Abstract
Exposing the brain to sublethal ischemia affects the response to a subsequent, otherwise injurious ischemia, resulting in transcriptional suppression and neuroprotection, a response called ischemic tolerance. Here, we show that the proteomic signature of the ischemic-tolerant brain is characterized by increased abundance of transcriptional repressors, particularly polycomb group (PcG) proteins. Knocking down PcG proteins precluded the induction of ischemic tolerance, whereas in an in vitro model, overexpressing the PcG proteins SCMH1 or BMI1 induced tolerance to ischemia without preconditioning. We found that PcG proteins are associated with the promoter regions of genes encoding two potassium channel proteins that show decreased abundance in ischemic-tolerant brains. Furthermore, PcG proteins decreased potassium currents in cultured neuronal cells and knocking down potassium channels elicited tolerance without preconditioning. These findings reveal a previously unknown mechanism of neuroprotection that involves gene repressors of the PcG family.
INTRODUCTION
Brain ischemia, resulting from either permanent or transient impaired blood flow, produces injury in the ischemic region of the brain. The clinical features of ischemic brain injury or “stroke” are pertinent to the region of ischemia (for instance, weakness of the face, arm, and leg on the side of the body contralateral to ischemia of the middle cerebral artery supplying parietal cortex). However, if it has previously been exposed to brief, noninjurious ischemia (preconditioning ischemia), the brain can undergo a prolonged ischemic insult without incurring injury (a phenomenon known as ischemic tolerance).
Identifying effectors of ischemic tolerance, as one approach to developing new strategies to treat stroke, has previously been undertaken at the transcriptional level (1).
The ischemic-tolerant brain shows an overall suppression of gene expression in response to prolonged ischemia (1). Genes down-regulated in ischemic-tolerant brains include those that encode proteins that have been implicated in regulation of metabolism, molecular transport, immune defense, and cell-cycle control. Such a profile of suppressed gene expression, which is not seen in ischemic-preconditioned or ischemic-injured brains but only in ischemic-tolerant brains, has been suggested to be a hibernation pattern (1). The molecular mechanism that underlies the ischemic-tolerant–specific suppression of gene expression is not known, although a repressive epigenetic mechanism can be assumed given the broad range of genes that are down-regulated.
The physiological characteristics of ischemic-tolerant brains remain to be described. A feature of ischemic-tolerant cortical neurons, as determined in an in vitro model of ischemia, is the suppression of outward potassium currents (1). Furthermore, genomic analyses have shown that the expression of Kcna5, the gene encoding voltage-gated potassium channel subunit 5 (KCNA5, also known as Kv1.5), is decreased in ischemic-tolerant brain (1). Thus, it can be anticipated that the target genes under the regulation of the repressive epigenetic mechanisms assumed above may include some that regulate channel activity, especially that of potassium channels, which provide the dominant contribution to outward currents.
Here, we performed an unbiased quantitative proteomic characterization of ischemic-preconditioned, ischemic-tolerant, and ischemic-injured mouse brains with the goal of identifying potential effectors of brain ischemic tolerance. We found that proteins that were uniquely up-regulated in ischemic-tolerant brain (proteins that increased in abundance only in ischemic-tolerant brain) included a number of gene repressor proteins, namely, histone H2A and H2B variants, and polycomb group (PcG) protein SCMH1, a mammalian homolog of the Drosophila PcG protein Sex comb on midleg (2–4). Bioinformatic analyses of proteins with increased abundance in ischemic-tolerant brains revealed an association of those proteins with chromatin remodeling processes, suggesting that these proteins might mediate repressive genomic reprogramming and, thereby, the induction of brain ischemic tolerance.
Other than a possible involvement in chromatin modification at the XY chromatin domain (2), the function of SCMH1 in mammalian cells is unknown. SCMH1 is a constituent of polycomb repressive complex 1 (PRC1) (2), which has several ring finger proteins and possesses histone H2A mono-ubiquitination activity (2, 5, 6), and operates in a repressive machinery that involves several PcG protein complexes [for general reviews on PcG protein–mediated transcriptional regulation, see (7–10)]. Although PcG proteins were initially thought to regulate the transcription of only a few genes, including the homeotic (Hox) genes, the list of PcG-targeted genes has expanded in recent years to include genes encoding proteins involved in electron transport and glucose transport, as well as endopeptidases, oxidoreductases, and G protein–coupled receptors (11, 12). A recent study by Lee and colleagues shows that the PRC1 protein BMI1 protects against chemical stress–induced cell death (13). Another study by Chatoo et al. suggests an antiapoptotic role for BMI1 through the regulation of the p53-mediated cell death pathway (14). The implication of the PRC1 proteins in antiapoptotic pathways, together with the increased abundance of SCMH1 in ischemic-tolerant brain, led us to investigate the potential involvement of PcG proteins, with a focus on PRC1 proteins, in the induction of brain ischemic tolerance.
To accommodate studies on molecular mechanisms that underlie PcG protein–mediated ischemic tolerance, we established an in vitro model of ischemic tolerance involving oxygen-glucose deprivation (OGD) of differentiated NS20Y cells, a mouse brain–derived neuroblastoma cell line that differentiates into a neuronal phenotype when treated with adenosine 3′,5′-monophosphate (cAMP) analogs (15, 16). Previous studies have investigated tolerance to OGD in primary cortical neuronal cultures (1); however, NS2OY cells provide an advantage over primary cultures in that their protein abundance can be readily manipulated through transfection of small interfering RNA (siRNA) or recombinant complementary DNA (cDNA). In this cell line, a 2-hour period of OGD results in protein degradation and blockade of normal protein biosynthesis (17). Here, we determined that 2-hour OGD results in a 40 to 50% decrease of cell viability (and is thus injurious); 30-min OGD can effectively induce OGD tolerance if performed 24 hours before the injurious 2-hour OGD, a paradigm essentially identical to that used for inducing tolerance in primary cortical neuron cultures (1).
Following leads from proteomic analyses and using both in vivo and in vitro models of ischemic tolerance, we found the following: (i) In addition to the changes in H2A and SCMH1, the abundance of BMI1 and RING2, two other PRC1 proteins, also increased in ischemic tolerance. (ii) Knocking down SCMH1 or BMI1 blocked tolerance both in vivo and in vitro, whereas overexpressing either of these proteins induced tolerance without preconditioning. (iii) Under- or overexpressing PcG proteins in cultured neuronal cells respectively increased or decreased the activity of voltage-gated potassium channels, known to be decreased in ischemic-tolerant neurons. (iv) PcG proteins interacted with the promoter regions of the genes encoding two potassium channel proteins (which showed decreased abundance in ischemic-tolerant brains; chosen according to results from the previous genomic study (1) and the current study), as determined by chromatin immunoprecipitation (ChIP) assays. And, (v), knocking down potassium channel protein abundance protected neuronal cells from ischemic injury. These findings collectively point to PcG proteins as being involved in acquiring or sustaining (or both) the ischemic-tolerant condition in brain. Broadly, this involves programmed transcriptional suppression; specifically, we have identified genes encoding potassium channel proteins as one pertinent target of this transcriptional suppression.
RESULTS
Proteomic characteristics of ischemic-tolerant brains
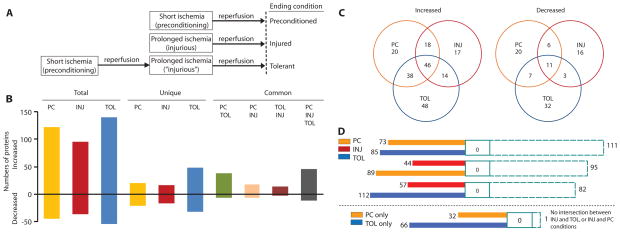
We used a middle cerebral artery occlusion (MCAO) experimental paradigm for studies of ischemic tolerance (Fig. 1A). Using a label-free, quantitative mass spectrometry (MS) system, we compared the abundance of 562 identified proteins in ischemic-preconditioned, ischemic-tolerant, and ischemic-injured brains, respectively, to that in sham-operated brains (Fig. 1B and tables S1 to S6). There was less overlap between ischemic-tolerant and ischemic-injured brains than between ischemic-tolerant and ischemic-preconditioned brains, or between ischemic-preconditioned and ischemic-injured brains for both proteins increased in abundance relative to that in sham-operated brain (up-regulated proteins) and those decreased in abundance (down-regulated proteins) (Fig. 1C). Bioinformatic analysis for possible intersections of cellular networks (see Materials and Methods), as presented by a data set, between or among data sets of proteins that were uniquely up-regulated under each brain ischemic condition, revealed little overlap between the conditions (Fig. 1D). This argues that proteins uniquely regulated in ischemic-tolerant brains are candidates for effectors of brain ischemic tolerance. Further bioinformatic analysis of proteins that increased in abundance only in ischemic-tolerant brains revealed a highly significant association of these proteins with processes involved in nucleosome assembly and chromatin assembly (Table 1; further details and P values are in table S7). Up-regulated proteins that contribute to these processes included several histone H2A and H2B variants and SCMH1 (Table 2). The specific and coordinate increase in the abundance of H2A and SCMH1 in ischemic-tolerant brains led us to investigate whether PcG proteins of PRC1 complex might be effectors of brain ischemic tolerance.
Fig. 1.
Proteomic signature of ischemic-tolerant brains. (A) Experimental paradigm. In mice, 15-min MCAO (preconditioning ischemia) followed by 72-hour reperfusion protects the cortex from ischemic injury when the preconditioned brain is subjected to 1-hour MCAO. Without preconditioning ischemia, 1-hour MCAO causes massive cortical and striatal infarction (1). (B) Numbers of cortical proteins showing changes in abundance under various ischemic conditions, as determined by highly reproducible, quantitative MS analyses (fig. S1). Most of the proteins analyzed (~400) did not show a change in abundance under any given condition. Complete lists of proteins and their quantities under various conditions are provided in tables S1 and S2 to S6. (C) Venn diagram of proteins that changed in abundance. (D) Intersection of data sets involving proteins that increased in abundance (top, including proteins that increased in abundance in more than one ischemic condition; bottom, involving proteins that increased in abundance only under a specific ischemic condition). This figure presents numbers of network objects (see Materials and Methods for definition) that are unique to a particular ischemic condition (color-filled), associated with two of three conditions (open) or all three conditions (dashed line). PC, ischemic-preconditioned; INJ, ischemic-injured; TOL, ischemic-tolerant.
Table 1.
Biological processes mediated by proteins with increased abundance in ischemic-tolerant brain. Results of bioinformatic analyses for all proteins that change in abundance under different brain ischemic conditions (with statistic relevance as indicated by calculated P values) are provided in table S7.
| Proteins increased in tolerant brain* | Proteins increased in tolerant brain only |
|---|---|
| Small GTPase-mediated signal transduction | Nucleosome assembly |
| Protein localization | Nucleosome organization |
| Establishment of protein localization | Chromatin assembly |
| Macromolecule localization | DNA packaging |
| Intracellular signaling cascade | Chromatin assembly or disassembly |
| Protein transport | Protein-DNA complex assembly |
| Establishment of localization | Cellular component assembly |
| Localization | Cellular macromolecular complex assembly |
| Transport | Localization |
| Vesicle-mediated transport | Protein transport |
Includes proteins with increased abundance in other ischemic conditions as well. Processes were defined by Gene Ontology (GO) terms. Proteins are matched to GO terms with the use of the MetaCore program. Processes listed in the table are those statistically most relevant (with the smallest P values).
Table 2.
Nuclear proteins that increased in abundance only in ischemic-tolerant brain.
| Genes | Protein names |
|---|---|
| Gadd45gip1 | Growth arrest and DNA-damage–inducible proteins–interacting protein 1 |
| Hist1h2ah | Histone H2A type 1-H |
| Hist2h2ab | Histone H2A type 2-B |
| Hist1h2ba | Histone H2B type 1-A |
| Hist2h2be | Histone H2B type 2-E |
| Hist3h2bb | Histone H2B type 3-B |
| Ran | GTP-binding nuclear protein Ran |
| Rhoc | Rho-related GTP-binding protein RhoC |
| Scmh1 | Polycomb protein Scmh1 |
| Sep10 | Septin-10 |
| Cct1 | T-complex protein 1 subunit alpha A and B |
Changes in abundance of PRC1 complex proteins under different brain ischemic conditions
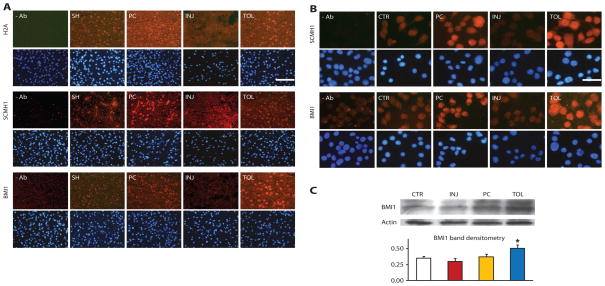
By immunohistochemical analyses, H2A and SCMH1 showed increased immunoreactivity in ischemic-preconditioned and ischemic-tolerant brains. BMI1 also demonstrated an increase in both ischemic-preconditioned and ischemic-tolerant brains, with changes in the latter more robust than in the former (Fig. 2A). H2A and PcG abundance in ischemic-tolerant brains increased primarily in cortical neurons (fig. S2), and neuronal H2A and PcG abundance increased primarily in the tolerant region (fig. S2). In ischemic-injured brains, however, the abundance of these proteins was greatly decreased (Fig. 2A).
Fig. 2.
Differential changes of representative gene repressor proteins under different ischemic conditions in brains and cultured neuronal cells. (A) Immunohistochemical analysis of H2A, SCMH1, and BMI1 showed increased immunoreactivity (red) in ischemic-preconditioned (PC) and ischemic-tolerant (TOL) brains, relative to that in sham brains (SH), but decreased immunoreactivity in injured brains (INJ). Mice were subjected to the MCAO treatment paradigm described in Fig. 1. Bottom rows show DAPI staining (blue) to reveal nuclei. At least two brains of each condition were analyzed with similar results. The scale bar represents 50 μm. (B and C) Increased immunoreactivity of SCMH1 and BMI1 in NS20Y cells under simulated ischemic-tolerant (OGD-tolerant) conditions. Four groups of differentiated NS20Y cells were treated with the following conditions: (1) control (CTR); (2) preconditioning (PC); (3) injurious (INJ); (4) tolerant (TOL) (details are in Materials and Methods). Cells were then fixed and examined by immunocytochemistry for SCMH1 or BMI1 (B). BMI1 was also analyzed by Western blot (Western blot of actin serves as a loading control), with parallel analysis of actin (C, bottom) as a loading control. Similar results were obtained in at least three independent cultures. Scale bar, 25 μm. *P < 0.05.
Figure 2, B and C, shows that, in NS20Y cells in vitro, the abundance of SCMH1 and BMI1 increased robustly in OGD-tolerant cells but decreased in OGD-injured cells.
Regulation of in vitro and in vivo induction of ischemic tolerance by PcG proteins
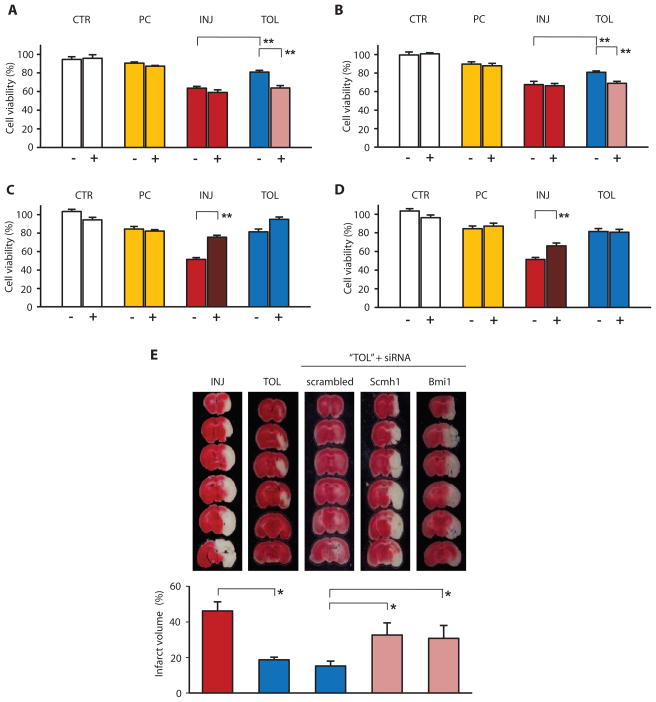
We investigated the requirement for PcG proteins in ischemic tolerance using siRNA directed against Scmh1 or Bmi1 messenger RNA (mRNA) to knock down the abundance of SCMH1 and BMI1. Knocking down SCMH1 (Fig. 3A) or BMI1 (Fig. 3B) ablated tolerance to OGD in NS20Y cells. In contrast, overexpressing SCMH1 or BMI1 proteins by transfecting cells with recombinant Scmh1 or Bmi1 protected NS20Y cells from OGD-induced injury even in the absence of preconditioning (Fig. 3, C and D). The essential role of SCMH1 and BMI1 in mediating ischemic tolerance was further demonstrated in vivo. Intracerebroventricular administration of siRNA directed against Scmh1 or Bmi1 mRNA resulted in a significant increase in brain tissue infarction in mice, in what would otherwise have been ischemic-tolerant brains (Fig. 3E). The similar outcomes of manipulating SCMH1 and BMI1 abundance, both in vitro and in vivo, suggest that PcG proteins of PRC1 complex are prominent effectors of brain ischemic tolerance.
Fig. 3.
Dependence of ischemic tolerance on PcG protein abundance in vitro and in vivo. Before differentiation and OGD, NS20Y cells were transiently transfected with scrambled RNA (−) or siRNA oligonucleotides (+) directed against Scmh1 (A) or Bmi1 (B), or stably transfected with plasmid encoding recombinant Scmh1 (C) or Bmi1 (D), respectively. OGD-induced injury was assessed by cell viability (percent to control cells). Data represent mean ± SE of at least three independent cultures. A 40 to 50% decrease in viability was caused by 2-hour OGD, which could be prevented by a preceding 30-min OGD. (E) Effects of knocking down PcG expression on ischemic tolerance in vivo. One-hour MCAO caused massive infarction of the mouse cortex (INJ), which was avoided when the brain was exposed to a 15-min preconditioning ischemia (TOL). Administration of siRNA directed against Scmh1 or Bmi1, but not scrambled RNA, significantly diminished the preconditioning ischemia-induced ischemic tolerance. Tissue infarction was revealed (top) and quantified (bottom; mean ± SE, n = 5 to 8 animals each group) by vital dye staining of fresh brain slices. Altered PcG protein abundance with the application of siRNA or ORF cDNA are demonstrated in fig. S3. *P < 0.05; **P < 0.005.
PcG proteins and regulation of potassium channels
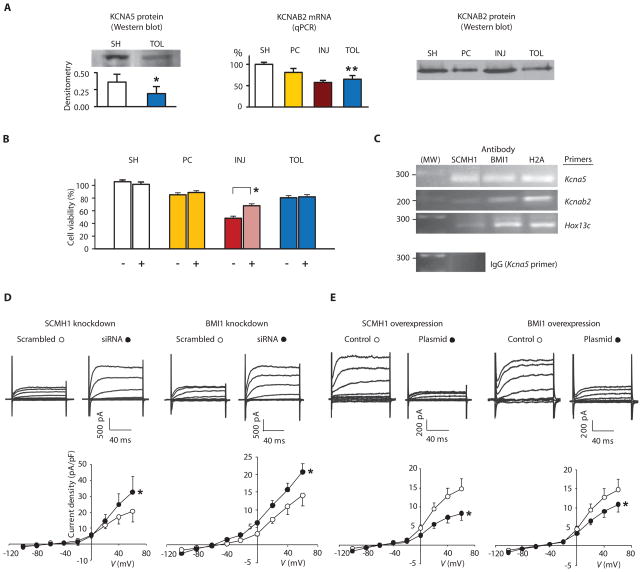
If an essential role for PcG proteins in brain ischemic tolerance exists, what are the target genes that PcG proteins regulate under ischemic-tolerant conditions? Although a complete answer will rely on a high-throughput screening, we pursued one possibility suggested by existing data. A bioinformatic analysis of potential PcG target genes indicated genes associated with potassium channel activity (8). Moreover, as previously noted, the expression level of Kcna5 is decreased in ischemic tolerance (1), and reduced outward potassium currents have been observed in ischemic-tolerant neurons in vitro (1). In the present work, we found decreased abundance of KCNA5 protein in ischemic-tolerant brains (Fig. 4A). Furthermore, quantitative polymerase chain reaction (qPCR) and Western blotting, respectively, revealed a decrease in mRNA and protein abundance of voltage-gated potassium channel subunit β2 (Kcnab2 and KCNAB2, respectively), in ischemic-tolerant brains as well (Fig. 4A). KCNAB2 is known to co-assemble with KCNA5 and can accelerate KCNA5 activation (18, 19).
Fig. 4.
Implication of potassium channel suppression in ischemic tolerance and inhibition of K+ current by PcG proteins. (A) Analyses of potassium channel abundance in brain. Left, KCNA5; middle and right, KCNAB2. SH, PC, INJ, and TOL: as noted in Fig. 1. (B) Effects of knocking down potassium channels on tolerance. NS20Y cells were transfected with scrambled RNA (−) or a mixture of siRNA oligos (+) against Kcna5 and Kcnab2, followed by differentiation and OGD treatments as described in Figs. 2B and 3, A and B. Data are presented as mean ± SE of at least three independent cultures. (C) Results of ChIP assays of NS20Y cells, with antibodies noted in the figure and PCR primers, also noted in the figure, for the promoter region of Kcna5, Kcnab2, or Hox13c. Predicted sizes for PCR fragments are 270, 184, and 271 base pairs for Kcna5, Kcnab2, and Hox13c, respectively. Antibody against H2A and Hox13c primers were included as positive controls and immunoglobulin G (IgG) as a negative control. The analyses were repeated twice on two independent cultures with similar results. (D and E) Effects of knocking down (D) or overexpressing (E) SCMH1 or BMI1 on potassium currents. NS20Y cells were transfected with siRNA oligos or plasmids as noted in the figure and differentiated (n = 6 to 8 each condition). *P < 0.05; **P < 0.005.
These observations prompted us first to determine whether the abundance of potassium channel proteins may play a role in ischemic tolerance. We found that when KCNA5 and KCNAB2 abundance was decreased through knockdown with siRNAs directed against Kcna5 and Kcnab2 mRNA, NS20Y cells showed increased viability under ischemic conditions (that is, tolerance to OGD) without preconditioning (Fig. 4B). We then determined whether PcG proteins interacted with the promoter region of Kcna5 or Kcnab2 and whether changes in PcG protein abundance can result in alteration of outward potassium currents. ChIP assays with antibodies against SCMH1 or BMI1 indicated that both SCMH1 and BMI1 bound to the promoter regions of Kcna5 and Kcnab2 (Fig. 4C). Finally, when SCMH1 or BMI1 in NS20Y cells were knocked down with siRNA or overexpressed through transfection of recombinant Scmh1 or Bmi1, opposing changes in potassium currents were observed (Fig. 4, D and E). SCMH1 or BMI1 knockdown increased potassium currents, whereas their overexpression decreased them.
DISCUSSION
Although ischemic tolerance in brain depends on protein synthesis, ischemic-tolerant brains are marked by an overall decrease in gene transcription (1). Our quantitative proteomic characterization of ischemic-preconditioned, ischemic-tolerant, and ischemic-injured brains has identified the proteomic signature of ischemic tolerance as an increase in the abundance of transcriptional repressors (Fig. 1 and Tables 1 and 2). Specifically, we found a coordinate increase in the abundance of several PcG proteins and histone proteins (Table 2 and Fig. 2) in ischemic-tolerant brains. These proteins work together to achieve epigenetic repression of the genome (7–10). Thus, the dependence of ischemic tolerance on PcG protein abundance (Fig. 3), PcG proteins’ actions on PcG protein targets that showed decrease in abundance in tolerance (Fig. 4), and the induction of tolerance by direct knocking down of potential PcG protein targets (Fig. 4) suggest that PcG protein–mediated transcriptional repression is a central effector mechanism of brain ischemic tolerance.
The current proteomic data agree with the genomic understanding of tolerance. At the genomic level, ischemic-tolerant brains are characterized by a global decrease in gene expression. Using full genome screening, we and others have shown that the expression of arrays of genes is up-regulated in ischemic brains. However, in brains preconditioned to produce tolerance, ischemia induces a global decrease in transcription (1). Thus, the neuroprotective phenotype of the tolerant brain is characterized by transcriptional suppression. How this transcriptional suppression is accomplished in tolerance has been unknown. Furthermore, the fact that transcriptional suppression results in tolerance despite tolerance being protein synthesis–dependent (20) has remained a paradox. Here, we demonstrate increased abundance of gene repressor proteins in the tolerant proteome. These proteomic findings, together with the published genomic data, are mutually supportive in describing a mechanism whereby endogenous neuroprotection (tolerance) is induced by transcriptional suppression. Thus, the combination of transcriptional suppression with the protein synthesis dependence of tolerance is no longer a paradox.
Our results uncover a previously unknown role for PcG proteins, the mediation of an endogenous neuroprotective mechanism that can be induced by preconditioning ischemia.
In both mouse brains and cultured neuronal cells, the changes in abundance of the PcG proteins SCMH1 and BMI1 appear to be ischemic condition–specific; their abundance increased under ischemic-preconditioned and ischemic-tolerant conditions (a change more robust in the latter) but decreased under conditions of ischemic injury (Fig. 2). PcG proteins play a role in genomic reprogramming during development and differentiation (7). In mouse brains, the abundance of PcG gene transcripts and proteins undergoes dynamic changes during development (21). These, together with our present results, suggest that the expression of PcG proteins is subject to change under both physiological and pathophysiological conditions. The temporal order of changes in PcG protein abundance, and that of their gene transcripts, under different brain ischemic conditions remains to be characterized. The abundance of the few PcG proteins that we studied here decreased in ischemic-injured brains or cells. Although the implications of such a decrease remain to be elucidated, the current results indicate that a sustained amount of PcG proteins is crucial to the survival of brain cells under ischemic conditions.
The molecular mechanism that underlies PcG protein–mediated brain ischemic tolerance remains to be fully characterized. PcG proteins were first described for their roles in pattern formation and differentiation during Drosophila melanogaster development in the 1970s (22, 23). Thereafter, a connection between PcG genes, including Bmi1, and cancer development has been observed and oncogenic roles have been proposed for PcG proteins [for reviews, see (24–26)]. Knocking down BMI1 results in cell death in several human cancer cell lines such as mammary epithelial adenocarcinoma, metastatic neuroblastoma, and ovary adenocarcinoma cells (27). PcG proteins may suppress p53-mediated apoptosis in neurons, as suggested by a study describing increased p19Arf/p53 abundance and increased vulnerability to oxidative stress in Bmi1-deficient mice and cultured Bmi1-deficient neurons (14). Our current results support the existence of a cell survival role for PcG protein–mediated transcriptional repression that occurs during tolerance to acute ischemic stress, with gene suppression resulting in the neuroprotection of ischemic tolerance.
Besides regulating known cell death pathways, such as p53-mediated pathway (mentioned above), PcG proteins may exert their protective role through suppressive regulation of potassium channels in brain and in cultured neurons. Channel arrest (suppression of cell membrane permeability to ions) is a phenomenon in ischemic tolerance of hibernation (1). Reduced potassium channel abundance results in decreased K+ efflux, thereby decreasing energy consumption necessary for restoring the normal potassium gradient across the plasma membrane [for instance, through the Na+- and K+-dependent adenosine triphosphatase (Na+,K+-ATPase)]. Such energy conservation during oxygen limitation has been suggested to be a mechanism underlying ischemic tolerance (1). A reciprocal relationship between abundance of the potassium channel subunit KCNA5 and apoptotic resistance exists in cancer cells (28). Furthermore, blocking potassium channels can attenuate hypoxia- or glutamate-induced apoptosis in cultured neurons in vitro (29, 30), or ischemia-induced brain injury in vivo (29, 30). Thus, it is not surprising that we found decreased gene expression (1) (Fig. 4), protein abundance (Fig. 4), and activity (1) for potassium channels under ischemic-tolerant conditions. A critical issue, then, is whether these potassium channel proteins are under the regulation of PcG proteins. As previously noted, bioinformatic analyses of the results of high-throughput genomic screens have identified potassium channel genes as possible PcG protein targets (8). Remarkably, genes encoding potassium channels constitute nearly 3% of potential PcG target genes. We found that potassium channel activity can be regulated, as anticipated, by regulating the abundance of PcG proteins (Fig. 4, D and E), and demonstrated an interaction between PcG proteins and the promoter regions of the genes encoding KCNA5 and KCNAB2 (Fig. 4C). Our demonstration that knocking down KCNA5 and KCNAB2 with siRNA produced OGD tolerance in the absence of preconditioning (Fig. 4B), just as overexpressing PcG proteins did (Fig. 3, C and D), argues that potassium channel activity plays a critical role in the induction of ischemic tolerance.
Much remains to be determined. What other proteins might be under the regulation of PcG proteins during the induction of brain ischemic tolerance? Are other potassium channel proteins also involved? A high-throughput genomic screen of PcG targets under different brain ischemic conditions could help to address these questions. Are other PcG proteins also involved in regulating the induction of brain ischemic tolerance? The current understanding of PcG protein–mediated gene repression involves the concerted action of multiple PRC complexes. In addition to SCMH1 and BMI1, our data show a robust increase in the abundance of RING2 (fig. S4), another PRC1 component with H2A ubiquitin E3 ligase activity (31, 32) in ischemic-tolerant brains and in cells in which mono-ubiquitinated H2A also increased. A number of reports have implicated PRC2 protein EZH2 in cancer cell apoptosis and have indicated a possible regulatory role of EZH2 on Bmi1 (33, 34), suggesting that EZH2 might be a candidate for investigation of a role in regulating brain ischemic tolerance. Hence, future studies will likely reveal the involvement of other PcG proteins in ischemic tolerance of brain.
In summary, the increase in PcG protein abundance in ischemic tolerance, which initiates transcriptional repression, reconciles ischemic-tolerant–specific changes at the genomic, proteomic, and physiological levels. This explains the paradox that ischemic tolerance in brain is characterized by transcriptional suppression but depends on translation.
MATERIALS AND METHODS
Cerebral ischemic tolerance in mice
Transient focal cerebral ischemia was induced by MCAO in mice (C57BL/6J, male, 20 to 25 g; Charles River Laboratories) with the suture method, as previously described (1). Four groups of animals (n = 7 each) were subjected to different periods of MCAO and reperfusion as follows: sham-operated: 0 hours of MCAO, 24 hours of reperfusion; ischemic-preconditioned: 15 min of MCAO, 72 hours of reperfusion; ischemic-injured: 60 min of MCAO, 24 hours of reperfusion; ischemic-tolerant: 15 min of MCAO, 72 hours of reperfusion, followed by another 60 min of MCAO and 24 hours of reperfusion. At the termination of reperfusion, under anesthesia, animals were decapitated and whole brains were removed. For proteomic analyses (described next), ipsilateral cortices from ischemic cortical regions of MCAO territory were dissected, frozen on dry ice, and kept at −80°C for further analyses.
Protein extraction and tryptic digestion
Individual cortices were homogenized with RIPA (radioimmunoprecipitation assay) buffer [50 mM tris (pH 8.0), 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, and a cocktail of protease inhibitors]. Homogenates were centrifuged at 10,000g for 10 min at 4°C, and protein concentrations of cleared supernatants were determined with the Bradford method. For each experimental group, supernatants containing equal amounts of proteins from each animal were pooled, and 30 μl of each pooled sample (~210 μg proteins) was exchanged with 50 mM ammonium bicarbonate five times with a 5-kD ultrafiltration device. Proteins were denatured (0.06% RapiGest at 60°C for 15 min) (35), reduced (5 mM dithiothreitol at 60°C for 30 min), and alkylated (10 mM iodoacetamide for 30 min at room temperature in the dark). Samples were then digested with sequencing-grade trypsin (Promega) (0.05 μg/μl, 37°C overnight). Digestion was terminated by the addition of 1 μl of 5% trifluoroacetic acid (TFA), which also destroyed RapiGest in the incubation.
Liquid chromatography and MS
Yeast alcohol dehydrogenase (ADH; P00330) was added to the digests (50 fmol/μl) as an internal standard for absolute quantitation (36). Nanoflow separations of tryptic peptides were performed on a nanoACQUITYultra performance liquid chromatography (UPLC) system (Waters) equipped with a 5-μl sample loop and a 75-μm by 150-mm analytical column packed with 1.7 μm bridged-ethyl hybrid (BEH) C18. Column temperature was maintained at 35°C. Partial loop injections were performed. Samples were injected in a randomized order with three 1-μl injections of each sample to establish an optimized loading amount, after which another three injections of the same sample were performed. Eventually, all samples were analyzed at ~500 ng per run. The UPLC system was equilibrated with 0.1% formic acid in water (mobile phase A) containing 5% mobile phase B [0.1% formic acid in acetonitrile (ACN)]. The separation of injected tryptic peptides was achieved by application of a gradient of mobile phase B, as detailed in the legend of fig. S1. A lock mass solution (200 fmol per microliter of Glu-fibrinopeptide in 25% ACN and 0.1% formic acid in water) was delivered via the auxiliary solvent manager at 500 nl/min into the reference sprayer of the NanoLockSpray source.
Analyses of nano-UPLC elutes were performed on a Q-Tof Premier mass spectrometer (Waters). Electrospray ionization was performed with an uncoated, pulled fused silica emitter (New Objective) with a potential of 3.0 kV. The Q-Tof was operated in the V mode with a typical mass resolution of 10,000. Alternating scans were used to detect precursor ions and then fragment ions. The masses of the precursors were detected with a 0.5-s scan with a relatively low collision energy (CE) of 4 eV (MS scan), which was followed by a 0.5-s scan during which the CE was ramped from 15 to 40 eV (MSE scan). The instrument was calibrated with 13 fragment ions of the MS/MS spectrum of Glu-fibrinopeptide. To correct any shifts in masses that occurred during instrument operation, a single-point calibration was performed against the lock mass compound, doubly-charged Glu-fibrinopeptide [mass/charge ratio (m/z) 785.8426], which was sampled every 30 s.
Analyses of MS data and bioinformatics
MS data were analyzed with ProteinLynx Global Server (PLGS) software version 2.3 (Waters). A database of mouse proteins was generated by searching the Swiss-Prot and TrEMBL databases (http://www.expasy.ch) for the keyword Mus musculus. At the time that samples were analyzed, the database contained 12,943 mouse protein sequences and an equal number of randomized sequences for the estimation of false-positive identifications. The algorithm used to identify proteins has been described previously (37). For database search and acceptance of a protein, the following criteria were imposed: three fragment ions per peptide and seven fragment ions per protein with at least one unique peptide per protein; within 10 and 20 parts per million (ppm) of the theoretical masses for precursor and fragment ions, respectively. The database search returned results for each injection until 4% of the identified proteins were false (random sequences). After all of the data were processed, a further requirement was made that, for acceptance, a protein must be identified in at least two of three replicate injections of the same loading amount of the same sample. Homologous proteins that were not identified with a highly confident, unique peptide were removed from the final list. Absolute quantitation of identified peptides was achieved with ion chromatographic peak areas relative to that of the internal standard (ADH). The amounts of protein in each sample were then normalized to femtomoles per microgram to take into account the variability in sample concentrations, as well as different loading amounts of the individual samples. For each accepted protein, relative standard deviation of femtomole numbers from three replicate runs was calculated. The femtomoles per microgram value was used to calculate protein ratios between brain conditions.
Bioinformatic analyses were performed for proteins that showed at least 30% difference in quantities between an experimental group and the sham group (referred as regulated proteins or proteins that change in abundance) with the assistance of the MetaCore program (Version 5.0; GeneGo, Inc) using their gene names as entries (gene names were the acceptable identifier by the MetaCore program at the time the analyses were performed). Intersections between or among data sets of up-regulated or down-regulated proteins under each brain ischemic condition were revealed with the Compare Experiments Workflow tool of the MetaCore program, by which “network objects” that were unique to a data set or shared by two or more data sets are determined. Network object is a term used by the MetaCore program, referring to a protein or its known attributes (for example, reaction, process) presented as an object in a network—a graphical representation of objects connected according to their attributes (for example, direction of interaction). Each protein may be involved in more than one cellular processes or pathways, and thus can be assigned more than one network object. Enriched presence of biological processes that were represented by proteins regulated under each brain condition was also determined with the “Functional Enrichment by Ontology” of the MetaCore, by which P values (statistic relevance) of found matches between a data set (by gene names) and a process were calculated according to the proteins and the number of proteins in the data set. Unless stated otherwise, the 10 processes with the smallest P values were shown in Results.
Fluorescent immunohistocytochemistry or immunocytochemistry and Western blot analyses
Both analyses, as well as preparation of frozen brain sections and tissue homogenates, were performed following standard protocols as previously described (17). Rabbit polyclonal antibody directed against H2A (used at 1:100), mouse monoclonal antibody directed against RING2 (1:200 for Western blotting, 1:250 for immunohistochemistry), and rabbit polyclonal antibody directed against actin (1:500) were obtained from Santa Cruz Biotechnology. Mouse monoclonal antibody directed against SCMH1 (1:250) was from Novus. Goat polyclonal antibody against BMI1 (1:500) was from Abcam. Mouse monoclonal antibody against KCNAB2 (1:250) was obtained from Antibodies Incorporated (National Institute of Neurological Disorders and Stroke–National Institute of Mental Health NeuroMab Facility at University of California in Davis). Rabbit polyclonal antibody for KCNA5 (1:400) was purchased from Millipore. Mouse monoclonal antibodies against NeuN (1:100) and against glial fibrillary acidic protein (GFAP) (1:400) were from Millipore and Sigma, respectively. For all immunohistochemical analyses in this study, fluorescein isothiocyanate (FITC)– or cyanine dye 3 (Cy3)–conjugated secondary antibodies from appropriate species were obtained from Jackson ImmunoResearch. Sections were mounted with a mounting fluid (Vector Co.) containing DAPI (4′,6-diamidino-2-phenylindole) to counterstain nuclei. The fluorescent images were examined and documented with an epifluorescence microscope (Leica Microsystems, Inc).
siRNA oligos and plasmids encoding full-length cDNAs for PcG proteins
Double-stranded naked oligonucleotides were designed and synthesized by Sigma. Sequences of primers used in this study are as follows (only sense strands are shown; two primers per target): scrambled RNA: 5′-GAUCAUACGUGCGAUCAGATT; Scmh1 siRNA: 5′-UUCAAGUA-GAUACAAACUG; 5′-CAGUUUGUAUCUACUUGAATT; Bmi1 siRNA: 5′-CACUUAAUGUGUGUCCUGU; 5′-GAAUCAAGAUCACUGAGCU; Kcna5 siRNA: 5′-CACCUAAAGGCCAAGAGCA; 5′-CUACUUCGAUCCCUUGAGA; Kcnab2 siRNA: 5′-GGUAUUAGGGAACAUCAUU; 5′-CGGAAUAUCACAUGUUCCA. Plasmid pCMV6 carrying the full-length open reading frame (ORF) of recombinant human Scmh1 or Bmi1 was obtained from OriGene.
Neuronal cell cultures, in vitro simulated ischemia (OGD), and transfection
Mouse brain–derived neuroblastoma NS20Y cells (16) were used to test the effects of under- or overexpressing selected PcG proteins on the induction of ischemic tolerance. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum and penicillin-streptomycin (100 U/ml to 100 μg/ml) (regular growth medium) under normoxic conditions in the presence of 5.1% CO2. Differentiation was induced by incubating cells in the regular growth medium with the addition of 1 mM 8-(4-chlorophenylthio) cAMP (ctp-cAMP) for 24 hours. OGD was achieved by incubating cells for various periods of time in glucose-free, serum-free, glutamate-free medium (OGD medium) containing 5 mM Hepes (pH 7.4), 2 mM CaCl2, 135 mM NaCl, 5 mM KCl, 1X essential amino acids (without L-glutamine), 25 mM 2-deoxyglucose (2-DG) and penicillin-streptomycin (100 U/ml to 100 μg/ml) in an anaerobic chamber (Forma Scientific, model 1025) equilibrated with 85% N2/5% CO2/10% H2. Time-matched control cells were incubated in the control medium (OGD medium without 2-DG but supplemented with 25 mM glucose and 1 mM L-glutamine) under normoxic conditions. The extent of OGD-induced injury was determined by measuring changes in cell viability with the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] method, following standard protocols. On a 96-well plate, cells were transfected with scrambled RNA or siRNA (5 nM) or plasmid (1 μg) to be tested, each in quadruplet wells, by the Lipofectamine2000 method (Invitrogen). Twenty-four hours after transfection, cells were differentiated with ctp-cAMP (16) and proceeded to OGD treatments.
Four plates of identically transfected and differentiated cells were subjected to the following treatment conditions: control (normoxic condition in the control medium, followed by 24-hour recovery incubation in the regular growth medium); OGD-preconditioned (30 min of OGD, 24 hours of recovery incubation in the regular growth medium); OGD-injured (2 hours of OGD, 24 hours of incubation in the regular growth medium); OGD-tolerant (30 min of OGD followed by 24 hours of recovery incubation in the regular growth medium, then another 2 hours of OGD followed by another 24 hours of recovery incubation in the regular growth medium). At the end of 24 hours of recovery incubations (for OGD-tolerant condition, at the end of the second 24-hour recovery incubation), MTT assay was performed and plates were read on a SpectraMax Plus plate reader (Molecular Devices) (testing wavelength, 570 nm; reference wavelength, 630 nm). A “master control” value (defined as 100% of viability) was determined on quadruplet wells of cells that were kept under normoxic conditions in the regular growth medium. For all other conditions, percentages of viable cells (living cells) were calculated by dividing their MTT readings with the master control value and then multiplied by 100.
Intracerebroventricular administration of siRNA
Intracerebroventricular injection was performed following previously described protocols (17). Briefly, in adult mice, under anesthesia, a 26-gauge cannula was implanted into the right lateral ventricle followed by the insertion of a 31-gauge cannula into the lumen of the guide to inject 1 μl of artificial cerebrospinal fluid (aCSF) or scrambled RNA or siRNA (0.2 to 0.5 nmol). The animals were then subjected to the same MCAO-reperfusion conditions as used for the ischemic-tolerant group. The effectiveness of siRNA in knocking down selected PcG proteins was verified by immunohistochemistry (fig. S3). At the termination of reperfusions, under anesthesia, whole brains were removed and sectioned coronally at 1-mm intervals. The freshly cut brain slices were stained with vital dye 2,3,5-triphenyltetrazolium chloride [TTC; 2% in phosphate-buffered saline (PBS)] (38) to visualize infarct tissues. Infarct volumes in each ipsilateral hemisphere were determined according to the infarct areas on each brain slice and the number of slices from each brain (39), and compared across experimental groups.
Isolation of RNA and protein
Total RNA was isolated from cortices with a SuperPrep RNA/DNA/Protein Purification kit from Fisher. Mouse brains from sham (n = 5), preconditioned (n = 6), injurious (n = 7), and tolerant (n = 6) conditions were processed according to the manufacturer’s instructions. Briefly, each frozen cortical tissue was weighed and homogenized in a lysis buffer (provided by the kit) using an overhead stirrer with a tapered tissue grinder (Wheaton Industries Inc.). After homogenization, additional lysis buffer was added to reach a final concentration of ~30 mg (wet weight)/ml. An equal volume of 70% ethanol was then added to each lysate and the mixture was vortexed for 10 s. The lysate was added to a spin column (provided with the kit) and centrifuged. The protein fraction (flow-through) was removed to a fresh tube for later purification. The column was then washed and the RNA was eluted with an elution buffer (provided). The column was washed again and the DNA was eluted with another elution buffer (provided). Finally, the column was regenerated and activated for protein binding. The pH of the protein flow-through was adjusted to <3.5 and the protein was added to the column. The column was then washed and the protein was eluted with the provided elution buffer into a tube containing the protein neutralizer to neutralize the pH of the solution. RNA yield and quality were assessed by A260/A280 (ratio of the absorbance at 260 nm to the absorbance at 280 nm) values for each sample. The DNA fraction was stored at −80°C for future use. The protein concentration was determined by Bradford assay.
Reverse transcription and real-time PCR
cDNA was prepared from 100 ng of total RNA with MuLV reverse transcriptase and random hexamers as primers (Applied Biosystems) and the following thermal profile: 25°C, 10 min; 37°C, 120 min; and 85°C, 5 min. Real-time PCR was performed with the 7500 Fast Real-time PCR system (Applied Biosystems) using Applied Biosystems assay-on-demand primers and probes (4352341E: Actin, endogenous control primer; Kcnab2: Mm01260263_m1) and the TaqMan Gene Expression Master Mix kit (Applied Biosystems) via the following thermal profile: 50°C, 2 min; 95°C, 10 min; followed by 40 cycles of 95°C, 15 s, and 60°C, 1 min. Three independent experiments were performed with the multiplex comparative CT method and results were analyzed with SDS 4.6 software from Applied Biosystems.
ChIP assay
ChIP assays were performed with the EZ-ChIP kit (Millipore), following the manufacturer’s instructions. Approximately 1.5 × 107 NS20Y cells were plated on 10-cm dishes, differentiated by ctp-cAMP, and cross-linked with 1% formaldehyde in PBS. The chromosomal DNAs in the lysates were sheared by sonication with a digital sonifier (Branson 450) at 30% of maximum power output for 8 × 10 s, yielding DNA fragments between 300 to 1000 base pairs. The protein-DNA complexes were immunoprecipitated with 10 μg of appropriate antibodies and protein G–agarose beads and de-cross-linked. Mouse monoclonal antibodies against SCMH1 or BMI1 and rabbit polyclonal antibody against H2A were from Novus, Abcam, and Santa Cruz Biotechnology, respectively. The DNA was purified and subjected to PCR analyses with the FastStart TaqDNA Polymerase kit (Roche). PCR reactions (0.26 μM each primer) were carried out with a PTC-100 Programmable Thermal Controller (MJ Research Inc) as follows: 95°C, 3 min; 40 × 95°C, 20 s/50°C, 30 s/72°C, 30 s, 72°C, 2 min. The primers for the promoter regions of mouse Kcna5 (F-ATTTACCGTTGTGGGGTTCA, R-TACCAGGAGCGCTGAAAACT); Kcnab2 (F-GATACCGCTCACTCAGCACA, R-TTCACGCCAGTCA-CAGTCTC); and Hox13c (F-TGCAGCGGAGCGAGCCCC, R-TCAA-CAGGGATGAGCGCGTCGTG) were designed with Invitrogen’s online primer designing tools and synthesized by Sigma. The PCR-generated DNA fragments were analyzed with 2% agarose gel in TBE [tris-borate-EDTA (ethylenediaminetetraacetic acid)] buffer.
Electrophysiology
NS20Y cells were transfected with scrambled RNA or siRNA or plasmid and differentiated. Plasmid pEGFP-N3 (CloneTech) was cotransfected for identifying positively transfected cells. Potassium currents were recorded with whole-cell patch-clamp at room temperature (20°C to 22°C) (1). In general, cells were voltage-clamped at −60 mV. Patch pipettes were pulled from borosilicate glass (1.5 mm diameter) on a two-stage puller, producing pipettes with a resistance of 2 to 4 MΩ when filled with the intra-cellular solution (see below). Membrane capacitance was recorded for each cell as a measure of cell size. Voltage-gated potassium currents were elicited by a 100-ms depolarizing potential from −100 mV to +80 mV with 20-mV increments. To ensure high-quality voltage-clamp, only recordings with an access resistance of less than 10 megohm and a leak current of less than 100 pA at −60 mV were included for data analysis. Standard extra-cellular solution (ECF) contained 140 mM NaCl, 5.4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 20 mM Hepes, and 10 mM glucose. The pH was adjusted to 7.4 with NaOH, and the osmolarity was adjusted to 320 to 335 mosM. Intracellular solution contained 140 mM KF, 11 mM EGTA, 1 mM CaCl2, 4 mM MgCl2, 10 mM Hepes (pH 7.3) adjusted with KOH; 290 to 300 mosM. Data were acquired with an AXOPATCH 200B amplifier and pCLAMP 8.1 software (Axon Instruments). Data were analyzed with Clampfit (Axon Instruments). Current-voltage relations of voltage-gated channels were plotted by Sigma plot 8.0 software.
Statistics
Statistical analyses were performed with either SPSS statistics v16 (SPSS Inc) or MATLAB (The Mathworks Inc). All tests were performed at the alpha level of significance of 0.05. Data were tested for significance with either one-way analysis of variance (ANOVA), followed by Tukey and Scheffe post hoc analysis, or two-way ANOVA followed by Bonferroni post hoc analysis, for electrophysiology experiments. Tests for factors included pairwise comparisons with Mann-Whitney U tests. All data are reported or graphed as mean ± SE from n individual measurements.
Supplementary Material
Fig. S1. Chromatographic peak area measurement.
Fig. S2. Immunohistochemistry of H2A and BMI1.
Fig. S3. Knocking down or overexpressing PcG proteins.
Fig. S4. Changes in abundance of RING2 and mUb-H2A in mouse brains and in NS20Y cells.
Table S1. Total identified and quantified proteins in mouse cortex.
Table S2. Numbers of proteins that change in abundance under each condition.
Table S3. Proteins that change in abundance under ischemic-preconditioned condition.
Table S4. Proteins that change in abundance under ischemic-tolerant condition.
Table S5. Proteins that change in abundance under ischemic-injured condition.
Table S6. Proteins that change in abundance under a unique brain ischemic condition or more than one condition.
Table S7. Biological processes presented by proteins that change in abundance under different conditions of brain ischemia.
Acknowledgments
We thank A. White for helping with processing of mouse brain samples, S. Crawford for administrative assistance, and S. Thompson for helpful discussion on technical details of some of the experiments.
Funding: The study was supported by grants from American Heart Association (0850129Z, A.Z.), NIH (1R01NS046560, A.Z.; 5P01NS035965, R.P.S.; R01NS47506, Z.G.X.), Legacy Good Samaritan Foundation (A.Z.), and by a Legacy Research RAC grant (A.Z.). C.S. was supported by Discoveries in Sight of Legacy Research.
Footnotes
Author contributions: A.Z. designed the entire project and most experiments; Z.G.X. designed electrophysiology experiments; J.L. directed the development of mass spectrometry tools; M.S., C.P., T.Y., M.L., C.S., J.Q.L., and J.A.S. performed the experiments; A.Z., M.S., Z.G.X., J.A.S., C.S., and S.G. analyzed the data; A.Z., R.P.S., Z.G.X., and J.A.S. interpreted results; and A.Z. and R.P.S. wrote the manuscript (M.S., Z.G.X., and J.A.S. wrote portions of Materials and Methods and C.P. assisted in figure preparation). None of the material has been published or is under consideration elsewhere, including the Internet. The MCAO and intracerebroventricular injection protocols used in this study were approved by the Institutional Animal Care and Use Committee of Legacy Research. All animal experiments were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
References
- 1.Stenzel-Poore MP, Stevens SL, Xiong Z, Lessov NS, Harrington CA, Mori M, Meller R, Rosenzweig HL, Tobar E, Shaw TE, Chu X, Simon RP. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: Similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362:1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- 2.Takada Y, Isono K, Shinga J, Turner JM, Kitamura H, Ohara O, Watanabe G, Singh PB, Kamijo T, Jenuwein T, Burgoyne PS, Koseki H. Mammalian Polycomb Scmh1 mediates exclusion of Polycomb complexes from the XY body in the pachytene spermatocytes. Development. 2007;134:579–590. doi: 10.1242/dev.02747. [DOI] [PubMed] [Google Scholar]
- 3.Berger J, Kurahashi H, Takihara Y, Shimada K, Brock HW, Randazzo F. The human homolog of Sex comb on midleg (SCMH1) maps to chromosome 1p34. Gene. 1999;237:185–191. doi: 10.1016/s0378-1119(99)00285-1. [DOI] [PubMed] [Google Scholar]
- 4.Tomotsune D, Takihara Y, Berger J, Duhl D, Joo S, Kyba M, Shirai M, Ohta H, Matsuda Y, Honda BM, Simon J, Shimada K, Brock HW, Randazzo F. A novel member of murine Polycomb-group proteins, Sex comb on midleg homolog protein, is highly conserved, and interacts with RAE28/mph1 in vitro. Differentiation. 1999;65:229–239. doi: 10.1046/j.1432-0436.1999.6540229.x. [DOI] [PubMed] [Google Scholar]
- 5.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Ohtsubo M, Yasunaga S, Ohno Y, Tsumura M, Okada S, Ishikawa N, Shirao K, Kikuchi A, Nishitani H, Kobayashi M, Takihara Y. Polycomb-group complex 1 acts as an E3 ubiquitin ligase for Geminin to sustain hematopoietic stem cell activity. Proc Natl Acad Sci USA. 2008;105:10396–10401. doi: 10.1073/pnas.0800672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breiling A, Sessa L, Orlando V. Biology of polycomb and trithorax group proteins. Int Rev Cytol. 2007;258:83–136. doi: 10.1016/S0074-7696(07)58002-2. [DOI] [PubMed] [Google Scholar]
- 8.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Kerppola TK. Polycomb group complexes—many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 13.Lee K, Adhikary G, Balasubramanian S, Gopalakrishnan R, McCormick T, Dimri GP, Eckert RL, Rorke EA. Expression of Bmi-1 in epidermis enhances cell survival by altering cell cycle regulatory protein expression and inhibiting apoptosis. J Invest Dermatol. 2008;128:9–17. doi: 10.1038/sj.jid.5700949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatoo W, Abdouh M, David J, Champagne MP, Ferreira J, Rodier F, Bernier G. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J Neurosci. 2009;29:529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito Y, Maruyama K, Saido TC, Kawashima S. Overexpression of a neuropeptide nociceptin/orphanin FQ precursor gene, N23K/N27K, induces neurite outgrowth in mouse NS20Y cells. J Neurosci Res. 1997;48:397–406. doi: 10.1002/(sici)1097-4547(19970601)48:5<397::aid-jnr2>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 16.Sirianni MJ, Fujimoto KI, Nelson CS, Pellegrino MJ, Allen RG. Cyclic AMP analogs induce synthesis, processing, and secretion of prepro nociceptin/orphanin FQ-derived peptides by NS20Y neuroblastoma cells. DNA Cell Biol. 1999;18:51–58. doi: 10.1089/104454999315619. [DOI] [PubMed] [Google Scholar]
- 17.Zhan S, Zhao H, White AJ, Minami M, Pignataro G, Yang T, Zhu X, Lan J, Xiong Z, Steiner DF, Simon RP, Zhou A. Defective neuropeptide processing and ischemic brain injury: A study on proprotein convertase 2 and its substrate neuropeptide in ischemic brains. J Cereb Blood Flow Metab. 2009;29:698–706. doi: 10.1038/jcbfm.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinemann SH, Rettig J, Graack HR, Pongs O. Functional characterization of Kv channel β-subunits from rat brain. J Physiol. 1996;493:625–633. doi: 10.1113/jphysiol.1996.sp021409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uebele VN, England SK, Chaudhary A, Tamkun MM, Snyders DJ. Functional differences in Kv1.5 currents expressed in mammalian cell lines are due to the presence of endogenous Kvβ2.1 subunits. J Biol Chem. 1996;271:2406–2412. doi: 10.1074/jbc.271.5.2406. [DOI] [PubMed] [Google Scholar]
- 20.Barone FC, White RF, Spera PA, Ellison J, Currie RW, Wang X, Feuerstein GZ. Ischemic preconditioning and brain tolerance: Temporal histological and functional outcomes, protein synthesis requirement, and interleukin-1 receptor antagonist and early gene expression. Stroke. 1998;29:1937–1950. doi: 10.1161/01.str.29.9.1937. [DOI] [PubMed] [Google Scholar]
- 21.Vogel T, Stoykova A, Gruss P. Differential expression of polycomb repression complex 1 (PRC1) members in the developing mouse brain reveals multiple complexes. Dev Dyn. 2006;235:2574–2585. doi: 10.1002/dvdy.20876. [DOI] [PubMed] [Google Scholar]
- 22.Puro J, Nygrén T. Mode of action of a homoeotic gene in Drosophila melanogaster. Localization and dosage effects of Polycomb. Hereditas. 1975;81:237–248. doi: 10.1111/j.1601-5223.1975.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 23.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 24.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 25.Bracken AP, Helin K. Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 26.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, Andrews LG, Tollefsbol TO. Loss of the human polycomb group protein BMI1 promotes cancer-specific cell death. Oncogene. 2006;25:4370–4375. doi: 10.1038/sj.onc.1209454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Bonnet S, Harry G, Hashimoto K, Porter CJ, Andrade MA, Thebaud B, Michelakis ED. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 29.Wei L, Yu SP, Gottron F, Snider BJ, Zipfel GJ, Choi DW. Potassium channel blockers attenuate hypoxia- and ischemia-induced neuronal death in vitro and in vivo. Stroke. 2003;34:1281–1286. doi: 10.1161/01.STR.0000065828.18661.FE. [DOI] [PubMed] [Google Scholar]
- 30.Zhao YM, Sun LN, Zhou HY, Wang XL. Voltage-dependent potassium channels are involved in glutamate-induced apoptosis of rat hippocampal neurons. Neurosci Lett. 2006;398:22–27. doi: 10.1016/j.neulet.2005.12.073. [DOI] [PubMed] [Google Scholar]
- 31.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring–Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, Kerkhoven RM, Voncken JW, Wessels LF, van Lohuizen M. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 34.Wu ZL, Zheng SS, Li ZM, Qiao YY, Aau MY, Yu Q. Polycomb protein EZH2 regulates E2F1-dependent apoptosis through epigenetically modulating Bim expression. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.162. [DOI] [PubMed] [Google Scholar]
- 35.Yu YQ, Gilar M, Lee PJ, Bouvier ES, Gebler JC. Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal Chem. 2003;75:6023–6028. doi: 10.1021/ac0346196. [DOI] [PubMed] [Google Scholar]
- 36.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 38.Bederson JB, Pitts LH, Germano SM, Nishimura MC, Davis RL, Bartkowski HM. Evaluation of 2,3,5-triphenyltetrazolium chloride as a stain for detection and quantification of experimental cerebral infarction in rats. Stroke. 1986;17:1304–1308. doi: 10.1161/01.str.17.6.1304. [DOI] [PubMed] [Google Scholar]
- 39.Yang Y, Shuaib A, Li Q. Quantification of infarct size on focal cerebral ischemia model of rats using a simple and economical method. J Neurosci Methods. 1998;84:9–16. doi: 10.1016/s0165-0270(98)00067-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Chromatographic peak area measurement.
Fig. S2. Immunohistochemistry of H2A and BMI1.
Fig. S3. Knocking down or overexpressing PcG proteins.
Fig. S4. Changes in abundance of RING2 and mUb-H2A in mouse brains and in NS20Y cells.
Table S1. Total identified and quantified proteins in mouse cortex.
Table S2. Numbers of proteins that change in abundance under each condition.
Table S3. Proteins that change in abundance under ischemic-preconditioned condition.
Table S4. Proteins that change in abundance under ischemic-tolerant condition.
Table S5. Proteins that change in abundance under ischemic-injured condition.
Table S6. Proteins that change in abundance under a unique brain ischemic condition or more than one condition.
Table S7. Biological processes presented by proteins that change in abundance under different conditions of brain ischemia.