Abstract
Porphyromonas gingivalis, an oral bacterium, might play a role in the pathogenesis or progression of adult periodontitis. In this study, we isolated from P. gingivalis a putative glycosyltransferase gene, designated gtfA, which had a consensus domain for glycosyltransferase in its N terminus. GtfA consisted of 248 amino acids and its predicted molecular mass was 28 kDa; however, as the molecular mass of endogenous GtfA protein was around 40 kDa, this suggested that GtfA had undergone some posttranslational modifications. To reveal the role of the gtfA gene in P. gingivalis, we established gtfA-deficient strains by allelic replacement. Morphologically, gtfA-deficient P. gingivalis lacked mature fimbriae. gtfA-deficient P. gingivalis also showed a very low ability for autoaggregation, and its ability to attach to epithelial cells was severely impaired. Thus, the results indicate that the gtfA gene is required for P. gingivalis autoaggregation as well as attachment to epithelial cells. These results suggest that GtfA might have an important role in the pathogenicity of P. gingivalis by regulating adhesion.
Oral bacterial communities, called biofilms, are composed of numerous genetically distinct types of bacteria that exist close to host surfaces such as enamel or oral epithelia (14). For example, more than 300 species of bacteria may exist in the subgingival area (18), which suggests the complexity of oral bacterial communities. Among the flora, several anaerobes are considered to be associated with human periodontitis. In particular, Porphyromonas gingivalis, a black-pigmented bacterium, has been implicated as playing an important role in the pathogenesis of periodontitis (19). P. gingivalis possesses a variety of virulence factors, such as secreted proteases, that can act directly on periodontal tissues. These can induce the degradation of host tissues and the activation of host proenzymes (18), and the virulence of these proteases has been revealed by using mutant strains (7, 28, 29). In addition, there are other factors that might have indirect effects by regulating host reactions. For example, P. gingivalis lipid A, a bioactive center of lipopolysaccharide, activates toll-like receptor 4- and MyD88-dependent pathway (26). Another study has shown that P. gingivalis lipopolysaccharide also activates toll-like receptor 2 (12). Thus, recent intensive studies have revealed the roles of these potent virulent factors.
Another major virulence factor of P. gingivalis is its ability to interact with a variety of surfaces (18). P. gingivalis can interact with various microbial cells (10, 11, 13, 17), saliva components (2), host epithelial cells (6), and extracellular matrices (22). Furthermore, P. gingivalis can enter epithelial cells (16); this ability might play an important part in the penetration of periodontal tissues. P. gingivalis is detected throughout human periodontal pockets (25). At the bottom of the periodontal pocket, the so-called plaque-free zone, P. gingivalis adheres to root surfaces and is covered with a glycocalyx-like structure (24). Recent studies have also indicated that P. gingivalis has the ability to form biofilms in vitro on various surfaces (4, 20). We believe that this might be an important aspect of the pathogenicity of P. gingivalis.
In several bacteria, sugar metabolites or glycoconjugates can act as bacterial adhesins, and glycosyltransferases participate in the synthesis of these products as well as in the adherence of bacteria to other surfaces. For example, it is well known that an open reading frame (ORF) in the ica locus, involved in the synthesis of polysaccharide intercellular adhesin, exhibits N-acetylglucosaminyltransferase activity in Staphylococcus epidermidis (9). In Escherichia coli K-12, rfaG, encoding a glycosyltransferase, is required for adhesion to polystyrene plates as well as for lipopolysaccharide synthesis (8). Thus, glycosyltransferases might have important roles in bacterial adhesion, but little is known about their role in P. gingivalis.
In this study, we isolated a putative glycosyltransferase gene, designated gtfA. The GtfA protein consists of 248 amino acids and has a glycosyltransferase domain at its N terminus. By homologous recombination, we established gtfA-deficient mutants, which lacked mature fimbriae and showed reduced abilities for autoaggregation and attachment to epithelial cells. In addition, the ability to attach to extracellular matrices, including type I collagen, was impaired in the gtfA-deficient mutants. Thus, the gtfA gene regulates attachment ability and might be involved in colonization of oral surfaces.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. gingivalis 381 was cultured anaerobically (90% N2, 5% CO2, 5% H2) at 37°C in GAM broth (Nissui Pharmaceutical, Tokyo, Japan) or on 5% sheep blood agar plates (Trypticase soy agar; Becton Dickinson) supplemented with hemin (5 μg/ml) and menadione (1 μg/ml). Escherichia coli DH5α and BL21 were cultured in Luria-Bertani (LB) broth or on LB agar plates containing ampicillin (100 μg/ml) at 37°C.
Isolation of the gtfA gene.
For isolation of putative glycosyltransferase genes, the P. gingivalis genomic database was searched with the BLAST program at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). As a query, we used pfam00535, a consensus sequence of type 2 glycosyltransferases, which transfer sugar from nucleotide-sugar conjugates to various substrates (3). We designated one of the genes encoding a putative glycosyltransferase gtfA. We found that the locus name for the gtfA ORF was PG0750 in the TIGR database (http://www.tigr.org). For cloning of genomic fragments containing the ORF for gtfA from P. gingivalis 381, PCR was performed with specific primers 5′-GCC TCT TTG TGC CGG TAT CGA C-3′ and 5′-TTT GTA GGA CTT TGT GAC CCG G-3′, based on the genomic sequence from P. gingivalis W83. The PCR product was subcloned into the pGEM-T easy vector (Promega) and designated pGEM-T-GtfA.
Generation of gtfA-deficient P. gingivalis.
For construction of a targeting vector, an ermF-ermAM cassette encoding an erythromycin resistance gene was excised from pYKP009 with SacI and PstI (15). Then, the fragment was blunted by T4 DNA polymerase and inserted into the EcoRV site of pGEM-T-GtfA, resulting in pGEM-T-GtfA-ermF-ermAM, which was linearized with SacI, and 2 μg of the linearized vector was introduced into P. gingivalis by electroporation as described previously (7). After electroporation, cells were incubated in GAM broth for 16 h, then plated onto 5% sheep blood agar plates containing erythromycin (10 μg/ml), and incubated for 7 days. Erythromycin-resistant colonies were inoculated into GAM broth, and genomic DNA was prepared from these clones. NcoI-digested DNA was subjected to Southern blotting to isolate the clones that had undergone homologous recombination, and these were designated P. gingivalis RE1. Two independent clones were used for the following experiments to confirm the phenotype. A digoxigenin-labeled probe was made with the EcoRI fragment of pGEM-T-GtfA and a digoxigenin High Prime kit (Roche) according to the manufacturer's recommendations. The hybridized probe was detected with an alkaline phosphatase-conjugated antidigoxigenin antibody and CSPD (Roche).
Preparation of antiserum against GtfA and Western blotting.
For construction of an expression vector that expresses GtfA protein with a 6x His tag in its N terminus, a fragment containing the ORF for GtfA was amplified by PCR with pGEM-T-GtfA as the template. The sequences of the primers used were 5′-GAG GAT CCG AAG AGA ACA AAC CTG TTT TTT CGA-3′ and 5′-TTT GTA GGA CTT TGT GAC CCG G-3′; the BamHI site is in italics. The PCR product was subcloned into pGEM-T-Easy, and then the BamHI-SalI fragment was ligated into the BamHI and SalI sites of pQE80L (Qiagen, Hilden, Germany), resulting in pQE80L-GtfA. E. coli BL21 was used as the host strain for pQE80L-GtfA, and the 6x His-tagged GtfA protein was purified with Ni-nitrilotriacetic acid-agarose (Qiagen) under denaturing conditions. After dialysis with phosphate-buffered saline (PBS), 6x His-tagged GtfA protein was used as the antigen to obtain polyclonal antiserum from rabbits. Immunization was performed at Medical & Biological Laboratories Co., Ltd., Nagoya, Japan.
For Western blotting, 108 P. gingivalis cells were suspended in Laemmli's sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and boiled for 5 min. The eluted proteins were separated on a 4 to 20% gradient SDS-polyacrylamide gel. After transfer to polyvinylidene difluoride membranes (Immobilon P; Millipore), they were incubated with 1:2,000-diluted anti-GtfA antiserum or 1:2,000-diluted antifimbria antiserum, which recognizes FimA (30), at 4°C overnight. Membranes were washed with Tris-buffered saline containing 0.1% Tween 20 and then incubated with 1:10,000-diluted horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Dako, Glostrup, Denmark). The immune complexes were visualized by a chemiluminescence system (ECL; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom), and representative data are shown.
Transmission electron microscopy.
P. gingivalis cells were washed three times with PBS and fixed with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer, followed by embedding in 3% agar. After postfixation by osmium tetroxide and dehydration, specimens were embedded in LR White (EM Polysciences, Warrington, Pa.). Thin sections were contrasted with uranyl acetate and lead citrate. Images were obtained with a Hitachi 7100 electron microscope at 75 kV.
Autoaggregation of P. gingivalis.
Autoaggregation assays of P. gingivalis cells were performed as previously described (29), except that a different wavelength was used to monitor the optical densities.
Briefly, P. gingivalis cells in the early to mid-log phase were collected by centrifugation, washed three times with PBS, and then resuspended in PBS to an optical density (OD) of 1.0 at 550 nm. The OD of each suspension was monitored for autoaggregation. Three independent experiments were performed, and representative data are shown.
Attachment to epithelial cells and extracellular matrices.
HEp-2 epithelial cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum in a humidified atmosphere with 5% CO2. HEp-2 cells were plated in 24-well plates (105 cells per well) and allowed to attach for 24 h. P. gingivalis cells were washed three times with PBS and resuspended in PBS at a density of 107 or 106 cells/ml. Prior to seeding of P. gingivalis cells, the culture medium of the HEp-2 cells was sucked up and washed three times with PBS. One milliliter of P. gingivalis suspensions was seeded on the HEp-2 cells and incubated for 90 min. After the incubation, nonadherent P. gingivalis cells were removed by washing three times with PBS. HEp-2 cells were lysed with 1 ml of sterile water, and attached or internalized P. gingivalis cells were recovered. One-hundred-microliter aliquots of P. gingivalis cell suspensions were plated onto 5% sheep blood agar plates, followed by colony counting. The averages and standard deviations of the CFU from five wells were calculated, and representative data from three independent experiments are shown.
To evaluate the ability to attach to extracellular matrices, 96-well plates coated with type I collagen, laminin, and fibronectin were used (Biocoat; Becton Dickinson). One-hundred-microliter aliquots of P. gingivalis suspensions in PBS (108 cells/ml) were plated into wells and incubated for 3 h. Nonadherent cells were removed by washing three times with PBS, and adherent cells were stained with 0.1% crystal violet. After being washed with distilled water, the plates were dried and the OD at 570 nm was measured. Averages and standard deviations of the ODs for five wells were calculated, and representative data from three independent experiments are shown.
RT-PCR.
Total RNA was purified from bacterial culture in mid-log phase. cDNA was synthesized by use of random primers and ReverTra Ace (Nacalai Tesque, Kyoto, Japan). To detect fimA expression, we used specific primers for type I fimA, because P. gingivalis 381 possesses the type I fimA gene (1). The sequences of the primers used were as follows: type I fimA, 5′-CTG TGT GTT TAT GGC AAA CTT C-3′ and 5′-AAC CCC GCT CCC TGT ATT CCG A-3′; and 16S rRNA, 5′-TGT AGA TGA CTG ATG GTG AAA ACC-3′ and 5′-ACG TCA TCC CCA CCT TCC TC-3′ (1). The PCR conditions are available on request.
RESULTS
Isolation of a putative glycosyltransferase gene from P. gingivalis.
To isolate a putative glycosyltransferase gene(s) from P. gingivalis, we carried out a BLAST search with the consensus amino acid sequence of glycosyltransferases as the query. We obtained some ORFs which had a consensus domain for glycosyltransferase from a genomic database for P. gingivalis W83. We designated one of the ORFs as gtfA (Fig. 1). The product GtfA consisted of 248 amino acids and had a consensus domain for glycosyltransferase at its N terminus. The glycosyltransferase domain in the N terminus showed 30% identity to pfam00535, and its product had a predicted molecular mass of approximately 28kDa. We isolated the gtfA gene from P. gingivalis 381 by genomic PCR, which indicated that the gtfA gene was also conserved in P. gingivalis 381. gtfA showed the highest identity to TIGR locus BT2947, an ORF for a putative glycosyltransferase in Bacteroides thetaiotaomicron (48%), although the function of the ORF remains unknown. From the BLAST search of the E. coli genome database, GtfA showed the highest identity to WcaE (25% identity), a putative glycosyltransferase that might be involved in extracellular polysaccharide synthesis in E. coli (27).
FIG. 1.
Amino acid sequences of GtfA and pfam00535, a consensus sequence for glycosyltransferases. Alignment of GtfA from P. gingivalis W83 (248 amino acids) and pfam00535 (168 amino acids) is shown. Asterisks indicate identical residues.
Generation of gtfA-deficient mutants.
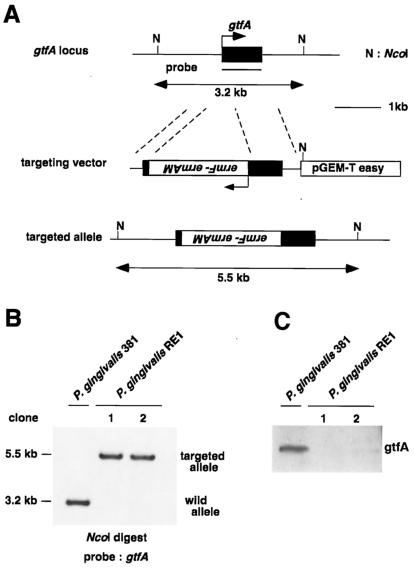
To reveal the role of the gtfA gene, we developed gtfA-deficient P. gingivalis by homologous recombination (Fig. 2A). The targeting vector was constructed by insertion of an erythromycin resistance cassette into the gtfA ORF. After electroporation of the targeting vector, erythromycin-resistant clones were obtained. Following the first screening of homologous recombinants by PCR (data not shown), we performed Southern blot analysis to identify clones that had undergone the correct recombination. In this analysis, homologous recombinants showed a 5.5-kb NcoI fragment due to an insertion of erythromycin resistant cassette, compared to the 3.2-kb NcoI fragment of the wild-type strain (Fig. 2B).
FIG. 2.
Targeted disruption of gtfA locus. (A) Structures of the gtfA locus, targeting vector, and targeted allele. N, NcoI. (B) Southern blot analysis of P. gingivalis clones after selection with erythromycin. NcoI-digested DNA was probed with the gtfA ORF. The 3.2-kb wild-type fragment and the 5.5-kb targeted fragment are indicated. (C) Western blotting with anti-GtfA antiserum. Whole extracts from P. gingivalis 381 and P. gingivalis RE1 were immunoblotted with rabbit anti-GtfA polyclonal antiserum.
The homologous recombinants, designated P. gingivalis RE1 and P. gingivalis 381, were subjected to Western blotting with anti-GtfA antiserum (Fig. 2C). The molecular mass of the endogenous GtfA protein was approximately 40 kDa, suggesting that GtfA had undergone some posttranslational modifications. GtfA protein could not be detected in P. gingivalis RE1, indicating that the gtfA gene was correctly disrupted in P. gingivalis RE1. The appearance or pigmentation of the colonies of P. gingivalis RE1 was similar to that of the wild type, and its growth was slightly slower than that of the wild type (data not shown). These results suggested that gtfA is not essential for cell survival, cell growth, or pigmentation.
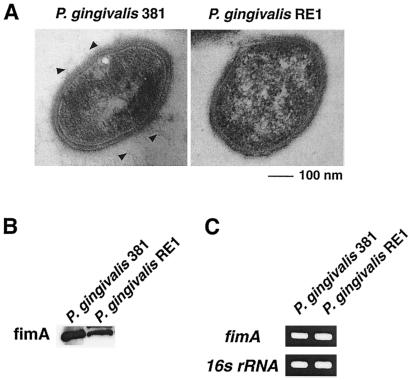
We observed the morphology of P. gingivalis RE1 by transmission electron microscopy. There was no significant difference in the thickness of the capsular layer between P. gingivalis 381 and P. gingivalis RE1. However, P. gingivalis RE1 lacked mature fimbria, which is a key virulence factor in the ability to attach (Fig. 3A). From this result, gtfA must be required for the development of mature fimbria. We examined the expression of FimA, a subunit of fimbriae, by Western blotting and RT-PCR and found no significant change in fimA expression in P. gingivalis RE1 (Fig. 3B and C), indicating that GtfA is not required for fimA expression. These results suggested that sugar transfer might be involved in fimbria development in P. gingivalis.
FIG. 3.
Transmission electron microscopic image and expression of fimA in gtfA-deficient mutant. (A) Images of P. gingivalis 381 (left panel) and P. gingivalis RE1 (right panel) are shown. Bar, 100 nm. Arrowheads indicate fimbriae of P. gingivalis 381. (B) Detection of FimA protein by Western blotting. (C) Detection of fimA mRNA expression by RT-PCR.
Autoaggregation activity of gtfA-deficient P. gingivalis.
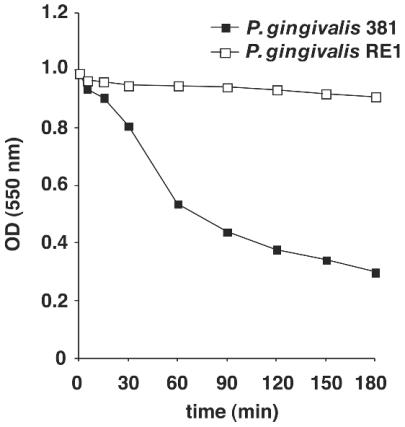
It has been reported that P. gingivalis shows autoaggregation and that this might be important for colonization of human oral surfaces (28, 29). Therefore, we examined whether the gtfA gene was involved in autoaggregation. The OD of cell suspensions was monitored to evaluate autoaggregation activity. In wild-type P. gingivalis, reduction of the OD was observed within several hours, whereas P. gingivalis RE1 showed only a slight reduction in OD (Fig. 4). This result indicated that P. gingivalis RE1 has low autoaggregation activity and that the gtfA gene is required for autoaggregation.
FIG. 4.
Autoaggregation activities of wild-type and gtfA-deficient P. gingivalis. The OD of each P. gingivalis suspension was measured at the indicated times. Representative data from three independent experiments are shown. Open squares, P. gingivalis 381; solid squares, P. gingivalis RE1.
Role of gtfA in attachment to epithelial cells and extracellular matrices.
It has been reported that P. gingivalis can adhere to cultured epithelial cells (6) and that this might be an important property in the colonization of P. gingivalis. Therefore, we also examined whether the gtfA gene is involved in the process of attachment to cultured epithelial cells. P. gingivalis suspensions were added to confluent HEp-2 epithelial cells and incubated for 90 min. Compared to P. gingivalis 381, P. gingivalis RE1 was defective in adherence to HEp-2 cells (Table 1). This result indicated that gtfA has an indispensable role in adherence to epithelial cells.
TABLE 1.
Attachment of wild-type and gtfA-deficient P. gingivalis to HEp-2 epithelial cellsa
| P. gingivalis strain | No. of cells/ml | Mean CFU (105) ± SD |
|---|---|---|
| 381 | 106 | 1.26 ± 0.063 |
| 107 | 2.12 ± 0.13 | |
| RE1 | 106 | No viable colonies |
| 107 | No viable colonies |
P. gingivalis suspensions were added to HEp-2 cells and allowed to adhere for 90 min. Adherent P. gingivalis cells were recovered, and viable cells were counted.
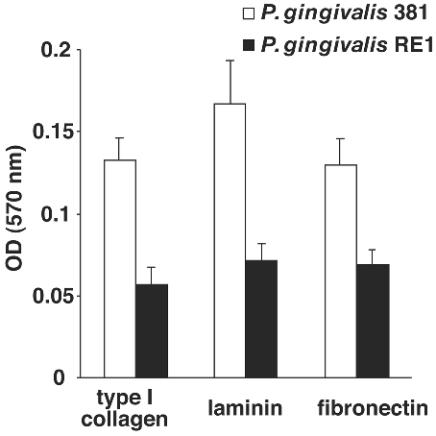
It has also been reported that P. gingivalis can attach to various proteins, including extracellular proteins such as collagen and salivary components (18). To investigate binding to extracellular matrices, we examined binding to type I collagen, laminin, and fibronectin. The ability of P. gingivalis RE1 to attach to extracellular matrices was reduced to approximately 40 to 60% of that of the wild type (Fig. 5). These results indicated that gtfA is involved, at least in part, in attachment to extracellular matrices, including type I collagen, laminin, and fibronectin. Thus, GtfA plays a role in attachment to various proteins as well as to living epithelial cells.
FIG. 5.
Attachment to extracellular matrices. Ninety-six-well plates with immobilized type I collagen, fibronectin, or laminin were used. After incubation with P. gingivalis suspension, nonadherent cells were removed by washing, followed by crystal violet staining. The means of the OD at 570 nm and standard deviations are shown.
DISCUSSION
In this report, we characterized the role of gtfA, a putative glycosyltransferase gene in P. gingivalis. Some glycosyltransferase genes have already been identified in P. gingivalis. The narH gene encodes β-N-acetylhexosaminidase, which is likely a lipoprotein (21). Duncan et al. identified an ORF for a putative glycosyltransferase from an expression screening to isolate ORFs related to attachment to epithelial cells (5). They indicated that overexpression of an ORF encoding a putative glycosyltransferase in E. coli could induce attachment to epithelial cells and that this result suggested that the glycosyltransferase might be involved in attachment to epithelial cells. However, the roles of these ORFs in P. gingivalis have remained unknown. Characterization of a loss-of-function mutant for these ORFs would be helpful in understanding their roles in P. gingivalis. A BLAST search of the P. gingivalis genomic database (data not shown) identified a number of ORFs for putative glycosyltransferases, and it might be of interest to determine the native role of each ORF.
P. gingivalis is a periodontopathogenic organism that has already been analyzed extensively, and the roles of virulence factors such as proteases and adhesion molecules have been characterized. In particular, P. gingivalis has the ability to interact with various surfaces, including other bacteria and epithelial cells, and also has the potential to be a virulence factor (19). Some reports have suggested that fimbriae have a central role in attachment to various surfaces; for example, a fimA-defective mutant showed loss of attachment ability as well as internalization into epithelial cells (23). Some proteases are also involved in the interaction between P. gingivalis and epithelial cells, and protease-defective mutants showed altered expression of fimA, suggesting that the expression of fimbriae might affect the ability to attach to epithelial cells (28, 29). In this study, we showed that P. gingivalis RE1 lacked mature fimbriae (Fig. 3A), even though expression of FimA protein or mRNA was not altered (Fig. 3B and 3C). This implies that a sugar transfer reaction might be involved in the production of fimbriae. The thickness of the capsular layer in P. gingivalis RE1 was similar to that of the wild type; however, the composition of the capsular layer remains unknown, and it is possible that the composition and function might differ.
We also showed that the gtfA gene is involved in autoaggregation; the interaction between P. gingivalis and epithelial cells, as well as binding to all the extracellular matrices used in this study. These phenotypes might be explained by the lack of fimbriae in P. gingivalis RE1. It is also possible that a surface matrix might have lost its potential to act as an adhesin. Both attachment and aggregation are important steps in biofilm formation (14). As P. gingivalis RE1 showed reduced autoaggregation and attachment to cell surfaces and several extracellular matrices, it is likely that the gtfA gene is involved in the development of P. gingivalis biofilms.
Our data showed that gtfA regulates autoaggregation and attachment to epithelial cells and extracellular matrices. The gtfA gene might regulate attachment ability in subgingival biofilms and act as a potent virulence factor.
Acknowledgments
We thank Ichiro Nakagawa for helpful advice. We also acknowledge Yumi Kumagai for plasmid pYKP009 and Fuminobu Yoshimura for antifimbria antiserum.
This study was supported by grants [A(2)14207080] from the Japan Society for the Promotion of Science and the 21st century COE entitled Origination of Frontier Biodentistry at Osaka University Graduate School of Dentistry from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Editor: V. J. DiRita
REFERENCES
- 1.Amano, A., I. Nakagawa, K. Kataoka, I. Morisaki, and S. Hamada. 1999. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J. Clin. Microbiol. 37:1426-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano, A., S. Shizukuishi, H. Horie, S. Kimura, I. Morisaki, and S. Hamada. 1998. Binding of Porphyromonas gingivalis fimbriae to proline-rich glycoproteins in parotid saliva via a domain shared by major salivary components. Infect. Immun. 66:2072-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326:929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, W., R. J. Palmer, and H. K. Kuramitsu. 2002. Role of polyphosphate kinase in biofilm formation by Porphyromonas gingivalis. Infect. Immun. 70:4708-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan, M. J., S. A. Emory, and E. C. Almira. 1996. Porphyromonas gingivalis genes isolated by screening for epithelial cell attachment. Infect. Immun. 64:3624-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan, M. J., S. Nakao, Z. Skobe, and H. Xie. 1993. Interactions of Porphyromonas gingivalis with epithelial cells. Infect. Immun. 61:2260-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fletcher, H. M., H. A. Schenkein, R. M. Morgan, K. A. Bailey, C. R. Berry, and F. L. Macrina. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172:1-8. [DOI] [PubMed] [Google Scholar]
- 9.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Gotz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 10.Goulbourne, P. A., and R. P. Ellen. 1991. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J. Bacteriol. 173:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grenier, D. 1992. Demonstration of a bimodal coaggregation reaction between Porphyromonas gingivalis and Treponema denticola. Oral Microbiol. Immunol. 7:280-284. [DOI] [PubMed] [Google Scholar]
- 12.Hirschfeld, M., J. J. Weis, V. Toshchakov, C. A. Salkowski, M. J. Cody, D. C. Ward, N. Qureshi, S. M. Michalek, and S. N. Vogel. 2001. Signaling by toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect. Immun. 69:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinder, S. A., and S. C. Holt. 1993. Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J. Bacteriol. 175:840-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 15.Kumagai, Y., K. Konishi, T. Gomi, H. Yagishita, A. Yajima, and M. Yoshikawa. 2000. Enzymatic properties of dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis and its participation in virulence. Infect. Immun. 68:716-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamont, R. J., S. Gil, D. R. Demuth, D. Malamud, and B. Rosan. 1994. Molecules of Streptococcus gordonii that bind to Porphyromonas gingivalis. Microbiology 140:867-872. [DOI] [PubMed] [Google Scholar]
- 18.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamont, R. J., and H. F. Jenkinson. 2000. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol. Immunol. 15:341-349. [DOI] [PubMed] [Google Scholar]
- 20.Larsen, T. 2002. Susceptibility of Porphyromonas gingivalis in biofilms to amoxicillin, doxycycline and metronidazole. Oral Microbiol. Immunol. 17:267-271. [DOI] [PubMed] [Google Scholar]
- 21.Lovatt, A., and I. S. Roberts. 1994. Cloning and expression in Escherichia coli of the nahA gene from Porphyromonas gingivalis indicates that beta-N-acetylhexosaminidase is an outer-membrane-associated lipoprotein. Microbiology 140:3399-3406. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura, T., A. Amano, I. Nakagawa, and S. Hamada. 1999. Specific interactions between Porphyromonas gingivalis fimbriae and human extracellular matrix proteins. FEMS Microbiol. Lett. 175:267-272. [DOI] [PubMed] [Google Scholar]
- 23.Njoroge, T., R. J. Genco, H. T. Sojar, N. Hamada, and C. A. Genco. 1997. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 65:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noiri, Y., and S. Ebisu. 2000. Identification of periodontal disease-associated bacteria in the “plaque-free zone.” J. Periodontol. 71:1319-1326. [DOI] [PubMed] [Google Scholar]
- 25.Noiri, Y., K. Ozaki, H. Nakae, T. Matsuo, and S. Ebisu. 1997. An immunohistochemical study on the localization of Porphyromonas gingivalis, Campylobacter rectus and Actinomyces viscosus in human periodontal pockets. J. Periodontal Res. 32:598-607. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa, T., Y. Asai, M. Hashimoto, O. Takeuchi, T. Kurita, Y. Yoshikai, K. Miyake, and S. Akira. 2002. Cell activation by Porphyromonas gingivalis lipid A molecule through Toll-like receptor 4- and myeloid differentiation factor 88-dependent signaling pathway. Int. Immunol. 14:1325-1332. [DOI] [PubMed] [Google Scholar]
- 27.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokuda, M., M. Duncan, M. I. Cho, and H. K. Kuramitsu. 1996. Role of Porphyromonas gingivalis protease activity in colonization of oral surfaces. Infect. Immun. 64:4067-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tokuda, M., T. Karunakaran, M. Duncan, N. Hamada, and H. Kuramitsu. 1998. Role of Arg-gingipain A in virulence of Porphyromonas gingivalis. Infect. Immun. 66:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimura, F., K. Takahashi, Y. Nodasaka, and T. Suzuki. 1984. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J. Bacteriol. 160:949-957. [DOI] [PMC free article] [PubMed] [Google Scholar]