Abstract
Recently, a consensus has emerged that cofilin severing activity can generate free actin filament ends that are accessible for F-actin polymerization and depolymerization without changing the rate of G-actin association and dissociation at either filament end. The structural basis of actin filament severing by cofilin is now better understood. These results have been integrated with recently discovered mechanisms for cofilin activation in migrating cells, which led to new models for cofilin function that provide insights into how cofilin regulation determines the temporal and spatial control of cell behaviour.
Most eukaryotic cells migrate by binding to the extra-cellular matrix using a motility cycle in which actin polymerization supplies the pushing force for protrusion of the leading edge to establish the direction of migration. During classic amoeboid cell migration, the motility cycle includes actin polymerization-driven formation of a protrusion in the new direction of migration, attachment to the substratum, generation of traction force and retraction of the tail. These events are dependent on actomyosin-mediated contractile force production and the actin cytoskeleton, which is dynamic and undergoes repeated cycles of actin polymerization and depolymerization in a spatially and temporally coordinated pattern (FIG. 1). Furthermore, the cytoskeleton has an essential role in establishing the internal cell architecture that controls cell migration. Other forms of movement such as blebbing1 and rolling2 have been described, but the importance of actin polymerization in these rare forms of motility is not well understood, and the involvement of cofilin in these types of cell motility has not been extensively studied. Therefore, our Review discusses the role of cofilin in cell locomotion and invasion in which actin polymerization is the pushing force for protrusion.
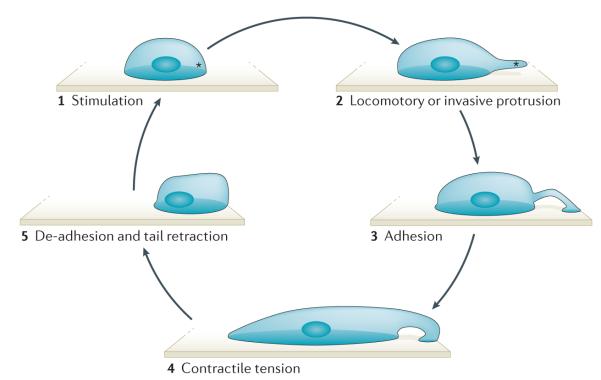
Figure 1. Steps of the cell motility cycle.
An external or internal signal stimulates the local and asymmetric polymerization of actin (depicted by an asterisk) (1). Polymerizing actin pushes against the cell membrane to form a locomotory or invasive protrusion (2). The attachment of the protrusion to the extracellular matrix initiates adhesion formation at the leading edge and signals to the contractile machinery of the cell to initiate contraction (3). Contractile tension distorts the cell into an elongated shape (4). Release of the rear adhesion allows tail retraction and locomotion of the cell (5).
The actin-binding proteins involved in each of the steps of the motility cycle include cofilin, integrin–talin complexes, the actin-related protein 2/3 (ARP2/3) complex, formins, profilin, capping protein and myosin, among others (reviewed in REFS 3–7). Cofilin functions in the first step to initiate asymmetric actin polymerization and in later steps to recycle actin filaments. The involvement of cofilin in controlling the temporal and spatial extent of actin dynamics is seen in processes as diverse as tumour metastasis in mice8, cytokinesis in yeast9, morphogenesis in flies10, neuronal plasticity in rats11, as well as during inflammation12. The ADF/cofilin protein family consists of ADF (actin depolymerizing factor), cofilin 1 and cofilin 2. Cofilin 1 (hereafter referred to as cofilin) is the most abundant and ubiquitous member of the family in vertebrate non-muscle tissues and the only one required for viability13. The development of new technologies and their application to measure cofilin activity in vivo (BOX 1; Supplementary information S1 (box)) has provided new insights into its regulation and challenged previous models of cofilin function in cell migration.
Box 1. High-resolution methods to study cofilin in migrating cells*.
Fluorescence resonance energy transfer (FRET)
The dynamics of the dissociation of active cofilin from the plasma membrane can be monitored by FRET between GFP–cofilin and mCherry-CAAX49 in living eukaryotic cells. The use of expressed GFP-tagged cofilin in eukaryotic cells has been validated: GFP-cofilin interacts with actin in cell extracts (using co-immunoprecipitation); it severs actin filaments in vitro (as shown by co-sedimentation assays); and has been observed to fragment individual actin filaments in an in vitro assay (using total internal reflection (TIRF) microscopy)121. However, in budding yeast122 and fission yeast9, the GFP–tagged cofilin fusion protein is not fully functional and had to be overexpressed to rescue cofilin deletion phenotypes.
These results suggest that it is important to validate the use of GFP–cofilin fusion proteins in each cell type by an independent method such as antibody-based FRET.
The binding of endogenous cofilin to F-actin can also be measured by antibody-based FRET (for example, between immunolabelled endogenous cofilin and F-actin46,49,113 in fixed cells). These two FRET signals, GFP and/or mCherry and antibody-based FRET, can be studied independently during cell protrusion even though they are within the diffraction limited spot of light microscopes. Both of these independent methods yield similar results in eukaryotic cells49,113.
Fluorescence loss in photobleaching (FLIP)
The mobility of plasma membrane- and F-actin-bound cofilin can be measured by bleaching cytosolic GFP–cofilin. Using appropriate conditions, mobility differences between the plasma membrane- and F-actin-bound fractions are quantified49,96. Fluorescence recovery after photobleaching (FRAP) experiments can be used to analyse the lateral mobility of plasma membrane-bound cofilin.
Bimolecular fluorescence complementation (BiFC)
Binding of cofilin to F-actin and G-actin can be monitored using Venus fluorescent protein (a derivative of YFP) that is split between actin and cofilin. It does not distinguish between F-actin- and G-actin-bound cofilin. The actin–cofilin binding induced by this approach is irreversible123.
Proximity ligation assay (PLA)
This method can be used to study F-actin and G-actin binding to endogenous cofilin at single-molecule resolution46. PLA uses antibodies conjugated to oligonucleotides that form complementary segments to a circularization probe, which are in situ ligated and circularly amplified. The amplified DNA can be detected by dye-labelled probe complementation. PLA has an extremely high signal-to-noise ratio for the detection of protein–protein binding in situ124.
Caged cofilin
A constitutively active S3A cofilin phosphorylation mimetic mutant (in which Ser3 was replaced with Asp) is chemically caged using a photolabile moiety. Upon photocleavage, activated cofilin is released. This technique can be used to initiate focal activation of cofilin in live cells and has been applied to validate the model presented in FIG. 4. The system is not reversible, and therefore the potential effects on cells from the accumulation of active cofilin must be considered97.
Cofilin-dependent barbed ends in situ
By labelling actin barbed ends with fluorescently tagged G-actin, cofilin severing and/or barbed end generation activity can be measured in situ113,117.
Recent reviews have highlighted the biochemistry and cell biology of the ADF/cofilin protein family in cell migration, chemotaxis and cancer8,13,14. Here, we focus on recent results describing mechanisms of cofilin activation at different subcellular locations that led to new models for cofilin function in migrating cells.
Functions of cofilin
Microscopy studies revealing the interaction of F-actin with cofilin have shown that cofilin severs actin filaments but does not enhance depolymerization from the pointed end of the filament9,15–17. Conclusions from early biochemical experiments that cofilin increases the dissociation rate of G-actin from the pointed end of the filament18,19 were incorrect, because they were not taking into account the number of filament ends created by cofilin severing9,16. Submicromolar concentrations of cofilin produce free barbed ends by severing existing filaments, and these new ends can be used to nucleate actin polymerization20. In addition, at micromolar concentrations, cofilin nucleates actin polymerization directly15. By creating new actin filaments through severing, cofilin supports ARP2/3 complex-mediated actin branching, as branches nucleated by the ARP2/3 complex are tenfold more stable on such recently polymerized filaments than on older filaments20,21. Cofilin also dissociates ARP2/3 complex-produced branches from older actin filaments22. Moreover, cofilin is involved in supporting contractility at the cell rear through local F-actin depolymerization23 and in regulating actomyosin assembly by inhibiting binding of myosin II to F-actin24.
Thus, cofilin has a central role in controlling actin dynamics, by catalysing actin polymerization and actin depolymerization through its severing activity, as well as by inducing dendritic nucleation and debranching.
Structural understanding of cofilin function
Several recent studies have advanced our understanding of the structural basis of cofilin-mediated severing — a process by which non-covalent bonds between actin molecules in F-actin are broken (FIG. 2). Cofilin increases the torsional dynamics of F-actin, and this can be propagated to regions of the filament that are not bound to cofilin25,26. A key finding is that cofilin severs F-actin at the junction between undecorated and cofilin-decorated regions of F-actin27. This observation, combined with a high-resolution structure of the cofilin-decorated actin filament25,28, revealed that cofilin binding to F-actin disrupts the longitudinal interface in the filament (double-stranded actin filaments are defined by two interfaces: longitudinal and diagonal) while forming a ‘bridge’ to maintain the connection between interfaces. However, the cofilin-induced change in one of the four subdomains of G-actin, subdomain 2, disorders it and increases the flexibility of the F-actin filament. These changes in subdomain 2 cooperatively propagate into the undecorated filament regions29. The absence of cofilin in these regions makes the filament unstable due to the absence of the cofilin-mediated longitudinal stabilization, resulting in filament severing25. However, the discovery in protozoan (for example, Toxoplasma gondii) of cofilin family members that sever F-actin to about the same degree as their mammalian homologues with no detectable stable F-actin binding suggests additional mechanisms for severing, possibly involving increased sequestration of G-actin monomers30. Further studies will be required to investigate whether there is a common severing mechanism in protozoa and higher organisms.
Figure 2. Domain structure of cofilin and its main functions.
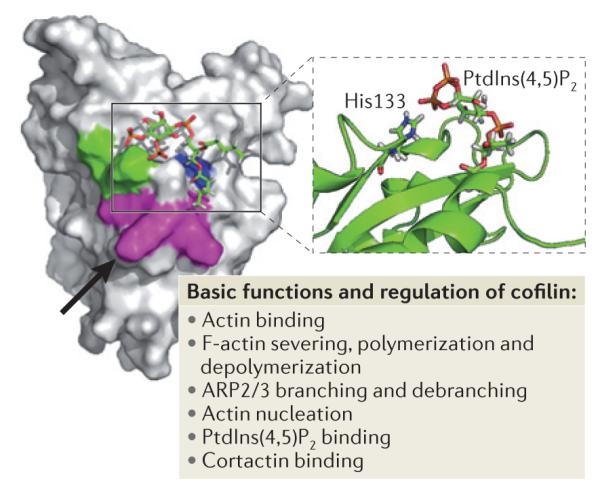
Key amino acids involved in the regulation of cofilin function are Asp122 and His133. The black arrow points to Asp122 located underneath Leu126 (in magenta). Asp122 regulates binding of cofilin to phosphatidylinositol-4,5-bisphophate (PtdIns(4,5)P2). A cofilin mutant in which Asp122 has been replaced with Ala has increased binding affinity for PtdIns (4,5)P2 and this results in the inhibition of actin polymerization, cell protrusion and migration. His133 decreases cofilin affinity for PtdIns(4,5)P2 when deprotonated. Substitution of His133 with Ala in cofilin causes loss of H+ binding and increases steady-state actin polymerization and cell protrusion. Lys132 (which is involved in actin binding) and His133 are shown in green, Lys125, Lys126, and Lys127 (which are alternative residues for PtdIns(4,5)P2 binding) are shown in magenta and Phe15 and Leu99 are shown in blue. The figure is modified, with permission, from REF. 102 © (2008) Rockefeller University Press.
Polymerization or depolymerization of F-actin?
A central question is what determines whether polymerization or depolymerization results from cofilin-mediated severing of the actin filament. Several studies have shown, both in vitro and in vivo, that the relative concentration of G-actin is a key determinant. Because dissociation of both ADP•G-actin and ATP•G-actin at the pointed end is slow, and the association rate for ATP•G-actin at the barbed end is 15–40 times higher, net polymerization occurs at sites of actin filament severing at physio logical ATP•G-actin concentrations31. Cyclase-associated protein (CAP) can accelerate the exchange of cofilin-associated ADP•G-actin with ATP•G-actin to help ensure the local supply of polymerization-competent G-actin and to support cofilin-induced actin polymerization32. However, ATP•G-actin concentrations in migrating cells are thought to be mostly maintained by profilin, which catalyses the exchange of nucleotides on G-actin and sustains free ATP•G-actin at a concentration of about 1 μM in vivo33. Recently, CAP has been shown to accelerate cofilin-dependent actin filament severing at a neutral pH34, providing a mechanism for increased actin filament dynamics without changes in intracellular pH. Under conditions of low G-actin concentrations (as, hypothetically, could occur in cells that undergo prolonged and continuous cell migration), severing by cofilin would cause net actin depolymerization. However, G-actin concentrations that are low enough to achieve net depolymerization are not found in the most commonly studied migrating cells, such as fibroblasts and epithelial cells, which suggests that net polymerization is the initial result from cofilin severing in vivo35. In addition, cofilin increases the concentration of G-actin by promoting actin depolymerization and thus contributes to the maintenance of physiological G-actin levels36,37.
Although the relative concentrations of active cofilin and ATP•G-actin help to determine the relative balance of actin polymerization and depolymerization that is likely to occur in vivo upon local cofilin activation, this balance can also be regulated by additional actin-modulating proteins such as AIP1 (actin interacting protein 1), MENA, RHOC, CAP, coronin and gelsolin (reviewed in REFS 13,14,38). AIP1 can tip the balance from polymerization towards net depolymerization39,40. In Caenorhabditis elegans, AIP-1 has been shown to potentiate cofilin severing41, but AIP 1 also favours net cofilin-induced actin filament disassembly by capping the barbed ends of severed filaments to prevent their elongation and reannealing in both Xenopus laevis and C. elegans42,43. However, ENA/VASP (enabled/vasodilator-stimulated phosphoprotein) proteins such as MENA, which have anticapping activity, have been reported to maintain free barbed ends that have been generated by cofilin at the leading edge of cell protrusions44. RHOC has been shown to regulate cofilin phosphorylation through ROCK (RHO-associated protein kinase) and LIMK (LIM-domain kinase) to determine the location and amount of cofilin activity and actin polymerization at invadopodium and lamellipodium compartments45,46. Gelsolin has actin filament severing and barbed end capping activity and can cause net depolymerization. Coronin can regulate cofilin activity in two ways: preventing its binding to new actin filaments and enhancing its binding to old ADP•G-actin filaments.
In addition to the functions of cofilin in actin remodelling, a role for cofilin in the regulation of phopholipase D1 (PLD1; which catalyses the hydrolysis of phosphatidilcholine to choline and phosphatidic acid) has been proposed47.
Beyond these recent insights into the functions of cofilin, progress has been made in our understanding of cofilin cellular localization and how it is locally activated in migrating cells.
The localization of cofilin
Cofilin is a small protein of 19 kDa that freely diffuses within cells and can be found in multiple cellular compartments, including the cytoplasm and nucleoplasm. Live-cell imaging of GFP–cofilin expressed in unstimulated carcinoma cells48,49 and HeLa cells50 suggests that cofilin is uniformly distributed. However, immunofluorescence microscopy of fixed carcinoma cells revealed that the two major forms of cofilin — cofilin and cofilin that is phosphorylated on Ser3 — are differentially distributed in the cell51. Ser3 is the major phosphorylation site involved in the regulation of cofilin, as phosphorylation at that residue inhibits all cofilin–actin interactions, including binding to G-actin, and it inhibits severing of F-actin. Phosphorylation of other cofilin residues also regulates its activity. For example, protein kinase Cα (PKCα)-dependent phosphorylation at Ser23 and Ser24 terminates histamine degranulation52, and phosphorylation at Tyr68 by constitutively active SRC can promote the ubiquitin-mediated degradation of cofilin53.
Antibodies specific to total cofilin and cofilin phosphorylated at Ser3, which allows the analysis of dephosphorylated cofilin51, revealed that these cofilin forms show differences in their cellular distribution. Non-phosphorylated cofilin is found in locomotory and invasive protrusions such as lamellipodia in motile epithelial cells and invadopodia in motile carcinoma cells, whereas phosphorylated cofilin is more uniformly distributed throughout the cytoplasm, except at the leading edge46,51,54. In general, cofilin is active at the tip of the leading edge protrusion of migrating cells and inactivated by Ser3 phosphorylation 1 μm behind the leading edge50. These results are in line with the findings that inhibition of cofilin activity causes defects in protrusion, cell polarity and chemotaxis, the latter of which is dependent on directional protrusion formation46,54–56.
Cofilin that is not phosphorylated at Ser3 is found in regions of the cell where it is involved in rapid, reversible binding interactions (for example, on the order of seconds for F-actin57 and milliseconds for G-actin58) that are associated with the cofilin activity cycle. These involve interactions with F-actin, phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) at the plasma membrane, cortactin in invadopodia and diffusible G-actin heterodimers in the cytoplasm59. The binding of cofilin to PtdIns(4,5)P2, cortactin and G-actin inhibits cofilin severing activity, and the binding of cofilin to F-actin is limited to tropomyosin-free filaments near the plasma membrane60. Overall, these binding interactions not only serve to regulate cofilin activity but also contribute to the positioning of cofilin once it is activated.
Cofilin is also found in the nucleus61,62, where it can form stable heteropolymers with F-actin. However, this property is not thought to be involved in rapid actin dynamics or the regulation of cofilin activity during cell motility13. To fully understand how the location and timing of cofilin activation are influenced by these interactions, we must understand how cofilin is activated.
The mechanisms of cofilin activation
There are three important mechanisms that regulate the activation of cofilin: its dephosphorylation at Ser3; its release from PtdIns(4,5)P2; and its release from cortactin (FIGS 3–5). These mechanisms have been studied in detail, and their relative effects on cofilin activity and the resulting behaviour of motile cells have been revealed. However, these activation mechanisms are not mutually exclusive, and how they are coordinated in motile cells is the subject of intense investigation.
Figure 3. Regulatory mechanisms of cofilin phosphorylation and dephosphorylation.
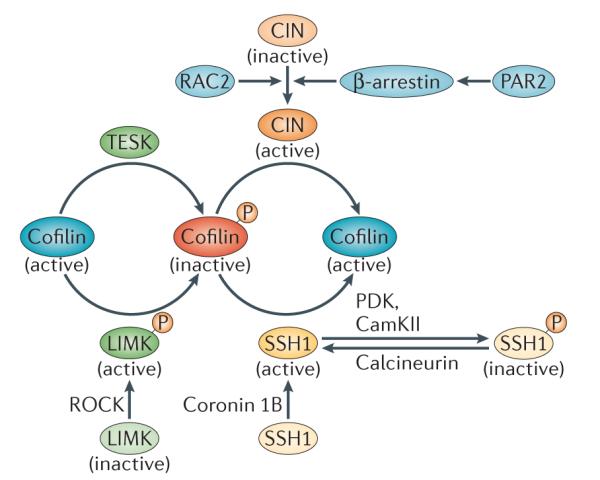
Cofilin phosphorylation at Ser3 is driven by LIM-domain kinase (LIMK) and TES kinase (TESK). LIMK is activated by ROH-associated protein kinase (ROCK). Dephosphorylation of cofilin at Ser3 is mediated by the phosphatases chronophin (CIN) and slinghshot 1 (SSH1).The localization and activation of SSH1 is mediated by coronin 1B. Phosphorylation of SSH1 by Ca2+/calmodulin-dependent protein kinase II (CaMKII) and phosphoinositide-dependent protein kinase (PDK) results in its inactivation. Calcineurin activates SSH1 by dephosphorylating it. CIN is activated by its interaction with β-arrestin, which is activated by protease-activated receptor 2 (PAR2). In neutrophils, RAC2 has been shown to induce cofilin activation in a CIN-dependent manner.
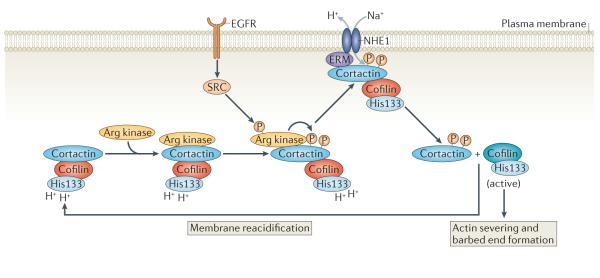
Figure 5. A new model for the activation of cofilin in invasive protrusions.
Tyr phosphorylation of cortactin by Arg kinase, which is activated downstream of epidermal growth factor receptor (EGFR), regulates the interaction between the Na+–H+ exchanger 1 (NHE1) and cortactin. The recruitment of phosphorylated cortactin to NHE1 may be mediated by ERM (ezrin–radixin–moesin) proteins. NHE1 increases the intracellular pH, which induces the release of cortactin-bound cofilin.
Cofilin Ser3 dephosphorylation
Dephosphorylation of cofilin Ser3 was the first activation mechanism to be well characterized63,64 (FIG. 3). Unphosphorylated cofilin, but not cofilin phosphorylated at Ser3, can bind actin and promote its polymerization and depolymerization in vitro65. In addition, S3D-cofilin (a phosphorylation mimic in which Ser3 was replaced with Asp) fails to bind to and sever F-actin, whereas S3A-cofilin (a mutant in which Ser3 was replaced with Ala) does64,65. Slingshot (SSH) was shown to be a major phosphatase responsible for dephosphorylating cofilin at Ser3 (REF. 66), and chronophin (CIN)67 was recently identified as a cofilin-specific phosphatase. Although the Ser phosphatases PP1 and PP2A68 can also dephosphorylate cofilin at Ser3, these phosphatases have broader substrate specificity than SSH or CIN.
Several recent studies have provided insights into the regulation of SSH1 activation, localization and scaffolding69. For example, coronin 1B was found to interact with SSH1 and target it to lamellipodia, thereby increasing cofilin activity at lamellipodia. A direct interaction may also take place between coronin 1B and cofilin, but further studies are needed to confirm this70. In neuregulin-stimulated breast carcinoma cells, the 14-3-3 family of regulatory proteins has been shown to inhibit F-actin-mediated activation of SSH1 through Ser978 dephosphorylation, which prevented the accumulation of SSH1 at the lamellipodium, thereby increasing the pool of inactive, phosphorylated cofilin71. Recently, protein kinase D (PKD) proteins72,73 were shown to phosphorylate SSH1 at Ser978, which leads to 14-3-3 recruitment and PAK4 (p21-activated kinase 4) activation73 and thus LIMK1 activation74. This suggests that PKD proteins can inhibit cofilin dephosphorylation by inactivating SSH1 and activating LIMK 1. Calcineurin has also been shown to trigger cofilin dephosphorylation through the activation of SSH1 (REF. 75), whereas Ca2+/calmodulindependent protein kinase II (CaMKII) negatively regulates SSH1 (REF. 76), which suggests that CaMKII and calcineurin act as a switch controlling Ca2+-dependent cofilin activation. Moreover, β-arrestin scaffolding has been shown to promote cofilin dephosphorylation following protease-activated receptor 2 (PAR2) stimulation, which activates CIN and antagonizes LIMK1 (REF. 77). These pathways are summarized in FIG. 3.
The rapid dephosphorylation of cofilin at Ser3 following stimulation with the G protein-coupled receptor ligands formylmethionine-Leu-phenylalanine (fMLF) and interleukin-8 (IL-8)78 is required to initiate chemotaxis towards these ligands in leukocytes79,80. It was demonstrated that the generation of free actin filament barbed ends in these cells requires cofilin activation79. This was regulated predominately through a RAC2-dependent pathway in mouse neutrophils, as cofilin was not dephosphorylated in response to fMLF in RAC2-knockout cells. Overexpression of CIN rescued cofilin dephosphorylation under these circumstances80. Similarly, dephosphorylation of cofilin downstream of fMLF can be induced by the activation of the phosphatase SSH2 through repression of glycogen synthase kinase 3 (GSK3)81. PI3K can promote the insulin-induced activation of SSH1 and cofilin dephosphorylation in breast carcinoma cells, whereas PTEN antagonizes this effect, indicating a role for PtdIns(3,4,5) P3 in this pathway82.
LIMK1 and LIMK2 (REFS 45,83) as well as TES kinase 1 (TESK1) and TESK2 (REFS 84,85) phosphorylate co filin at Ser3 in vivo. In crawling cells, LIMKs are the most well-studied kinases and have been proposed to be the dominant kinase in the regulation of actin dynamics69. TESKs are thought to be involved in integrin signalling during focal adhesion formation69. LIMK1 and LIMK2 are activated by phosphorylation at Thr508 and Thr505, respectively, by various kinases, including ROCK86 PAK1, PAK2, PAK4 (REFS 87,88), MRCKα89 and MAPK-activated protein kinase 2 (REF. 90). The balance of activity between kinases and phosphatases that target cofilin is crucial in determining the amount and location of co filin activity in migrating cells91. However, it is not always accurate to describe this balance, as we have in FIG. 3, by simply illustrating that LIMKs inhibits cofilin, whereas phosphatases activate it. Mathematical simulations based on known rate constants of reactions in the cofilin activity cycle indicate that LIMK-dependent cofilin phosphorylation in the context of other regulatory interactions, including cofilin dephosphorylation and actin monomer and PtdIns(4,5)P 2 binding, can actually amplify cofilin activity locally. Modelling predicts that increasing LIMK activity in a spatially enclosed system, such as the leading edge, can amplify the cofilin severing response and cofilin-dependent barbed end production in the presence of these other regulatory events, even though LIMKs are generally assumed to deactivate cofilin59.
Although dephosphorylation at Ser3 is required for cofilin activity, it is not always sufficient for cofilin activation in a closed system in which other key regulatory events of the cofilin activity cycle can take place51. Thus, the amount of cofilin that is not phosphorylated on Ser3 does not directly reflect the level of cofilin activity. Other mechanisms that regulate cofilin activation, including its release from its inhibitory interaction with PtdIns(4,5)P2 and cortactin, must be considered.
Dissociation of the cofilin–PtdIns(4,5)P2 inhibitory complex
Cofilin is known to be inactivated by its interaction with PtdIns(4,5)P2 at the plasma membrane92,93. This follows a general mechanism whereby membrane lipids have been shown to bind various actin regulatory proteins such as profilin33 and gelsolin94 that also interact with PtdIns(4,5)P2 to inhibit their function95. In migrating cells, the hydrolysis of PtdIns(4,5)P2 by phospholipase C (PLC) to form inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG) can release cofilin from its inhibitory interaction with the lipid, resulting in the local activation of F-actin filament severing, protrusion and cell polarity49,96 (FIG. 4). In addition, the rapamycin-induced targeting of phosphoinositide 5-phosphatase to the plasma membrane, which results in dephosphorylation of PtdIns(4,5)P2, is sufficient to release cofilin from the plasma membrane and initiate the formation of cellular protrusions49. This indicates that PtdIns(4,5) P2 binding to cofilin directly inhibits cofilin activity at the plasma membrane. Moreover, new protrusions are initiated by the local photoactivation of caged S3C-cofilin (a mutant in which Ser3 was replaced with Cys), and these protrusions determine the direction of cell migration97. This suggests that the local release of cofilin from PtdIns(4,5)P2 binding does not only serve as an activation switch for protrusion initiation, but that cofilin locally activated in response to external stimuli causing hydrolysis of PtdIns(4,5)P2 can drive chemotaxis and determine the direction of cell migration49,55,56,96.
Figure 4. A new model for the activation of cofilin at the leading edge of locomotory protrusions.
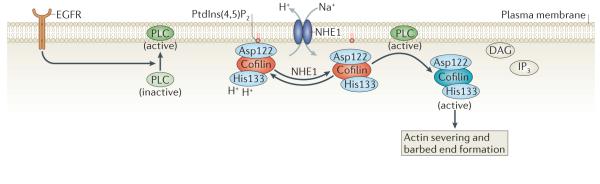
Inactive cofilin is bound to phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) at the plasma membrane through its Asp122 residue. Na+–H+ exchanger 1 (NHE1) can increase the intracellular pH by exchanging intracellular H+ for extracellular Na+. This causes the deprotonation of cofilin at His133, which alters its binding affinity for PtdIns(4,5)P2. An increased pH facilitates phospholipase C (PLC)-mediated hydrolysis of PtdIns(4,5)P2 to diacylglycerol (DAG) and inositol-1,4,5,-trisphosphate (IP3) and the release of cofilin from PtdIns(4,5)P2. Epidermal growth factor receptor (EGFR) stimulates PLC activity, which in turn increases cofilin activation. See BOX 1 for methods describing how to study cofilin mobility as well as cofilin binding to the plasma membrane and F-actin.
An additional regulatory overlay in the activation of cofilin by release from PtdIns(4,5)P2 is the involvement of local pH changes. Cofilin has long been recognized as a pH-sensitive actin-binding protein. Cofilin activity in vitro increases in the range of physiological pH from pH 6.8 to 7.2 (REFS 98–100) and, in fibroblasts, a rise in intracellular pH is necessary for cofilin-regulated actin dynamics101. His133 of cofilin is unprotonated at higher pH, which induces structural changes that decrease its binding affinity for other proteins, including cortactin102,103 (FIG. 2). The intracellular pH is regulated by the Na+–H+ exchanger 1 (NHE1), an ubiquitously expressed transmembrane protein that exchanges extracellular Na+ for intracellular H+ (REF. 104). The local increase in pH at the cytoplasmic side of the plasma membrane, which results from the recruitment and activation of NHE1, decreases cofilin-dependent clustering of PtdIns(4,5)P2 (REF. 105). This could lead to changes in PtdIns(4,5)P2 density and PLC-dependent lipid hydrolysis in vivo (FIG. 4). In this context, the pH-regulated clustering interaction between cofilin and PtdIns(4,5)P2 would act as a pH biosensor, connecting changes in intracellular pH to local protrusion and motility in crawling cells in response to growth factor stimulation102. This pH-dependent cofilin activation pathway may contribute to the spread of metastatic tumours106, which require the activation of cofilin for tumour cell motility and chemotaxis8,107,108. However, the release of cofilin from PtdIns(4,5)P2 by lipid hydrolysis is not absolutely dependent on increasing pH, as the ectopic delivery of an active phosphoinositide 5-phosphatase to the plasma membrane is sufficient for the activation of cofilin and membrane protrusion49. A new signalling pathway for the activation of cofilin that involves release from PtdIns(4,5)P2 is summarized in FIG. 4.
Dissociation of the cofilin–cortactin inhibitory complex
The binding of cofilin to cortactin also negatively regulates cofilin activity, and this mechanism seems to be specific to invasive protrusions, such as invadopodia109–112 (FIG. 5). Initially, the release of cofilin from cortactin in invadopodia has been correlated with cortactin phosphorylation113. More recently, it was shown that cortactin phosphorylation is catalysed by Arg kinase in tumour cells114, but this does not directly regulate co filin–cortactin binding, as both the phosphorylated and dephosphorylated forms of cortactin bind equally well to cofilin103. Further analysis demonstrated that cortactin phosphorylation results in the recruitment of NHE1 to the invadopodium core, causing a local increase in the intracellular pH. This increase in pH releases cofilin from its inhibitory binding to cortactin, resulting in the local activation of cofilin severing and actin polymerization in invadopodia. This mechanism has been shown to regulate the oscillatory protrusion of invadopodia and subsequently tumour cell invasion in two dimensions and three dimensions, as the submembrane compartment undergoes oscillatory changes in the intracellular pH103. Interestingly, cofilin activation at the leading edge of locomotory protrusions (for example, lamellipodia) and in invasive protrusions (invadopodia) requires local pH changes to enable the release of cofilin from its inhibitory binding partners, PtdIns(4,5)P2 and cortactin, respectively. Whether this common pattern extends to cofilin in other cellular compartments, and how these different compartments are coordinated in their activation of cofilin during cell migration, remains to be investigated.
The array treadmilling protrusion model
A well-known model for actin polymerization during protrusion in migrating cells is the array treadmilling protrusion model (FIG. 6a). This model was proposed on the basis of in vitro biochemical data and suggests that the ARP2/3 complex is the central organizer of actin dynamics during protrusion115,116.
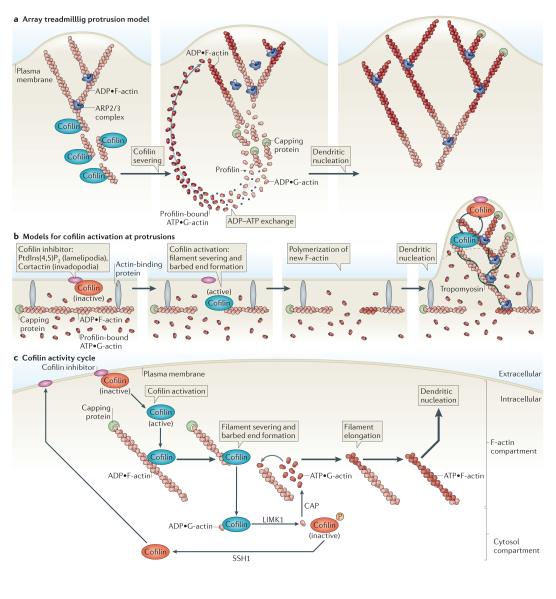
Figure 6. Models for cofilin function.
a ∣ Array treadmilling protrusion model. Cofilin severs and depolymerizes actin filaments at the base of the lamellipodium, thereby supplying G-actin monomers for steady-state actin polymerization in conditions of G-actin depletion. Dendritic nucleation is mediated by the actin-related protein 2/3 (ARP2/3) complex. b ∣ Models for cofilin activation at protrusions. Release of cofilin from its inhibitor (phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) at lamellipodia or cortactin at invadopodia) at the plasma membrane increases severing of actin filaments, generating free barbed ends that define the sites of dendritic nucleation by the ARP2/3 complex. G-actin monomers are supplied from an abundant pre-existing G-actin pool. The ARP2/3 complex mediates dendritic nucleation. Tropomyosin limits cofilin action, as it inhibits binding of cofilin to F-actin. This confines cofilin severing to the tip of protrusions (dotted black line), where cofilin continues its cycles of activation and deactivation due to the local cofilin activity cycle. c ∣ Cofilin activity cycle. Following its dephosphorylation or its release from inhibitory binding partners, activated cofilin binds to and severs actin filaments, generating barbed ends that are used for actin polymerization. LIM kinase 1 (LIMK1) and cyclase-associated protein (CAP) accelerate the dissociation of ADP•G-actin–cofilin complexes, and this increases the concentration of ATP•G-actin monomers available for filament elongation at the newly formed barbed ends. The capping of new barbed ends by actin interacting protein 1 (AIP1) (not shown) promotes cofilin-dependent filament disassembly. LIMK1 phosphorylates cofilin and thus inactivates it, leading to the release of bound G-actin. Upon the activation of slingshot 1 (SSH1) , cofilin is dephosphorylated and recycled to the inhibitory compartment. Figures are not drawn to scale. See BOX 1 for methods describing how to study cofilin activation.
One interpretation of this model is that cofilin functions exclusively as an actin depolymerization factor that depolymerizes actin filaments at the back of the protrusion to yield G-actin. In this way, cofilin functions only to generate G-actin to sustain steady-state ARP2/3 complex-mediated dendritic nucleation of actin polymerization at the leading edge of the cell69. Cofilin-dependent actin depolymerization coupled to ARP2/3 complex activity may be required for the extension of the dendritic actin network to power protrusion, the first step of the motility cycle of migrating cells (FIG. 1).
The array treadmilling protrusion model should be revised to incorporate recent observations about the dynamics of actin polymerization in protrusions and cofilin functions in particular.
The treadmilling protrusion model should include cofilin severing and cofilin nucleation that together produce free actin filament barbed ends at the leading edge for actin polymerization and protrusion initiation. For example, cofilin itself functions in early polymerization that precedes ARP2/3 complex-mediated dendritic nucleation117 (FIG. 6b).
Furthermore, the different shapes of protrusions, ranging from flat lamellipodia to needle-shaped invadopodia, are not explained by the array treadmilling protrusion model. The local release of cofilin from PtdIns(4,5)P2 at membranes and from cortactin within the invadopodium core, which leads to its activation, can define the boundaries of actin polymerization into complex shapes. For example, during invadopodium formation, a RHOC–ROCK dependent pathway increases LIMK1 activity surrounding the invadopodium core.
Activation of p190 RHO guanine nucleotide exchange factor (p190 RHO GEF; which activates RHOC) outside the core and of p190 RHO GTPase-activating protein (GAP; which deactivates RHOC) within the core restricts RHOC activity to a ring surrounding the invadopodium core. As RHOC activates LIMK1, this concentrates active cofilin (that is, not phosphorylated on Ser3) within the core and inactive Ser3 phosphorylated cofilin outside the core. The same mechanism has been shown to regulate spatial confinement of cofilin at the leading edge of locomotory protrusions. The restriction of cofilin activity to the core of the invadopodium45 or at the tip of the leading edge46 confines actin polymerization and F-actin turnover to this region. As a result, optimized focal invadopodial and lamellipodial protrusions and retraction oscillations are achieved during tumour cell invasion38,45,103.
Evidence shows that a fraction of active cofilin that functions in filament severing is found adjacent to the cytoplasmic surface of the plasma membrane, whereas Ser3 phosphorylated cofilin is more distant from the plasma membrane46,49,51,56,96,102. This adds an additional dimension to the location of cofilin activity in the array treadmilling protrusion model, placing cofilin activity at the front of the extending protrusion (FIG. 6b).
The current array treadmilling protrusion model does not take into account the synergistic interaction between cofilin and the ARP2/3 complex, in which cofilin supplies newly polymerizing actin filaments on which the ARP2/3 complex-nucleated branches are more stable thereby placing these branches at the cell membrane20,35,118.
In addition, the inhibitory effect of tropomyosin on cofilin activity at the base of locomotory protrusions, where tropomyosin is bound to actin filaments, prevents the severing and depolymerization of actin filaments at a region where the severing activity of cofilin has been proposed to occur in the array treadmilling protrusion model60 (FIG. 6b).
Notably, cofilin, but not the ARP2/3 complex, is required for the early actin polymerization response to growth factor stimulation and the resulting initiation of protrusions in migrating cells49,96,102,117. These results further highlight the importance of cofilin as an actin polymerization factor at the leading edge of locomotory protrusions.
Finally, cofilin, but not the ARP2/3 complex, is involved in directional protrusion during chemotaxis in cancer cells55,97,117, which is consistent with studies in fibroblasts showing that the ARP2/3 complex is not required for chemotaxis119. These results are consistent with the finding that ARP2/3 complex-dependent array treadmilling is not part of the early actin polymerization phase involved in chemosensing at the leading edge55,117.
New models for cofilin function in migrating cells
New models for the initiation of cofilin activity in locomotory and invasive protrusions, are shown in FIGS 4–6. The cofilin activity cycle (FIG. 6c) is a common feature of both invasive and locomotory protrusions and is common to cell types as diverse as leukocytes and tumour cells120.
Two major additions have been made to the array treadmilling protrusion model. First, the model now suggests that a significant fraction of active cofilin associates with plasma membrane-adjacent actin filaments at the leading edge of locomotory protrusions49,55,59 (FIGS 4,6b) and not only at the back of the lamellipodium as previously suggested (FIG. 6a). This is consistent with a role of cofilin in initiating protrusion activity. In addition, cofilin is localized at the cortactin-containing core of invasive protrusions (FIGS 5,6b).
Second, an activation step that spatially determines cofilin activation is now central to new models for cofilin function during protrusion. In locomotory protrusions, this mechanism includes the local action of NHE1 and PLC leading to the release of active cofilin from the plasma membrane (FIG. 4). In invasive protrusions, cortactin and NHE1 regulate the localized activation of cofilin at the invadopodium core. Cortactin phosphorylation by SRC and Arg kinase recruits NHE1 (a step thought to involve ERM (ezrin–radixin–moesin) proteins), and the resulting rise in pH releases cofilin from its inhibitory interaction with cortactin and local cofilin activation (FIG. 5).
On the basis of these new models, it is now possible to clarify inconsistencies that have complicated our understanding of cofilin function in vivo. A common cofilin activity cycle that describes cofilin activity in crawling cells (FIG. 6c) explains how results obtained from various cell types that begin the cycle at different starting points can suggest that there are distinct mechanisms of cofilin regulation depending on cell type. This confusion largely stems from the relatively low resolution of methods that are used to analyse cofilin activity in vivo. Due to the rapid movement of cofilin between the cytosol, plasma membrane and cortactin- and F-actin-rich compartments it seems that cofilin activity is uncoupled from these regulatory events in each compartment. Cofilin localization to these subcellular compartments cannot be distinguished using standard light microscopy, as the distance between these compartments is within the diffraction limited spot of conventional light microscopes. Determining the localization and movement of cofilin in these different compartments and where cofilin is active requires high-resolution imaging methods as described in BOX 1 (see also Supplementary information S1 (box).
For example, because increased cofilin phosphorylation at Ser3 accompanies the initiation of its activity cycle, it has been proposed that cofilin phosphorylation is required for actin polymerization and that dephosphorylated cofilin is only involved in actin depolymerization. However, the new models derived from high-resolution imaging reveal that increased levels of cofilin phosphorylation are a result of cofilin activation by dephosphorylation-independent mechanisms51 (FIGS 4–6), which can lead to actin polymerization as described above. It has been proposed that cofilin phosphorylation is involved in recycling cofilin back to the initial starting point in its activity cycle (FIG. 6c) and in spatially restricting cofilin activity to determine the shape of protrusions45,59.
Manipulating the expression or activity of a component of the cofilin activity cycle without measuring the corresponding change in output (that is, the cofilin-dependent actin filament free barbed end transient) can also lead to inconsistent data. For example, increasing LIMK activity in a crawling cell may inactivate cofilin globally. If an increase in cell protrusion activity is observed after increasing LIMK, it is sometimes mistakenly concluded that cofilin inhibits protrusion. However, cofilin activity at the leading edge can increase following a rise in LIMK activity as cofilin is restricted to this location, leading to increased protrusion. This relationship can only be detected by directly measuring cofilin-generated barbed end transients59,91 (BOX 1, Supplementary information S1 (box)).
Conclusions
Recent studies have confirmed that cofilin is a central component of actin dynamics in migrating cells because it catalyses either actin polymerization or depolymerization through its severing activity and due to its ability to catalyse dendritic nucleation stability and debranching. We now understand the structural basis of cofilin-mediated severing, which explains how cofilin can lead to both polymerization and depolymerization in a single severing event. Cofilin is active at the leading edge of locomotory protrusions of migrating cells, and inhibition of its activity causes defects in protrusion, cell polarity and chemotaxis. This conclusion is supported by the discovery that there are three main mechanisms by which cofilin is activated: the dephosphorylation of cofilin at Ser3; the release of cofilin from PtdIns(4,5)P2; and the release of cofilin from cortactin. In particular, the PtdIns(4,5)P2-dependent regulatory mechanism, which is governed by stimulated PLC activation, is important for the spatial and temporal control of cofilin activation required for the initiation of the locomotory protrusion, cell polarity and chemotaxis. The integration of these regulatory mechanisms within the cofilin activity cycle has allowed us to add refinements to the array treadmilling model of protrusion dynamics and presents an opportunity, as illustrated in the new models of cofilin function, to understand the temporal and spatial shaping of cell protrusions during cell migration.
Supplementary Material
Acknowledgements
The authors would like to thank members of the Condeelis and Hodgson laboratories for helpful discussions. They apologize to those whose work could not be cited owing to space limitations. The authors’ research is funded by grants GM093121 (to L.H. and J.J.B.-C.) and CA150344 (to J.C., R.E. and J.J.B.-C.).
Glossary
- Motility cycle
A series of steps that enables cell movement. The first step is the formation of a directional protrusion driven by actin polymerization, followed by adhesion, contractile tension and tail retraction.
- Amoeboid cell migration
A common type of motility whereby cells extend actin polymerization-dependent protrusions in the direction of migration.
- Chemotaxis
Polarized cell migration in response to extracellular, soluble cues. Chemotaxis is characterized by the directional extension of a locomotory protrusion towards the source of a chemoattractant.
- Pointed end
The end of the actin filament that is characterized by a slow growing rate.
- Barbed ends
The fast growing ends of actin filaments that are characterized by a fast ‘on rate’ and high affinity for ATP•G-actin.
- Invadopodium
Invasive protrusion that is extended by actin polymerization and is involved in extracellular matrix degradation.
- Lamellipodium
A common locomotory protrusion that is extended by actin polymerization.
- Histamine degranulation
Release of the pro-inflammatory molecule histamine from intracellular granules.
- Crawling cells
Cells that use the motility cycle for locomotion.
- Focal adhesion
Macromolecular complexes involved in cell–extracellular matrix interactions.
Footnotes
Images for this box are available in Supplementary information S1 (box).
Competing interests statement The authors declare competing financial interests: see Web version for details.
SUPPLEMENTARY INFORMATION See online article: S1 (box)
References
- 1.Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nature Rev. Mol. Cell Biol. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 2.Daubon T, Rochelle T, Bourmeyster N, Genot E. Invadopodia and rolling-type motility are specific features of highly invasive p190bcr-abl leukemic cells. Eur. J. Cell Biol. 2012;91:978–987. doi: 10.1016/j.ejcb.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J. Cell Sci. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nature Rev. Cancer. 2011;11:177–187. doi: 10.1038/nrc3003. [DOI] [PubMed] [Google Scholar]
- 5.Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nature Rev. Mol. Cell Biol. 2012;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 7.Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells — over and over and over again. Nature Cell Biol. 2002;4:e97–e100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- 8.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nature Rev. Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Pollard TD. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J. Cell Biol. 2011;195:485–498. doi: 10.1083/jcb.201103067. Demonstrates that cofilin severing activity is crucial for the assembly and contractility of the contractile ring during cytokinesis.
- 10.Zhang L, et al. Regulation of cofilin phosphorylation and asymmetry in collective cell migration during morphogenesis. Development. 2011;138:455–464. doi: 10.1242/dev.046870. [DOI] [PubMed] [Google Scholar]
- 11.Gu J, et al. ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nature Neurosci. 2010;13:1208–1215. doi: 10.1038/nn.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bamburg JR, Wiggan OP. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 2002;12:598–605. doi: 10.1016/s0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poukkula M, Kremneva E, Serlachius M, Lappalainen P. Actin-depolymerizing factor homology domain: a conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton (Hoboken) 2011;68:471–490. doi: 10.1002/cm.20530. Reviews the different proteins that contain ADF homology domains.
- 15.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Ichetovkin I, Han J, Pang KM, Knecht DA, Condeelis JS. Actin filaments are severed by both native and recombinant dictyostelium cofilin but to different extents. Cell Motil. Cytoskeleton. 2000;45:293–306. doi: 10.1002/(SICI)1097-0169(200004)45:4<293::AID-CM5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J. Mol. Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlier MF, et al. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlier MF, Pantaloni D. Control of actin dynamics in cell motility. J. Mol. Biol. 1997;269:459–467. doi: 10.1006/jmbi.1997.1062. [DOI] [PubMed] [Google Scholar]
- 20.Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- 21.Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys. J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mseka T, Cramer LP. Actin depolymerizationbased force retracts the cell rear in polarizing and migrating cells. Curr. Biol. 2011;21:2085–2091. doi: 10.1016/j.cub.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Wiggan O, Shaw AE, DeLuca JG, Bamburg JR. ADF/cofilin regulates actomyosin assembly through competitive inhibition of myosin II binding to F-actin. Dev. Cell. 2012;22:530–543. doi: 10.1016/j.devcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galkin VE, et al. Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl Acad. Sci. USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. Describes the mechanism of actin filament severing by cofilin.
- 26.Prochniewicz E, Janson N, Thomas DD, De La Cruz EM. Cofilin increases the torsional flexibility and dynamics of actin filaments. J. Mol. Biol. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 27.Suarez C, et al. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCullough BR, Blanchoin L, Martiel JL, De La Cruz EM. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. J. Mol. Biol. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bobkov AA, et al. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J. Mol. Biol. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 30.Mehta S, Sibley LD. Toxoplasma gondii actin depolymerizing factor acts primarily to sequester G-actin. J. Biol. Chem. 2010;285:6835–6847. doi: 10.1074/jbc.M109.068155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriyama K, Yahara I. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J. Cell Sci. 2002;115:1591–1601. doi: 10.1242/jcs.115.8.1591. [DOI] [PubMed] [Google Scholar]
- 33.Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev. Physiol. Biochem. Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- 34.Normoyle KP, Brieher WM. Cyclase-associated protein (CAP) acts directly on F-actin to accelerate cofilin-mediated actin severing across the range of physiological pH. J. Biol. Chem. 2012;287:35722–35732. doi: 10.1074/jbc.M112.396051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DesMarais V, Ghosh M, Eddy R, Condeelis J. Cofilin takes the lead. J. Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- 36.Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiuchi T, Ohashi K, Kurita S, Mizuno K. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J. Cell Biol. 2007;177:465–476. doi: 10.1083/jcb.200610005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J. Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin MC, Galletta BJ, Sept D, Cooper JA. Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J. Cell Sci. 2010;123:1329–1342. doi: 10.1242/jcs.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohri K, Ono K, Yu R, Yamashiro S, Ono S. Enhancement of actin-depolymerizing factor/cofilindependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol. Biol. Cell. 2006;17:2190–2199. doi: 10.1091/mbc.E05-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okada K, et al. Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 2002;277:43011–43016. doi: 10.1074/jbc.M203111200. [DOI] [PubMed] [Google Scholar]
- 43.Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/Cofilin-bound actin filaments. J. Biol. Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- 44.Philippar U, et al. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev. Cell. 2008;15:813–828. doi: 10.1016/j.devcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bravo-Cordero JJ, et al. A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr. Biol. 2011;21:635–644. doi: 10.1016/j.cub.2011.03.039. Shows that confinement of cofilin activity at the invadopodium core involves RHOC-mediated cofilin phosphorylation outside the invadopodium, concentrating activated cofilin inside the invadopodium core.
- 46.Bravo-Cordero JJ, et al. Spatial regulation of RhoC activity defines protrusion formation in migrating cells. J. Cell Sci. 2013 May 23; doi: 10.1242/jcs.123547. (doi:10.1242/jcs.123547) Demonstrates how RHOC spatially restricts cofilin activity at the leading edge to drive polarized protrusions and chemotaxis.
- 47.Han L, et al. Direct stimulation of receptor-controlled phospholipase D1 by phospho-cofilin. EMBO J. 2007;26:4189–4202. doi: 10.1038/sj.emboj.7601852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DesMarais V, Macaluso F, Condeelis J, Bailly M. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J. Cell Sci. 2004;117:3499–3510. doi: 10.1242/jcs.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Rheenen J, et al. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 2007;179:1247–1259. doi: 10.1083/jcb.200706206. Shows how PLC releases the inhibitory interaction of cofilin with PtdIns(4,5)P2 at the plasma membrane, resulting in the local activation of cofilin severing activity.
- 50.Kim JS, Huang TY, Bokoch GM. Reactive oxygen species regulate a Slingshot–cofilin activation pathway. Mol. Biol. Cell. 2009;20:2650–2660. doi: 10.1091/mbc.E09-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song X, et al. Initiation of cofilin activity in response to EGF is uncoupled from cofilin phosphorylation and dephosphorylation in carcinoma cells. J. Cell Sci. 2006;119:2871–2881. doi: 10.1242/jcs.03017. [DOI] [PubMed] [Google Scholar]
- 52.Sakuma M, et al. Novel PKCα-mediated phosphorylation site(s) on cofilin and their potential role in terminating histamine release. Mol. Biol. Cell. 2012;23:3707–3721. doi: 10.1091/mbc.E12-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoo Y, Ho HJ, Wang C, Guan JL. Tyrosine phosphorylation of cofilin at Y68 by v-Src leads to its degradation through ubiquitin–proteasome pathway. Oncogene. 2010;29:263–272. doi: 10.1038/onc.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delorme V, et al. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev. Cell. 2007;13:646–662. doi: 10.1016/j.devcel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mouneimne G, et al. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 2006;16:2193–2205. doi: 10.1016/j.cub.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 56.Sidani M, et al. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 2007;179:777–791. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin–actin filament interactions. J. Mol. Biol. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 58.Ressad F, Didry D, Egile C, Pantaloni D, Carlier MF. Control of actin filament length and turnover by actin depolymerizing factor (ADF/cofilin) in the presence of capping proteins and ARP2/3 complex. J. Biol. Chem. 1999;274:20970–20976. doi: 10.1074/jbc.274.30.20970. [DOI] [PubMed] [Google Scholar]
- 59.Tania N, Prosk E, Condeelis J, Edelstein-Keshet L. A temporal model of cofilin regulation and the early peak of actin barbed ends in invasive tumor cells. Biophys. J. 2011;100:1883–1892. doi: 10.1016/j.bpj.2011.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 2002;115:4649–4660. doi: 10.1242/jcs.00147. [DOI] [PubMed] [Google Scholar]
- 61.Munsie LN, Desmond CR, Truant R. Cofilin nuclear–cytoplasmic shuttling affects cofilin–actin rod formation during stress. J. Cell Sci. 2012;125:3977–3988. doi: 10.1242/jcs.097667. [DOI] [PubMed] [Google Scholar]
- 62.Obrdlik A, Percipalle P. The F-actin severing protein cofilin-1 is required for RNA polymerase II transcription elongation. Nucleus. 2011;2:72–79. doi: 10.4161/nucl.2.1.14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arber S, et al. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 64.Nagaoka R, Minami N, Hayakawa K, Abe H, Obinata T. Quantitative analysis of low molecular weight G-actin-binding proteins, cofilin, ADF and profilin, expressed in developing and degenerating chicken skeletal muscles. J. Muscle Res. Cell Motil. 1996;17:463–473. doi: 10.1007/BF00123362. [DOI] [PubMed] [Google Scholar]
- 65.Moriyama K, Iida K, Yahara I. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells. 1996;1:73–86. doi: 10.1046/j.1365-2443.1996.05005.x. [DOI] [PubMed] [Google Scholar]
- 66.Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- 67.Gohla A, Birkenfeld J, Bokoch GM. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nature Cell Biol. 2005;7:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- 68.Ambach A, et al. The serine phosphatases PP1 and PP2A associate with and activate the actin-binding protein cofilin in human T lymphocytes. Eur. J. Immunol. 2000;30:3422–3431. doi: 10.1002/1521-4141(2000012)30:12<3422::AID-IMMU3422>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 69.Mizuno K. Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell. Signal. 2012;25:457–469. doi: 10.1016/j.cellsig.2012.11.001. Reviews the mechanisms regulating the activity of LIMK and SSH and how these proteins influence cofilin activity.
- 70.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagata-Ohashi K, et al. A pathway of neuregulininduced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J. Cell Biol. 2004;165:465–471. doi: 10.1083/jcb.200401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eiseler T, et al. Protein kinase D1 regulates cofilinmediated F-actin reorganization and cell motility through slingshot. Nature Cell Biol. 2009;11:545–556. doi: 10.1038/ncb1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spratley SJ, Bastea LI, Doppler H, Mizuno K, Storz P. Protein kinase D regulates cofilin activity through p21-activated kinase 4. J. Biol. Chem. 2011;286:34254–34261. doi: 10.1074/jbc.M111.259424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterburs P, et al. Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 2009;69:5634–5638. doi: 10.1158/0008-5472.CAN-09-0718. [DOI] [PubMed] [Google Scholar]
- 75.Wang Y, Shibasaki F, Mizuno K. Calcium signalinduced cofilin dephosphorylation is mediated by Slingshot via calcineurin. J. Biol. Chem. 2005;280:12683–12689. doi: 10.1074/jbc.M411494200. [DOI] [PubMed] [Google Scholar]
- 76.Zhao JW, et al. Regulation of cofilin activity by CaMKII and calcineurin. Am. J. Med. Sci. 2012;344:462–472. doi: 10.1097/MAJ.0b013e318244745b. [DOI] [PubMed] [Google Scholar]
- 77.Zoudilova M, et al. β-arrestin-dependent regulation of the cofilin pathway downstream of proteaseactivated receptor-2. J. Biol. Chem. 2007;282:20634–20646. doi: 10.1074/jbc.M701391200. [DOI] [PubMed] [Google Scholar]
- 78.Hirayama A, Adachi R, Otani S, Kasahara T, Suzuki K. Cofilin plays a critical role in IL-8-dependent chemotaxis of neutrophilic HL-60 cells through changes in phosphorylation. J. Leukoc. Biol. 2007;81:720–728. doi: 10.1189/jlb.0506314. [DOI] [PubMed] [Google Scholar]
- 79.Boldt K, Rist W, Weiss SM, Weith A, Lenter MC. FPRL-1 induces modifications of migration-associated proteins in human neutrophils. Proteomics. 2006;6:4790–4799. doi: 10.1002/pmic.200600121. [DOI] [PubMed] [Google Scholar]
- 80.Sun CX, Magalhaes MA, Glogauer M. Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J. Cell Biol. 2007;179:239–245. doi: 10.1083/jcb.200705122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang W, et al. A PLCβ/PI3Kγ–GSK3 signaling pathway regulates cofilin phosphatase slingshot2 and neutrophil polarization and chemotaxis. Dev. Cell. 2011;21:1038–1050. doi: 10.1016/j.devcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nishita M, et al. Phosphoinositide 3-kinase-mediated activation of cofilin phosphatase Slingshot and its role for insulin-induced membrane protrusion. J. Biol. Chem. 2004;279:7193–7198. doi: 10.1074/jbc.M312591200. [DOI] [PubMed] [Google Scholar]
- 83.Scott RW, et al. LIM kinases are required for invasive path generation by tumor and tumorassociated stromal cells. J. Cell Biol. 2010;191:169–185. doi: 10.1083/jcb.201002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toshima J, et al. Cofilin phosphorylation by protein kinase testicular protein kinase 1 and its role in integrin-mediated actin reorganization and focal adhesion formation. Mol. Biol. Cell. 2001;12:1131–1145. doi: 10.1091/mbc.12.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toshima J, Toshima JY, Takeuchi K, Mori R, Mizuno K. Cofilin phosphorylation and actin reorganization activities of testicular protein kinase 2 and its predominant expression in testicular Sertoli cells. J. Biol. Chem. 2001;276:31449–31458. doi: 10.1074/jbc.M102988200. [DOI] [PubMed] [Google Scholar]
- 86.Maekawa M, et al. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIMkinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 87.Dan C, Kelly A, Bernard O, Minden A. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J. Biol. Chem. 2001;276:32115–32121. doi: 10.1074/jbc.M100871200. [DOI] [PubMed] [Google Scholar]
- 88.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nature Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 89.Sumi T, Matsumoto K, Shibuya A, Nakamura T. Activation of LIM kinases by myotonic dystrophy kinase-related Cdc42-binding kinase α. J. Biol. Chem. 2001;276:23092–23096. doi: 10.1074/jbc.C100196200. [DOI] [PubMed] [Google Scholar]
- 90.Kobayashi M, Nishita M, Mishima T, Ohashi K, Mizuno K. MAPKAPK-2-mediated LIM-kinase activation is critical for VEGF-induced actin remodeling and cell migration. EMBO J. 2006;25:713–726. doi: 10.1038/sj.emboj.7600973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nature Rev. Cancer. 2007;7:429–440. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gorbatyuk VY, et al. Mapping the phosphoinositidebinding site on chick cofilin explains how PIP2 regulates the cofilin–actin interaction. Mol. Cell. 2006;24:511–522. doi: 10.1016/j.molcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 93.Yonezawa N, Nishida E, Iida K, Yahara I, Sakai H. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 1990;265:8382–8386. [PubMed] [Google Scholar]
- 94.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J. Biol. Chem. 1999;274:33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 95.Yin HL, Janmey PA. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- 96.Leyman S, et al. Unbalancing the phosphatidylinositol-4,5-bisphosphate–cofilin interaction impairs cell steering. Mol. Biol. Cell. 2009;20:4509–4523. doi: 10.1091/mbc.E09-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ghosh M, et al. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 98.Chen H, et al. In vitro activity differences between proteins of the ADF/cofilin family define two distinct subgroups. Biochemistry. 2004;43:7127–7142. doi: 10.1021/bi049797n. [DOI] [PubMed] [Google Scholar]
- 99.Hawkins M, Pope B, Maciver SK, Weeds AG. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- 100.Maciver SK, Pope BJ, Whytock S, Weeds AG. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur. J. Biochem. 1998;256:388–397. doi: 10.1046/j.1432-1327.1998.2560388.x. [DOI] [PubMed] [Google Scholar]
- 101.Bernstein BW, et al. Intracellular pH modulation of ADF/cofilin proteins. Cell Motil. Cytoskeleton. 2000;47:319–336. doi: 10.1002/1097-0169(200012)47:4<319::AID-CM6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 102.Frantz C, et al. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 2008;183:865–879. doi: 10.1083/jcb.200804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Magalhaes MA, et al. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 2011;195:903–920. doi: 10.1083/jcb.201103045. Shows that the recruitment of NHE1 to invadopodia locally increases the pH, which releases the inhibitory interacting of cofilin with cortactin and leads to the activation of cofilin severing activity.
- 104.Kemp G, Young H, Fliegel L. Structure and function of the human Na+/H+ exchanger isoform 1. Channels (Austin) 2008;2:329–336. doi: 10.4161/chan.2.5.6898. [DOI] [PubMed] [Google Scholar]
- 105.Zhao H, Hakala M, Lappalainen P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP2-density sensor. Biophys. J. 2010;98:2327–2336. doi: 10.1016/j.bpj.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nature Rev. Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 107.Wang W, et al. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang W, et al. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007;67:3505–3511. doi: 10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- 109.Buccione R, Caldieri G, Ayala I. Invadopodia: specialized tumor cell structures for the focal degradation of the extracellular matrix. Cancer Metastasis Rev. 2009;28:137–149. doi: 10.1007/s10555-008-9176-1. [DOI] [PubMed] [Google Scholar]
- 110.Linder S, Wiesner C, Himmel M. Degrading devices: invadosomes in proteolytic cell invasion. Annu. Rev. Cell Dev. Biol. 2011;27:185–211. doi: 10.1146/annurev-cellbio-092910-154216. [DOI] [PubMed] [Google Scholar]
- 111.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nature Rev. Mol. Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weaver AM. Invadopodia: specialized cell structures for cancer invasion. Clin. Exp. Metastasis. 2006;23:97–105. doi: 10.1007/s10585-006-9014-1. References 109–112 review invasive protrusions.
- 113.Oser M, et al. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 2009;186:571–587. doi: 10.1083/jcb.200812176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mader CC, et al. An EGFR–Src–Arg–cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71:1730–1741. doi: 10.1158/0008-5472.CAN-10-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 116.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 117.Mouneimne G, et al. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 2004;166:697–708. doi: 10.1083/jcb.200405156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shao D, Forge A, Munro PM, Bailly M. Arp2/3 complex-mediated actin polymerisation occurs on specific pre-existing networks in cells and requires spatial restriction to sustain functional lamellipod extension. Cell Motil. Cytoskeleton. 2006;63:395–414. doi: 10.1002/cm.20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu C, et al. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Rheenen J, Condeelis J, Glogauer M. A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J. Cell Sci. 2009;122:305–311. doi: 10.1242/jcs.031146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lai FP, et al. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ohashi K, Kiuchi T, Shoji K, Sampei K, Mizuno K. Visualization of cofilin-actin and Ras–Raf interactions by bimolecular fluorescence complementation assays using a new pair of split Venus fragments. Biotechniques. 2012;52:45–50. doi: 10.2144/000113777. [DOI] [PubMed] [Google Scholar]
- 124.Soderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.