. Sequencing reads (8219) from the strain 86-028NP clones were assembled into 576 contigs. Seventy-five of the eighty-eight contigs of ≥5,000 bp showed significant similarity via BLASTN to genes in H. influenzae strain Rd. To visualize the relationship between the gene order in H. influenzae strain 86-028NP and H. influenzae strain Rd, the strain 86-028NP threefold-contig set and the Rd gene set were compared by BLAST using tricross. The results were plotted by sorting the contigs based on gene coordinates of the Rd genes hit, then anchoring each contig at the smallest Rd coordinate found (Fig. 2) (43). When compared in this fashion, if the 86-028NP genome had the same gene order as strain Rd and the contig assemblies were completely correct, the plot would appear as an increasing stairstep. At the current level of resolution, a high level of synteny is observed between the two genomes. This is in contrast to a comparison of the H. ducreyi and H. influenzae genomes, where little synteny is observed (Munson, Ray, and coworkers, unpublished data).
FIG. 2.
Contigs larger than 5 kb were aligned to the strain Rd genomic sequence as described in the text. Each point represents a hit to a strain Rd gene or several adjacent hits to closely linked strain Rd genes. The scatter plot shown here indicates that there are some differences between the gene arrangement in the strain 86-028NP genome (or assembly errors) compared to the gene arrangement in strain Rd, though an analysis of the data on a contig-by-contig basis supports the hypothesis that rearrangements represent a small portion of the genome.
BLASTX was used to identify 161 genes that had homologues in GenBank but were not found in H. influenzae strain Rd. We identified sequences related to known Haemophilus genes not present in strain Rd. The genes that encode the high-molecular weight-adhesin proteins and their associated processing proteins, the lav gene, the tnaA gene, the hicAB genes and restriction systems, among others, were identified in this data set (Table 2). Sequences with homology to transposases, phage genes and hypothetical genes were also identified.
TABLE 2.
Protein similarities identified in the strain 86-028NP three-fold contig seta
| Contig | E value | Hit | Database reference | Organism |
|---|---|---|---|---|
| 153 | 1.00E-129 | Adhesin | gb|AAA20524.1| | Haemophilus influenzae strain 12 |
| 502 | 1.00E-104 | Adhesin | gb|AAA20524.1| | Haemophilus influenzae strain 12 |
| 153 | 2.00E-07 | Outer membrane protein A | pir||T30852| | Rickettsia conorii |
| 270 | 0 | HmwA | gb|AAD56660.1| | Haemophilus influenzae A950006 |
| 1 | 1.00E-30 | HmwC | gb|AAF00477.1| | Haemophilus influenzae A950006 |
| 502 | 0 | HmwC | gb|AAF00477.1| | Haemophilus influenzae A950006 |
| 545 | 0 | HmwC | gb|AAF00477.1| | Haemophilus influenzae A950006 |
| 1 | 5.00E-37 | Putative accessory processing protein | gb|AAA20525.1| | Haemophilus influenzae strain 12 |
| 502 | 0 | Putative accessory processing protein | gb|AAA20525.1| | Haemophilus influenzae strain 12 |
| 421 | 2.00E-57 | HicB | gb|AAC35810.1| | Haemophilus influenzae R3001 |
| 421 | 3.00E-39 | HicAb | gb|AAC35809.1| | Haemophilus influenzae R3001 |
| 505 | 0 | Lavb | gb|AAK76425.1| | Haemophilus influenzae Int1/R2866 |
| 201 | 1.00E-20 | Peroxiredoxin | ref|NP_759448.1| | Vibrio vulnificus CMCP6 |
| 363 | 6.00E-05 | Putative protein phosphatase | ref|NP_711609.1| | Leptospira interrogans serovar lai strain 56601 |
| 398 | 1.00E-23 | Putative cytoplasmic protein | ref|NP_463063.1| | Salmonella enterica Typhimurium LT2 |
| 417 | 1.00E-05 | Repressor protein-related protein | ref|NP_745902.1| | Pseudomonas putida KT2440 |
| 442 | 0 | TnaAb | sp|O07674| | Haemophilus influenzae strain b (Eagan) |
| 118 | 1.00E-07 | ATP-dependent DNA helicase recG | ref|NP_813441.1| | Bacteroides thetaiotaomicron VPI-5482 |
| 501 | 7.00E-31 | ATP-dependent DNA helicase recG | ref|NP_813441.1| | Bacteroides thetaiotaomicron VPI-5482 |
| 110 | 5.00E-12 | Type I restriction-modification system methyltransferase subunit | gb|EAA25229.1| | Fusobacterium nucleatum subsp. vincentii ATCC 49256 |
| 118 | 8.00E-45 | DNA methylase HsdM, putative | ref|NP_231404.1| | Vibrio cholerae |
| 118 | 1.00E-18 | Probable restriction-modification system protein | ref|NP_251425.1| | Pseudomonas aeruginosa PA01 |
| 138 | 1.00E-148 | Type II restriction enzyme HaeII (endonuclease HaeII) (R. HaeII) | sp|O30869| | Haemophilus aegyptius |
| 240 | 8.00E-27 | Modification methylase Bepl (cytosine-specific methyltransferase BepI) (M. BepI) | sp|P10283| | Brevibacterium epidermidis |
| 297 | 1.00E-158 | DNA methylase HsdM, putative | ref|NP_231404.1| | Vibrio cholerae |
| 417 | 5.00E-06 | Adenine-specific DNA methylase | ref|NP_393798.1| | Thermoplasma acidophilum |
| 452 | 2.00E-98 | Type I restriction-modification system methylation subunit | gb|EAA23174.1| | Fusobacterium nucleatum subsp. vincentii ATCC 49256 |
| 501 | 0 | Type I restriction enzyme HsdR, putativeb | ref|NP_231400.1| | Vibrio cholerae |
Sequences identified in this study with homology to transposons, bacteriophage, and hypothetical genes are not shown in the print version but are available on our Web site (http://www.microbial-pathogenesis.org/hinf/files/20030917_Table2a.html). The complete alignment of each database hit as well as a link to the homologue in the NCBI database is also available on our Web site. In the online table, database hits to sequences also identified in the strain 1885MEE dataset are highlighted.
Database hits to sequences also identified in strain 1885MEE (see Table 1)
Further characterization of genes identified in both data sets but absent from strain Rd.
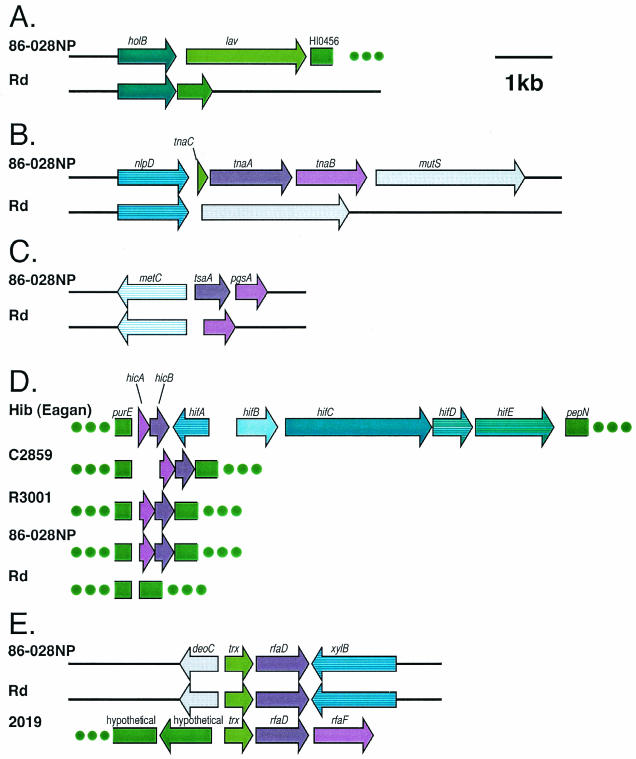
Several potential genes of interest were observed in data sets from both isolates. The lav gene encodes a member of the AIDA-I/VirG/PerT family of virulence-associated autotransporters and was reported to be restricted in distribution to a small number of pathogenic strains, including H. influenzae biogroup aegyptius and Brazilian purpuric fever isolates. It has previously been reported as absent from strain Rd (12). We completed the sequence for this gene from strain 86-028NP (GenBank accession number AY559035). The derived amino acid sequence of the strain 86-028NP Lav protein was 88% identical to the derived amino acid sequence of lav from INT1. Interestingly, lav from INT1, H. influenzae biogroup aegyptius, N. meningitides and N. gonorrhoeae have a series of GCAA repeats 10 bp downstream of the start codon, which are thought to be involved in slip-strand mispairing during DNA replication, giving rise to phase variation of this surface-exposed molecule. The coding sequence of lav in strain 86-026NP has 20 GCAA repeats in the same location. The comparative arrangement of the genes in the lav region for 86-028NP and strain Rd are shown in Fig. 3A.
FIG. 3.
(A) The lav gene of strain 86-028NP was sequenced and the gene arrangement in the lav region was compared to that of strain Rd. (B) The tnaA gene of strain 86-028NP was sequenced and the gene arrangement in the tna region was compared to that of strain Rd. (C) The tsaA gene of strain 86-028NP was sequenced and the gene arrangement in the tsaA region was compared to that of strain Rd. (D) The LKP gene region in a panel of Haemophilus isolates. The strain 86-028NP sequence is identical in this region to the sequence in this region of NTHI strain R3001, an NTHI isolate characterized by Mhlanga-Mutangadura and coworkers (37). Both strains 86-028NP and R3001 lack the hif gene cluster encoding the hemagglutinating pilus. The format of this figure is consistent with the format used by Mhlanga-Mutangadura and coworkers who characterized the LKP pilus gene cluster from a panel of Haemophilus isolates including strains C2859, R3001, and H. influenzae serotype b strain Eagan (37). (E) The rfaD region in a panel of Haemophilus isolates. The gene arrangement in the rfaD region of the strain 86-028NP genome is similar to that of the strain Rd genome, but different than the arrangement of these genes found in the genome of most NTHI strains examined including strain 2019, a strain characterized by Nichols et al. (40).
The tnaA gene encodes tryptophanase and is part of the tna gene cluster. This gene cluster has been widely studied in E. coli. In both E. coli and H. influenzae serotype b strain Eagan, the other components of the gene cluster are tnaB, which encodes a low-affinity tryptophan permease, and tnaC, which encodes a small peptide involved in regulation of expression of tnaAB. The tnaA gene cluster was located between the nlpD and mutS genes and is absent from the strain Rd genome (34). In the 86-028NP threefold-contig set, fragments of tnaA were identified spanning the ends of two contigs with the same organization as that found in strain Eagan. We completed the sequence from strain 86-028NP (GenBank accession number AY559033). The derived amino acid sequence of tnaA from strain 86-028NP was 99% identical to the sequence from strain Eagan. The comparative arrangement of the genes in the tna region for 86-028NP and strain Rd are shown in Fig. 3B.
Nearly all pathogenic isolates of H. influenzae test positive for indole production, one of the phenotypes used in the H. influenzae biotyping scheme (29). Tryptophanase catabolizes tryptophan to indole, pyruvate and ammonia, allowing tryptophan to be used as both a carbon and a nitrogen source (39). Tryptophanase may also play a role in both buffering against extremes in pH and in epithelial cell adherence and biofilm formation (15, 47).
Although not initially found in our BLAST analysis of the strain 86-028NP contig set, the tsaA gene was identified in strain 86-028NP contig set when the strain 1885MEE sequence was used to query the threefold database. The sequence of the strain 86-028NP gene was completed (GenBank accession number AY559033). The derived amino acid sequence was 89% identical to the P. multocida homologue. The comparative arrangement of the genes in the tsaA region for strains 86-028NP and Rd are shown in Fig. 3C.
The hicAB genes were identified in the strain 86-028NP data set and the hicA gene was identified in the strain 1885MEE data set. In serotype b organisms, these genes are in close proximity to the hemagglutinating pilus (LKP) genes. Mhlanga-Mutangadura and coworkers characterized the LKP pilus gene cluster from a panel of Haemophilus isolates (37). The pilus gene cluster is located between the purE and pepN genes, as depicted in Fig. 3D. The serotype b strain Eagan contains the hifABCDE gene cluster and produces hemagglutinating pili. Strain Rd lacks the hicAB genes as well as the hifABCDE gene cluster. In general, the nontypeable strains examined by Mhlanga-Mutangadura and coworkers contained the hicAB genes but not the hif genes that encode the hemagglutinating pilus. The strain 86-028NP sequence was identical in this region to the sequence in NTHI strain R3001, a strain that lacks the hif gene cluster (Fig. 3D).
The rfaD gene encodes an enzyme involved in the biosynthesis of endotoxin. Nichols and coworkers characterized the rfaD gene from NTHI strain 2019 (40). In strain 2019, the rfaD gene is immediately upstream of the rfaF gene, a gene that encodes an additional enzyme involved in endotoxin biosynthesis. The gene arrangement in strain Rd is different; the rfaD and rfaF genes are separated by approximately 11 kb of sequence. Nichols and coworkers noted that most nontypeable strains examined contained the gene arrangement found in strain 2019. In contrast, strain 86-028NP had a gene arrangement identical to that found in strain Rd (Fig. 3E).
Both strains had genes homologous to the HaeII restriction system found in H. influenzae biogroup aegyptius. A number of additional sequences with homology to different restriction systems were identified in both gene sets (Tables 1 and 2).
In summary, using microarray analysis, the genome of a NTHI isolate was analyzed and several genes were identified that were not present in strain Rd. We also partially sequenced the genome of a second strain. The assembly currently contains 576 contigs, many with regions of low coverage. A global analysis of the current assembly indicated that the gene content and order were similar to that in strain Rd. A more detailed analysis revealed that there were a substantial number of genes not previously found in the Pasteurellaceae and some regions where the gene content and order were different than that found in strain Rd. These data suggested that the strain 86-028NP genome would contain a complex mosaic of strain Rd and non-strain-Rd-like features.
Both approaches to comparative genome analysis identified genes present in NTHI strains relative to strain Rd, demonstrating the complementary nature of these approaches, as well as the genetic similarity of the two strains under investigation. These approaches are both survey methods, limited in precision by the physical process of microarray analysis and by the statistical parameters chosen for the bioinformatics analysis. Nonetheless, as survey methods they have identified a number of verifiable differences between the genomes under investigation, while not forecasting an unmanageable level of false positives. They require neither exhaustive sequencing of the genomes nor massive replication of the microarray experiments. Moreover, the specificity and selectivity of the methods were both sufficiently good that putative virulence genes were identified in the NTHI strains, genes that may be important in understanding the pathogenesis of OM and perhaps other NTHI-induced diseases of the respiratory tract.
The threefold-contig data are available on our Web site at http://www.microbial-pathogenesis.org. Updated data will be made available as the genome sequence is completed.
Acknowledgments
We thank Huachun Zhong for excellent technical assistance.
This work was supported by National Institutes of Health grants R01 DC03915 (to L.O.B.), R01 P20-RR15564 (to D.W.D.), and P20-RR16478 (to D.W.D.). The DNA Sequencing Core at Columbus Children's Research Institute was supported in part by the National Institutes of Health grant HD34615.
Editor: J. N. Weiser
REFERENCES
- 1.Alsarraf, R., C. J. Jung, J. Perkins, C. Crowley, N. W. Alsarraf, and G. A. Gates. 1999. Measuring the indirect and direct costs of acute otitis media. Arch. Otolaryngol. Head Neck Surg. 125:12-18. [DOI] [PubMed] [Google Scholar]
- 2.Bakaletz, L. O., E. R. Leake, J. M. Billy, and P. T. Kaumaya. 1997. Relative immunogenicity and efficacy of two synthetic chimeric peptides of fimbrin as vaccinogens against nasopharyngeal colonization by nontypeable Haemophilus influenzae in the chinchilla. Vaccine 15:955-961. [DOI] [PubMed] [Google Scholar]
- 3.Bakaletz, L. O., B. M. Tallan, T. Hoepf, T. F. DeMaria, H. G. Birck, and D. J. Lim. 1988. Frequency of fimbriation of nontypable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect. Immun. 56:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin, R. L. 1993. Effects of otitis media on child development. Am. J. Otol. 14:601-604. [PubMed] [Google Scholar]
- 5.Barenkamp, S. J., R. S. Munson, Jr., and D. M. Granoff. 1982. Outer membrane protein and biotype analysis of pathogenic nontypable Haemophilus influenzae. Infect. Immun 36:535-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergman, N. H., and B. J. Akerley. 2003. Position-based scanning for comparative genomics and identification of genetic islands in Haemophilus influenzae type b. Infect. Immun. 71:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunny, K., J. Liu, and J. Roth. 2002. Phenotypes of lexA mutations in Salmonella enterica: evidence for a lethal lexA null phenotype due to the Fels-2 prophage. J. Bacteriol. 184:6235-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassell, G. H. 1997. New and reemerging infectious diseases: a global crisis and immediate threat to the nation's health. The role of research. American Society for Microbiology, Washington, D.C.
- 9.Cassell, G. H., G. L. Archer, T. R. Beam, G. M. J., D. Goldmann, D. C. Hooper, R. N. Jones, S. H. Kleven, J. Lederberg, S. B. Levy, D. H. Lein, R. C. Moellering, T. F. O'Brien, B. Osburn, M. Osterholm, D. M. Shlaes, M. Terry, S. A. Tolin, and A. Tomasz. 1994. Report of the ASM Task Force of Antibiotic Resistance. American Society for Microbiology, Washington, D.C.
- 10.Chissoe, S. L., Y. F. Wang, S. W. Clifton, N. Ma, H. J. Sun, J. S. Lobsinger, S. M. Kenton, J. D. White, and B. A. Roe. 1991. Strategies for rapid and accurate DNA sequencing. Methods Companion Methods Enzymol. 3:55-65. [Google Scholar]
- 11.Daines, D. A., L. A. Cohn, H. N. Coleman, K. S. Kim, and A. L. Smith. 2003. Haemophilus influenzae Rd KW20 has virulence properties. J. Med. Microbiol. 52:277-282. [DOI] [PubMed] [Google Scholar]
- 12.Davis, J., A. L. Smith, W. R. Hughes, and M. Golomb. 2001. Evolution of an autotransporter: domain shuffling and lateral transfer from pathogenic Haemophilus to Neisseria. J. Bacteriol. 183:4626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Beccaro, M. A., P. M. Mendelman, A. F. Inglis, M. A. Richardson, N. O. Duncan, C. R. Clausen, and T. L. Stull. 1992. Bacteriology of acute otitis media: a new perspective. J. Pediatr. 120:81-84. [DOI] [PubMed] [Google Scholar]
- 14.DeMaria, T. F., D. M. Murwin, and E. R. Leake. 1996. Immunization with outer membrane protein P6 from nontypeable Haemophilus influenzae induces bactericidal antibody and affords protection in the chinchilla model of otitis media. Infect. Immun. 64:5187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Martino, P., A. Merieau, R. Phillips, N. Orange, and C. Hulen. 2002. Isolation of an Escherichia coil strain mutant unable to form biofilm on polystyrene and to adhere to human pneumocyte cells: involvement of tryptophanase. Can. J. Microbiol. 48:132-137. [DOI] [PubMed] [Google Scholar]
- 16.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 18.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 19.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 20.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, and et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 21.Francis, K. P., P. D. Taylor, C. J. Inchley, and M. P. Gallagher. 1997. Identification of the ahp operon of Salmonella typhimurium as a macrophage-induced locus. J. Bacteriol. 179:4046-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giebink, G. S. 1994. Immunology: promise of new vaccines. Pediatr. Infect. Dis. J. 13:1064-1068. [PubMed] [Google Scholar]
- 23.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 24.Holmes, K. A., and L. O. Bakaletz. 1997. Adherence of non-typeable Haemophilus influenzae promotes reorganization of the actin cytoskeleton in human or chinchilla epithelial cells in vitro. Microb. Pathog. 23:157-166. [DOI] [PubMed] [Google Scholar]
- 25.Hunter, L. L., R. H. Margolis, and G. S. Giebink. 1994. Identification of hearing loss in children with otitis media. Ann. Otol. Rhinol. Laryngol. Suppl. 163:59-61. [DOI] [PubMed] [Google Scholar]
- 26.Infante-Rivard, C., and A. Fernandez. 1993. Otitis media in children: frequency, risk factors, and research avenues. Epidemiol. Rev. 15:444-465. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan, B., T. L. Wandstrat, and J. R. Cunningham. 1997. Overall cost in the treatment of otitis media. Pediatr. Infect. Dis. J. 16:S9-11. [DOI] [PubMed] [Google Scholar]
- 28.Karma, P. H., L. O. Bakaletz, G. S. Giebink, G. Mogi, and B. Rynnel-Dagoo. 1995. Immunological aspects of otitis media: present views on possibilities of immunoprophylaxis of acute otitis media in infants and children. Int. J. Pediatr. Otorhinolaryngol. 32(Suppl.):S127-S134. [DOI] [PubMed] [Google Scholar]
- 29.Kilian, M. 1976. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 93:9-62. [DOI] [PubMed] [Google Scholar]
- 30.Kilpi, T., E. Herva, T. Kaijalainen, R. Syrjanen, and A. K. Takala. 2001. Bacteriology of acute otitis media in a cohort of Finnish children followed for the first two years of life. Pediatr. Infect. Dis. J. 20:654-662. [DOI] [PubMed] [Google Scholar]
- 31.Klein, J. O. 2000. The burden of otitis media. Vaccine 19(Suppl. 1):S2-S8. [DOI] [PubMed] [Google Scholar]
- 32.Klein, J. O. 1997. Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr. Infect. Dis. J. 16:S5-S8. [DOI] [PubMed] [Google Scholar]
- 33.Li, M. S., J. L. Farrant, P. R. Langford, and J. S. Kroll. 2003. Identification and characterization of genomic loci unique to the Brazilian purpuric fever clonal group of H. influenzae biogroup aegyptius: functionality explored using meningococcal homology. Mol. Microbiol. 47:1101-1111. [DOI] [PubMed] [Google Scholar]
- 34.Martin, K., G. Morlin, A. Smith, A. Nordyke, A. Eisenstark, and M. Golomb. 1998. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J. Bacteriol. 180:107-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Master, S. S., B. Springer, P. Sander, E. C. Boettger, V. Deretic, and G. S. Timmins. 2002. Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology 148:3139-3144. [DOI] [PubMed] [Google Scholar]
- 36.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mhlanga-Mutangadura, T., G. Morlin, A. L. Smith, A. Eisenstark, and M. Golomb. 1998. Evolution of the major pilus gene cluster of Haemophilus influenzae. J. Bacteriol. 180:4693-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton, W. A., and E. F. Snell. 1964. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc. Natl. Acad. Sci. USA 51:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nichols, W. A., B. W. Gibson, W. Melaugh, N. G. Lee, M. Sunshine, and M. A. Apicella. 1997. Identification of the ADP-L-glycero-D-manno-heptose-6-epimerase (rfaD) and heptosyltransferase II (rfaF) biosynthesis genes from nontypeable Haemophilus influenzae 2019. Infect. Immun. 65:1377-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paap, C. M. 1996. Management of otitis media with effusion in young children. Ann. Pharmacother. 30:1291-1297. [DOI] [PubMed] [Google Scholar]
- 42.Pettigrew, M. M., B. Foxman, Z. Ecevit, C. F. Marrs, and J. Gilsdorf. 2002. Use of pulsed-field gel electrophoresis, enterobacterial repetitive intergenic consensus typing, and automated ribotyping to assess genomic variability among strains of nontypeable Haemophilus influenzae. J. Clin. Microbiol. 40:660-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray, W. C., R. S. Munson, Jr., and C. J. Daniels. 2001. Tricross: using dot-plots in sequence-id space to detect uncataloged intergenic features. Bioinformatics 17:1105-1112. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Smoot, L. M., D. D. Franke, G. McGillivary, and L. A. Actis. 2002. Genomic analysis of the F3031 Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius by PCR-based subtractive hybridization. Infect. Immun. 70:2694-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spinola, S. M., J. Peacock, F. W. Denny, D. L. Smith, and J. G. Cannon. 1986. Epidemiology of colonization by nontypable Haemophilus influenzae in children: a longitudinal study. J. Infect. Dis. 154:100-109. [DOI] [PubMed] [Google Scholar]
- 47.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki, K., and L. O. Bakaletz. 1994. Synergistic effect of adenovirus type 1 and nontypeable Haemophilus influenzae in a chinchilla model of experimental otitis media. Infect. Immun. 62:1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teele, D. W., J. O. Klein, C. Chase, P. Menyuk, B. A. Rosner, et al. 1990. Otitis media in infancy and intellectual ability, school achievement, speech, and language at age 7 years. J. Infect. Dis. 162:685-694. [DOI] [PubMed] [Google Scholar]
- 50.Williams, B. J., M. Golomb, T. Phillips, J. Brownlee, M. V. Olson, and A. L. Smith. 2002. Bacteriophage HP2 of Haemophilus influenzae. J Bacteriol. 184:6893-6905. [DOI] [PMC free article] [PubMed] [Google Scholar]