Abstract
The opportunistic pathogen Legionella pneumophila alternates between two states: replication within phagocytes and transmission between host amoebae or macrophages. In broth cultures that model this life cycle, during the replication period, CsrA inhibits expression of transmission traits. When nutrients become limiting, the alarmone (p)ppGpp accumulates and the sigma factors RpoS and FliA and the positive activators LetA/S and LetE promote differentiation to the transmissible form. Here we show that when cells enter the postexponential growth phase, RpoS increases expression of the transmission genes fliA, flaA, and mip, factors L. pneumophila needs to establish a new replication niche. In contrast, in exponential (E)-phase cells whose (p)ppGpp levels are low, rpoS inhibits expression of transmission traits, on the basis of three separate observations. First, rpoS RNA levels peak in the E phase, suggestive of a role for RpoS during replication. Second, in multiple copies, rpoS decreases the amounts of csrA, letE, fliA, and flaA transcripts and inhibits the transmission traits of motility, infectivity, and cytotoxicity. Third, rpoS blocks expression of cytotoxicity and motility by E-phase bacteria that have been induced to express the LetA activator ectopically. The data are discussed in the context of a model in which the alarmone (p)ppGpp enables RpoS to outcompete other sigma factors for binding to RNA polymerase to promote transcription of transmission genes, while LetA/S acts in parallel to relieve CsrA posttranscriptional repression of the transmission regulon. By coupling transcriptional and posttranscriptional control pathways, intracellular L. pneumophila could respond to stress by rapidly differentiating to a transmissible form.
Legionella pneumophila has the remarkable ability to replicate within two distantly related host cells, freshwater amoebae and alveolar macrophages. However, infection of the human lung is likely fortuitous. Disease outbreaks that occur when L. pneumophila contaminates water sources can affect large numbers of people, but no secondary cases due to person-to-person transmission have been reported. Accordingly, the ability of L. pneumophila to cause pneumonia in humans is likely the consequence of selective pressures to disrupt bactericidal activities of amoebae that are common to macrophages (reviewed in reference 59).
When ingested by a macrophage or an amoeba, L. pneumophila blocks its immediate delivery to lysosomes (6, 31). Instead, its phagosome transiently associates with mitochondria and endoplasmic reticulum, and then the microbe replicates to high numbers in an acidic vacuole that acquires lysosomal traits (18, 30, 58, 60). When it has exhausted the nutrient supply, L. pneumophila must exit the host cell and search for new prey. To do so, after the replication period, the progeny not only increase their resistance to extracellular stresses (4, 20, 22, 42) but also express a pore-forming activity to escape the host (2, 8, 35), assemble a unipolar flagellum to disseminate (8, 51), and use a type IV secretion system to inhibit phagosome maturation in the new host cell (5, 52, 56, 62), where the cycle repeats.
From studies of broth cultures that model the L. pneumophila life cycle, several features of the regulatory circuit that governs bacterial differentiation have been deduced. Exponential (E)-phase bacteria respond to nutrient depletion or other stress by producing the alarmone (p)ppGpp (21). Subsequently, as the bacteria transition to the postexponential (PE) phase, the sigma factor RpoS induces motility, sodium sensitivity, and evasion of the endocytic pathway, and it also promotes subsequent replication of the intracellular bacteria (3, 20). In parallel, the LetA/S two-component regulator system also induces motility, sodium sensitivity, and evasion of lysosomes, and it activates cytotoxicity, pigmentation, resistance to heat and osmotic stress, and cell shortening (17, 22, 42). LetA/S likely does so by relieving translational repression by CsrA, thereby activating the transmission phenotype by pathways that are both dependent on and independent of the flagellar sigma factor FliA (17, 44). By an unknown mechanism, the LetE protein enhances expression of LetA/S-dependent transmission traits (22). Thus, by integrating the activity of multiple sigma factors, a two-component regulatory system, and a repressor of translation, L. pneumophila can efficiently alternate between an intracellular replicative state and a form fit to spread among populations of amoebae or macrophages.
Among gram-negative bacteria, RpoS, LetA/S, and CsrA are highly conserved global regulators of stationary-phase physiology that act by distinct mechanisms. The RpoS sigma factor directs RNA polymerase to transcribe genes important for survival in stationary phase and under other stressful conditions (24). In contrast, the LetA/S homologues known as GacA/S or BarA/SirA act posttranscriptionally (23). Specifically, these two-component regulatory systems relieve repression by the mRNA binding protein CsrA by inducing the expression of the regulatory RNA csrB, which binds CsrA protein to derepress translation of mRNAs (reviewed in reference 50). In a number of human pathogens, including L. pneumophila, Salmonella enterica serovar Typhimurium, and Pseudomonas aeruginosa, a subset of genes in both the RpoS and the LetA/S (GacA/S, BarA/SirA) regulons encode virulence effectors (1, 3, 15, 17, 20, 22, 34, 39, 42, 44, 49). By using both transcriptional and translational control mechanisms, bacteria could respond quickly to environmental fluctuations by altering their composition.
In a number of bacterial species, the RpoS sigma factor and the LetA/S posttranscriptional regulatory system directly interact, although to different effect. In Escherichia coli and Pseudomonas fluorescens, LetA/S homologues upregulate rpoS transcript levels beginning in E phase and are required for RpoS-dependent traits in PE phase (46, 63). Conversely, Erwinia carotovora RpoS functionally opposes the LetA/S homologues GacA/S by positively activating RsmA (CsrA), the mRNA binding protein that destabilizes targeted transcripts (45). In L. pneumophila, phenotypic analysis of bacteria that lack or overexpress rpoS has predicted not only unexpected roles for RpoS but also both cooperative and antagonistic interactions with LetA/S. RpoS is dispensable for L. pneumophila to become resistant to oxidative, osmotic, and acidic stress in the PE phase (20). Instead, the rpoS locus promotes resistance to osmotic stress during the E phase, whereas LetA/S activates stress resistance in the PE phase (3, 20, 22, 42, 44). Conversely, when present in multiple copies, wild-type rpoS inhibits L. pneumophila replication in Acanthamoeba castellanii (20) and also motility and cytotoxicity of PE-phase L. pneumophila, two traits induced by LetA/S (3). On the basis of these paradoxical results, we tested the hypothesis that, depending on the growth phase, RpoS either positively or negatively modulates traits that are induced by LetA/S.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
L. pneumophila Lp02, a virulent thymine auxotroph (5); MB379 (rpoS::kan; 3); and mariner Tn mutant strains (Kanr; 22) MB414 (letA-22), MB417 (letS-36), and MB420 (letE-121) derived from Lp02 were cultured in N-(2-acetamido)-2-aminoethanesulfonic acid (ACES; Sigma)-buffered yeast extract broth supplemented with 100 μg of thymidine per ml (AYET) at 37°C. Bacteria were plated on ACES-buffered charcoal-yeast extract agar supplemented with 100 μg of thymidine per ml (CYET). Where indicated, media was supplemented with gentamicin to a final concentration of 10 μg/ml or with kanamycin to 25 μg/ml.
The RSF1010-based plasmids used in this study were digested with AgeI to delete mobA, since conjugation function has been demonstrated to interfere with L. pneumophila virulence (57). Plasmids pMB384 (pKB5ΔmobA Thy+), pLrpoS (pMB391, pKB5ΔmobA containing a 6.8-kb rpoS genomic locus), and pLrpoSΔ (3) were transformed into Lp02 by electroporation and selected on CYE, which lacks thymidine. Likewise, pLetA (pMMB-GentΔmobA with a 1.4-kb letA-containing fragment colinear with the PTAC promoter) was transformed into Lp02 or MB379 and selected with gentamicin. As previously reported, LetA homologues can inhibit growth in laboratory culture (14), a trait we observed as heterogeneity in the colony size of transformants, regardless of their rpoS allele. Inocula from small colonies grew poorly in gentamicin-supplemented AYET and displayed heterogeneous colony morphology when colony purified or plated from broth cultures. These small isolates were not studied further. Approximately 1 to 10% of total transformants, large isolates exhibited a stable colony size when colony purified or plated from broth cultures. Inocula from three large-colony isolates of each strain containing plasmid-borne letA replicated in broth, reached PE phase, and became motile and cytotoxic with kinetics similar to that of wild-type Lp02 (data not shown), and these clones were used for subsequent experiments.
RNA preparation and Northern analysis.
For mutant analysis, overnight cultures were quantified by optical density at 600 nm (OD600) and then diluted to 0.3 or 0.03 to generate PE-phase and E-phase cultures after an additional 16 h of incubation. Time course experiments were performed by diluting overnight cultures to an OD600 of 0.015 in 25 ml and, to allow for equal aeration, dividing them into five 5-ml aliquots. After an additional overnight incubation, cultures were in mid-E phase (time = 0 h). Subsequently, RNA was prepared by collecting one aliquot every 3 h for a 12-h period, at which point the cultures had entered the PE phase. Bacteria from 4.5 ml of E-phase cultures and 1.5 ml of PE-phase cultures were collected by centrifugation, resuspended in 1 ml of Trizol reagent, and purified as directed by the manufacturer (Invitrogen). RNA samples were quantified by OD260, and 10 μg of total RNA per sample was electrophoresed on a 1.2% formaldehyde gel. The gel was prepared, electrophoresed, and transferred to BrightStar BioDetect nylon membranes in accordance with the NorthernMax kit protocol (Ambion, Inc.).
For each gene of interest, a segment within the open reading frame was amplified by PCR with the primers listed in Table 1. Biotin-labeled, antisense, single-stranded DNA (ssDNA) probes were synthesized with the Strip EZ PCR kit (Ambion, Inc.) from 10 ng of each PCR product by using primer 2 of each pair and 200 nM biotin 14-ATP (Invitrogen). Membranes were hybridized with NorthernMax Ultrahyb and a final probe concentration of ∼1 ng/ml. Probes were initially quantified, and mRNA was subsequently detected with the BrightStar BioDetect kit (Ambion) and Kodak ML chemiluminescence film. The net intensity of hybridization was measured with Kodak Digital Science 1D software and expressed as arbitrary units (data not shown). Probes were stripped by incubation for 10 min at room temperature in probe degradation buffer, 10 min at 68°C, and 10 min in blot reconstitution buffer at 68°C (Strip EZ PCR kit; Ambion).
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| rpoS-int1 | 5′-CCA AGC GAG GAT TCC GTT TTT C-3′ |
| rpoS-int2a | 5′-AAA ACG TCT TGC GAT AAC CTG-3′ |
| mip-int1 | 5′-AAA TCT CTA TCG CTA ACG CAC AAG-3′ |
| mip-int2a | 5′-AAC TCA CTC ACC CCA CCA CGA C-3′ |
| fliA-int1 | 5′-GGT GCC GCG TTC TGT TTA TC-3′ |
| fliA-int2a | 5′-CGG TGC GTT GCT TGA CTT-3′ |
| flaA-int1 | 5′-CCA ATT TTA CCG GGG CAC TA-3′ |
| flaA-int2a | 5′-AAC GGC GCT GAT TGA TAA AG-3′ |
| csrA-int1b | 5′-AGC TGC TAT GTT AGG GAG-3′ |
| csrA-int2a,c | 5′-TAC TGC TTG TTC CGA ATC-3′ |
| letETn1 | 5′-TAC ATG CAC TAA AAA GCG GTT CTG-3′ |
| letETn2a | 5′-TGA TCA TGC TAC GAG CTC AAG TAA-3′ |
Contact-dependent cytotoxicity.
Quantification of contact-dependent cytotoxicity of PE-phase cultures was performed as previously described (21). For LetA induction experiments, overnight cultures were diluted to an OD600 of ∼0.007, divided in two, and incubated overnight with or without 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG). On the following morning, culture density was quantified by OD600, motility was assessed by phase microscopy, cultures were diluted to equivalent concentrations in RPMI medium plus 10% fetal bovine serum (R10), and A/J mouse bone marrow macrophages were infected with each sample in triplicate. Inocula were diluted and plated on CYET containing gentamicin. To quantify CFU, after a 1-h incubation with macrophages, bacteria were removed by repeated washing with R10 and macrophage viability was quantified by incubation with 10% Alamar blue. Reduction of the colorimetric dye by viable macrophages was measured by the ratio of OD570 to OD600 in a Spectramax 250 plate reader (Molecular Devices).
Bone marrow-derived macrophage infectivity.
Enumeration of L. pneumophila bacteria that bind, enter, and survive within A/J mouse bone marrow-derived macrophages during a 2-h incubation was performed as previously described (8), with the following modifications. Macrophages were infected with L. pneumophila at a multiplicity of infection of 0.2 to avoid cytotoxicity, and gentamicin treatment was omitted since control studies indicate that this antibiotic is only weakly bactericidal against PE-phase Legionella (S. Sturgill-Koszycki, unpublished results). Instead, the majority of extracellular bacteria were removed by washing the infected monolayers with 3 × 0.5 ml of R10 immediately before lysis by trituration with 2 × 0.5 ml of phosphate-buffered saline. Infectivity was expressed as (cell-associated CFU at 2 h/CFU added at 0 h) × 100.
RESULTS
L. pneumophila expresses rpoS RNA maximally in E phase by a mechanism independent of the LetA/S pathway.
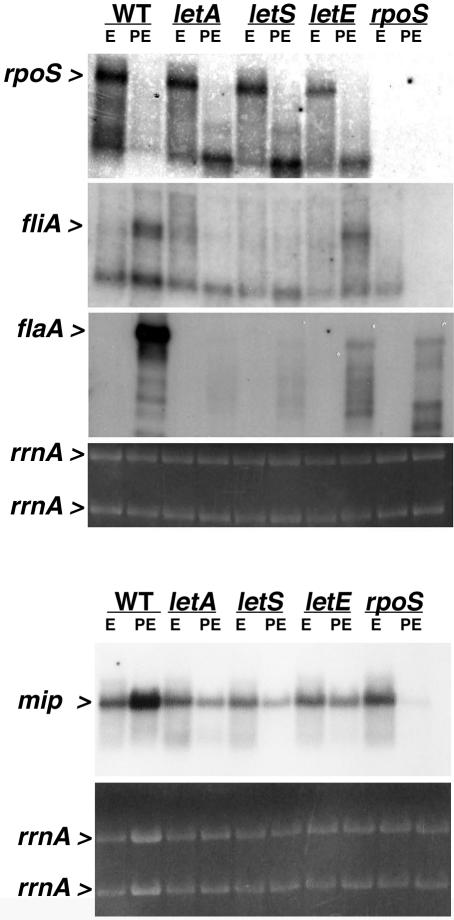
Consistent with the primary role of RpoS as a stationary-phase sigma factor, E. coli expresses rpoS RNA and protein maximally in early PE phase (24, 46). As a first step to investigate how RpoS regulates growth phase-dependent differentiation of L. pneumophila, we compared rpoS expression by wild-type bacteria cultured to either the E or the PE phase. Although the L. pneumophila RpoS protein is most abundant in the PE phase (20), its rpoS transcripts were maximal in the E phase and undetectable in the PE phase (Fig. 1), a pattern generally consistent with a previous genetic study of rpoS promoter activity (42). To a first approximation, its pattern of RNA abundance suggests an atypical E-phase function and/or mode of regulation for L. pneumophila RpoS.
FIG. 1.
Accumulation of rpoS, fliA, flaA, and mip mRNAs by E- and PE-phase wild-type (WT) and mutant L. pneumophila. Northern analysis was performed on 10 μg of total RNA collected from E-phase (OD600, 0.8 to 1.2) and PE-phase (OD600, 3.1 to 3.6) cultures of wild-type Lp02 bacteria and letA-22, letS-36, letE-121, and rpoS mutant bacteria with a biotin-labeled ssDNA probe. Hybridization was detected with streptavidin conjugated to alkaline phosphatase and visualized by CDP-Star (Tropix). The hybridization pattern shown is representative of results obtained from at least two independent sets of RNA samples. The relative amount of total RNA in each sample is demonstrated by ethidium bromide staining of rrnA from the formaldehyde gel prior to membrane transfer. Greater hybridization to mip in the wild-type PE-phase sample is due to a higher concentration of total RNA, as demonstrated by ethidium bromide staining of rrnA.
In E. coli, rpoS transcription is induced to a low level in the E phase by BarA, a tripartite sensor kinase that is homologous to LetS (22, 46). Therefore, we examined whether L. pneumophila activates rpoS expression in the E phase via its cognate LetA/S two-component regulatory system. The effect of a panel of regulatory mutations on rpoS RNA accumulation was determined by Northern analysis (Fig. 1). In the E phase, the amount of rpoS RNA in letA and letS mutants approached that of wild-type L. pneumophila. Likewise, LetE, which is predicted to cooperate with LetA/S to induce transmission traits (22), had only a modest effect on rpoS RNA accumulation by E-phase L. pneumophila (Fig. 1). Thus, rpoS RNA accumulates during the replication period by a mechanism that is largely independent of LetA/S and LetE, putative posttranscriptional activators of the PE transmission phenotype (22, 23).
RpoS and LetA/S induce transmission genes by independent mechanisms.
As another approach to investigate how RpoS and LetA/S cooperate to control L. pneumophila differentiation, their effects on the expression of three genes known to promote phagocyte infection were compared. The macrophage infectivity potentiator (Mip) is a peptidyl-prolyl cis-trans isomerase that enhances L. pneumophila invasion and replication within both amoebae and macrophages (10, 11). As expected, the mip RNA was present in both E- and PE-phase wild-type cells (36, 64). The LetA/S and LetE proteins are minor inducers of mip RNA expression in the PE phase since, compared to the wild type, bacteria that lack letA, letS, and letE contained less but still appreciable levels of mip transcripts (Fig. 1). In comparison, RpoS is critical for the accumulation of mip RNA in the PE phase, since its transcripts were barely detectable in PE rpoS mutant cells (Fig. 1). The observation that mip PE-phase expression is controlled primarily by RpoS may account for the similar lag in growth initiation that is characteristic of both mip and rpoS mutants that have infected either macrophages or amoebae (3, 10, 11).
In addition to Mip, L. pneumophila uses two genes of the flagellar regulon to infect phagocytes efficiently. The flagellar sigma factor FliA is required for expression of the three PE-phase transmission traits of motility, infectivity, and cytotoxicity and also for replication in amoebae (22, 27, 44). FliA activates several genes needed for the terminal stages of flagellum development, including flaA, the structural gene for flagellin which itself is required for efficient infection of phagocytes (12). Therefore, we next used these two genes of the flagellar regulon as tools to investigate how the RpoS and the LetA/S and LetE pathways cooperate to regulate differentiation of PE-phase L. pneumophila.
Replicating wild-type L. pneumophila contained a small amount of full-length fliA RNA; upon entry into the PE phase, the quantity increased dramatically (Fig. 1). Results of previous studies predict that fliA induction in the PE phase requires LetA/S to relieve CsrA repression (17, 44). Indeed, the growth phase regulation of fliA expression required LetA/S, as well as RpoS. Full-length fliA RNA was rare in E- and PE-phase letA and letS mutants, diminished in letE mutants, and undetectable in rpoS mutants (Fig. 1). Therefore, the robust induction of fliA RNA levels in the PE phase appears to be accomplished by two mechanisms: the sigma factor RpoS likely regulates transcription initiation, whereas the LetA/S pathway is predicted to relieve posttranscriptional repression mediated by CsrA (17).
Consistent with the paradigm of FliA-directed transcription of the flaA flagellin gene and genetic assays of its promoter activity (17, 21, 26, 28), flaA RNA levels were induced in the PE phase by LetA/S-, LetE-, and RpoS-dependent pathways (Fig. 1). Compared to wild-type L. pneumophila, PE-phase letA and letS mutants have dramatically reduced amounts of flaA RNA, whereas letE and rpoS mutants contained a low level of flaA RNA (Fig. 1), a pattern consistent with the degree of their motility defects (3, 22).
It was notable, however, that the abundance of flaA RNA in the letE and the rpoS mutant strains did not correspond directly to their levels of fliA transcripts. Relative to PE-phase letA or letS mutants, rpoS mutants reproducibly harbored less RNA encoding the FliA sigma factor yet accumulated similar amounts of RNA encoding flagellin. In contrast, PE-phase letE mutants contained a substantial amount of fliA RNA but little flaA RNA (Fig. 1). These results are also consistent with a model in which the steady-state level of flaA RNA is controlled by two mechanisms: the sigma factor RpoS regulates transcription initiation, whereas LetE and LetA/S act posttranscriptionally as stabilizers of effector mRNAs (23, 24). Accordingly, letE mutants contain ample RpoS and FliA sigma factors to transcribe flaA, but the mRNA product is destabilized by the constitutive CsrA posttranscriptional repressor. Conversely, an rpoS mutant transcribes fliA poorly, but the mRNA yield is sufficient for flaA expression since LetA/S and LetE relieve mRNA destabilization by CsrA (17, 44, 50).
In multiple copies, rpoS represses transcription of transmission genes.
Previous studies documented that in multiple copies the wild-type rpoS locus inhibits L. pneumophila replication in amoebae (20) and also represses two fliA-dependent PE transmission traits, motility and cytotoxicity (3). Therefore, as an independent approach to analyze how RpoS functionally interacts with other regulators of L. pneumophila differentiation, the effect of multiple copies of the wild-type rpoS locus on the transcription of known transmission genes was examined. For this purpose, RNA samples from wild-type bacteria that contained either the rpoS plasmid pLrpoS or the control vector pMB384 were collected at 3-h intervals between the E phase and the PE phase for Northern analysis (Fig. 2A). As expected, compared to bacteria containing the control vector, L. pneumophila transformed with pLrpoS had increased levels of both full-length and shorter rpoS transcripts throughout the E phase (Fig. 2B).
FIG. 2.
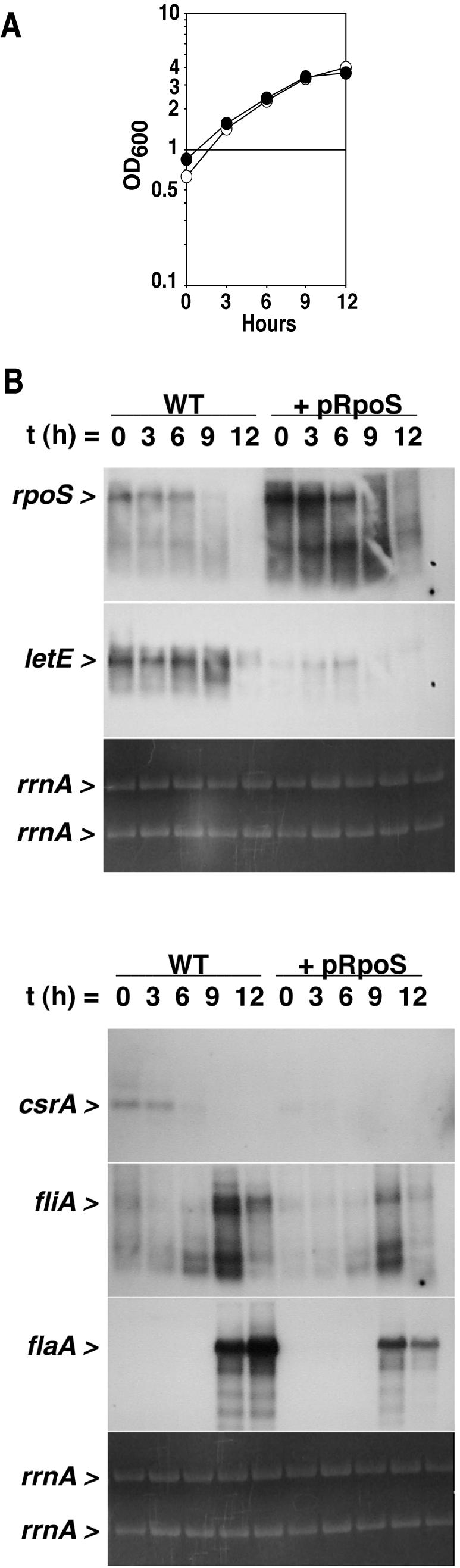
RpoS overexpression represses letE, csrA, fliA, and flaA expression by a mechanism that does not affect L. pneumophila growthin broth. (A) OD600 of cultures of wild-type Lp02 containing the vector pMB384 (open circles) or pLrpoS (pMB391, closed circles). At each time point shown, aliquots were removed, centrifuged, permeabilized with Trizol reagent, extracted for total RNA as directed by the manufacturer (Invitrogen), and analyzed as described for panel B. (B) Northern analysis was performed on 10 μg of total RNA collected as described for panel A. The same membrane was stripped and reprobed for each transcript. The relative amount of total RNA in each sample is demonstrated by ethidium bromide staining of rrnA from the formaldehyde gel prior to membrane transfer. The results shown are representative of two sets of RNA samples collected from independent cultures at similar optical densities. WT, wild type.
When rpoS RNA was excessive, the expression of a number of transmission genes was inhibited. L. pneumophila transformed with pLrpoS contained less RNA for two regulators of transmission that are transcribed predominantly in the E phase, namely, the positive activator letE and the repressor csrA (17, 44). A similar effect was observed for two genes expressed primarily in the PE phase: multiple copies of rpoS significantly decreased the magnitude, but not the timing, of fliA and flaA expression (Fig. 2B). The observation that excess rpoS can inhibit the expression of four different genes, letE, csrA, fliA, and flaA, is consistent with the work of Nystrom and colleagues demonstrating that E. coli sigma factors compete for binding to core RNA polymerase. By influencing the competition among sigma factors, growth or experimental conditions can alter the cellular pattern of gene expression (16, 33, 40, 43).
RpoS overexpression represses LetA/S-dependent virulence traits.
To test more rigorously whether RpoS can antagonize LetA/S-dependent activation of the transmission phenotype, the effect of pLrpoS on the expression of three transmission traits by wild-type L. pneumophila was analyzed. Whereas ∼75% of PE-phase wild-type cells that contained only the chromosomal rpoS locus were motile, motility was rare or absent when L. pneumophila contained multiple copies of rpoS, consistent with the inhibitory effects of pLrpoS on fliA and flaA expression (3).
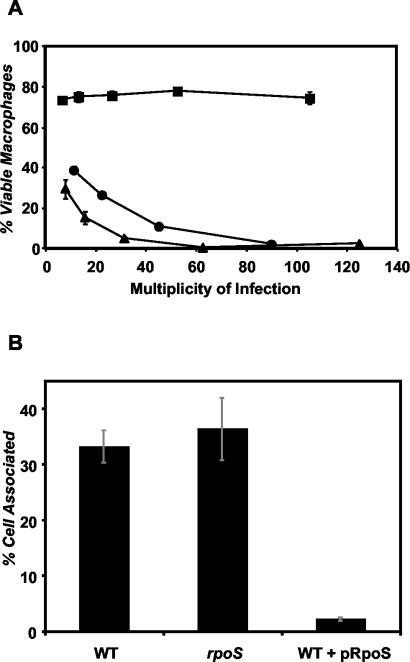
In the PE phase, L. pneumophila expresses a contact-dependent pore-forming activity that is toxic to macrophages and is thought to promote bacterial escape from phagocytes when nutrients become scarce (2, 8, 35). Induction of cytotoxicity in the PE phase requires LetA/S, LetE, and FliA, but not RpoS (3, 22). Nevertheless, when wild-type L. pneumophila was transformed with pLrpoS, cytotoxicity was completely repressed (Fig. 3), a phenotype consistent with the negative effect of pLrpoS on fliA expression (Fig. 2B). To verify that the pLrpoS-mediated repression was conferred by the rpoS locus and not other plasmid sequences, the effect of its derivative pLrpoSΔ, which contains an rpoS::kan null allele, was examined. Strains transformed with this control plasmid were at least as cytotoxic as the wild type that contained only the parent vector pMB384, demonstrating rpoS specificity (Fig. 3).
FIG. 3.
Multicopy expression of rpoS eliminates cytotoxicity and infectivity. (A) Contact-dependent cytotoxicity for macrophages. Macrophages were incubated for 1 h with samples of PE-phase wild-type Lp02 containing the vector pMB384 (circles), pLrpoS harboring a 6.8-kb rpoS locus (pMB391, squares), or pLrpoS::kan carrying an rpoS::kan mutation in the same locus (pMB392, triangles). Macrophage viability was assessed by quantifying reduction of the colorimetric dye Alamar blue. Shown are means and standard errors of triplicate samples from one representative experiment of three performed. (B) Efficiency of macrophage infection. Macrophages were infected at a multiplicity of infection of 0.2 with the following PE-phase strains: wild-type (WT) strain Lp02 containing the vector pMB384, rpoS mutant strain MB379 (rpoS::kan) containing pMB384 and the wild type plus pLrpoS. The percentage of each L. pneumophila inoculum that was cell associated after a 2-h infection with 2.5 × 105 macrophages is shown. The results shown are means of macrophage samples infected in triplicate; error bars represent standard deviations.
When ingested by macrophages, L. pneumophila can evade lysosomal degradation (31), provided the LetA/S regulator is active (22). In contrast, RpoS is dispensable for lysosome evasion, but it is necessary to block fusion of the bacterial phagosome with earlier, nonbactericidal endosomal compartments (3). To test whether multicopy rpoS also inhibited lysosome evasion, macrophage infection by rpoS mutant and wild-type cells with or without pLrpoS was assessed. After a 2-h incubation, similar percentages of wild-type and rpoS bacteria were viable and cell associated (Fig. 3), a result that reflects the persistence of rpoS mutants in vacuoles that contain LAMP-1 but not lysosomal contents (3). In contrast, carriage of pLrpoS by wild-type L. pneumophila lowered infectivity from ∼33% to ∼2% of the inoculum, a phenotype shared with letA and fliA mutants (22). The virulence defects conferred by plasmid-borne rpoS were not a simple consequence of slowing growth, as judged by replication of bacteria in broth (Fig. 2A) or in macrophages (3). In summary, multiple copies of rpoS caused poor motility, cytotoxicity, and infectivity in the PE phase, a spectrum of defects reminiscent of the pattern conferred by mutations in letA, letS, letE, or fliA (22). Therefore, in excess, rpoS antagonizes activators of the transmission phenotype.
RpoS prevents LetA induction of transmission traits in the E phase.
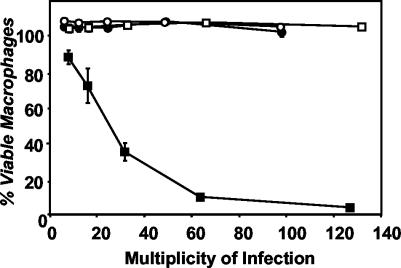
On the basis of its E-phase RNA expression and the inhibitory effects of pLrpoS, we postulated that during the replication period, RpoS protein represses transmission directly or indirectly. This hypothesis predicts that genetic inactivation of rpoS will allow LetA to induce transmission traits prematurely during the replication period. To test this supposition, wild-type and rpoS mutant bacteria were transformed with pLetA, a plasmid that encodes letA under the control of an inducible pTac promoter, and then their ability to respond to LetA by expressing cytotoxicity during the E phase was assessed (Fig. 4).
FIG. 4.
RpoS represses LetA-induced cytotoxicity in the E phase. After overnight growth in the presence (closed symbols) or absence (open symbols) of 200 μM IPTG to induce expression from pLetA, samples of E-phase wild-type Lp02 (circles) or rpoS mutant MB379 (squares) were incubated for 1 h with macrophages. Macrophage viability was assessed by quantifying reduction of the colorimetric dye Alamar blue. Shown are means and standard errors of triplicate samples from one representative experiment of six performed.
As demonstrated previously, wild-type bacteria were not cytotoxic in the E phase. Likewise, IPTG induction of letA was not sufficient to induce cytotoxicity in E-phase cultures of wild-type L. pneumophila. However, as predicted, in the absence of rpoS, E-phase cells responded to activating signals by LetA. Indeed, E-phase rpoS mutant cells induced to express letA resembled PE-phase L. pneumophila cells in their ability to kill macrophages (Fig. 4). Likewise, RpoS also repressed the expression of motility during the E phase, since induction of letA expression triggered flagellar motility only in the absence of rpoS function (data not shown). RpoS repression was relieved as cells entered the PE phase; in the presence or absence of pLetA, wild-type cells became fully cytotoxic, as expected (8). Therefore, the low level of RpoS protein normally present in E-phase L. pneumophila (20) blocks expression of transmission traits, even when the letA activator is expressed ectopically.
DISCUSSION
In L. pneumophila, RpoS function and regulation are atypical of its homologues in several other species of gram-negative bacteria, on the basis of four observations. First, although L. pneumophila substantially increases its resistance to oxidative, osmotic, and acidic stress in the PE phase, none of these fortifications require RpoS (3, 20). Second, RpoS induces osmotic resistance of replicating L. pneumophila (20). Third, transcription of E. coli rpoS increases ∼5- to 10-fold upon transition to the stationary phase (25); in L. pneumophila, abundant E-phase transcripts disappear in the PE phase (Fig. 1 and 2). Last, P. fluorescens and E. coli LetA/S homologues induce rpoS transcription (46), but L. pneumophila rpoS RNA species appear to be destabilized in the PE phase by a LetA/S-dependent pathway (Fig. 1). Together, these observations suggest that, in L. pneumophila, RpoS is used in pathways or under conditions beyond coordination of stationary-phase physiology.
Studies of E. coli and S. enterica serovar Typhimurium provide precedents for a role for RpoS during the L. pneumophila replication period. Although E. coli RpoS protein levels are low under nutrient-rich conditions, this alternate sigma factor directs the transcription of xthA and katG, genes that prevent UV and oxidative damage (32, 41, 53). When macrophages ingest S. enterica serovar Typhimurium, another gram-negative bacterium that replicates within intracellular vacuoles, its RpoS expression increases dramatically, approaching levels typical of stationary phase (9). RpoS is required for full virulence of S. enterica serovar Typhimurium in mice, as it activates the katE-encoded catalase and other spv-dependent and -independent factors (15). The biphasic life cycle of L. pneumophila may account for the diverse roles postulated for RpoS. To persist as a free-living aquatic microbe, L. pneumophila must tolerate nutrient limitation and other environmental stresses. To replicate efficiently within phagocytes, L. pneumophila must adapt to another harsh environment, i.e., acidic lysosomal compartments (58). To thrive within this stressful but nutrient-rich site, L. pneumophila may have evolved a regulatory circuit in which RpoS confers on replicating bacteria tolerance to lysosomal stresses while also preventing induction of the transmission phenotype.
The inverse pattern of L. pneumophila RpoS RNA and protein levels (20) likely indicates that RpoS is subject to posttranscriptional control, which is well documented in E. coli (25). A stockpile of rpoS mRNA may enable replicating L. pneumophila to respond quickly to deteriorating conditions. As L. pneumophila differentiates to the transmissible form, a LetA-dependent increase in rpoS promoter activity, predicted by a transcriptional rpoS-lacZ reporter (42), may further increase template levels. However, in the PE phase, full-length rpoS transcripts disappear (Fig. 1 and 2) while RpoS protein accumulates (20). LetA/S and LetE are attractive candidates to couple rpoS RNA translation to decay, given their predicted role as antagonists of the posttranscriptional repressor CsrA (23, 44). Interestingly, PE-phase letA, letS, and letE mutant cells contained a large quantity of smaller rpoS RNAs, which were rare in wild-type PE cells (Fig. 1). Similar rpoS species have been observed in other gram-negative bacteria under conditions of rpoS message excess and are thought to be stable degradation intermediates (38, 46). Further studies are required to determine whether the L. pneumophila LetA/S regulatory pathway mediates rpoS RNA decay after its translation in the PE phase.
This study provides insight into the organization of the L. pneumophila RpoS regulon, showing that this sigma factor is required for PE-phase expression of mip, the flagellar sigma factor fliA, and its target flaA, three well-described virulence factors that promote L. pneumophila infection of amoebae and macrophages (19, 22, 27). Both mip and rpoS mutants exhibit a characteristic lag in growth initiation after infection of phagocytes (3, 10, 11). However, unlike a mip mutant, an rpoS mutant cannot replicate at all in amoebae (20). Its more severe phenotype is consistent with the observation that RpoS regulates additional genes, including fliA and flaA, factors that together with Mip are required for efficient infection and growth inside protozoa (12, 27).
Several lines of genetic and molecular evidence indicate that RpoS and LetA/S activate the expression of transmission genes by cooperative but distinct mechanisms. In comparison to an rpoS mutation, mutations in letA or letS have a less extreme effect on the levels of mip and fliA transcripts, yet they impair flaA expression more severely (Fig. 1). Thus, a LetA/S- and LetE-dependent mechanism may compensate for low fliA message in an rpoS mutant to allow expression of FliA-dependent traits. Consistent with their more extreme effects on fliA and flaA expression, LetA/S and FliA are required for cytotoxicity and infectivity in the PE phase, whereas RpoS is not (3, 22). Furthermore, when L. pneumophila is engulfed by a phagocyte, RpoS and LetA/S promote two different activities: immediate arrest of phagosome maturation and subsequent bacterial replication. letA or letS mutants infect macrophages and amoebae poorly (22, 42). However, even though 50% of internalized letA mutant bacteria are degraded by macrophage lysosomes, the survivors replicate as efficiently as the wild type (22). rpoS mutants display the opposite pattern: the majority of rpoS mutant bacteria remain intact in an intermediate endosomal compartment, but they fail to multiply (3). Also, the requirement for LetA can be bypassed by genetic inactivation of the CsrA repressor, whereas the need for RpoS cannot (44). Therefore, RpoS and LetA/S have specialized roles in the transmissive phase, as postulated previously (3, 42). By analogy to their demonstrated activity in E. coli and other gram-negative bacteria, RpoS is expected to control transcription initiation by RNA polymerase, whereas LetA/S is likely to act posttranscriptionally by relieving CsrA repression to stabilize fliA and/or flaA mRNA and promote synthesis of these and other transmission effectors (23, 24, 50).
By examining bacteria that lack or overexpress rpoS, we found that, depending on the growth phase, RpoS either negatively or positively influences expression of LetA/S-dependent transmission traits. In replicating L. pneumophila, RpoS not only promotes osmotic resistance (20) but also prevents LetA-induced transmission traits, since ectopically expressed LetA induces cytotoxicity and motility only when rpoS is absent (Fig. 4). Likewise, when overexpressed by PE cells, rpoS disrupts LetA/S-dependent motility, cytotoxicity, and infectivity (Fig. 3, data not shown) by inhibiting expression of fliA, flaA, and letE (Fig. 2). Consistent with this experimental condition, in wild-type L. pneumophila cultures, fliA and flaA transcripts are scarce when rpoS RNA is abundant (Fig. 1 and 2). Together, the data indicate that, by some direct or indirect mechanism, RpoS prevents activation of the transmission phenotype by replicating L. pneumophila.
During the replication period, the low level of RpoS protein (20) could inhibit the transmission phenotype indirectly by inducing a repressor. In E. coli and S. enterica serovar Typhimurium, RpoS-dependent repression of pilus phase variation is attributed to transcriptional activation of a repressor gene (13, 48). In E. coli, RpoS is also known to cooperate with Hfq to repress expression of mutH (61) and with Fis to repress a collection of genes at various points in the growth cycle (65). Because E. carotovora RpoS induces the posttranscriptional repressor RsmA (45), we tested whether L. pneumophila csrA RNA levels are regulated by RpoS. To the contrary, our data indicate that RpoS represses transmission genes along with csrA, and an rpoS mutation had little effect on csrA expression (data not shown).
By analogy to E. coli, L. pneumophila RpoS could also inhibit transmission gene expression by competing with other sigma factors for binding to RNA polymerase (16, 33, 40). For example, when overexpressed, RpoS inhibits RpoD-directed transcription by PE-phase E. coli by competing for binding to core RNA polymerase (16). In L. pneumophila, the observation that overexpression of rpoS inhibits multiple genes, including loci normally transcribed in either the E or the PE phase (Fig. 2), is in keeping with such a mechanism. Furthermore, the observation that, depending on the experimental conditions, RpoS can either repress or activate the expression of the same gene, fliA (Fig. 1 and 2B), is consistent with the detailed genetic and biochemical studies of Nystrom and colleagues demonstrating that sigma factors compete for binding to core RNA polymerase (16, 33, 40).
By taking into account the paradigm of sigma factor competition, known transcriptional and posttranscriptional control mechanisms of RpoS activity (25), the nucleotide sequence and genetic analysis of the L. pneumophila flagellar regulon by Heuner and colleagues (29), and genetic evidence from our laboratory and others, the following working model of L. pneumophila regulation of differentiation can be proposed. During the replication period, the rpoS gene is transcribed at high levels (Fig. 1); although most of the RpoS protein is presumably degraded (20, 25), there must be sufficient protein bound to RNA polymerase to mediate osmotic resistance (20) and promote intracellular replication (3, 20). At the same time, although sigma factor RpoN may direct some transcription of fliA (29), the posttranscriptional repressor CsrA targets any mRNA encoded by the flagellar regulon for degradation (17). Consequently, when amino acids are abundant and (p)ppGpp levels are low, fliA and flaA RNAs are rare and L. pneumophila cells are not motile, cytotoxic, or infectious. When the basal level of E-phase RpoS protein is eliminated genetically, sufficient RpoN may bind the freed RNA polymerase to induce the flagellar regulon, provided posttranscriptional repression by CsrA is also alleviated by ectopic expression of LetA/S (29, 44).
When conditions deteriorate, we postulate that accumulation of (p)ppGpp (21) increases the amount of RpoS protein (20, 25) and also promotes both RpoS- and RpoN-mediated transcription by RNA polymerase (33, 37, 40). By some mechanism, RpoS promotes transcription initiation in the flagellar regulon (Fig. 1), perhaps indirectly by elevating the expression of an RpoN coactivator protein (7). According to this model, excess RpoS could inhibit PE expression of the flagellar regulon (Fig. 2) by outcompeting RpoN for binding to RNA polymerase. In wild-type L. pneumophila, (p)ppGpp (21, 22) or some other PE-phase signal (66) concomitantly activates the LetA/S two-component regulatory system to relieve CsrA repression of fliA mRNA translation (17, 44). In this scenario, a rapid and robust induction of the flagellar and transmission regulons occurs when (p)ppGpp coordinates a parallel increase in RpoS- and RpoN-dependent transcription initiation and a LetA/S-dependent decrease in posttranscriptional repression by CsrA. Consequently, by coupling both transcriptional and translational controls, intracellular L. pneumophila can respond to deteriorating conditions by efficiently differentiating to a cytotoxic, motile, and infectious form that is fit to be transmitted to a new host.
The demand theory of gene regulation states that one can predict a positive mode of control for genes that are in high demand in an organism's natural environment and negative control for those genes in low demand (47, 54, 55). For organisms that alternate between different environments, the mode of regulation is predicted to change in accordance with the current demand for the regulated genes. Consistent with the predictions of the demand theory, when conditions are favorable for growth, L. pneumophila transmission genes are in low demand and thus are repressed by CsrA and RpoS. Conversely, when the amino acid supply becomes limiting, transmission genes are in high demand and are thus under a positive mode of control, mediated by LetA/S, LetE, FliA, and RpoS (Table 2 and Fig. 1 to 4) (17, 44). Knowledge of the regulatory circuit that controls L. pneumophila differentiation can be used to identify the effectors of the replicative and transmission phenotypes. Once the effector genes are identified, detailed analysis of their transcription initiation and mRNA stability can test current genetic models of how RpoS and the LetA/S-and-LetE system cooperate to govern bacterial physiology to allow L. pneumophila to tolerate or exploit its surroundings.
TABLE 2.
Demand theorya of L. pneumophila differentiation
Acknowledgments
We gratefully acknowledge Victor DiRita, N. Cary Engleberg, Alex Ninfa, and Ari Molofsky for insight and critical analysis and also advice on the manuscript.
This project was funded by NIH grant AI 44212-01 and the University of Michigan President's Initiative Fund for Graduate Training in Microbial Pathogenesis.
Editor: J. N. Weiser
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Alli, O. A. T., L.-Y. Gao, L. L. Pedersen, S. Zink, M. Radulic, M. Doric, and Y. Abu Kwaik. 2000. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect. Immun. 68:6431-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 4.Bandyopadhyay, P., and H. M. Steinman. 1998. Legionella pneumophila catalase-peroxidases: cloning of the katB gene and studies of KatB function. J. Bacteriol. 180:5369-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 6.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64:668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck, M., M.-T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. Y., L. Eckmann, S. J. Libby, F. C. Fang, S. Okamoto, M. F. Kagnoff, J. Fierer, and D. G. Guiney. 1996. Expression of Salmonella typhimurium rpoS and rpoS-dependent genes in the intracellular environment of eukaryotic cells. Infect. Immun. 64:4739-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cianciotto, N. P., R. Long, B. I. Eisenstein, and N. C. Engleberg. 1989. A Legionella pneumophila gene encoding a species-specific surface protein potentiates the initiation of intracellular infection. Infect. Immun. 57:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dove, S. L., S. G. Smith, and C. J. Dorman. 1997. Control of Escherichia coli type 1 fimbrial gene expression in stationary phase: a negative role for RpoS. Mol. Gen. Genet. 254:13-20. [DOI] [PubMed] [Google Scholar]
- 14.Duffy, B. K., and G. Defago. 2000. Controlling instability in gacA-gacS regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 17.Fettes, P. S., V. Forsbach-Birk, D. Lynch, and R. Marre. 2001. Overexpression of the Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int. J. Med. Microbiol. 291:353-360. [DOI] [PubMed] [Google Scholar]
- 18.Fields, B. S., J. M. Barbaree, E. B. Shotts, Jr., J. C. Feeley, W. E. Morrill, G. N. Sanden, and M. J. Dykstra. 1986. Comparison of guinea pig and protozoan models for determining virulence of Legionella species. Infect. Immun. 53:553-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer, G., H. Bang, B. Ludwig, K. Mann, and J. Hacker. 1992. Mip protein of Legionella pneumophila exhibits peptidyl-prolyl-cis/trans isomerase (PPIase) activity. Mol. Microbiol. 6:1375-1383. [DOI] [PubMed] [Google Scholar]
- 20.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer, B. K., and M. S. Swanson. 1999. Coordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 22.Hammer, B. K., E. Tateda, and M. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 23.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 24.Hengge-Aronis, R. 1996. Regulation of gene expression during entry into stationary phase, p. 1497-1512. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 25.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heuner, K., L. Bender-Beck, B. C. Brand, P. C. Luck, K.-H. Mann, R. Marre, M. Ott, and J. Hacker. 1995. Cloning and genetic characterization of the flagellum subunit gene (flaA) of Legionella pneumophila serogroup 1. Infect. Immun. 63:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heuner, K., C. Dietrich, C. Skriwan, M. Steinert, and J. Hacker. 2002. Influence of the alternative σ28 factor on virulence and flagellum expression of Legionella pneumophila. Infect. Immun. 70:1604-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heuner, K., J. Hacker, and B. C. Brand. 1997. The alternative sigma factor σ28 of Legionella pneumophila restores flagellation and motility to an Escherichia coli fliA mutant. J. Bacteriol. 179:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heuner, K., and M. Steinert. 2003. The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int. J. Med. Microbiol. 293:133-143. [DOI] [PubMed] [Google Scholar]
- 30.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanova, A., C. Miller, G. Glinsky, and A. Eisenstark. 1994. Role of rpoS (katF) in oxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol. Microbiol. 12:571-578. [DOI] [PubMed] [Google Scholar]
- 33.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 35.Kirby, J. E., J. P. Vogel, H. L. Andrews, and R. R. Isberg. 1998. Evidence of pore-forming ability by Legionella pneumophila. Mol. Microbiol. 27:323-336. [DOI] [PubMed] [Google Scholar]
- 36.Köhler, R., A. Bubert, W. Goebel, M. Steinert, J. Havker, and B. Bubert. 2000. Expression and use of the green fluorescent protein as a reporter system in Legionella pneumophila. Mol. Gen. Genet. 262:1060-1069. [DOI] [PubMed] [Google Scholar]
- 37.Kvint, K., A. Farewell, and T. Nystrom. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σs. J. Biol. Chem. 275:14795-14798. [DOI] [PubMed] [Google Scholar]
- 38.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 40.Laurie, A. D., L. M. Bernardo, C. C. Sze, E. Skarfstad, A. Szalewska-Palasz, T. Nystrom, and V. Shingler. 2003. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J. Biol. Chem. 278:1494-1503. [DOI] [PubMed] [Google Scholar]
- 41.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 42.Lynch, D., N. Rieser, K. Gloggler, V. Forsbach-Brik, and R. Marre. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219:241-248. [DOI] [PubMed] [Google Scholar]
- 43.Magnusson, L. U., T. Nystrom, and A. Farewell. 2003. Underproduction of sigma 70 mimics a stringent response. A proteome approach. J. Biol. Chem. 278:968-973. [DOI] [PubMed] [Google Scholar]
- 44.Molofsky, A., and M. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50:445-461. [DOI] [PubMed] [Google Scholar]
- 45.Mukherjee, A., Y. Cui, Y. Liu, A. Ishihama, A. Eisenstark, and A. K. Chatterjee. 1998. RpoS (sigma-S) controls expression of rsmA, a global regulator of secondary metabolites, harpin, and extracellular proteins in Erwinia carotovora. J. Bacteriol. 180:3629-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukhopadhyay, S., J. P. Audia, R. N. Roy, and H. E. Schellhorn. 2000. Transcriptional induction of the conserved alternative sigma factor RpoS in Escherichia coli is dependent on BarA, a probable two-component regulator. Mol. Microbiol. 37:371-381. [DOI] [PubMed] [Google Scholar]
- 47.Neidhardt, F. C., and M. A. Savageau. 1996. Regulation beyond the operon, p. 1310-1324. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 48.Nicholson, B., and D. Low. 2000. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol. Microbiol. 35:728-742. [DOI] [PubMed] [Google Scholar]
- 49.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 50.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 51.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 52.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 53.Sak, B. D., A. Eisenstark, and D. Touati. 1989. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc. Natl. Acad. Sci. USA 86:3271-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savageau, M. A. 1977. Design of molecular control mechanisms and the demand for gene expression. Proc. Natl. Acad. Sci. USA 74:5647-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savageau, M. A. 1983. Regulation of differentiated cell-specific functions. Proc. Natl. Acad. Sci. USA 80:1411-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Segal, G., and H. A. Shuman. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol. Microbiol. 30:197-208. [DOI] [PubMed] [Google Scholar]
- 58.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 60.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsui, H. T., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by Hfq and RpoS global regulators of E. coli K-12. J. Bacteriol. 179:7476-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 63.Whistler, C. A., N. A. Corbell, A. Sarniguet, W. Ream, and J. E. Loper. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor σS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wieland, H., M. Faiglea, F. Lang, H. Northoffa, and B. Neumeister. 2002. Regulation of the Legionella mip-promotor during infection of human monocytes. FEMS Microbiol. Lett. 212:127-132. [DOI] [PubMed] [Google Scholar]
- 65.Xu, J., and R. C. Johnson. 1995. Identification of genes negatively regulated by Fis: Fis and RpoS comodulate growth-phase-dependent gene expression in Escherichia coli. J. Bacteriol. 177:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]