Abstract
Background
Recent genomic evidence suggests frequent phosphatidylinositide 3-kinase (PI3K) pathway activation in human papillomavirus (HPV) positive oropharyngeal squamous cell carcinoma. Mutations/amplification of the gene encoding p110α catalytic subunit of phosphoinositide 3-kinase (PIK3CA), loss of phosphatase and tensin homolog (PTEN) and HRAS mutations are known to activate PI3K pathway.
Methods and results
PIK3CA mutations were identified by Sanger sequencing in 23 of 75 (31%) HPV-positive oropharyngeal carcinomas, including exon 9 (p.E545K [n = 10] and p.E542K [n = 5]) or exon 20 (p.H1047Y, n = 2) mutations. Five rare and one novel (p.R537Q) PIK3CA mutations were identified. HRAS mutation (p.Q61L) was detected in 1 of 62 tested cases. PIK3CA amplification by fluorescence in situ hybridization (FISH) was identified in 4 cases (4/21, 20%), while PTEN loss was seen in 7 (7/21, 33%) cases (chromosome 10 monosomy [n = 4], homozygous deletion [n = 3]).
Conclusions
Overall, genetic alterations that likely lead to PI3K pathway activation were identified in 34 of 75 cases (45%) and did not correlate with disease specific survival. These findings offer a molecular rationale for therapeutic targeting of PI3K pathway in patients with HPV-positive oropharyngeal carcinoma.
Keywords: Oropharyngeal squamous carcinoma, PIK3CA, HRAS, PTEN, HPV
Background
The phosphatidylinositide 3-kinase (PI3K) pathway is activated in about half of head and neck squamous cell carcinomas (SCC) by a number of mechanisms, including mutation or amplification of the gene encoding p110α catalytic subunit of phosphoinositide 3-kinase (PIK3CA)[1-4]. The higher incidence of PI3K pathway activation in oropharyngeal SCC was previously reported [5]. Oropharyngeal SCC are increasingly associated with human papillomavirus (HPV) infection [6,7] and the higher prevalence of PI3K pathway abnormalities in these tumors was eventually linked to HPV [8,9].
Most recent characterization of the mutational landscape of head and neck SCC showed that the genetic profile of HPV-positive SCC is distinct from that of HPV-negative SCC. For instance, HPV-positive oropharyngeal SCC harbor fewer mutations overall (e.g., no TP53 mutations) and more PIK3CA mutations. Specifically, of the 15 HPV-positive SCC with known PIK3CA status reported in the literature, 4 tumors harbored PIK3CA mutation (4/15, 27%) [10,11]. In contrast, PIK3CA mutations are present in about 5% (5/91) of HPV-negative head and neck SCC. The higher incidence of PIK3CA mutations in HPV-positive SCC suggests a new therapeutic option, as PI3K pathway is targeted by multiple drugs in development: PX-866 [12], and MK-2066 [13], and RAD001 [14]. Indeed, our most recent findings demonstrated that HPV-positive SCC tumorgrafts with activating PIK3CA mutation were highly responsive to PI3K-targeted therapy [15].
Increased PI3K signaling can also result from mutations in other genes in the PI3K pathway such as HRAS[16,17]. In addition to PIK3CA mutations and/or amplification, PI3K pathway may also be activated due to phosphatase and tensin homolog (PTEN) deletion, a known negative regulator of the PI3K signaling pathway [18].
The aim of the present study was to elucidate the molecular basis for therapeutic targeting of PI3K pathway in HPV-positive oropharyngeal SCC by characterizing the prevalence and prognostic significance of PIK3CA and HRAS mutations, PIK3CA amplification, and PTEN loss in 75 patients with HPV-positive oropharyngeal SCC.
Methods
Patients
This study was approved by the Institutional Review Board of the University of Pittsburgh Medical Center (IRB# PRO11010195). Seventy five cases of HPV-positive oropharyngeal SCC were identified from 1983 to 2007 and satisfied the following inclusion criteria: availability of formalin fixed paraffin embedded tissue, p16 immunohistochemistry and HPV in situ hybridization positivity, presence of tumor areas with >50% represented by cancer cells, and extraction of adequate DNA.
HPV in situ hybridization and p16 immunohistochemistry
HPV detection by in-situ hybridization was performed using probes targeting 37 distinct HPV subtypes, including 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, and 52 (Y1404; Dako, Carpinteria, CA). Five-micrometer tissue sections were deparaffinized and digested with proteinase K (Roche Diagnostics, Indianapolis, IN). Cases with punctate nuclear signal were considered positive [19].
For p16 analysis, five-micrometer sections were deparaffinized. Heat-induced epitope retrieval was then performed in a citrate buffer. Immunohistochemistry for p16 (G175-405; BD Pharmingen, San Diego, CA) was performed as per the manufacturer’s protocol. Cases were considered positive if >70% of tumor cells showed diffuse and strong cytoplasmic and nuclear staining [19].
PIK3CA and HRAS mutation analysis
Tissue cores from tumor targets were obtained as previously described [20]. DNA was isolated from tissue cores using the DNeasy tissue kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. For the detection of mutations, DNA was amplified with primers flanking exon 3 of the HRAS gene (forward primer 5′- GTC CTC CTG CAG GAT TCC TA -3′ and reverse primer 5′- CGG GGT TCA CCT GTA CT -3′), exon 9 of the PIK3CA gene (forward primer 5′- TGA AAA TGT ATT TGC TTT TTC TGT -3′ and reverse primer 5′- TGT AAA TTC TGC TTT ATT TAT TCC -3′) and exon 20 of the PIK3CA gene (forward primer 5′- TTT GCT CCA AAC TGA CCA A -3′ and reverse primer 5′- GCA TGC TGT TTA ATT GTG TGG -3′). PCR products were sequenced in both sense and antisense directions using the BigDye Terminator v3.1 cycle sequencing kit on ABI 3730 (Applied Biosystems, Inc., Foster City, CA) according to the manufacturer’s instructions (Additional file 1: Figure S1 and Additional file 2: Figure S2). The sequences were analyzed using Mutation Surveyor software (SoftGenetics, LLC., State College, PA).
The presence of most common PIK3CA mutations (p.E545K and p.E542K) was confirmed by SNaPshot PCR as per the manufacturer’s manual and as previously described [21] (Additional file 3: Figure S3). Briefly, primers for exon 9 (forward 5′-AGTAACAGACTAGCTAGAGA-3′ and reverse 5′-ATTTTAGCACTTACCTGTGAC-3′) and exon 20 (forward 5′-GACCCTAGCCTTAGATAAAAC-3′ and reverse 5′-GTGGAAGATCCAATCCATTT-3′) were used for amplification. Denatured products were analyzed on an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA).
PTEN and PIK3CA fluorescence in situ hybridization (FISH)
Cases with known wild type PIK3CA and HRAS and available tissue were tested for PIK3CA and PTEN copy number changes by FISH (n = 22) (Additional file 4: Figure S4). Sixty to 130 cells were analyzed. PTEN (SpectrumOrange) and chromosomal enumeration probe 10 (CEP10, Spectrum Green) FISH was performed as per manufacturer’s recommendations (Abbot Molecular, Des Plaines, IL, USA) and as previously described [22]. Results were interpreted using previously established thresholds [23,24]: PTEN homozygous deletion was defined as >20% of cells without PTEN locus signal and the presence of ≥2 CEP10 signals. Hemizygous PTEN deletion was defined as >30% of cells with only one PTEN signal and the presence of ≥2 CEP10 signals. As previously suggested, cases with >50% of cells with a single CEP10 signal were categorized as “Chromosome 10 monosomy”, an additional mechanism of PTEN loss [22]. Despite repeated hybridization attempts, no data were obtained in one case.
PIK3CA (Spectrum Green) and CEP3 (Spectrum Orange) (Abbot Molecular, Des Plaines, IL, USA) FISH was performed as per manufacture’s recommendation. PIK3CA amplification was defined as PIK3CA/CEP3 >2. Despite repeated hybridization attempts, no data were obtained in one case.
Statistical analysis
The primary endpoint was disease-specific survival (DSS) defined as time elapsed from the date of diagnosis until death from cancer. DSS was chosen as a primary endpoint over the overall survival due to the predominance of patients deceased from causes unrelated to the oropharyngeal SCC (n = 21). DSS was assessed only for patients with > 2 months of follow-up (n = 72). Patients who were alive at last follow-up or had died from other causes were censored. Survival data were presented as Kaplan-Meier plots. The log rank test was used to test survival equality. Covariates examined for association with survival included age, gender, smoking (ever versus never), site (tonsil and base of tongue), T and N classification, AJCC clinical stage, adjuvant chemotherapy or radiotherapy. Cross-tabulated categorical data were tested for independence with Fisher’s exact test.
Results
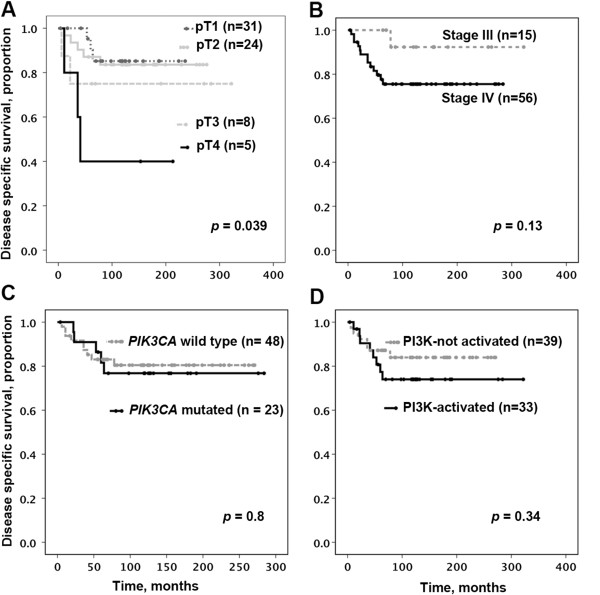
The clinicopathologic characteristics of 75 HPV-positive oropharyngeal SCC patients are summarized in Table 1. The mean follow-up was 122 months (minimum – maximum, 4–322). While 21 patients deceased of causes unrelated to oropharyngeal SCC, 14 patients died of disease. The 3-year disease specific survival (DSS) was 85%. In this clinicopathologically uniform group of patients only pathologic tumor stage (pT) correlated with DSS (Figure 1A). For instance, the DSS at 5 years was 87% for pT1 and 40% for pT4 (p = 0.039). There was a trend toward a worse DSS for patients with clinical stage IV disease; however, it did not reach statistical significance, most likely due to the low number of events in stage III patients (Figure 1B). It is noteworthy that patients’ gender, age, smoking history, oropharyngeal sub-site, pN, and treatment modality was not associated with survival in these individuals.
Table 1.
Clinicopathologic features of patients with human papillomavirus positive oropharyngeal SCC, overall and by PIK3CA mutational status
| Feature | Total 1 , n = 75 |
PIK3CA
wild type |
PIK3CA
mutant |
Notes | |
|---|---|---|---|---|---|
| n = 51 | n = 23 | ||||
| Men/women |
64/11 |
43/8 |
20/3 |
n.s. |
|
| Average age, years (range) |
56 (34–78) |
56 |
55.3 |
n.s. |
|
| Smoking2 |
Never |
22 |
15 |
7 |
n.s. |
| Ever |
40 |
26 |
14 |
||
| Site3 |
BOT |
31 |
24 |
7 |
n.s. |
| Tonsil |
43 |
27 |
15 |
||
| pT4 |
1 |
34 |
23 |
11 |
n.s. |
| 2 |
25 |
17 |
8 |
||
| 3 |
7 |
4 |
3 |
||
| 4 |
5 |
4 |
1 |
||
| pN6 |
0 |
2 |
2 |
0 |
n.s. |
| 1 |
16 |
11 |
5 |
||
| 2 |
51 |
35 |
15 |
||
| 3 |
5 |
3 |
2 |
||
| Resection Margins5 |
Positive |
10 |
7 |
3 |
n.s. |
| Negative |
54 |
36 |
18 |
||
| Clinical Stage6 |
I |
1 |
1 |
0 |
n.s. |
| III |
15 |
10 |
4 |
||
| IV |
58 |
40 |
18 |
||
| Therapy7 |
RT |
43 |
32 |
11 |
n.s. |
| CRT |
21 |
14 |
7 |
||
| Non-keratinizing |
52 |
35 |
17 |
n.s. |
|
| Mixed |
23 |
17 |
5 |
||
| Deceased of disease | 14 | 9 | 5 | ||
1For one patient with HRAS mutation PIK3CA status was indeterminate. The clinicopathologic features of the case with indeterminate PIK3CA status are not presented in the columns 3 and 4 of the Table.
2Smoking history was not available for 13 patients.
3The oropharyngeal sub-site for one case was unknown.
4pT was unknown for 4 cases with biopsies only.
5Margin status was not assessed in 11 cases (biopsies, fragmented specimens).
6pN and clinical stage were unknown for 1 patient.
7Seven patients received no postoperative therapy; for 4 patients these data were not available.
BOT base of tongue, RT radiotherapy, CT chemotherapy, CRT chemoradiotherapy, mixed keratinizing and non-keratinizing, n.s. not statistically significant.
Figure 1.
Disease specific survival (DSS) of patients with p16 and human papillomavirus positive oropharyngeal squamous cell carcinoma. DSS was assessed only for patients with > 2 months of follow-up (n = 72). A. pT stage and disease specific survival. B. Clinical stage and disease specific survival. C. PIK3CA mutational status and disease specific survival. D. Presumed activation of PI3K pathway and disease specific survival. Cases with PI3K pathway activating alteration with < 2 months of follow-up were excluded from the DSS analysis.
PIK3CA, HRAS, and PTEN alterations
PIK3CA mutations were identified in 23 of 75 patients with oropharyngeal SCC (31%), including exon 9 (p.E545K [n = 10] and p.E542K [n = 5]) or exon 20 (p.H1047Y, n = 2) mutations. Five cases with rare mutations and one case with novel mutation are presented in Table 2. Patients’ gender, age, smoking history, oropharyngeal sub-site, pT, pN, clinical stage, and treatment modality were similar between cases with wild type and mutated PIK3CA. Disease specific survival (DSS) of the patients in these two groups was not significantly different (Figure 1C).
Table 2.
Rare and novel PIK3CA mutations in HPV positive oropharyngeal SCC
| Mutation | Clinicopathologic features and number of cases in this study, n | Prior reports and References |
|---|---|---|
| Exon 9, c.1571G > A, p.R524K and c.1573G > A, p.E525K |
53 year old man, NK, pT1 N2, no postoperative therapy, NED at 132 mo, n = 1 |
None. While individually both mutations were previously reported, the combination of the two was not. |
| Exon 9, c.1610G > A, p.R537Q |
60 year old man, NK, pT1 N2, CRT, DOD in 22 months, n = 1 |
None |
| Exon 20, c.2975G > A, p.R992Q |
68 year old man, NK, pT2 N1, radiotherapy, DOC at 70 months, n = 1 |
Glioblastoma multiforme, n = 1 [25] |
| Exon 20, c.3103G > A, p.A1035T |
54 year old man, mixed K& NK pT2 N2, radiotherapy, DOD at 53 months, n = 1 |
Endometrial adenocarcinoma, n = 1 [26] |
| Urothelial carcinoma of the bladder, n =1 [27] | ||
| Exon 20, c.3129G > C, |
48 year old man, mixed K & NK, pT3 N1, radiotherapy, NED at 284 months, n = 1 |
Carcinoma of the breast, n = 2 [28] |
| p.M1043I | ||
| Exon 20, c.3153G > A, p.W1051* | 71 year old man, NK, pT1 N1, treated with radiotherapy, DOC at 182 months, n = 1 | Ovarian clear cell carcinoma, n = 1; conventional colorectal carcinoma, n = 1; [29,30] |
NK non-keratinizing, K keratinizing, DOC deceased of other causes, DOD deceased of disease, CRT chemoradiotherapy, NED no evidence of disease.
HRAS mutation (c.182A > T, p.Q61L) was identified in 1 of 62 tested cases (or in 1 of 58 successfully tested cases; in 4 cases the status of HRAS was indeterminate). In the only case with HRAS mutation, the mutational status of PIK3CA was indeterminate.
PIK3CA amplification was identified in 4 of 21 cases (20%). PTEN loss was identified in 7 of 21 cases (33%) (chromosome 10 monosomy [n = 4), homozygous deletion [n = 3]; note, for one of the cases with homozygous deletion clinical follow-up was not available).
Assuming that PIK3CA mutation or amplification, HRAS mutation, or loss of PTEN lead to PI3K pathway activation, patients with tumors harboring one of these events were combined into a “PI3K-activated” group and compared to patients whose tumors did not harbor any of the above genetic alterations. PI3K pathway activation did not correlate with DSS (Figure 1D).
Discussion
The clinical and pathologic characteristics of our HPV-positive oropharyngeal SCC population and the performance of conventional pathologic prognosticators (e.g., pT, pN) are consistent with prior reports [31].
To our knowledge, this is the largest HPV-positive oropharyngeal SCC cohort to undergo evaluation for PIK3CA and HRAS mutation and PIK3CA and PTEN amplification/loss. Our findings suggest that mutation or amplification of PIK3CA may represent the most common alteration in HPV-positive oropharyngeal SCC. It is noteworthy that recent mutational analyses of head and neck SCC also found PIK3CA alterations, albeit at lower rates [10,11,15]. The variation in PIK3CA mutation incidence is most likely due to the relative underrepresentation of HPV-positive oropharyngeal SCC in other cohorts, use of oropharyngeal site as a surrogate marker for HPV status, and the use of different techniques to assess for PIK3CA mutations. The recently published data [11,15] highlighted an interesting phenomenon that even though HPV-positive SCC harbored fewer mutations on average, as high as 20% of HPV-positive SCC (3/15 cases [15]) harbored PIK3CA mutation as the only cancer gene mutation, indicating that PI3K pathway mutations are enriched in HPV-positive tumors despite the lower rate of gene mutations in general. The higher prevalence of PI3K pathway abnormalities in oropharyngeal SCC was previously linked to HPV [8,9].
All mutations found in the samples of HPV-positive oropharyngeal SCC were heterozygous with mutant allelic frequency that appeared to range from 20% to 50% of alleles (corresponding to 40%– 100% of cancer cells with a heterozygous mutation). None of the cases showed mutant allelic frequency of more than 50% suggesting that loss of the wild type PIK3CA allele or amplification of the mutant PIK3CA allele in cancer cells is exceedingly rare.
Although HRAS mutations have been reported to modulate signaling through the PI3K pathway [32,33], the role of the mutation found in a single HPV-positive oropharyngeal SCC in this study remains unclear.
PTEN is generally understood to function as a tumor suppressor gene and to negatively regulate PI3K pathway. Therefore, loss of PTEN should lead to PI3K pathway activation. The incidence of PTEN alterations in head and neck SCC varies in the literature and there is little indication that PTEN loss has an independent prognostic value [34,35]. We found that PTEN loss (as assessed by FISH) was relatively common in HPV-positive oropharyngeal SCC.
Activation of the PI3K pathway, generally by virtue of PIK3CA gene amplification, has been previously reported to represent a poor prognostic biomarker in head and neck SCC [36]. Others have reported that phosphorylation of AKT, a downstream target of PIK3CA, is associated with poor clinical outcome in oropharyngeal SCC, specifically [37]. Although HPV status was not specifically assessed in this cohort of oropharyngeal SCC, it is reasonable to presume that it was enriched for HPV-positive SCC. Our analysis showed no association between the genetic alterations we assessed for (combined into a “PI3K activated” group) and clinical outcome. Prior reports have generally focused on a single alteration or biomarker assessment. It is possible that some of the alterations we detected in HPV-positive oropharyngeal SCC do not activate the pathway as predicted. Or, more likely, each alteration modulates PI3K oncogenic signaling. Further functional studies in relevant preclinical models are needed to decipher the precise contribution of each mutation, amplification and/or loss to PI3K pathway status in HPV-positive oropharyngeal SCC.
One of the technical limitations of this study is that we restricted our assessment to exons 9 and 20 of PIK3CA gene and we have likely underestimated the frequency of PIK3CA mutation in this cohort. Similarly, we only assessed codon 61 of HRAS and did not perform codon 12/13 testing. Therefore, the actual mutation frequency of both PIK3CA and HRAS could be higher than reported here.
The variety of potential mechanisms leading to PI3K pathway activation underscores the complexity of the potential implications of our findings. It is possible, as reported by others and us, that head and neck SCC harboring “driver” PIK3CA mutations demonstrate enhanced response to PI3K pathway inhibitors [15,38,39]. Similar findings have been reported in clinical trials of patients with breast or gynecologic malignancies [40]. PI3K pathway inhibitors are under early investigation in head and neck SCC and clinical results are not yet available.
The EGFR monoclonal antibody cetuximab is FDA-approved in both newly diagnosed head and neck SCC as well as in the recurrent or metastatic setting [41]. We previously reported that PI3K pathway activation correlates with clinical resistance to cetuximab in head and neck SCC patients and targeting the PI3K pathway enhanced the antitumor effects of EGFR inhibitors in head and neck SCC preclinical models [42-44]. Therefore, molecular determinants of PI3K activation may identify individuals who may benefit from co-targeting of EGFR in conjunction with PI3K pathway inhibition.
Conclusion
In conclusion, we report an analysis of a large HPV-positive oropharyngeal SCC cohort and demonstrate distinct, but perhaps functionally homologous, mechanisms of PI3K pathway activation: PIK3CA mutations/amplification, HRAS mutation, or PTEN loss. We provide evidence, for the first time, of potentially activating genetic alterations of the PI3K signaling pathway in about 45% (34/75) of HPV-positive oropharyngeal SCC. The significance of the affected PIK3CA exon or specific PIK3CA mutation types, mechanism of PTEN loss, and the association with alternative mechanisms of PI3K signaling remain incompletely understood. Our findings offer a molecular basis for future studies of therapeutic targeting of PI3K pathway in HPV-positive oropharyngeal SCC.
Competing interests
P50CA97190 and the American Cancer Society (to JRG) and the Patricia L. Knebel Fund of the Pittsburgh Foundation (to VWYL).
Authors’ contributions
MM and AL carried out molecular genetic studies. SC, JRG, VWYL drafted the manuscript. SC, MNN conceived, designed the study, and acquired the data. SC performed the statistical analysis. All authors were involved in analysis and interpretation of data and critical revision of the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Representative cases of human papillomavirus-positive oropharyngeal squamous cell carcinoma, with HRAS (A) or PIK3CA mutations (B through I), sequencing electropherograms (SE). Portions of the SE surrounding the point mutation were scanned. A. HRAS c.182A>T, p.Q61L. B. PIK3CA, exon 9, c.1571G>A, p.R524K and c.1573G>A, p.E525K. C. PIK3CA, exon 20, c.3103G>A, p.A1035T. D. PIK3CA, exon 20, c.3129G>C, p.M1043I. E. PIK3CA, exon 20, c.3139C>T, p.H1047Y. F. PIK3CA, exon 20, c.3153G>A, p.W1051*.G. PIK3CA, exon 9, c.1633G>A, p.E545K. H. PIK3CA, exon 9, c.1610G>A, p.R537Q. I. PIK3CA, exon 20, c.2975G>A, p.R992Q.
Representative sequencing electropherograms (SE) illustrating PIK3CA mutations. The entirely scanned SE are presented to illustrate low background.
SNaPshot detection of common hotspot mutations in the exon 9 of the PIK3CA gene. SNaPshot result illustrating c. 1633G>A, p.E545K electropherogram pattern (mutant peak is indicated by a grey arrow). The bases are color-coded: “A” – green, while “G” – blue. The red trigon-shaped peaks represent internal size standards.
Representative images of PTEN and PIK3CA fluorescence in situ hybridization, FISH, original magnification 1000x. All cases included in this figure are oropharyngeal squamous cell carcinoma that are human papillomavirus-positive, HRAS wild-type, PIK3CA wild-type, without PIK3CA amplification. The nuclei were counterstained with DAPI/Antifade 1 (blue) (Vysis, Inc., Downers Grove, IL). A. PTEN FISH, representative field: most nuclei are characterized by two PTEN signals (orange) and two chromosomal enumeration probe 10 (CEP10) signals (green), consistent with normal PTEN copy number. B. PTEN FISH, representative field: 57.1% of cells (36/63) showed only one pair of PTEN and CEP10 signals, consistent with chromosome 10 monosomy. C. PTEN FISH, representative field: 90.2% (55/61) of analyzed cells showed no PTEN signal, consistent with homozygous PTEN loss. Also, 55.7% (34/61) of analyzed cells showed only one CEP 10 signal consistent with chromosome 10 monosomy. The PTEN/CEP10 ratio was 0.11. D. PIK3CA (green) and CEP3 (orange) FISH, representative field. The PIK3CA/CEP3 ratio is 2.5.
Contributor Information
Simion I Chiosea, Email: chioseasi@upmc.edu.
Jennifer R Grandis, Email: grandisjr@upmc.edu.
Vivian W Y Lui, Email: luivw@upmc.edu.
Brenda Diergaarde, Email: diergaardeb@upmc.edu.
Jessica H Maxwell, Email: maxwelljh@upmc.edu.
Robert L Ferris, Email: ferrisrl@upmc.edu.
Seungwon W Kim, Email: kimsw@upmc.edu.
Alyssa Luvison, Email: luvisona@upmc.edu.
Megan Miller, Email: millerm@upmc.edu.
Marina N Nikiforova, Email: nikiforovamn@upmc.edu.
Acknowledgements
The authors wish to thank members of the Molecular Anatomic Pathology and Developmental laboratories of the Department of Pathology, University of Pittsburgh, for excellent technical support and Robyn Roche for outstanding secretarial support. Dr. Chiosea would like to dedicate this paper to his parents, Ivan Chiosea and Feodora Chiosea, who are the source of determination and discipline.
References
- Woenckhaus J, Steger K, Werner E, Fenic I, Gamerdinger U, Dreyer T, Stahl U. Genomic gain of PIK3CA and increased expression of p110alpha are associated with progression of dysplasia into invasive squamous cell carcinoma. J Pathol. 2002;13(3):335–342. doi: 10.1002/path.1207. [DOI] [PubMed] [Google Scholar]
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;13(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- Murugan AK, Hong NT, Fukui Y, Munirajan AK, Tsuchida N. Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. Int J Oncol. 2008;13(1):101–111. [PubMed] [Google Scholar]
- Murugan AK, Munirajan AK, Tsuchida N. Ras oncogenes in oral cancer: the past 20 years. Oral Oncol. 2012;13(5):383–392. doi: 10.1016/j.oraloncology.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Qiu W, Tong GX, Manolidis S, Close LG, Assaad AM, Su GH. Novel mutant-enriched sequencing identified high frequency of PIK3CA mutations in pharyngeal cancer. Int J Cancer. 2008;13(5):1189–1194. doi: 10.1002/ijc.23217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W. et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;13(32):4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert J, Chiosea S. Incidence of human papillomavirus in oropharyngeal squamous cell carcinomas: now and 50 years ago. Hum Pathol. 2012;13(1):17–22. doi: 10.1016/j.humpath.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Pimkhaokham A, Hasegawa S, Tsuda H, Omura K, Inazawa J. PIK3CA mutation is an oncogenic aberration at advanced stages of oral squamous cell carcinoma. Cancer Sci. 2006;13(12):1351–1358. doi: 10.1111/j.1349-7006.2006.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won HS, Jung CK, Chun SH, Kang JH, Kim YS, Sun DI, Kim MS. Difference in expression of EGFR, pAkt, and PTEN between oropharyngeal and oral cavity squamous cell carcinoma. Oral Oncol. 2012;13(10):985–990. doi: 10.1016/j.oraloncology.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J. et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;13(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A. et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;13(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek V, Fu S, Piha-Paul SA, Lee JJ, Luthra R. et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early phase clinical trials. Cancer Res. 2013;13(1):276–284. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TA, Yan L, Patnaik A, Fearen I, Olmos D, Papadopoulos K, Baird RD, Delgado L, Taylor A, Lupinacci L. et al. First-in-man clinical trial of the oral pan-AKT inhibitor MK-2206 in patients with advanced solid tumors. J Clin Oncol. 2011;13(35):4688–4695. doi: 10.1200/JCO.2011.35.5263. [DOI] [PubMed] [Google Scholar]
- Nakabayashi M, Werner L, Courtney KD, Buckle G, Oh WK, Bubley GJ, Hayes JH, Weckstein D, Elfiky A, Sims DM. et al. Phase II trial of RAD001 and bicalutamide for castration-resistant prostate cancer. BJU Int. 2012;13(11):1729–1735. doi: 10.1111/j.1464-410X.2012.11456.x. [DOI] [PubMed] [Google Scholar]
- Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, Lu Y, Zhang Q, Du Y, Gilbert B. et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;13(7):761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Vanhaesebroeck B, Waterfield MD, Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. Embo J. 1996;13(10):2442–2451. [PMC free article] [PubMed] [Google Scholar]
- Espada J, Perez-Moreno M, Braga VM, Rodriguez-Viciana P, Cano A. H-Ras activation promotes cytoplasmic accumulation and phosphoinositide 3-OH kinase association of beta-catenin in epidermal keratinocytes. J Cell Biol. 1999;13(5):967–980. doi: 10.1083/jcb.146.5.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1998;13(26):15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;13(9):2166–2173. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]
- Prince ME, Ubell ML, Castro J, Ogawa H, Ogawa T, Narayan A, Paulino A, Cole A, Wolf GT, Rubin MA. et al. Tissue-preserving approach to extracting DNA from paraffin-embedded specimens using tissue microarray technology. Head Neck. 2007;13(5):465–471. doi: 10.1002/hed.20547. [DOI] [PubMed] [Google Scholar]
- Hurst CD, Zuiverloon TC, Hafner C, Zwarthoff EC, Knowles MA. A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res Notes. 2009;13:66. doi: 10.1186/1756-0500-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack IF, Hamilton RL, James CD, Finkelstein SD, Burnham J, Yates AJ, Holmes EJ, Zhou T, Finlay JL. Rarity of PTEN deletions and EGFR amplification in malignant gliomas of childhood: results from the Children’s Cancer Group 945 cohort. J Neurosurg. 2006;13(5 Suppl):418–424. doi: 10.3171/ped.2006.105.5.418. [DOI] [PubMed] [Google Scholar]
- Ettl T, Baader K, Stiegler C, Muller M, Agaimy A, Zenk J, Kuhnel T, Gosau M, Zeitler K, Schwarz S. et al. Loss of PTEN is associated with elevated EGFR and HER2 expression and worse prognosis in salivary gland cancer. Br J Cancer. 2012;13(4):719–726. doi: 10.1038/bjc.2011.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sircar K, Yoshimoto M, Monzon FA, Koumakpayi IH, Katz RL, Khanna A, Alvarez K, Chen G, Darnel AD, Aprikian AG. et al. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol. 2009;13(4):505–513. doi: 10.1002/path.2559. [DOI] [PubMed] [Google Scholar]
- Derakhshandeh-Peykar P, Alivi J, Hossein-nezhad A, Rautenstrauss B, Vesal RE, Doriani A. High frequency of mutations in the PIK3CA gene helical and kinase coding regions in a group of Iranian patients with high-grade glioblastomas: five novel mutations. J Neurogenet. 2011;13(4):189–194. doi: 10.3109/01677063.2011.623202. [DOI] [PubMed] [Google Scholar]
- Catasus L, Gallardo A, Cuatrecasas M, Prat J. PIK3CA mutations in the kinase domain (exon 20) of uterine endometrial adenocarcinomas are associated with adverse prognostic parameters. Mod Pathol. 2008;13(2):131–139. doi: 10.1038/modpathol.3800992. [DOI] [PubMed] [Google Scholar]
- Korkolopoulou P, Levidou G, Trigka EA, Prekete N, Karlou M, Thymara I, Sakellariou S, Fragkou P, Isaiadis D, Pavlopoulos P. et al. A comprehensive immunohistochemical and molecular approach to the PI3K/AKT/mTOR (phosphoinositide 3-kinase/v-akt murine thymoma viral oncogene/mammalian target of rapamycin) pathway in bladder urothelial carcinoma. BJU Int. 2012;13(11 Pt C):E1237–E1248. doi: 10.1111/j.1464-410X.2012.11569.x. [DOI] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;13(7308):869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- Kuo KT, Mao TL, Jones S, Veras E, Ayhan A, Wang TL, Glas R, Slamon D, Velculescu VE, Kuman RJ. et al. Frequent activating mutations of PIK3CA in ovarian clear cell carcinoma. Am J Pathol. 2009;13(5):1597–1601. doi: 10.2353/ajpath.2009.081000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Solano J, Conesa-Zamora P, Carbonell P, Trujillo-Santos J, Torres-Moreno DD, Pagan-Gomez I, Rodriguez-Braun E, Perez-Guillermo M. Colorectal serrated adenocarcinoma shows a different profile of oncogene mutations, MSI status and DNA repair protein expression compared to conventional and sporadic MSI-H carcinomas. Int J Cancer. 2012;13(8):1790–1799. doi: 10.1002/ijc.27454. [DOI] [PubMed] [Google Scholar]
- Sinha P, Lewis JS Jr, Piccirillo JF, Kallogjeri D, Haughey BH. Extracapsular spread and adjuvant therapy in human papillomavirus-related, p16-positive oropharyngeal carcinoma. Cancer. 2012;13(14):3519–3530. doi: 10.1002/cncr.26671. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Bifeld E, Stabley DL, Hopkins E, Meien S, Vinette K, Sol-Church K, Rosenberger G. A novel HRAS substitution (c.266C>G; p.S89C) resulting in decreased downstream signaling suggests a new dimension of RAS pathway dysregulation in human development. Am J Med Genet A. 2012;13(9):2106–2118. doi: 10.1002/ajmg.a.35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juanpere N, Agell L, Lorenzo M, de Muga S, Lopez-Vilaro L, Murillo R, Mojal S, Serrano S, Lorente JA, Lloreta J. et al. Mutations in FGFR3 and PIK3CA, singly or combined with RAS and AKT1, are associated with AKT but not with MAPK pathway activation in urothelial bladder cancer. Hum Pathol. 2012;13(10):1573–1582. doi: 10.1016/j.humpath.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mavros A, Hahn M, Wieland I, Koy S, Koufaki ON, Strelocke K, Koch R, Haroske G, Schackert HK, Eckelt U. Infrequent genetic alterations of the tumor suppressor gene PTEN/MMAC1 in squamous cell carcinoma of the oral cavity. J Oral Pathol Med. 2002;13(5):270–276. doi: 10.1034/j.1600-0714.2002.310504.x. [DOI] [PubMed] [Google Scholar]
- Lee JI, Soria JC, Hassan KA, El-Naggar AK, Tang X, Liu DD, Hong WK, Mao L. Loss of PTEN expression as a prognostic marker for tongue cancer. Arch Otolaryngol Head Neck Surg. 2001;13(12):1441–1445. doi: 10.1001/archotol.127.12.1441. [DOI] [PubMed] [Google Scholar]
- Suda T, Hama T, Kondo S, Yuza Y, Yoshikawa M, Urashima M, Kato T, Moriyama H. Copy number amplification of the PIK3CA gene is associated with poor prognosis in non-lymph node metastatic head and neck squamous cell carcinoma. BMC Cancer. 2012;13:416. doi: 10.1186/1471-2407-12-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Weinberger PM, Sasaki C, Egleston BL, Speier WF, Haffty B, Kowalski D, Camp R, Rimm D, Vairaktaris E. et al. Phosphorylation of Akt (Ser473) predicts poor clinical outcome in oropharyngeal squamous cell cancer. Cancer Epidemiol Biomarkers Prev. 2007;13(3):553–558. doi: 10.1158/1055-9965.EPI-06-0121. [DOI] [PubMed] [Google Scholar]
- Erlich RB, Kherrouche Z, Rickwood D, Endo-Munoz L, Cameron S, Dahler A, Hazar-Rethinam M, de Long LM, Wooley K, Guminski A. et al. Preclinical evaluation of dual PI3K-mTOR inhibitors and histone deacetylase inhibitors in head and neck squamous cell carcinoma. Br J Cancer. 2012;13(1):107–115. doi: 10.1038/bjc.2011.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles JA, Golden B, Yan L, Carroll WR, Helman EE, Rosenthal EL. Disruption of the AKT pathway inhibits metastasis in an orthotopic model of head and neck squamous cell carcinoma. Laryngoscope. 2011;13(11):2359–2365. doi: 10.1002/lary.22180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F, Wheler JJ, Westin SN, Moulder SL, Naing A, Tsimberidou AM, Fu S, Falchook GS, Hong DS, Garrido-Laguna I. et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;13(8):777–782. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandis JR, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, Drenning SD, Tweardy DJ. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;13(11):824–832. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- Zhang X, Guo A, Yu J, Possemato A, Chen Y, Zheng W, Polakiewicz RD, Kinzler KW, Vogelstein B, Velculescu VE. et al. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc Natl Acad Sci USA. 2007;13(10):4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhola NE, Thomas SM, Freilino M, Joyce S, Sahu A, Maxwell J, Argiris A, Seethala R, Grandis JR. Targeting GPCR-mediated p70S6K activity may improve head and neck cancer response to cetuximab. Clin Cancer Res. 2011;13(15):4996–5004. doi: 10.1158/1078-0432.CCR-10-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhola NE, Freilino ML, Joyce SC, Sen M, Thomas SM, Sahu A, Cassell A, Chen CS, Grandis JR. Antitumor mechanisms of targeting the PDK1 pathway in head and neck cancer. Mol Cancer Ther. 2012;13(6):1236–1246. doi: 10.1158/1535-7163.MCT-11-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative cases of human papillomavirus-positive oropharyngeal squamous cell carcinoma, with HRAS (A) or PIK3CA mutations (B through I), sequencing electropherograms (SE). Portions of the SE surrounding the point mutation were scanned. A. HRAS c.182A>T, p.Q61L. B. PIK3CA, exon 9, c.1571G>A, p.R524K and c.1573G>A, p.E525K. C. PIK3CA, exon 20, c.3103G>A, p.A1035T. D. PIK3CA, exon 20, c.3129G>C, p.M1043I. E. PIK3CA, exon 20, c.3139C>T, p.H1047Y. F. PIK3CA, exon 20, c.3153G>A, p.W1051*.G. PIK3CA, exon 9, c.1633G>A, p.E545K. H. PIK3CA, exon 9, c.1610G>A, p.R537Q. I. PIK3CA, exon 20, c.2975G>A, p.R992Q.
Representative sequencing electropherograms (SE) illustrating PIK3CA mutations. The entirely scanned SE are presented to illustrate low background.
SNaPshot detection of common hotspot mutations in the exon 9 of the PIK3CA gene. SNaPshot result illustrating c. 1633G>A, p.E545K electropherogram pattern (mutant peak is indicated by a grey arrow). The bases are color-coded: “A” – green, while “G” – blue. The red trigon-shaped peaks represent internal size standards.
Representative images of PTEN and PIK3CA fluorescence in situ hybridization, FISH, original magnification 1000x. All cases included in this figure are oropharyngeal squamous cell carcinoma that are human papillomavirus-positive, HRAS wild-type, PIK3CA wild-type, without PIK3CA amplification. The nuclei were counterstained with DAPI/Antifade 1 (blue) (Vysis, Inc., Downers Grove, IL). A. PTEN FISH, representative field: most nuclei are characterized by two PTEN signals (orange) and two chromosomal enumeration probe 10 (CEP10) signals (green), consistent with normal PTEN copy number. B. PTEN FISH, representative field: 57.1% of cells (36/63) showed only one pair of PTEN and CEP10 signals, consistent with chromosome 10 monosomy. C. PTEN FISH, representative field: 90.2% (55/61) of analyzed cells showed no PTEN signal, consistent with homozygous PTEN loss. Also, 55.7% (34/61) of analyzed cells showed only one CEP 10 signal consistent with chromosome 10 monosomy. The PTEN/CEP10 ratio was 0.11. D. PIK3CA (green) and CEP3 (orange) FISH, representative field. The PIK3CA/CEP3 ratio is 2.5.