Abstract
The type III secretion system (T3SS) encoded by the Salmonella pathogenicity island 2 (SPI2) has a central role in systemic infections by Salmonella enterica and for the intracellular phenotype. Intracellular S. enterica uses the SPI2-encoded T3SS to translocate a set of effector proteins into the host cell, which modify host cell functions, enabling intracellular survival and replication of the bacteria. We sought to determine whether specific functions of the SPI2-encoded T3SS can be transferred to heterologous hosts Salmonella bongori and Escherichia coli Mutaflor, species that lack the SPI2 locus and loci encoding effector proteins. The SPI2 virulence locus was cloned and functionally expressed in S. bongori and E. coli. Here, we demonstrate that S. bongori harboring the SPI2 locus is capable of secretion of SPI2 substrate proteins under culture conditions, as well as of translocation of effector proteins under intracellular conditions. An SPI2-mediated cellular phenotype was induced by S. bongori harboring the SPI2 if the sifA locus was cotransferred. An interference with the host cell microtubule cytoskeleton, a novel SPI2-dependent phenotype, was observed in epithelial cells infected with S. bongori harboring SPI2 without additional effector genes. S. bongori harboring SPI2 showed increased intracellular persistence in a cell culture model, but SPI2 transfer was not sufficient to confer to S. bongori systemic pathogenicity in a murine model. Transfer of SPI2 to heterologous hosts offers a new tool for the study of SPI2 functions and the phenotypes of individual effectors.
During the evolution of bacterial pathogenesis, the horizontal transfer of virulence genes has been a driving force for the emergence of new species and the adaptation of pathogens to new hosts and specific sites in these hosts. It has been concluded that large clusters of virulence genes are exchanged between various bacteria, and the acquisitions of so-called “pathogenicity islands” are considered as quantum leaps in the evolution of bacterial pathogenesis (13, 35).
Salmonella spp. form a complex group of gastrointestinal pathogens of animals and humans. Traditionally, a large number of isolates of Salmonella enterica have been defined as species due to distinct serological differences. However, molecular analyses indicated that most of these isolates represent serotypes of the species S. enterica, for example, S. enterica subspecies I serotype Typhimurium, and current taxonomy defines two species of Salmonella. Human infections are most frequently caused by various serovars of S. enterica. Disease outcomes associated with S. enterica can range from self-limiting localized gastrointestinal infections to life-threatening systemic infections such as typhoid fever (9). S. bongori, a second Salmonella species, is only rarely observed in human infections and is found as a commensal of cold-blooded animals (4, 38). Molecular analyses indicated that S. bongori represents a phylogenetically older separation from the genus Salmonella (4, 6).
Besides their clinical relevance, Salmonella spp. are interesting organisms for study of the interaction between pathogen and host cell, as well as events during the evolution of a pathogen. A large number of virulence genes have been identified in S. enterica. Two key virulence traits are involved in the interaction of bacteria and host cells. S. enterica is able to invade nonphagocytic cells such epithelial cells of the gastrointestinal mucosa. S. enterica is also a facultative intracellular pathogen that can survive phagocytosis by macrophages and replicate inside eukaryotic host cells. These two hallmarks of pathogenesis are linked to the presence of two pathogenicity islands (reviewed in reference 14). Salmonella pathogenicity island 1 (SPI1) encodes a type III secretion system (T3SS) that has a function for bacterial invasion (11) and further interactions between Salmonella and enterocytes (45). SPI2 encodes a second T3SS with a role in subsequent phases of pathogenesis (18). Mutant strains deficient in SPI2 genes are severely attenuated in virulence and are deficient in intracellular survival and proliferation. SPI2 genes are induced by intracellular bacteria, and it has been demonstrated that SPI2 functions modify several host cell functions such as intracellular transport (42) and antimicrobial defense mechanisms involving reactive oxygen intermediates (44), as well as reactive nitrogen intermediates (5). SPI2 is also important for maintaining the Salmonella-containing vesicle, and the SPI2 effector protein SifA makes a significant contribution to this phenotype (3).
Both S. enterica and S. bongori harbor SPI1, but the SPI2 locus is only present in S. enterica and is considered a more recent evolutionary acquisition (20, 34). The gain of SPI2 is thought to be a major step toward successful systemic colonization of host organisms (1).
The SPI1- and SPI2-encoded T3SSs both mediate the contact-dependent translocation of bacterial effector proteins into eukaryotic target cells. Interestingly, these effector proteins are not only encoded by genes within SPI1 and SPI2 but also by various distinct loci that represent independent acquisitions. With respect to the translocated effector proteins of the SPI2-encoded T3SS, a group of effector proteins referred to as STE proteins has been defined by the presence of a N-terminal secretion signal (30). There are also the translocated effector proteins SseF and SseG encoded within the SPI2 locus that colocalize with the endosomal membrane compartments after translocation by intracellular Salmonella (25). All of the SPI2-specific effector proteins currently known are absent in S. bongori (16, 24, 31). The total number of genes encoding translocated effectors is not known, and the function of the identified effector proteins is only partially understood. Sequence similarity of various effector proteins may indicate a redundancy in the function of these proteins. This situation makes it difficult to examine the contribution of an individual effector to the intracellular phenotype of S. enterica.
In the experimental setup described here, we investigated whether SPI2-functions can be transferred into SPI2-negative host organisms. We observed that the SPI2-system can be functionally transferred to S. bongori and that SPI2-mediated phenotypes are expressed if eukaryotic host cells are infected with S. bongori harboring the SPI2 locus. This system, when used with a heterologous host organism, allows the dissection of the roles of individual effector proteins.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains used in the present study are listed in Table 1. S. enterica subspecies I serotype Typhimurium NCTC 12023 (S. enterica serotype Typhimurium) was used as a wild-type strain. S. bongori was retrieved from Salmonella reference collection C (SARC) of the Salmonella Genetic Stock Center (Calgary, Alberta, Canada). The E. coli strains DH5α (Gibco-BRL) and XL1-Blue MR (Stratagene) were used for the propagation of plasmids, and the probiotic E. coli strain Mutaflor was used as a host for transfer of the SPI2 locus.
TABLE 1.
Bacteria strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source and/or referenceb |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| NCTC 12023 | Wild type | NCTC, Colindale, United Kingdom |
| MvP371 | ΔSPI2, 25-kb deletion; Kanr | This study |
| S. bongori SARC12 | Wild type | 4; SGSC, Calgary, Alberta, Canada |
| E. coli | ||
| XL-1 MR | Stratagene | |
| Mutaflor (Nissle 1917) | Wild type, serotype O6:K5:H1 | DSM strain 6601 (provided by S. Schubert, Munich, Germany) |
| Plasmids | ||
| pSuperCos1 | Cosmid vector; Cmr | Stratagene |
| pBeloBac11 | BAC vector, single copy; Cmr | 22 |
| pWSK29 | Low-copy vector; Ampr | 46 |
| pFPV25.1 | Prorpsm GFP | 43 |
| p1-H4 | SPI2 in pSuperCos1 | This study |
| pB6 | SPI2 in pBeloBac11 | This study |
| p2104 | ProsifA sifA::M45 in pWSK29 | 17 |
| p2684 | ProsseJ sseJ::HA in pWSK29 | This study |
| p2777 | Constitutive GFP in p2684 | This study |
| p2812 | Constitutive GFP in p2104 | This study |
Kanr, Kanamycin resistance; Cmr, chloramphenicol resistance; Ampr, ampicillin resistance.
SGSC, Salmonella Genetic Stock Center; NCTC, National Collection of Type Cultures.
Construction of a cosmid bank of S. enterica serotype Typhimurium.
Chromosomal DNA of S. enterica serotype Typhimurium was partially digested with SauIIIA and cloned into the BamHI restriction site of the cosmid pSuperCos1 (Stratagene). The cosmids were packed by phage λ by using a Gigapack III Gold Packaging Kit (Stratagene) and transferred into E. coli XL1-Blue MR (Stratagene). The cosmid library consisting of about 1,000 clones was examined by Southern blot analysis for the presence of the SPI2-encoded T3SS with a hybridization probe corresponding to ssaT. Positive clones were further analyzed by hybridization with probes corresponding to orf242 and ssaU, the most distal genes of the portion of SPI2 relevant for virulence.
Culture conditions.
For infection experiments, bacterial strains were grown in Luria-Bertani (LB) broth containing 50 μg of carbenicillin/ml or 34 μg of chloramphenicol/ml if required for the maintenance of plasmids. Bacterial cultures were grown in glass test tubes with agitation.
For in vitro studies of expression, strains were grown in minimal medium composed of N-salts medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 100 mM Bis-Tris-HCl (pH 7.0), 38 mM glycerol, 0.1% Casamino Acids] containing low (30 μM MgCl2) or high (10 mM MgCl2) concentrations of Mg2+. Minimal medium containing high (PCN) or low (PCN-P) concentrations of phosphate were used as described previously (8, 17). For PCN-P medium at pH 5.8, 80 mM morpholinepropanesulfonic acid was replaced by 80 mM morpholineethanesulfonic acid. All minimal media were prepared with double-distilled H2O.
Strain construction.
Plasmids were introduced into the respective strains by electroporation or by P22 transduction (26) (in case of S. enterica serotype Typhimurium) according to standard procedures. Strain MvP 371 with a replacement of the SPI2-encoded T3SS by the kanamycin resistance cassette was constructed by the Red deletion technique (7). The primers SPI2-Red-Del-for (5′-CCATACGTAACAAGGCTGCAACGGGTTCAAATAACGTTTCAGGGTGTAGGCTGGAGCTGCTTC-3′) and SPI2-Red-Del-rev (5′-GTAGCTTTTCGCTGATCCCATGATTGGTATACCCCTCGTCCATATGAATATCCTCCTTAG-3′) were used to amplify the aph gene from pKD4 (7). The resulting PCR product was purified and used to transform S. enterica serotype Typhimurium to kanamycin resistance as previously described (15). The deletion in the resulting strain was confirmed by Southern hybridization and DNA sequencing.
Construction of plasmids.
Plasmid p2777 for the expression of SseJ with a C-terminal fusion to the hemagglutinin (HA) epitope tag and the constitutively expression of green fluorescent protein (GFP) was constructed as follows. Primers SseJ-Pro-For (5′-CCGGAATTCACATAAAACACTAGCAC-3′) and SseJ-HA-Rev2 (5′-GCATCTAGATTAAGCGTAGTCTGGGACGTCGTATGGGTATTCAGTGGAATAATGATGAGC-3′) were used to amplify the sseJ gene with its promoter from genomic DNA of S. enterica serotype Typhimurium. The resulting product was digested with EcoRI and XbaI and subcloned into low-copy-number vector pWSK29 to yield p2684.
Primers GFP-Pro-For-SacI (5′-GATGAGCTCGTTCATGCGTGATGCAATG-3′) and GFP-Rev-SacI (5′-GATGAGCTCCATTTATTTGTATAGTTC-3′) were used to amplify the GFP gene and a constitutive promoter from plasmid pFPV25.1. The product was digested with SacI and cloned into the SacI site of p2684 to yield p2777 or cloned into p2104 to yield p2812. Plasmids were introduced into S. enterica serotype Typhimurium strains by electroporation. Regulated expression of the SseJ-HA fusion protein was analyzed under in vitro conditions and was identical to previously described fusion proteins with the M45 epitope tag (data not shown).
Protein secretion in vitro.
Preparation and analyses of proteins secreted by the SPI2-encoded T3SS in vitro was performed as described before (32). Briefly, bacteria were grown with agitation for 16 h at 37°C in 200 ml of PCN-P media adjusted to pH 5.8. Bacteria were pelleted by centrifugation, and the culture supernatant was passed through a 0.2-μm-pore-size filter membrane to remove residual bacteria. Protein secreted into the culture medium was concentrated by precipitation with 10% trichloroacetic acid. Secreted protein located on the bacterial cells surface was detached by vigorous mixing of the concentrated bacterial suspension and concentrated by trichloroacetic acid precipitation.
Cell culture.
The murine monocytic cell line RAW 264.7 and the human epithelial cell line HeLa were cultured in Dulbecco modified Eagle medium (DMEM; Invitrogen) containing 10% fetal calf serum (FCS; Invitrogen) and 2 mM glutamine at 37°C in an atmosphere of 5% CO2. Cells were cultured in the absence of antibiotics.
Bacterial infection of HeLa cells and invasion assay.
For the infection of HeLa cells, bacteria were grown at 37°C with agitation to stationary phase. The cultures were then diluted 1:30 with fresh LB broth and incubated for further 3.5 h at 37°C with agitation to reach late logarithmic phase. The optical density at 600 nm (OD600) of the cultures was adjusted with phosphate-buffered saline (PBS) to 0.2. The bacteria were diluted in DMEM containing FCS and glutamine and added to the cells seeded in 24-well tissue culture plates at a multiplicity of infection of 1 (for S. enterica serotype Typhimurium) to 50 (for S. bongori). The bacteria were centrifuged onto the cells at 500 × g for 5 min and then incubated for 25 min at 37°C in an atmosphere of 5% CO2. After infection, the macrophages were washed three times with PBS and then incubated in DMEM containing FCS, glutamine, and 100 μg of gentamicin/ml (Sigma). After 1 h of incubation, the medium was replaced with DMEM containing FCS, glutamine, and 10 μg of gentamicin/ml for the remainder of the experiment.
To assess the number of bacteria that actively entered the cells, serial dilutions of the inocula were plated onto Mueller-Hinton agar plates. HeLa cells were washed three times with PBS at 2 h after infection and lysed with 0.1% Triton X-100 in PBS for 10 min at 37°C. Serial dilutions were plated on Mueller-Hinton agar plates.
Bacterial infection of macrophages, invasion assay, and survival assay.
For infection of RAW 264.7 cells, bacteria were grown to stationary phase, and the OD600 values of the cultures were adjusted with PBS to 0.2. The bacteria were diluted in DMEM containing FCS and glutamine and then added to the cells seeded in 24-well tissue culture plates at a multiplicity of infection of 1 (for S. enterica serotype Typhimurium) to 50 (for S. bongori). The infected cells were further treated as described for the infection of HeLa cells.
For enumeration of intracellular bacteria, macrophages were washed three times with PBS and lysed with 0.1% Triton X-100 in PBS for 10 min at 37°C, and serial dilutions were plated on Mueller-Hinton agar plates.
Infection of mice.
Groups of 3 to 4 female BALB/c mice (6 to 8 weeks of age) were infected by orogastral application of a suspension containing mixture of 5 × 109 each of S. bongori(p2104) and S. bongori(pB6/p2104) or of S. bongori(p2777) and S. bongori(pB6/p2777). At day 2, 3, or 4 after infection, animals were sacrificed. Peyer's patches, mesenteric lymph nodes, spleens, and ceca were removed and homogenized in sterile PBS. Serial dilutions of the homogenates were plated onto LB agar containing carbenicillin to determine the total number of bacteria and onto LB agar containing chloramphenicol to determine the number of S. bongori(pB6/p2104) or S. bongori(p2777).
Immunofluorescence.
For immunofluorescence analyses, cells were grown in 24-well tissue culture plates on glass coverslips. After infection and incubation for different times, the cells were fixed with 3% paraformaldehyde in PBS for 15 min at room temperature and then washed three times with PBS. The antibodies were diluted in a blocking solution consisting of 10% normal goat serum, 1% bovine serum albumin, and 0.1% saponin (Sigma) in PBS. The coverslips were incubated with various antibodies as detailed below and then washed three times with PBS after each incubation step. The coverslips were mounted with Fluoprep (bioMèrieux) and sealed with Entellan (Merck). Samples were analyzed by using a laser-scanning confocal microscope (Leica TCS-NT).
Antibodies.
If not otherwise stated, all antibody incubations were performed for 1 h at room temperature. S. enterica serotype Typhimurium was detected by using rabbit α-Salmonella O4 test sera (1:1,000; Difco) as a primary antibody and a goat anti-rabbit Cy2-conjugate (1:1,000; Jackson). S. bongori was detected by using mouse α-Salmonella O48 sera (1:1,000; Sifin) as a primary antibody and goat anti-mouse Cy2-conjugate (1:1,000; Jackson). Rat monoclonal antibody anti-HA epitope (Roche) was used at a dilution of 1:500 and detected with goat α-rat conjugated to Cy3 (1:1,000). For immunostaining of microtubules, a mouse anti-β-tubulin Cy3-conjugate (Sigma) was used at a dilution of 1:200.
LAMP-1 of human cells was detected with mouse anti-human LAMP-1 (clone A4H3, 1:1,000; DSHB) as a primary antibody and goat anti-mouse Cy3-conjugate (1:1,000; Jackson) as the secondary antibody. Cellular F-actin was detected by using Texas red-conjugated phalloidin (1:200; Molecular Probes).
Polyclonal rabbit antiserum against the recombinant heat shock protein Hsp60 was used at a dilution of 1:400 (33). Rabbit antisera against recombinant SseB have been described previously (2).
RESULTS
Virulence characteristics of S. bongori and E. coli Mutaflor.
We used the bacterial species S. bongori and E. coli Mutaflor as heterologous hosts in the present study. We observed that various virulence factors of S. enterica are absent in S. bongori, most importantly the SPI2 locus (20, 34) and various loci encoding effector proteins of the SPI2-encoded T3SS (16, 24, 31). Mutaflor is an E. coli strain that is nonpathogenic but, in contrast to common lab strains, fully capable of colonizing the intestine. This strain is used as probiotic for the regeneration of the human intestinal flora.
In order to define the intracellular characteristics of S. bongori and E. coli Mutaflor, we applied cell culture models for invasion and intracellular replication. We used S. bongori strains from SARC (4), as well as clinical isolates from human infections of children in southern Italy (37). Analysis of invasion of HeLa cells indicated that E. coli Mutaflor and several of the clinical S. bongori isolates were noninvasive under the conditions applied. We found that the S. bongori strains SARC 11 and SARC 12 are able to invade HeLa cells with 2 to 5% of the invasiveness observed for S. enterica serotype Typhimurium strain NCTC 12023. Further analyses indicated that S. bongori was efficiently taken up by the murine monocyte cell line RAW 264.7. In contrast to intracellular S. enterica serotype Typhimurium that replicated rapidly, the number of intracellular S. bongori organisms decreased within 16 h of the experiment (data not shown).
Cloning of SPI2-encoded T3SS.
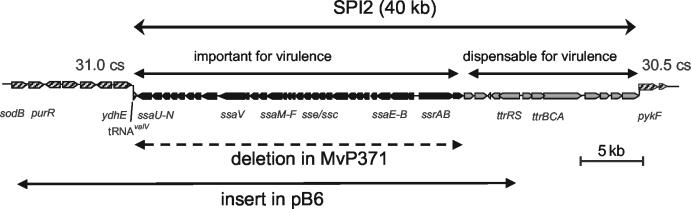
DNA fragments obtained by partial SauIIIA digest of genomic S. enterica serotype Typhimurium DNA were cloned into cosmid pSuperCos1 (Stratagene), and the resulting library was maintained in E. coli. Colony hybridization allowed the identification of clone p1-H4 that harbored the portion of SPI2 encoding subunits of the T3SS, secreted proteins, and the regulatory system SsrAB (Fig. 1). To circumvent artifacts due to the copy number of 7 to 10 reported for pSuperCos1, the insert of p1-H4 was recovered after digestion with NotI and subcloned into the bacterial artificial chromosome (BAC) vector pBeloBac11 (22), resulting in BAC clone pB6. pBeloBac11 is present as a single copy per cell.
FIG. 1.
Cloning of the SPI2-encoded T3SS. The SPI2 locus of S. enterica is 40 kb in size and is divided into two functional entities. A portion of 25 kb important for virulence contains the T3SS and several translocated effectors and is only present in S. enterica subspecies. In contrast, the 15-kb portion encoding the tetrathionate reductase complex and proteins of unknown function is not required for virulence and can be detected in S. enterica, as well as in S. bongori (19). The extent of the deletion of SPI2 genes in strain ΔSPI2 is indicated.
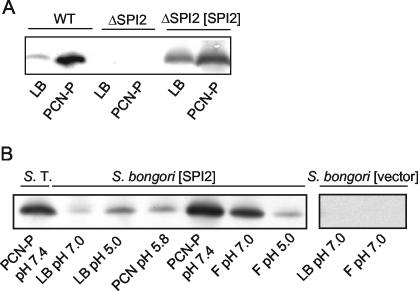
In order to verify the function of the pB6-encoded SPI2 translocation system, a SPI2 deletion strain of S. enterica serotype Typhimurium was constructed. A portion of 25 kb of SPI2 required for virulence was deleted from the S. enterica serotype Typhimurium chromosome by using a one-step inactivation system (7). The resulting strain failed to synthesize SPI2-encoded proteins, as shown for the secreted translocator protein SseB (Fig. 2A). The ΔSPI2 strain was attenuated in systemic infections in mice to a similar extent as previously described transposon mutant strains (40). Complementation of the ΔSPI2 strain with pB6 harboring the cloned SPI2 locus restored expression of SPI2-encoded proteins (Fig. 2A), as well as intracellular replication (data not shown). These experiments indicated that plasmid pB6 contains a functional set of genes encoding the T3SS and its regulatory system.
FIG. 2.
Expression of sseB by the cloned SPI2 locus. (A) S. enterica serotype Typhimurium wild type (WT), the SPI2 deletion mutant strain (ΔSPI2), and the SPI2 deletion mutant strain complemented with the plasmid-borne SPI2 locus (ΔSPI2 [SPI2]) were grown under conditions resulting in low (LB) or high (PCN-P) levels of the SPI2-encoded protein SseB. After growth for 16 h, lysates were prepared from equal amounts of bacteria, as determined from the OD600, and protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto nitrocellulose membranes. SseB was detected by using a polyclonal antibody as previously described (32). (B) The plasmid-borne SPI2 locus was introduced into S. bongori. S. enterica serotype Typhimurium wild type (S. T.) and S. bongori harboring the vector pBeloBac11 or the SPI2-encoding plasmid pB6 were grown for 16 h in various media as indicated. LB broth was adjusted to pH 7.0 or 5.0, PCN was adjusted to pH 5.8, PCN-P was adjusted to pH 7.4, and F minimal medium containing limiting amounts of Mg2+ (30 μM) was adjusted to pH 7.0 or 5.0. The amounts of SseB in total bacterial lysates were detected as described above.
Analyses of SPI2 gene expression and T3SS function in heterologous hosts.
Cosmid p1-H4 and BAC clone pB6 were introduced into various E. coli strains and S. bongori SARC 12 by electroporation, and the function of SPI2 genes was further analyzed. Previous observations revealed that the expression of SPI2 genes is induced during growth in medium containing limiting amounts of Mg2+ or phosphate. To investigate expression of SPI2 genes in heterologous hosts, strains harboring p1-H4 or pB6 were grown in various rich and minimal media, and the levels of the SPI2-encoded protein SseB were assayed by Western blot analyses of total cell fractions. E. coli strains did not grow under medium conditions required to induce SPI2 gene expression and were not further investigated. Western blot analyses indicated that high levels of SseB were present after growth of S. bongori(p1-H4) in rich medium but not after growth in SPI2-inducing minimal medium with phosphate limitation (i.e., PCN-P) or limiting amounts of Mg2+ (F medium) (data not shown). In contrast, analysis of S. bongori(pB6) showed that high levels of SseB were only observed after growth in PCN-P or F medium (Fig. 2B). The effects of medium composition on S. bongori(pB6) were similar to the effects on wild-type S. enterica serotype Typhimurium described previously (8).
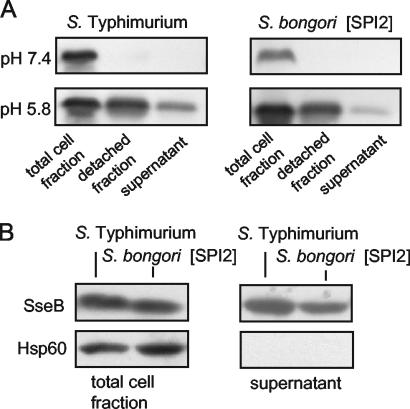
We next analyzed whether the SPI2-encoded T3SS is functioning in protein secretion in the heterologous host S. bongori. Previous studies showed that growth of S. enterica serotype Typhimurium in minimal medium at acidic pH induced the secretion of several SPI2-encoded proteins (2, 23, 32) and further effector proteins of the SPI2-encoded T3SS (17). These studies also revealed that large amounts of the secreted protein remain on the bacterial cell surface after secretion. To investigate SPI2-dependent protein secretion, cultures of S. enterica serotype Typhimurium and S. bongori(pB6) were grown in PCN-P at pH 7.4 or pH 5.8. Western blot analyses indicated the presence of SseB in the total cell fraction, the fraction detached from the bacterial cell surface (detached fraction) and in the culture supernatant (secreted protein fraction) if S. enterica serotype Typhimurium or S. bongori(pB6) was grown in PCN-P at pH 5.8 but not after growth in PCN-P at pH 7.4 (Fig. 3). To control whether the protein in the detached or supernatant fractions results from release of cytoplasmic protein due to cell lysis, we examined by Western blot the presence of Hsp60, a cytoplasmic heat shock protein that is not secreted. Hsp60 was neither detected in the supernatant fraction of S. enterica serotype Typhimurium nor in the supernatant fraction of S. bongori(pB6), indicating that growth conditions did not result in the release of detectable amounts of protein from bacterial cells.
FIG. 3.
Secretion of SseB by wild-type S. enterica serotype Typhimurium and S. bongori expressing the SPI2-encoded T3SS. S. enterica serotype Typhimurium or S. bongori(pB6) organisms were grown in PCN-P minimal medium adjusted to pH 7.4 or 5.8. After growth overnight, secreted protein was recovered from the bacterial cell surface (detached fraction) or the growth medium (supernatant) as previously described (32). (A) The amounts of SseB in total cells, in the detached fraction, and in the supernatant were analyzed by Western blotting as described for Fig. 2. (B) To control the integrity of S. enterica serotype Typhimurium and S. bongori(pB6) grown in PCN-P at pH 5.8, the presence of the Hsp60 was analyzed in the total cell and supernatant fractions. Hsp60 is a cytoplasmic protein that is not secreted.
Taken together, these experiments demonstrate that the effects of environmental stimuli on chromosomal SPI2 genes in S. enterica serotype Typhimurium and episomal SPI2 genes in S. bongori are very similar and that the SPI2-encoded T3SS is functional in protein secretion in S. bongori as a heterologous host.
Translocation of SPI2 proteins by S. bongori harboring plasmid-encoded SPI2.
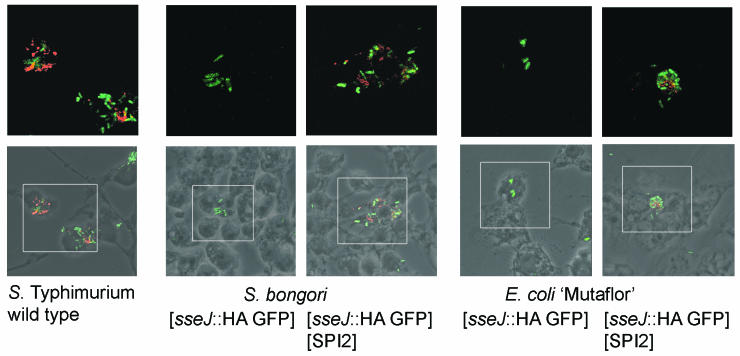
We next examined whether intracellular S. bongori harboring SPI2 was able to translocate effector proteins into the host cell. Using an epitope-tagging approach, we and others have previously demonstrated that the SPI2-encoded effector SseJ is secreted under in vitro conditions (17) and translocated by intracellular S. enterica serotype Typhimurium (10, 25). To analyze translocation by heterologous host species, plasmid p2777 for the expression of a fusion protein of SseJ and the HA epitope tag or both p2777 and pB6 were electroporated into S. bongori and E. coli Mutaflor.
RAW 264.7 cells were infected with S. enterica serotype Typhimurium harboring p2777 or S. bongori or E. coli Mutaflor, each harboring either p2777 or pB6/p2777, and translocated protein was detected by immunostaining (Fig. 4). As described before (25), we observed staining for translocated SseJ-HA within host cells after infection with S. enterica serotype Typhimurium harboring p2777. Only a lower number of intracellular E. coli was detected, indicating that the majority of the phagocytosed bacteria were rapidly degraded. No protein translocation was detected after infection with S. bongori(p2777) or E. coli(p2777). In contrast, after infection with S. bongori(pB6/p2777) or E. coli(pB6/p2777) we detected translocated SseJ-HA. These observations demonstrate that, in heterologous hosts, the SPI2-encoded T3SS is functional in translocating an effector protein.
FIG. 4.
Translocation of a SPI2 effector protein by intracellular S. enterica serotype Typhimurium and by S. bongori and E. coli harboring the SPI2-encoded type III secretion system. RAW 264.7 cells were grown on glass coverslips and infected with S. enterica serotype Typhimurium, S. bongori, or E. coli Mutaflor. All strains harbored plasmid p2777 for SPI2-regulated expression of sseJ::HA and constitutive expression of GFP. In addition, S. bongori or E. coli Mutaflor harbored pB6 containing the SPI2 locus, if indicated. At 16 h after infection, the cells were fixed and immunostained for SseJ-HA. Representative micrographs show merged phase contrast, intracellular bacteria (green), and translocated SseJ-HA (red).
Induction of SPI-dependent phenotypes by S. bongori harboring SPI2.
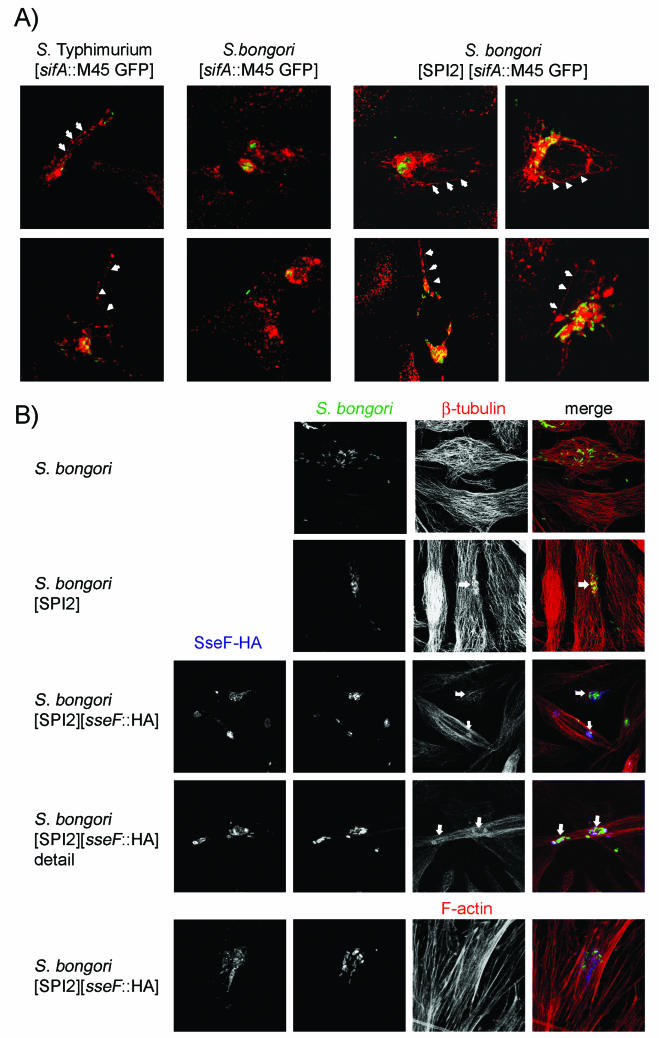
We then investigated whether an effector protein translocated by S. bongori harboring SPI2 can also induce a cellular phenotype within infected host cells. The specific effects of most SPI2 effector proteins on the host cell are not known in detail. However, a strong host cell phenotype is related to the translocation of SifA, a member of the STE family of SPI2 effectors (30). SifA is required for the formation of tubular aggregates of host cell endosomes containing lysosomal glycoproteins, such as LAMP-1 (12, 41), as well as for the maintenance of the Salmonella-containing vesicles (SCV) (3).
Plasmid p2812 encoding epitope-tagged SifA and constitutively expressed GFP was introduced in wild-type S. bongori and S. bongori(pB6). In S. enterica serotype Typhimurium, expression of sifA::M45 and T3SS-dependent secretion of SifA-M45 has been previously demonstrated (17). HeLa cells were infected with wild-type S. enterica serotype Typhimurium, S. bongori, or S. bongori(pB6), each harboring p2812. As observed previously, the formation of tubular endosomal aggregates, or Salmonella-induced filaments, was obvious in S. enterica serotype Typhimurium-infected host cells (Fig. 5). No SIF formation was observed in HeLa cells infected with S. bongori (data not shown) or with S. bongori(p2812) (Fig. 5). The formation of SIF was also observed in host cells infected with S. bongori(pB6/p2812), with endosomal aggregates of extend and frequency similar to those observed in S. enterica serotype Typhimurium-infected cells (Fig. 5). We were not able to analyze SIF induction by E. coli Mutaflor(pB6/p2812), since this did not invade HeLa cells. The induction of SIF by intracellular S. bongori(pB6/p2812) shows that an SPI2-dependent virulence function can be transferred to a heterologous host.
FIG.5.
Host cell phenotypes induced by S. bongori harboring SPI2. (A) Aggregation of host cell endosomes by S. bongori harboring the SPI2 locus and sifA. HeLa cells were infected with S. enterica serotype Typhimurium, S. bongori, or S. bongori harboring the plasmid-borne SPI2 locus. All strains harbored p2812 for the SPI2-dependent expression of sifA::M45 and the constitutive expression of GFP. At 16 h after infection, cells were fixed and immunostained for the late endosomal-lysosomal membrane protein LAMP-1. Representative micrographs show the organization of endosomal-lysosomal compartments (red) and intracellular Salmonella spp. Salmonella-induced filaments are indicated by arrowheads. (B) Accumulation of host cell tubulin. HeLa cells were infected with S. bongori, S. bongori harboring the plasmid-borne SPI2 locus, or S. bongori harboring the SPI2 locus and plasmid p2643 for the expression of sseF::HA. HeLa cells were fixed 16 h after infection and immunostaining was performed for S. bongori lipopolysaccharide (green), β-tubulin (red), and translocated SseF-HA (blue). Alternatively, F-actin was visualized by staining with Texas red-phalloidin (red). Representative cells with accumulation of tubulin at the SCV (indicated by arrows) were shown. Such tubulin accumulations were absent in cells infected with S. bongori or S. bongori harboring only plasmid p2643 (not shown). No accumulation of F-actin at the SCV was observed.
We next investigated the effect of S. bongori harboring SPI2 on the host cell cytoskeleton. Previous studies indicated SPI2-dependent modifications of the actin cytoskeleton (28, 29). Infection of HeLa cells with S. bongori harboring SPI2 did not result in detectable alterations in the organization of the actin cytoskeleton (Fig. 5). In contrast, infection with S. bongori harboring SPI2 affected the appearance of the microtubule cytoskeleton (Fig. 5). Accumulation of β-tubulin was detected around SCV containing replicating S. bongori(pB6) but was not observed in HeLa cells infected with S. bongori. We next sought to determine whether known SPI2-encoded effector proteins could be involved in this phenotype. A plasmid for the expression sseF::HA was introduced into S. bongori(pB6). In contrast to the pattern observed in infected RAW cells (Fig. 4), translocated SseF in HeLa cells was predominantly localized in the vicinity of the SCV. We observed frequent colocalization of SseF-HA translocated by intracellular S. bongori harboring SPI2 with β-tubulin accumulations at the SCV. The observations indicate that SPI2-dependent modifications of the actin cytoskeleton were not induced by S. bongori(pB6) in the absence of additional effector. However, SPI2-encoded effectors such as SseF and SseG were sufficient to induce alterations of the microtubule cytoskeleton by intracellular S. bongori.
Transfer of SPI2 to S. bongori confers an intracellular virulence phenotype.
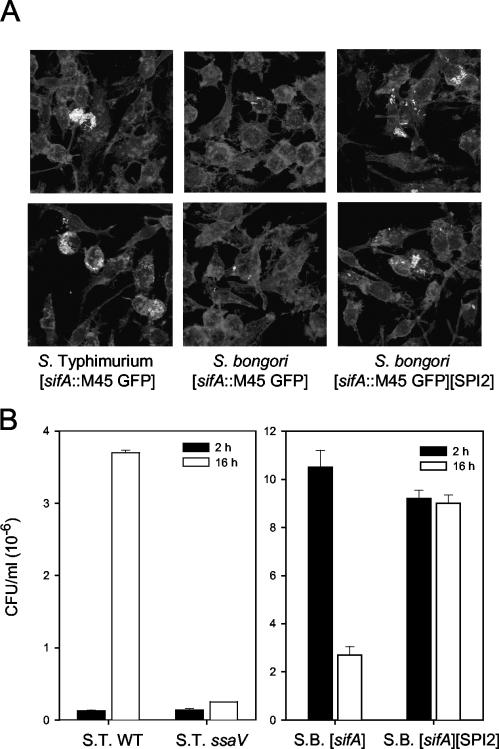
To assess the effect of the pB6-encoded T3SS on the intracellular phenotype of S. bongori, the intracellular replication rate was determined by using the gentamicin protection assay. RAW 264.7 cells were infected with S. enterica serotype Typhimurium or S. bongori as indicated in Fig. 6 and intracellular replication was examined by immuno-fluorescence microscopy. In host cells infected with S. enterica serotype Typhimurium(p2812), clusters of intracellular replicating bacteria were detected. Similar observations were made for S. bongori(pB6/p2812), but the frequency of clusters of replicating bacteria was lower. In contrast, only very few intracellular S. bongori(p2812) organisms were observed (Fig. 6A). For the quantification of intracellular proliferation, infected RAW 264.7 cells were lysed 2 h or 16 h after infection, and CFU counts were determined by plating lysates onto agar plates (Fig. 6B). In accordance with previous observations, strong intracellular proliferation of S. enterica serotype Typhimurium was dependent on the function of SPI2 genes. The numbers of viable S. bongori decreased within the experimental interval, whereas a constant number of intracellular S. bongori organisms harboring SPI2 was observed. The observation of clusters of replicating bacteria of S. bongori(pB6/p2812), but constant numbers of viable intracellular bacteria, indicates that a certain portion of S. bongori harboring SPI2 is able to replicate intracellularly but that a large number of the internalized bacteria is killed by the host cell mechanisms.
FIG. 6.
Intracellular proliferation of S. enterica serotype Typhimurium, S. bongori, and S. bongori(pB6). (A) RAW 264.7 macrophages were infected with S. enterica serotype Typhimurium wild type, S. bongori, or S. bongori harboring pB6 encoding the SPI2 locus. All strains harbored p2812 for SPI2-dependent expression of sifA::M45 and constitutive expression of GFP. Host cells were fixed 16 h after infection, and F-actin was stained by Texas red-phalloidin. Representative micrographs demonstrating the extent of intracellular proliferation are shown. (B) Macrophages were infected at a multiplicity of infection of ∼1 (for S. enterica serotype Typhimurium wild type [S. T. WT]) and 50 (for S. bongori [S. B.] strains), respectively. At 2 and 16 h after infection, macrophages were lysed by the addition of 0.1% Triton X-100, and serial dilutions were plated onto Mueller-Hinton agar for the enumeration of intracellular bacteria. The intracellular CFU was determined for both time points, and the rates of intracellular replication were expressed as the fold increase in CFU. Experiments were performed in triplicate on three different occasions.
We finally tested whether the transfer of SPI2 to S. bongori results in increased virulence in the murine model of salmonellosis. Mice were orally infected with mixtures of S. bongori(p2777) and S. bongori(pB6/p2777) or of S. bongori(p2104) and S. bongori(pB6/p2104). The bacterial burdens with both strains were determined 2, 3, and 4 days after infection in the ceca, Peyer's patches, mesenteric lymph nodes, and spleens (data not shown). S. bongori was rarely detected in the Peyer's patches and mesenteric lymph nodes. We did not observe an increased ability of S. bongori(pB6/p2104) or S. bongori(pB6/p2777) to colonize host organs. These results suggest that the transfer of SPI2 and a single effector locus is not sufficient to confer systemic virulence to S. bongori and that further pathogen or host factors determine the ability to initiate a systemic infection.
DISCUSSION
We reported here the cloning of the SPI2 virulence locus of S. enterica serotype Typhimurium and the functional transfer into the SPI2-negative species S. bongori and E. coli. Our experiments show that a T3SS of a pathogen of animals and humans can be functionally transferred to other bacterial species and genera.
We demonstrated that the expression of SPI2 genes is regulated in S. bongori and that the T3SS is functional, as indicated by the translocation of the SPI2-encoded effector SseJ into host cell. These observations indicate that all components required for regulated expression and assembly of a functional T3SS are present in S. bongori(pB6). In hosts other than Salmonella spp. or E. coli, the expression of episomal SPI2 genes was not properly regulated, suggesting that additional regulatory genes are required that are absent in these species (data not shown).
We observed here that the functions of the SPI2-encoded T3SS could be transferred to the heterologous species S. bongori and E. coli and enabled S. bongori to elicit SPI2-mediated phenotypes in infected host cells. However, the transfer of the SPI2 locus and a single effector locus did not confer the ability to cause systemic infections to S. bongori. This deficiency can be explained by the absence of further members of the group of STE proteins that are encoded by gene loci outside of the SPI2 locus. Although the evolution of bacterial pathogenesis is mediated by the transfer of large blocks of virulence genes, a combination of several virulence genes is required for a specific phenotype. It can be speculated that during the evolution of S. enterica and adaptation to new hosts, effector genes were sequentially acquired. The function of known effector proteins has only been partially described. In addition to the SPI2 virulence genes, this set of effector proteins has to be present in order to confer the full phenotype of intracellular virulence. Alternatively, factors other than SPI2 might be required for the complex virulence trait of systemic pathogenesis. Among other factors, the Salmonella virulence plasmid is not present in S. bongori, as indicated by hybridization experiments (unpublished observations) and by analysis of data from the genome sequencing project of this species. Furthermore, host restriction has been observed for several serotypes of S. enterica in cell culture systems. For example, S. enterica serotype Typhi is not able to proliferate in murine macrophages despite the presence of SPI2 and SPI2 effector genes. A similar host restriction has to be taken into account for cell culture models with S. bongori. Finally, in addition to virulence determinants of Salmonella, recent observations suggest that host factors such as antimicrobial peptides (39) can also dramatically influence the disease outcome in specific host-pathogen combinations.
Previous studies demonstrated the functional transfer of T3SSs of other pathogens. The ability to induce hypersensitive response in infected plants was transferred from plant-pathogenic Pseudomonas syringae pathovar syringae to P. fluorescens and E. coli by transfer of a cloned chromosomal locus (21). The LEE locus of enteropathogenic E. coli was transferred to an E. coli laboratory strain, and the LEE-dependent phenotypes were observed in cell culture models (27). Furthermore, pathogenicity islands encoding functions other than T3SS were transferred, e.g., the “high pathogenicity island” of a highly virulent strain of Yersinia enterocolitica to a low-virulence strain (36). Increased virulence in the mouse model was observed for the recipient strain.
A rather complex situation is given for effector proteins of the T3SS encoded by SPI1 and SPI2 of S. enterica. Both loci encode T3SS and a subset of effector proteins, but a large number of effector proteins of both systems are encoded by loci outside the SPI1 and SPI2 loci. At least 3 secreted and 13 translocated effector proteins of the SPI2-encoded T3SS had been identified thus far, but the total number of substrate proteins in this system is not known. The study of the functions of the individual effector proteins has been complicated by the large number and the possible redundancy in function. S. bongori containing the SPI2 locus represents an interesting novel tool for the dissection of functions of effector protein of the SPI2-encoded T3SS. Previous reports showed that genes encoding effector proteins are absent in S. bongori (16, 24, 31). We have demonstrated that individual effector genes can be transferred to S. bongori harboring episomal SPI2 and that the corresponding proteins are translocated and functional in the host cell. Furthermore, combinations of two effector proteins were translocated by S. bongori harboring the SPI2 locus (data not shown), and this feature will be useful for the study of the interaction of SPI2 effector protein after translocation.
Using S. bongori harboring SPI2, we have been able to detect a previously unknown host cell phenotype, the modification of the microtubule cytoskeleton (Fig. 5). This phenotype was induced by S. bongori harboring SPI2 without additional effector genes, suggesting that SPI2-encoded effectors such as SseF and SseG are involved in this phenotype. We performed further studies with S. enterica serovar Typhimurium-infected cells and observed that both SseF and SseG are targeted to microtubules and affect the organization of the microtubule cytoskeleton (V. Kuhle, D. Jäckel, and M. Hensel, unpublished observations).
Future studies will analyze the subcellular localization of various translocated effector proteins and their specific targets in the host cell. Furthermore, the S. bongori system will allow the study of the interaction of translocated effector proteins, for example, in the modulation of the Salmonella-containing phagosome, or the interference with specific host cell functions.
Acknowledgments
The study was supported by the DFG Priority Program grants HE 1964/4-2 and HE 1964/4-3. D.C. was supported by a research fellowship of the Humboldt foundation.
We thank Daniela Jäckel for excellent technical support and Volker Kuhle for help with confocal microscopy and image analysis. The contribution of Nina Kaiser to the construction of plasmids p2684 and p2777 is gratefully acknowledged.
Editor: B. B. Finlay
REFERENCES
- 1.Bäumler, A. J. 1997. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 5:318-322. [DOI] [PubMed] [Google Scholar]
- 2.Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33:806-816. [DOI] [PubMed] [Google Scholar]
- 3.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of sifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, E. F., F. S. Wang, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravortty, D., I. Hansen-Wester, and M. Hensel. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 195:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, H., S. Nordentoft, and J. E. Olsen. 1998. Phylogenetic relationships of Salmonella based on rRNA sequences. Int. J. Syst. Bacteriol. 48:605-610. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1764. [DOI] [PubMed] [Google Scholar]
- 9.Fierer, J., and D. G. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman, J. A., M. E. Ohl, and S. I. Miller. 2003. The Salmonella enterica serovar Typhimurium translocated effectors SseJ and SifB are targeted to the Salmonella-containing vacuole. Infect. Immun. 71:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-del Portillo, F., M. B. Zwick, K. Y. Leung, and B. B. Finlay. 1993. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. USA 90:10544-10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 14.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 15.Hansen-Wester, I., and M. Hensel. 2002. Genome-based identification of chromosomal regions specific for Salmonella spp. Infect. Immun. 70:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen-Wester, I., B. Stecher, and M. Hensel. 2002. Analyses of the evolutionary distribution of Salmonella translocated effectors. Infect. Immun. 70:1619-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen-Wester, I., B. Stecher, and M. Hensel. 2002. Type III secretion of Salmonella enterica serovar typhimurium translocated effectors and SseFG. Infect. Immun. 70:1403-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 19.Hensel, M., T. Nikolaus, and C. Egelseer. 1999. Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol. Microbiol. 31:489-498. [DOI] [PubMed] [Google Scholar]
- 20.Hensel, M., J. E. Shea, A. J. Bäumler, C. Gleeson, F. R. Blattner, and D. W. Holden. 1997. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J. Bacteriol. 179:1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, H. C., R. Schuurink, T. P. Denny, M. M. Atkinson, C. J. Baker, I. Yucel, S. W. Hutcheson, and A. Collmer. 1988. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J. Bacteriol. 170:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, U. J., B. W. Birren, T. Slepak, V. Mancino, C. Boysen, H. L. Kang, M. I. Simon, and H. Shizuya. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213-218. [DOI] [PubMed] [Google Scholar]
- 23.Klein, J. R., and B. D. Jones. 2001. Salmonella pathogenicity island 2-encoded proteins SseC and SseD are essential for virulence and are substrates of the type III secretion system. Infect. Immun. 69:737-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knodler, L. A., J. Celli, W. D. Hardt, B. A. Vallance, C. Yip, and B. B. Finlay. 2002. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 43:1089-1103. [DOI] [PubMed] [Google Scholar]
- 25.Kuhle, V., and M. Hensel. 2002. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell. Microbiol. 4:813-824. [DOI] [PubMed] [Google Scholar]
- 26.Maloy, S. R., V. L. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 28.Meresse, S., K. E. Unsworth, A. Habermann, G. Griffiths, F. Fang, M. J. Martinez-Lorenzo, S. R. Waterman, J.-P. Gorvel, and D. W. Holden. 2001. Remodeling of the actin cytoskeleton is essential for replication of intravacuolar Salmonella. Cell. Microbiol. 3:567-577. [DOI] [PubMed] [Google Scholar]
- 29.Miao, E. A., M. Brittnacher, A. Haraga, R. L. Jeng, M. D. Welch, and S. I. Miller. 2003. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol. Microbiol. 48:401-415. [DOI] [PubMed] [Google Scholar]
- 30.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Bäumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 32.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schröder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noll, A., A. Roggenkamp, J. Heesemann, and I. B. Autenrieth. 1994. Protective role for heat shock protein-reactive alpha beta T cells in murine yersiniosis. Infect. Immun. 62:2784-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochman, H., and E. A. Groisman. 1996. Distribution of pathogenicity islands in Salmonella spp. Infect. Immun. 64:5410-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 36.Pelludat, C., M. Hogardt, and J. Heesemann. 2002. Transfer of the core region genes of the Yersinia enterocolitica WA-C serotype O:8 high-pathogenicity island to Y. enterocolitica MRS40, a strain with low levels of pathogenicity, confers a yersiniabactin biosynthesis phenotype and enhanced mouse virulence. Infect. Immun. 70:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pignato, S., G. Giammanco, C. Santangelo, and G. M. Giammanco. 1998. Endemic presence of Salmonella bongori 48:z35:− causing enteritis in children in Sicily. Res. Microbiol. 149:429-431. [DOI] [PubMed] [Google Scholar]
- 38.Reeves, M. W., G. M. Evins, A. A. Heiba, B. D. Plikaytis, and J. J. 3. Farmer. 1989. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 27:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salzman, N. H., D. Ghosh, K. M. Huttner, Y. Paterson, and C. L. Bevins. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522-526. [DOI] [PubMed] [Google Scholar]
- 40.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein, M. A., K. Y. Leung, M. Zwick, F. Garcia del Portillo, and B. B. Finlay. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20:151-164. [DOI] [PubMed] [Google Scholar]
- 42.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 44.Vazquez-Torres, A., Y. Xu, J. Jones-Carson, D. W. Holden, S. M. Lucia, M. C. Dinauer, P. Mastroeni, and F. C. Fang. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655-1658. [DOI] [PubMed] [Google Scholar]
- 45.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritidis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 46.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing, and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]