Abstract
The tuberculin skin test for diagnosing Mycobacterium tuberculosis infection suffers from antigenic cross-reactivity of purified protein derivative with BCG, resulting in poor specificity in BCG-vaccinated populations. Comparative genomics has identified several genetic regions in M. tuberculosis and M. bovis that are deleted in M. bovis BCG. Proteins encoded in these regions will form the basis of new specific T-cell-based blood tests that do not cross-react with BCG, but only two, early secretory antigen target 6 and culture filtrate protein 10, have been studied in detail in humans. We investigated four novel gene products, encoded by RD2 (Rv1989c) and RD1 (Rv3873, Rv3878, and Rv3879c), that are absent from most or all of the vaccine strains of BCG, respectively. Sixty-seven overlapping peptides were tested in ex vivo gamma interferon enzyme-linked immunospot assays in 49 patients with culture-confirmed tuberculosis and 38 healthy BCG-vaccinated donors. Forty-five percent (95% confidence interval [CI], 31 to 57%) and 53% (95% CI, 39 to 67%) of the tuberculosis patients responded to Rv3879c and Rv3873, respectively, identifying these proteins as major M. tuberculosis T-cell antigens in humans, while 35 and 25% of the patients responded to Rv3878 and Rv1989c, respectively. Of the 38 BCG-vaccinated donors, 1 (2.6%) responded to peptides from Rv3878 and Rv3879c, 3 (7.9%) responded to Rv3873, and none responded to Rv1989c. Exclusion of cross-reactive peptides encoded in conserved motifs of Rv3873, a PPE family member, increased its specificity to 97.4%. The high specificity of Rv3879c peptides and nonconserved Rv3873 sequences, together with their moderate sensitivity in tuberculosis patients, identifies these peptides as candidates for inclusion in new T-cell-based tests for M. tuberculosis infection.
Accurate diagnosis of tuberculosis (TB) infection is essential for the treatment, prevention, and control of this resurgent disease (1). Since Mycobacterium tuberculosis is often difficult to culture from patients with active TB, and impossible to culture from healthy latently infected people, an immune-based diagnostic test indicating the presence or absence of M. tuberculosis infection would be very useful for the diagnosis of active TB and screening for latent M. tuberculosis infection. The only widely used test is the century-old tuberculin skin test (TST), which has many drawbacks. Foremost among these is its poor specificity (21). This results from the broad antigenic cross-reactivity of purified protein derivative (PPD), a crude mixture of more than 200 M. tuberculosis proteins widely shared among M. tuberculosis, M. bovis Bacillus Calmette-Guérin (BCG), and most environmental mycobacteria (22). Hence, false-positive results are common in people with environmental mycobacterial exposure and previous BCG vaccination. Since most of the world's population is BCG vaccinated and since the confounding effect of BCG persists for up to 15 years after vaccination (41), this is a significant problem. Thus, there has been an intensive search for specific M. tuberculosis antigens that are not cross-reactive with BCG (3, 8, 27). Since humoral immune responses are generally weak in latent M. tuberculosis infection, the search for specific proteins has focused increasingly on antigens that are targets of cell-mediated immune (CMI) responses (5, 19). A cocktail of a small number of such defined T-cell antigens could form the basis of new T-cell-based in vitro blood tests for M. tuberculosis infection that do not cross-react with BCG (28). Such tests promise to improve the detection of latent M. tuberculosis infection and could help with the rapid diagnosis of active TB in low-prevalence regions. Like the TST, T-cell-based tests do not distinguish between active TB and latent M. tuberculosis infection, so their diagnostic utility for active TB would be limited in populations with a high prevalence of latent infection.
By screening eluted fractions of antigens from M. tuberculosis and M. bovis culture filtrates for recognition by T cells from infected humans and cattle, respectively, Andersen and coworkers identified several low-molecular-mass antigens that are major targets of CMI responses (2, 12, 32, 33). Subtractive DNA hybridization of pathogenic M. bovis and BCG (27) and comparative genome-wide DNA microarray analysis of M. tuberculosis H37Rv and BCG (9) identified several regions of difference, designated RD1 to RD16, between M. tuberculosis and M. bovis on the one hand and BCG on the other (9). All represent parts of the M. bovis genome deleted during prolonged in vitro culture (13). RD1 was deleted before 1921, when BCG was first disseminated internationally for use as a vaccine. RD1 is thus absent from all vaccine strains of BCG, as well as most environmental mycobacteria, but is still present in the M. tuberculosis complex, including all clinical isolates of M. tuberculosis and M. bovis (9). There are nine open reading frames (ORFs) in the RD1 gene region (Rv3871 to Rv3879c) (9).
Two of the low-molecular-mass potent T-cell antigens identified by Andersen and colleagues, early secretory antigen target 6 (ESAT-6) and culture filtrate protein 10 (CFP10), are encoded in RD1 (2, 10) and have been intensively investigated in animal models and humans over the last few years (4, 7, 15, 16, 26, 30, 32, 33, 36, 40). Studies using several different assays, including lymphoproliferation, gamma interferon (IFN-γ) enzyme-linked immunosorbent assay (ELISA), and IFN-γ enzyme-linked immunospot (ELISPOT) assay, have shown that they are strong targets of the cellular immune response in animal models, TB patients, and contacts (4, 11, 24, 28, 30, 35, 37-39). However, we know very little about the remaining seven RD1 gene products. In particular, there is no published data on the cellular immune response to these antigens in humans, except for a recent report on Rv3873 (29). Hence, although they are of great interest as future candidate molecules for specific T-cell-based diagnosis of M. tuberculosis infection, they have not been evaluated for this purpose.
Recently, Cockle and colleagues investigated cellular immune responses to gene products from RD1, RD2, and RD14 in M. bovis-infected and BCG-vaccinated cattle (17). Five hundred thirty-six overlapping peptides spanning 13 ORFs were screened with a whole-blood IFN-γ ELISA. Eight antigens were recognized by peripheral blood mononuclear cells (PBMC) from more than 40% of M. bovis-infected cattle (n = 22) and were therefore deemed to be potent T-cell antigens (Rv1983, Rv1986, Rv3872, Rv3873, Rv3878, Rv3879c, Rv1979c, and Rv1769). Combining the T-cell responses to the most frequently recognized peptides from three RD1-encoded gene products, Rv3873, Rv3878, and Rv3879c, gave a 95% sensitivity for detection of infection in cattle (17). This screening thus identified certain gene products and, in particular, defined regions within these gene products that are of interest for evaluation in human TB. Since M. bovis has greater than 99.9% DNA sequence identity with M. tuberculosis, antigens thus identified in cattle will have amino acid sequences almost identical to those of their M. tuberculosis counterparts and can therefore be directly tested in humans (17, 20). Moreover, the observation that the major T-cell targets from short-term culture filtrate in M. bovis-infected cattle are also the immunodominant T-cell antigens in human TB provides a basis for postulating that newly identified strong T-cell antigens in infected cattle may prove to be similarly potent targets of CMI responses in TB patients (20, 33).
For this study, we selected four gene products on the basis of the following criteria. First, they were all strong targets of CMI responses in a high proportion of M. bovis-infected cattle (17). Second, they are present in M. tuberculosis, as well as M. bovis. Third, they are absent from most (Rv1989c from RD2) or all (Rv3873, Rv3879c, and Rv3878 from RD1) strains of BCG (9). This study was designed to evaluate T-cell responses to peptides from these four proteins in TB patients and healthy controls; our goal was to identify defined peptides with sufficient diagnostic sensitivity and specificity to warrant incorporation into new CMI response-based diagnostic tests for human TB. Peptides from those regions of Rv3873, Rv3878, Rv3879c, and Rv1989c that were the strongest and most frequent targets of T-cell responses in M. bovis-infected cattle were selected. Sixty-seven overlapping peptides were tested by ex vivo IFN-γ ELISPOT assay in 87 participants (6, 17, 23). Sensitivity was assessed in 49 culture-confirmed TB patients, and specificity was assessed in 38 healthy BCG-vaccinated donors with no known TB exposure.
MATERIALS AND METHODS
Study participants.
All participants were recruited prospectively in London and Oxford over a 14-month period from June 2002 through July 2003. Ethical approval for the study was granted by the Harrow and Central Oxford Research Ethics Committees. The diagnoses of all 49 TB patients were bacteriologically confirmed by positive cultures for M. tuberculosis from one or more clinical specimens. Patients were untreated or had received less than 2 weeks of therapy at the time of venipuncture for ELISPOT assay. Control participants were healthy, BCG-vaccinated laboratory personnel from regions with a low prevalence of TB and with no known exposure to M. tuberculosis. All had recently tested negative by IFN-γ ELISPOT assay with 35 overlapping 15-mer peptides spanning the lengths of ESAT-6 and CFP10, as previously described (24). Epidemiological data regarding place of birth, any period of residence in higher-prevalence regions, and absence of TB contact were collected from these volunteers at the point of venipuncture. Health care workers were not recruited owing to the risk of occupational TB exposure.
Peptides.
Sixty-seven synthetic peptides spanning selected regions of four ORFs were designed and purchased (Research Genetics, Huntsville, Ala.). The regions represented by the peptides were selected because they were the sequences most frequently recognized by whole-blood IFN-γ ELISA in M. bovis-infected cattle (17). In all four molecules except Rv3873, these sequences were at the amino terminus. For the exact regions represented by the peptides for each molecule, see Table 2. Each peptide was 15 residues long and overlapped the adjacent peptide by 10 amino acids. This approach has previously been shown to be effective for detecting HLA class I-restricted CD8, as well as HLA class II-restricted CD4, T-cell responses (31). We arranged the 67 peptides into 11 pools containing between five and seven peptides. See Table 2 for the pools in relation to the antigens they represent. For all peptides, identity was confirmed by mass spectrometry and purity was more than 70%.
TABLE 2.
Antigens and peptide pools evaluated and number of donors who responded to each peptide pool by IFN-γ-ELISPOT assay
| Region of difference and designation | Size (amino acids) | Putative functionc | Peptide pool (no. of constituent peptides) | Region of molecule represented by peptide pools (amino acid position) | No. of TB patients responding (n = 49) | No. of unexposed, BCG- vaccinated donors responding (n = 38) |
|---|---|---|---|---|---|---|
| RD1 | ||||||
| Rv 3873 | 368 | M. tuberculosis PPE family member | 1 (6) | 89-128 | 8 | 0 |
| 2 (6) | 129-158 | 25 | 2 | |||
| 3 (6) | 159-188 | 18 | 1 | |||
| Rv 3878 | 280 | Unknown alanine-rich protein | 1 (7) | 1-45 | 16 | 1 |
| 2 (7) | 36-80 | 14 | 1 | |||
| Rv 3879c | 729 | Unknown alanine- and proline- | 1 (6) | 1-40 | 12a | 1 |
| rich protein | 2 (6) | 31-70 | 18a | 0 | ||
| 3 (5) | 61-95 | 9b | 0 | |||
| RD2, Rv 1989c | 186 | Unknown | 1 (6) | 1-40 | 10b | 0 |
| 2 (6) | 31-70 | 7b | 0 | |||
| 3 (6) | 61-100 | 8b | 0 |
Ex vivo IFN-γ ELISPOT assays.
ELISPOT assays were performed as previously described (23, 26). IFN-γ ELISPOT assay plates (Mabtech AB, Stockholm, Sweden) were seeded with 2.5 × 105 PBMC per well: duplicate wells contained no antigen (negative control), phytohemagglutinin (positive control; ICN Biomedical) at 5 μg/ml, streptokinase-streptodornase (Varidase; Cyanamid, Hampshire, United Kingdom) at 100 U/ml, PPD (Statens Serum Institut, Copenhagen, Denmark) at 20 μg/ml, and 1 of 11 peptide pools, such that the final concentration of each peptide was 10 μg/ml. After overnight incubation at 37°C in 5% CO2, the plates were developed with preconjugated detector antibody and the chromogenic substrate 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium chloride (Moss Inc., Pasadena, Md.). For unexposed, BCG-vaccinated donors who responded to any of the pools, PBMC were retested against all 67 peptides individually in single ELISPOT assay wells at a final concentration of 10 μg/ml.
Assays were scored by an automated ELISPOT assay counter (AID-GmbH, Strassberg, Germany). For wells containing peptide pools, responses were scored as positive if the test well contained at least five more IFN-γ spot-forming cells (SFC) than negative control wells and this number also had to be at least twice the frequency found in the negative control wells. These predefined cutoff points translate into a detection threshold of 20 peptide-specific T cells per 106 PBMC. The person performing the assays was blind to personal identifiers of participants.
Bioinformatics.
The DNA sequence of M. tuberculosis H37Rv was visualized with the TubercuList database (http://genolist.pasteur.fr/TubercuList/). Basic Local Alignment Search Tool (BLAST) searches for protein sequence homology in available mycobacterial genomes were performed with TubercuList, the Sanger Centre (Cambridge, United Kingdom) server for the incomplete M. bovis BCG genome sequence (http://www.sanger.ac.uk/Projects/M_bovis/), and the National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/BLAST).
RESULTS
Demographic characteristics of study participants.
Demographic characteristics of the 49 culture-confirmed TB patients are shown in Table 1. Forty-two patients had pulmonary TB, and 23 of them were sputum smear positive. The seven patients with extrapulmonary TB comprised three patients with pleural TB, one with lymphadenitis, two with miliary TB, and one with urinary tract TB. The patients were from a broad range of ethnicities. The demographic characteristics of the BCG-vaccinated donors are shown in Table 1. All donors were born in regions with a low prevalence of TB (Europe or Australia). None had a history of known TB contact, and none had resided for more than 3 months in high-prevalence regions.
TABLE 1.
Demographic characteristics of TB patients and unexposed, BCG-vaccinated donors
| Parameter | Patients with tuberculosis (n = 49) | BCG-vaccinated donors (n = 38) |
|---|---|---|
| Mean age [yr (range)] | 34.0 (17-78) | 33.3 (20-50) |
| Sex (male/female) | 31/18 (63/37) | 22/16 (58/42) |
| Ethnicity (no. [%] of patients) | ||
| Indian subcontinent | 24 (49) | 1 |
| African | 18 (37) | 0 |
| Asian | 4 (8) | 0 |
| Caucasian | 3 (6) | 37 |
IFN-γ ELISPOT assay responses to peptides from Rv3873, Rv3878, Rv3879c, and Rv1989c in culture-confirmed TB patients.
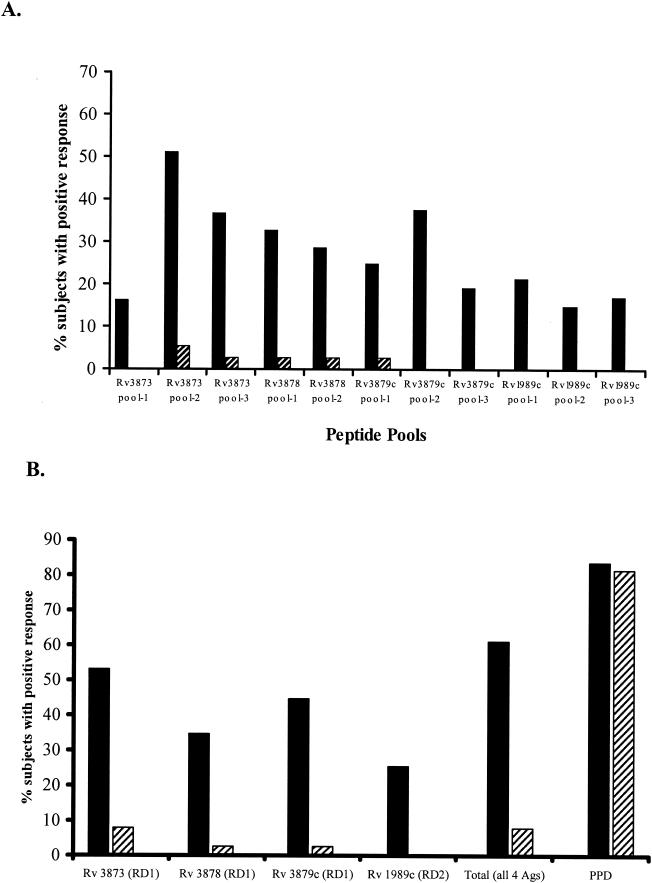
The IFN-γ ELISPOT assay responses of PBMC from all 49 TB patients to the 11 peptide pools from the four antigens are summarized in Fig. 1A. The percentages of responding patients varied between 25.5 and 53.1% for the different antigens (Fig. 1B). The proportions of patients responding to peptides from each of the antigens (Rv3873, Rv3879c, Rv3878, and Rv1989c) were 53.1% (95% confidence interval [CI], 39 to 67%), 44.7% (95% CI, 31 to 57%), 34.7% (95% CI, 22 to 48%), and 25.5% (95% CI, 13 to 39%), respectively (Fig. 1B). Combining these responses, 30 of 49 TB patients responded to peptide pools from one or more antigens, giving a diagnostic sensitivity of 61.2% (95% CI, 46.2 to 74.8%) for all of the peptides used together.
FIG. 1.
Proportions of culture-confirmed TB patients (n = 49) and healthy, unexposed BCG vaccinees (n = 38) responding in an IFN-γ ELISPOT assay to peptide pools from the four RD region gene products. PBMC from each participant were tested by the IFN-γ ELISPOT assay with peptide pools of between five and seven peptides representing different antigens from RD1 (Rv3873, Rv3878, and Rv3879c) and RD2 (Rv1989c). (A) Percentages of culture-confirmed TB patients and unexposed BCG vaccinees who responded to each peptide pool in an IFN-γ ELISPOT assay. (B) Percentages of culture-confirmed TB patients and unexposed BCG vaccinees who responded to one or more peptide pools from each of the RD1 and RD2 gene products. Total, percentage of donors who responded to 1 or more of the 11 peptide pools from the four antigens PPD, percentage of donors who responded to PPD; solid columns, response rates of TB patients; hatched columns, response rates of unexposed, BCG-vaccinated donors.
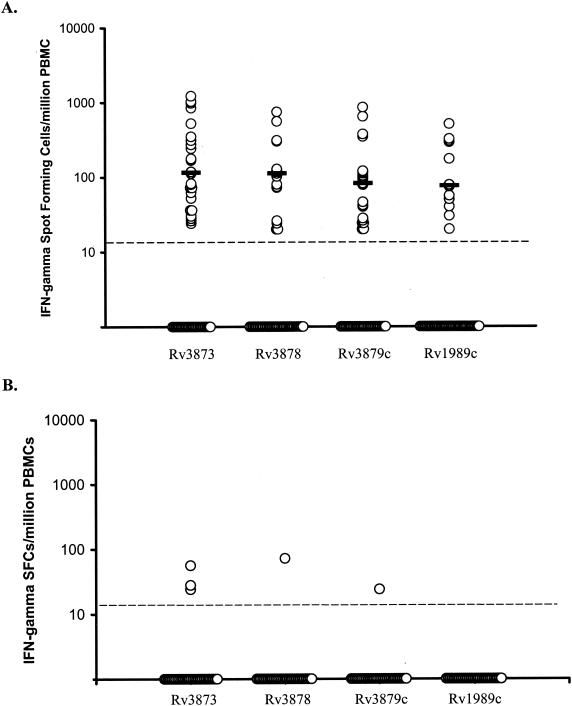
The frequencies of Rv3873, Rv3878, Rv3879c, and Rv1989c peptide-specific IFN-γ-secreting T cells for all responder patients were (median response and interquartile range) 115 (52 to 310), 112 (72 to 128), 82 (28 to 116), and 76 (45 to 296) per 106 PBMC, respectively (Fig. 2A).
FIG. 2.
Magnitudes of IFN-γ ELISPOT assay responses to RD region antigens in 49 culture-confirmed TB patients (A) and 38 healthy, unexposed BCG vaccinees (B). Frequencies of peptide-specific IFN-γ-secreting SFC summated for each of the constituent peptide pools for each antigen, enumerated by ex vivo ELISPOT assay in patients with TB (A) and healthy, unexposed, BCG-vaccinated donors (B), are shown. Each horizontal bar represents the median response for each antigen. Points on the baseline represent individuals with no response to a given antigen (i.e., fewer than five SFC above the negative control for each of the constituent peptides of each pool of the given antigen). The broken horizontal line represents the predefined cutoff point (five SFC per 2.5 × 105 PBMC, which translates into a threshold of detection of 20 peptide-specific T cells per 106 PBMC).
Comparison of proportions of patients responding to each antigen according to the clinical type of TB.
The TB patients were stratified by clinical type of TB, i.e., pulmonary (n = 42) versus extrapulmonary (n = 7). The proportions of patients from each group that responded to peptides from each different protein were then compared. Although there was no significant difference between the proportions of pulmonary and extrapulmonary patients that responded to Rv3873, Rv3878, and Rv1989c, significantly more extrapulmonary patients (6 [86%] of 7) responded to Rv3879c than did pulmonary patients (14 [33%] of 42) (P = 0.014).
IFN-γ ELISPOT assay responses in BCG-vaccinated healthy donors.
Rv3873 peptide pools elicited responses in 3 (7.9%) of 38 BCG-vaccinated, unexposed donors; Rv3878 and Rv3879c each elicited a response in 1 donor (2.6%); and Rv1989c elicited no responses. Two donors, donors 20 and 31, each responded to a different peptide from pool 2 of Rv3873, and one, donor 25, responded to pools from Rv3873, Rv3878, and Rv3879c (Table 2 and Fig. 1). Donors 20 and 31 responded to peptides 119-133 (LTATNFFGINTIPIA) and 139-153 (YFIRMWNQAALAMEV), respectively, both from pool 2 of Rv3873. Donor 25 responded to peptide 174-188 (LDPGASQSTTNPIFG) from Rv3873, peptides 16-30 (AAKLAGLVFPQPPAP) and 61-75 (ESLVSDGLPGVKAAL) from Rv3878, and peptide 26-40 (DTFYDRAQEYSQVLQ) from Rv3879c. Combining all of these responses, 3 (7.9%) of 38 BCG-vaccinated, healthy donors responded to one or more antigens while 81.6% responded to PPD.
The frequencies of peptide-specific IFN-γ SFC seen in BCG-vaccinated, unexposed donors were much lower than in the TB patients (Fig. 2B). The median frequencies of peptide-specific T cells (and interquartile range) were 28 (24 to 56), 72 (72), and 20 (20) per 106 PBMC for Rv3873, Rv3878, and Rv3879c, respectively.
BLAST searches of cross-reactive peptide sequences.
BLAST searches for protein sequences highly homologous to the six 15-mer peptides that gave a response in BCG-vaccinated donors were performed. Peptide 119-133 (LTATNFFGINTIPIA) had the greatest homology, with 93% identity to other mycobacterial proteins (14 out of 15 amino acids identical). This peptide is from pool 2 of Rv3873, a member of the PPE family of proteins, and is encoded within a 52-amino-acid-long motif that is highly conserved throughout the PPE family (Fig. 3). Consequently, it displays high levels of homology with many M. tuberculosis, M. bovis, and M. leprae PPE proteins (Table 3) that are encoded in the deleted and undeleted regions of the genomes of M. tuberculosis, M. bovis, and other mycobacteria. Peptide 139-153 (YFIRMWNQAALAMEV), which is also encoded within the 52-amino-acid conserved motif of Rv3873 (Fig. 3), also showed homology with sequences from many PPE proteins (Table 3), although the level of identity was considerably lower at 47% (7 out of 15 identical residues). In contrast, peptide 174-188 (LDPGASQSTTNPIFG) from Rv3873, which lies outside the conserved-motif region, had no significant homology with PPE family members. The two cross-reactive peptides from Rv3878 and the single cross-reactive peptide from Rv3879c had no significant sequence homology with any other mycobacterial proteins.
FIG. 3.
Location and homology of PPE protein family motif (as described at http://genolist.pasteur.fr/TubercuLIST/mast/P210.1.html) within the partial amino acid sequence of Rv3873 (amino acid residues 100 to 160). Amino acid residues are shown in the one-letter code. Underlined residues indicate the given peptide sequence. Identical residues are indicated with a cross.
TABLE 3.
Homologya between peptides 119-133 and 139-153 from Rv3873 with sequences from other proteins in the mycobacterial PPE family
| Peptide and designation(s)b | Amino acid sequencec |
|---|---|
| 119-133 | |
| Rv3873 | LTATNFFGINTIPIA |
| Rv3021c, Rv3018c, Rv0280, Rv1387 | LVATNFFGINTIPIA |
| Rv0256c | LMATNFFGINTIPIA |
| Rv0453 | MVATNFFGINTIPIA |
| 139-153 | |
| Rv3873 | YFIRMWNQAALAMEV |
| Rv2768c, Rv1039c | HYGEMWAQDALAMYG |
| Rv0286 | DYVRMWLQAAAVMGL |
| Rv1807 | QYAEMWSQDAMAMYG |
The homology search was performed with the BLAST program.
Designations of M. tuberculosis proteins are as previously described (18). The sequences of all of the related proteins described are also present in the M. bovis BCG genome (http://www.sanger.ac.uk/Projects/M_bovis/). Nonidentical residues are underlined. The putative function of each protein is membership in the M. tuberculosis PPE family.
Amino acid residues are shown in the one-letter code.
DISCUSSION
We have evaluated human cellular immune responses to peptide mixtures of four M. tuberculosis proteins encoded in regions of difference RD1 and RD2. This is the first such report for Rv3879c, Rv3878, and Rv1989c; for Rv3873, cellular immune responses were also recently described by Okkels et al. (29). Peptides from each protein were recognized by T cells from >25% of the TB patients in IFN-γ ELISPOT assays. Peptide pools from two RD1-encoded gene products were recognized in approximately half of the TB patients tested, i.e., Rv3879c (45%) and PPE family member Rv3873 (53%). This study thus identified these two proteins as major M. tuberculosis T-cell antigens in infected humans. IFN-γ ELISPOT assay responses to the peptides were rare in BCG-vaccinated donors, giving a specificity of 97.4% or more for all of the antigens except Rv3873, which, on account of cross-reactive peptides from conserved sequences, had a lower specificity of 92.1%. The high specificity of the Rv3879c peptides (97.4%), together with the fact that they are recognized in the IFN-γ ELISPOT assay by almost half of TB patients, identifies this molecule as a potentially useful T-cell antigen for inclusion in novel T-cell-based diagnostic tests of M. tuberculosis infection.
Antibody responses to Rv3873 and Rv3878 have previously been studied in humans (14). Of 75 TB patients, none (0%) had antibodies to Rv3873 and only 5 (6.7%) had detectable antibody responses to Rv3878 (14), considerably lower than the rates of response to these antigens of our series of 49 culture-confirmed TB patients in IFN-γ ELISPOT assays. Thus, the potential of these two RD1-encoded gene products to contribute to an improved diagnostic test for TB could only be recognized by evaluation of T-cell responses in TB patients.
CMI responses to the antigens in this study have previously been assessed in cattle (17). Despite being encoded in RD1, peptides derived from Rv3873 and Rv3879c elicited IFN-γ responses in a whole-blood ELISA in 17 and 33% of BCG-vaccinated cattle, respectively (17). However, the responses were only borderline positive, and the number of vaccinated cattle tested was low (n = 6). Fortunately, in our larger series of BCG-vaccinated humans, we have shown that the level of cross-reactivity of these antigens with BCG is far lower than was previously anticipated on the basis of a small number of cattle. Moreover, three of the five responses observed were borderline positive (Fig. 2B).
Compared to our findings in TB patients, Cockle and colleagues found that a significantly higher proportion of M. bovis-infected cattle responded to peptides derived from the four antigens. Response rates were 82, 77, 59, and <40% for Rv3873, Rv3878, Rv3879c, and Rv1989c, respectively (n = 22) (17). With combined responses to each of the most widely recognized peptides from the three most immunodominant antigens, 95% of the cattle responded, suggesting that these three antigens together might suffice to form the basis of a T-cell-based test for TB in cattle (17). Our results, obtained with peptides from the most immunogenic regions of these antigens, indicate that although they are prominent targets of T-cell responses in TB patients, these antigens would not be sufficient for the diagnosis of human M. tuberculosis infection unless supplemented by other antigens, such as ESAT-6 and CFP10. Combining responses to peptide pools from all four antigens would give an overall response rate of 61%, which is too low to be diagnostically useful. It is not clear why Cockle and coworkers, with peptides from the same regions of the same antigens used in this study, found higher response rates. One potential explanation might relate to the fact that the cattle were inbred (Friesians), and it is possible that the major histocompatibility complex background or other genetic factors in Friesians are associated with frequent cellular immune responses to these antigens, whereas the TB patients in our study were a genetically heterogeneous population. Although the two studies used different assays of antigen-specific T-cell responses, the IFN-γ ELISPOT assay is recognized as being more sensitive than the whole-blood IFN-γ ELISA, so this could not account for the higher response rates in the study of Cockle et al. One other study has assessed CMI responses to Rv3873 in humans; with a PBMC-based IFN-γ ELISA, Okkels and coworkers found that only 10 (42%) of 24 TB patients responded to recombinant protein, compared with 53% of the TB patients in this study who responded to Rv3873-derived peptides in an IFN-γ ELISPOT assay. Despite the generally lower response rates in humans, it is interesting that the rank order in the hierarchy of immunodominance for these four antigens was the same as in cattle (17). This observation lends further support to the notion that, from an immunological perspective, M. bovis-infected cattle represent a good model for human TB (20).
Why did a few BCG-vaccinated, healthy donors respond to peptides from Rv3873 (n = 3), Rv3878 (n = 1), and Rv3879c (n = 1), given that these proteins are encoded in RD1? It is unlikely that these individuals were latently infected with M. tuberculosis, as all were epidemiologically at a very low risk for M. tuberculosis exposure. Moreover, although these volunteers did not undergo a TST, all were negative by the IFN-γ ELISPOT assay with peptides spanning the lengths of ESAT-6 and CFP10. Given that the ESAT-6-CFP10-based ELISPOT assay appears to be at least as sensitive as the TST for detecting latent M. tuberculosis infection (19, 25), it is very unlikely that the responses to Rv3873, Rv3878, and Rv3879c in these unexposed donors were a result of latent M. tuberculosis infection. However, given the lack of a “gold standard” test for latent infection, this possibility cannot be completely excluded. Although the ORFs encoding the three proteins are deleted in BCG and do not have close homologues elsewhere in the BCG genome, it is still possible that the rare responses in the BCG-vaccinated donors reflect antigenic cross-reactivity. This is because the unit of cross-reactivity is the epitope, and genes encoding large molecules may contain limited sequences containing T-cell epitopes that are homologous to genes outside the deleted region. Rv3873 is a member of the PPE family, and two of the cross-reactive peptides (from pool 2) are from a highly conserved motif that characterizes PPE family members (Fig. 3). BLAST searches for sequences with substantial homology to the six cross-reactive peptides from Rv3873, Rv3878, and Rv3879c found several regions with high identity to one of the two peptides from the conserved motif of Rv3873; the second peptide from this motif was also homologous with several regions, but the level of identity was lower (Table 3). All of the regions of homology were in PPE family members encoded within undeleted regions of the M. tuberculosis, M. bovis, and M. leprae genomes. Homologous sequences were also found throughout the BCG genome, but since this sequence is not complete, the ORFs with homology are unannotated. The third cross-reactive peptide from Rv3873, the two cross-reactive peptides from Rv3878, and the single cross-reactive peptide from Rv3879c had no significant sequence homology with any other known mycobacterial proteins. Hence, we cannot explain the cross-reactivity to these three peptides on the basis of known sequence homology; interestingly, the responses to each of these peptides were all observed in the same single donor. Thus, cross-reactivity to the peptides we studied (except for those in the conserved motifs of Rv3873) is unlikely to be a major problem in BCG-vaccinated individuals. Our findings thus exemplify a significant advantage of using peptides rather than recombinant antigens, i.e., the ability to screen out cross-reactive sequences following systematic testing in BCG-vaccinated donors.
The very high specificity of the Rv3879c peptides and the non-cross-reactive sequences of Rv3873, together with their sensitivity of around 50% in culture-confirmed TB patients, identify these peptides as candidates for inclusion in a cocktail of peptides for a T-cell-based diagnostic test. T-cell responses to peptides spanning the lengths of ESAT-6 and CFP10 have been detected in 70 to 80% of TB patients by IFN-γ ELISA and in around 90% of TB patients by IFN-γ ELISPOT assay (3, 4, 6, 26, 28). We have identified peptides that may further enhance the sensitivity of T-cell-based assays with ESAT-6 and CFP10 without compromising specificity. The clinical impact of these new peptides on improving the diagnosis of TB will depend on what proportion of the patients who fail to respond to ESAT-6 and CFP10 respond to Rv3873 or Rv3879c. Therefore, we have recently begun to quantify the additional diagnostic sensitivity that could be achieved by using these novel peptides in parallel with ESAT-6- and CFP10-derived peptides in the IFN-γ ELISPOT assay in a blinded, prospective study of the most clinically relevant target population, i.e., patients presenting with clinically suspected active TB in whom the diagnosis is unproven at the time of enrollment and testing (34). After unblinding, ELISPOT assay results for peptides from ESAT-6, CFP10, Rv3873, and Rv3879c for each antigen individually and in combination will be compared with the final microbiological and clinical diagnoses.
Acknowledgments
This work was supported by The Wellcome Trust. X.Q.L. is a visiting Research Fellow partly funded by a grant to Peking Union Medical College from the China Medical Board (New York, N.Y.). A.L. is a Wellcome Senior Research Fellow in Clinical Science.
We are grateful to Martin Vordermeier and Glyn Hewinson for sharing raw data from their studies of M. bovis-infected cattle and also for invaluable advice and helpful discussions. We thank the TB patients and healthy donors for participating. We are grateful to Rob Wilkinson and Rob Davidson for valuable support in the recruitment of TB patients. We thank Rubamalar Gunatheesan for help in preparing some of the data for the manuscript.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.American Thoracic Society and Centers for Disease Control and Prevention. 2000. Targeted tuberculin testing and treatment of latent tuberculosis infection. Am. J. Respir. Crit. Care Med. 161:S221-S247. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, P., A. B. Andersen, A. L. Sorensen, and S. Nagai. 1995. Recall of long-lived immunity to Mycobacterium tuberculosis infection in mice. J. Immunol. 154:3359-3372. [PubMed] [Google Scholar]
- 3.Andersen, P., M. E. Munk, J. M. Pollock, and T. M. Doherty. 2000. Specific immune-based diagnosis of tuberculosis. Lancet 356:1099-1104. [DOI] [PubMed] [Google Scholar]
- 4.Arend, S. M., P. Andersen, K. E. van Meijgaarden, R. L. Skjot, Y. W. Subronto, J. T. van Dissel, and T. H. Ottenhoff. 2000. Detection of active tuberculosis infection by T cell responses to early-secreted antigenic target 6-kDa protein and culture filtrate protein 10. J. Infect. Dis. 181:1850-1854. [DOI] [PubMed] [Google Scholar]
- 5.Arend, S. M., A. C. Engelhard, G. Groot, K. de Boer, P. Andersen, T. H. Ottenhoff, and J. T. van Dissel. 2001. Tuberculin skin testing compared with T-cell responses to Mycobacterium tuberculosis-specific and nonspecific antigens for detection of latent infection in persons with recent tuberculosis contact. Clin. Diagn. Lab. Immunol. 8:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arend, S. M., A. Geluk, K. E. van Meijgaarden, J. T. van Dissel, M. Theisen, P. Andersen, and T. H. Ottenhoff. 2000. Antigenic equivalence of human T-cell responses to Mycobacterium tuberculosis-specific RD1-encoded protein antigens ESAT-6 and culture filtrate protein 10 and to mixtures of synthetic peptides. Infect. Immun. 68:3314-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arend, S. M., T. H. Ottenhoff, P. Andersen, and J. T. van Dissel. 2001. Uncommon presentations of tuberculosis: the potential value of a novel diagnostic assay based on the Mycobacterium tuberculosis-specific antigens ESAT-6 and CFP-10. Int. J. Tuberc. Lung Dis. 5:680-686. [PubMed] [Google Scholar]
- 8.Arend, S. M., K. E. van Meijgaarden, K. de Boer, E. C. de Palou, D. van Soolingen, T. H. Ottenhoff, and J. T. van Dissel. 2002. Tuberculin skin testing and in vitro T cell responses to ESAT-6 and culture filtrate protein 10 after infection with Mycobacterium marinum or M. kansasii. J. Infect. Dis. 186:1797-1807. [DOI] [PubMed] [Google Scholar]
- 9.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 10.Berthet, F. X., P. B. Rasmussen, I. Rosenkrands, P. Andersen, and B. Gicquel. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144(Pt 11):3195-3203. [DOI] [PubMed] [Google Scholar]
- 11.Black, G. F., R. E. Weir, S. D. Chaguluka, D. Warndorff, A. C. Crampin, L. Mwaungulu, L. Sichali, S. Floyd, L. Bliss, E. Jarman, L. Donovan, P. Andersen, W. Britton, G. Hewinson, K. Huygen, J. Paulsen, M. Singh, R. Prestidge, P. E. Fine, and H. M. Dockrell. 2003. Gamma interferon responses induced by a panel of recombinant and purified mycobacterial antigens in healthy, non-mycobacterium bovis BCG-vaccinated Malawian young adults. Clin. Diagn. Lab. Immunol. 10:602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boesen, H., B. N. Jensen, T. Wilcke, and P. Andersen. 1995. Human T-cell responses to secreted antigen fractions of Mycobacterium tuberculosis. Infect. Immun. 63:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brusasca, P. N., R. Colangeli, K. P. Lyashchenko, X. Zhao, M. Vogelstein, J. S. Spencer, D. N. McMurray, and M. L. Gennaro. 2001. Immunological characterization of antigens encoded by the RD1 region of the Mycobacterium tuberculosis genome. Scand. J. Immunol. 54:448-452. [DOI] [PubMed] [Google Scholar]
- 15.Buddle, B., N. A. Parlane, D. L. Keen, F. E. Aldwell, J. M. Pollock, K. Lightbody, and P. Andersen. 1999. Differentiation between Mycobacterium bovis BCG-vaccinated and M. bovis infected cattle using recombinant mycobacterial antigens. Clin. Diagn. Lab. Immunol. 6:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardoso, F. L., P. R. Antas, A. S. Milagres, A. Geluk, K. L. Franken, E. B. Oliveira, H. C. Teixeira, S. A. Nogueira, E. N. Sarno, P. Klatser, T. H. Ottenhoff, and E. P. Sampaio. 2002. T-cell responses to the Mycobacterium tuberculosis-specific antigen ESAT-6 in Brazilian tuberculosis patients. Infect. Immun. 70:6707-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cockle, P. J., S. V. Gordon, A. Lalvani, B. M. Buddle, R. G. Hewinson, and H. M. Vordermeier. 2002. Identification of novel Mycobacterium tuberculosis antigens with potential as diagnostic reagents or subunit vaccine candidates by comparative genomics. Infect. Immun. 70:6996-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 19.Ewer, K., J. Deeks, L. Alvarez, G. Bryant, S. Waller, P. Andersen, P. Monk, and A. Lalvani. 2003. Comparison of T-cell-based assay with tuberculin skin test for diagnosis of Mycobacterium tuberculosis infection in a school tuberculosis outbreak. Lancet 361:1168-1173. [DOI] [PubMed] [Google Scholar]
- 20.Hewinson, R. G., H. M. Vordermeier, and B. M. Buddle. 2003. Use of the bovine model of tuberculosis for the development of improved vaccines and diagnostics. Tuberculosis (Edinburgh) 83:119-130. [DOI] [PubMed] [Google Scholar]
- 21.Huebner, R. E., M. F. Schein, and J. B. Bass, Jr. 1993. The tuberculin skin test. Clin. Infect. Dis. 17:968-975. [DOI] [PubMed] [Google Scholar]
- 22.Jasmer, R. M., P. Nahid, and P. C. Hopewell. 2002. Clinical practice. Latent tuberculosis infection. N. Engl. J. Med. 347:1860-1866. [DOI] [PubMed] [Google Scholar]
- 23.Lalvani, A., R. Brookes, S. Hambleton, W. J. Britton, A. V. Hill, and A. J. McMichael. 1997. Rapid effector function in CD8+ memory T cells. J. Exp. Med. 186:859-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalvani, A., P. Nagvenkar, Z. Udwadia, A. A. Pathan, K. A. Wilkinson, J. S. Shastri, K. Ewer, A. V. Hill, A. Mehta, and C. Rodrigues. 2001. Enumeration of T cells specific for RD1-encoded antigens suggests a high prevalence of latent Mycobacterium tuberculosis infection in healthy urban Indians. J. Infect. Dis. 183:469-477. [DOI] [PubMed] [Google Scholar]
- 25.Lalvani, A., A. A. Pathan, H. Durkan, K. A. Wilkinson, A. Whelan, J. J. Deeks, W. H. Reece, M. Latif, G. Pasvol, and A. V. Hill. 2001. Enhanced contact tracing and spatial tracking of Mycobacterium tuberculosis infection by enumeration of antigen-specific T cells. Lancet 357:2017-2021. [DOI] [PubMed] [Google Scholar]
- 26.Lalvani, A., A. A. Pathan, H. McShane, R. J. Wilkinson, M. Latif, C. P. Conlon, G. Pasvol, and A. V. Hill. 2001. Rapid detection of Mycobacterium tuberculosis infection by enumeration of antigen-specific T cells. Am. J. Respir. Crit. Care Med. 163:824-828. [DOI] [PubMed] [Google Scholar]
- 27.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munk, M. E., S. M. Arend, I. Brock, T. H. Ottenhoff, and P. Andersen. 2001. Use of ESAT-6 and CFP-10 antigens for diagnosis of extrapulmonary tuberculosis. J. Infect. Dis. 183:175-176. [DOI] [PubMed] [Google Scholar]
- 29.Okkels, L. M., I. Brock, F. Follmann, E. M. Agger, S. M. Arend, T. H. Ottenhoff, F. Oftung, I. Rosenkrands, and P. Andersen. 2003. PPE protein (Rv3873) from DNA segment RD1 of Mycobacterium tuberculosis: strong recognition of both specific T-cell epitopes and epitopes conserved within the PPE family. Infect. Immun. 71:6116-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathan, A. A., K. A. Wilkinson, P. Klenerman, H. McShane, R. N. Davidson, G. Pasvol, A. V. Hill, and A. Lalvani. 2001. Direct ex vivo analysis of antigen-specific IFN-γ-secreting CD4 T cells in Mycobacterium tuberculosis-infected individuals: associations with clinical disease state and effect of treatment. J. Immunol. 167:5217-5225. [DOI] [PubMed] [Google Scholar]
- 31.Pathan, A. A., K. A. Wilkinson, R. J. Wilkinson, M. Latif, H. McShane, G. Pasvol, A. V. Hill, and A. Lalvani. 2000. High frequencies of circulating IFN-γ-secreting CD8 cytotoxic T cells specific for a novel MHC class I-restricted Mycobacterium tuberculosis epitope in M. tuberculosis-infected subjects without disease. Eur. J. Immunol. 30:2713-2721. [DOI] [PubMed] [Google Scholar]
- 32.Pollock, J. M., and P. Andersen. 1997. The potential of the ESAT-6 antigen secreted by virulent mycobacteria for specific diagnosis of tuberculosis. J. Infect. Dis. 175:1251-1254. [DOI] [PubMed] [Google Scholar]
- 33.Pollock, J. M., and P. Andersen. 1997. Predominant recognition of the ESAT-6 protein in the first phase of interferon with Mycobacterium bovis in cattle. Infect. Immun. 65:2587-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Small, P. M., and M. D. Perkins. 2000. More rigour needed in trials of new diagnostic agents for tuberculosis. Lancet 356:1048-1049. [DOI] [PubMed] [Google Scholar]
- 35.Smith, S. M., M. R. Klein, A. S. Malin, J. Sillah, K. Huygen, P. Andersen, K. P. McAdam, and H. M. Dockrell. 2000. Human CD8+ T cells specific for Mycobacterium tuberculosis secreted antigens in tuberculosis patients and healthy BCG-vaccinated controls in The Gambia. Infect. Immun. 68:7144-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorensen, A. L., S. Nagai, G. Houen, P. Andersen, and A. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 63:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulrichs, T., P. Anding, S. Porcelli, S. H. Kaufmann, and M. E. Munk. 2000. Increased numbers of ESAT-6- and purified protein derivative-specific gamma interferon-producing cells in subclinical and active tuberculosis infection. Infect. Immun. 68:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulrichs, T., R. Anding, S. H. Kaufmann, and M. E. Munk. 2000. Numbers of IFN-γ-producing cells against ESAT-6 increase in tuberculosis patients during chemotherapy. Int. J. Tuberc. Lung Dis. 4:1181-1183. [PubMed] [Google Scholar]
- 39.Ulrichs, T., M. E. Munk, H. Mollenkopf, S. Behr-Perst, R. Colangeli, M. L. Gennaro, and S. H. Kaufmann. 1998. Differential T cell responses to Mycobacterium tuberculosis ESAT6 in tuberculosis patients and healthy donors. Eur. J. Immunol. 28:3949-3958. [DOI] [PubMed] [Google Scholar]
- 40.Vordermeier, H. M., A. Whelan, P. J. Cockle, L. Farrant, N. Palmer, and R. G. Hewinson. 2001. Use of synthetic peptides derived from the antigens ESAT-6 and CFP-10 for differential diagnosis of bovine tuberculosis in cattle. Clin. Diagn. Lab. Immunol. 8:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, L., M. O. Turner, R. K. Elwood, M. Schulzer, and J. M. FitzGerald. 2002. A meta-analysis of the effect of Bacille Calmette Guérin vaccination on tuberculin skin test measurements. Thorax 57:804-809. [DOI] [PMC free article] [PubMed] [Google Scholar]