SUMMARY
Microglia are phagocytic cells that form the basis of the brain’s immune system. They derive from primitive macrophages that migrate into the brain during embryogenesis, but the genetic control of microglia development remains elusive. Starting with a genetic screen in zebrafish, we show that the non-canonical NOD-like receptor (NLR) nlrc3-like is essential for microglia formation. Although most NLRs trigger inflammatory signaling, nlrc3-like acts cell autonomously in microglia precursor cells to suppress unwarranted inflammation in the absence of overt immune challenge. In nlrc3-like mutants, primitive macrophages initiate a systemic inflammatory response with increased pro-inflammatory cytokines, and actively aggregate instead of migrating into the brain to form microglia. NLRC3-like requires both its pyrin and NACHT domains, and it can bind the inflammasome component ASC. Our studies suggest that NLRC3-like may regulate the inflammasome and other inflammatory pathways. Together, these results demonstrate that NLRC3-like prevents inappropriate macrophage activation, thereby allowing normal microglia development.
INTRODUCTION
As the only immune cells dedicated to the defense of the central nervous system, microglia have unique functions and developmental origins (Aguzzi et al., 2013, Ransohoff and Cardona, 2010). Microglia are extremely sensitive to signs of infection, injury, and disease of the brain, and they can respond rapidly, depending on the nature of the stimulus (Ransohoff and Perry, 2009, Perry et al., 2010). After infection, for example, microglia engulf pathogens and initiate an immune response, and they can clear cell corpses and promote healing after injury to the CNS. Microglia also interact with and eliminate neuronal synapses, and they may therefore modulate neuronal connectivity (Paolicelli et al., 2011, Peri and Nusslein-Volhard, 2008, Schafer et al., 2012, Ransohoff and Cardona, 2010). Unlike other CNS cell types, which derive from the neuroectoderm, microglia arise from a subset of primitive macrophages that migrate from the yolk sac into the brain during embryogenesis (Ginhoux et al., 2010, Herbomel et al., 2001). Primitive macrophages from the yolksac in mouse (Ginhoux et al., 2010) and zebrafish (Herbomel et al., 1999, Herbomel et al., 2001) enter the brain prior to definitive hematopoiesis, and differentiate into microglia that remain in the brain thereafter (Ginhoux et al., 2010). Because microglia in the adult brain derive from the early embryonic macrophage population, early disruption of microglia in the embryo may impair the immune system of the developing and mature CNS. The cellular and molecular mechanisms, however, that mediate the specification, migration, and differentiation of developing microglia remain elusive.
NOD-like receptors (NLRs) are intracellular pattern recognition receptors that regulate innate immunity and inflammatory processes (Chen et al., 2009, Davis et al., 2011, Mason et al., 2012). They can respond to pathogens and cellular stress by triggering caspase-1-dependent inflammatory signaling or by activating NF-kB to promote production of pro-inflammatory cytokines (Chen et al., 2009, Davis et al., 2011). Upon recognizing various ligands, putatively through their C-terminal leucine rich repeats (LRRs)(Chen et al., 2009, Davis et al., 2011, Mason et al., 2012), NLRs form large multi-protein complexes by self-oligomerization at their central NACHT domain. Activated NLRs recruit other proteins through homotypic interactions with their N-terminal domains, typically either a pyrin or a CARD domain, to activate NF-kB through the NODosome pathway or caspase-1 through the inflammasome mechanism (Chen et al., 2009, Davis et al., 2011). Mutations causing aberrant NLR signaling are linked to numerous human inflammatory disorders, such as inflammatory bowel disease, vitiligo, sarcoidosis, and cryopyrin-associated periodic syndromes (Chen et al., 2009, Davis et al., 2011, Mason et al., 2012), underscoring the importance of curbing inflammatory signaling mediated by the NLRs. Recent studies have shown that NLRC3 and NLRP12 can suppress inflammation after challenge by LPS or infection (Schneider et al., 2012, Allen et al., 2012, Zaki et al., 2011). Despite the far-reaching roles of the NLRs in regulating innate immunity, the mechanisms that regulate their functions and maintain homeostasis in the absence of infection are not well understood.
Starting with a genetic screen for mutations that disrupt microglia in zebrafish, we identified a non-canonical NOD-like receptor, nlrc3-like, that is essential for microglia formation in zebrafish. Our analysis demonstrates that nlrc3-like prevents runaway inflammation during embryogenesis and thereby allows the development of microglia. In nlrc3-like mutants, primitive macrophages adopt an inflammatory phenotype instead of migrating into the brain to differentiate as microglia. In addition, nlrc3-like mutants have systemic inflammation, as evidenced by elevation of pro-inflammatory cytokines and recruitment of neutrophils into the brain and circulation. Transgenic expression of the wildtype nlrc3-like gene in macrophages rescued microglia in the mutants, indicating that the gene acts autonomously in macrophages. Our results demonstrate that nlrc3-like serves as an essential, cell-autonomous checkpoint in primitive macrophages that prevents unwarranted inflammation in the absence of overt immune challenges and allows the normal development of microglia.
RESULTS
nlrc3-like is essential for microglia development
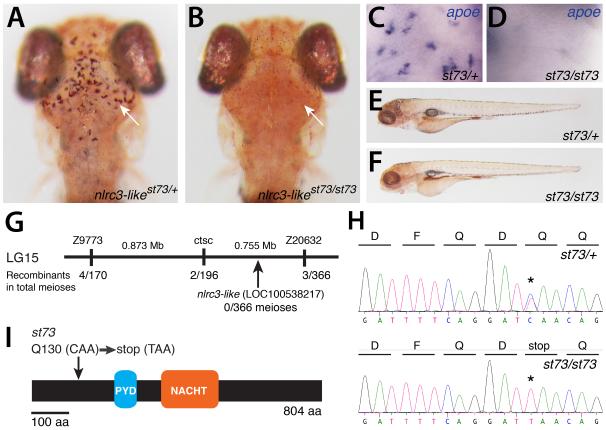
In a genetic screen in zebrafish, we identified st73 as a recessive mutation that eliminated microglia (Fig. 1A,B). At 5 days postfertilization (dpf), homozygous st73 mutant larvae lacked microglial cells (Fig. 1A-D), but did not have any apparent anatomical defects (Fig. 1E,F). High-resolution meiotic mapping localized the st73 mutation to a 1.63 Mb region of linkage group 15 (LG15) (Fig. 1G). Among the genes in this interval is nlrc3-like (LOC100538217) (Fig. 1G), which encodes an atypical member of the NLR family that contains the canonical pyrin (PYD) and NACHT domains, but lacks the common leucine-rich repeats thought to be important for ligand sensing and self-inhibition (Davis et al., 2011, Mason et al., 2012). NLRC3-like belongs to an expanded subfamily of teleost NLRs that shares significant similarity to the human NLRC3 (or Nod3), based on sequence comparisons of the NACHT domain (Laing et al., 2008, Hughes, 2006). Sequence analysis identified a nonsense mutation in nlrc3-like near the 5′ end of the open reading frame (Fig.1 H,I). All st73 mutants analyzed were homozygous for this lesion (n=300). Injection of synthetic mRNA encoding the wildtype NLRC3-like protein can fully rescue microglia in st73 mutants and had no effect on wildtype siblings (Fig. S1). Based on mapping, the identification of the nonsense lesion, and RNA rescue experiments, we conclude that st73 is a loss-of-function mutation in the nlrc3-like gene.
Figure 1. Lack of microglia in nlrc3-like mutants.
(A-B) Live 5 dpf zebrafish larvae stained by neutral red (Herbomel et al., 2001) to visualize microglia in the brain. (A) Heterozygous st73/+ larvae (arrow, microglia around optic tectum). (B) Homozygous st73 larvae have no microglia (arrow). (C,D) Microglial marker apoe expression at 5 dpf in the brain. (E-F) Neutral red stained heterozygous (E) and homozygous mutant (F) showing normal whole animal morphologies at 5 dpf. (G) Genetic and physical map of the st73 locus. (H) Sequence chromatogram showing the lesion (asterisk) in the coding sequence of nlrc3-like (LOC100538217). (I) The st73 mutation introduces a premature stop codon near the 5′ end of the open reading frame of nlrc3-like. (A, B) Dorsal views; anterior is to the top. (E,F) Lateral views; anterior is to the left. See also Figure S1.
Abnormal migration and inflammatory activation of macrophages in nlrc3-like mutants
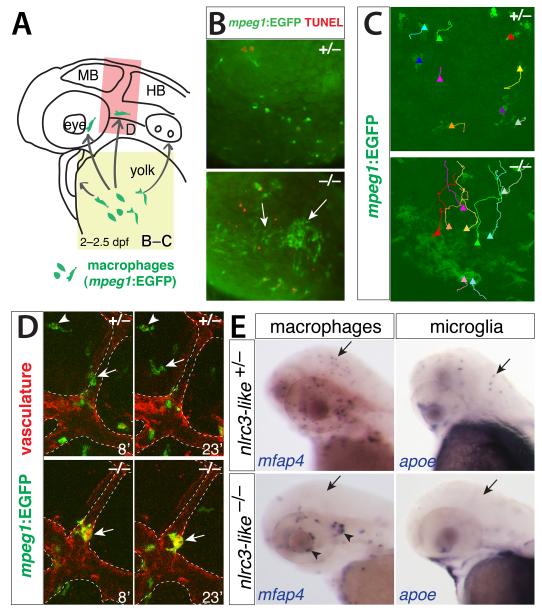
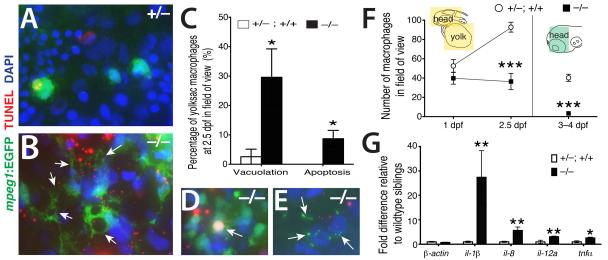
To define the role of nlrc3-like in microglia development, we examined different stages of microglia development starting from the formation of primitive macrophages to differentiation of microglia (20-60 hours postfertilization, hpf) using markers specific to macrophages (microfibrillar-associated protein 4, mfap4; macrophage expressed gene 1, mpeg1)(Ellett et al., 2011, Zakrzewska et al., 2010) and all leukocytes (leukocyte specific plastin, l-plastin)(Meijer and Spaink, 2011). We detected no difference in macrophage, neutrophil, or overall leukocyte number and distribution, vasculature formation, or blood circulation between mutants and their wildtype siblings at early time points up to 24 hpf (Fig. S2 and data not shown). By 2 dpf, however, mutant macrophages aggregated in large clusters on the yolk (Fig. 2A-C, and Fig. S2). Time-lapse imaging revealed that macrophages actively migrated toward and joined these aggregates in the mutant, in clear contrast to wildtype siblings, in which no aggregation occurred (Fig. 2C, and Movies S1 and S2). Other macrophages in mutants migrated from the yolk, but they often still formed small clusters (Fig. 2E, arrowheads). These macrophages typically did not enter the brain parenchyma in the mutants, but instead remained associated with the vasculature (Fig. 2D, Movies S3 and S4). Macrophage and microglia markers (mfap4 and apoe, respectively) confirmed the absence of brain macrophages and microglia through 2.5 dpf, even though peripheral macrophages were present (Fig. 2E). This aberrant migration and aggregation is characteristic of macrophages activated by infection or injury (Renshaw and Trede, 2012). We also observed that macrophages in mutants formed large cytoplasmic vacuoles, often several per cell (Fig. 3A-C), and that a higher fraction of macrophages were TUNEL-positive at 2.5 dpf in mutants than in sibling controls (Fig. 3C-E). TUNEL positive macrophages in the mutants exhibited condensed apoptotic nuclei (Fig. 3D) or cellular breakdown (Fig. 3E), two characteristics of an inflammatory form of apoptosis called pyroptosis (Lamkanfi and Dixit, 2010, Lage et al., 2013). The number of macrophages in the mutants declined significantly by 3-4 dpf (Fig. 3F), consistent with an increase in apoptotic macrophages. The active aggregation, vacuolation, and pyroptotic death of macrophages in nlrc3-like mutants resemble phenotypes previously described for macrophages responding to infection (Mujawar et al., 2006, Siracusa et al., 2008, Lage et al., 2013, Renshaw and Trede, 2012), suggesting that nlrc3-like mutant macrophages are activated despite the absence of overt infection.
Figure 2. Aberrant migration of primitive macrophages in nlrc3-like−/− mutants.
(A) Diagram showing the stereotypical migration route of macrophages from yolk sac to embryo proper (arrows). (B) Fluorescent images showing abnormal active coalescence of yolk sac macrophages (mpeg1:EGFP+ in green, arrows) that overlap with apoptotic marker TUNEL in red in mutants (bottom panel; n=12/12), which does not occur in wildtype (top panel; n=11/11). Region of imaging is indicated by yellow box in A. (C) Trajectories of individual yolk sac macrophages are shown using the MTrackJ cell tracking tool, where the end point is indicated by a solid triangle marker. See Movies S1 and S2 for the time-lapse series. (D) Time-lapse imaging of the head region (red box in A) where macrophages are expected to migrate into the brain, using transient transgenesis of the macrophage mpeg1:EGFP construct in green and the stable transgene kdrl:mCherry-CAAX to visualize the vasculature in red. Imaging shows macrophages entering the brain wildtype (top panels) but not in mutants (bottom panels). The numbers indicate the time in minutes. See Movies S3 and S4. (E) Whole mount in situ hybridization at 2.5 dpf showing macrophage mfap4+ and microglial apoe+ cells in wildtype heterozygous embryos (arrows) but not in mutants (arrows). Aberrant macrophage clusters form near cranial vasculature in mutants (arrowheads). MB, midbrain; HB, hindbrain; dpf, day postfertilization. See also Figure S2.
Figure 3. Inappropriate inflammatory activation of macrophages in nlrc3-like−/− mutants.
High magnification images showing striking cell morphology differences between yolksac macrophages of wildtype (A) and mutant (B). Macrophages in the mutants have multiple, large vacuoles (arrows). (C) Plot showing significantly higher percentages of primitive macrophages (mpeg1-EGFP+) exhibiting vacuolation (≥ 1 large cytoplasmic vacuole; p=0.047) and cell death (overlap of TUNEL staining with macrophage DAPI staining; p=0.037) in nlrc3-like−/− mutants (n=5 embryos) compared with siblings (n= 6 embryos). Statistical significance was determined by the two-tailed student’s t-test. Examples of dying mutant macrophages exhibiting apoptosis (D) and cellular breakdown (E). (F) Plot of macrophage number over time; diagram indicates the region of quantification (colored boxes); n=5 for each genotype at 1 dpf and 2.5 dpf, p = 0.0009 at 2.5 dpf; at 3–4 dpf, n=7 siblings and n= 3 mutants, p= 2E–05. (G) Graph shows relative mRNA levels of pro-inflammatory cytokines by qPCR comparing mutants (n=6) to wildtype siblings (n=6) at 3 dpf; p= 0.70 (β-actin); 0.002 (il-1β); 0.004 (il-8); 0.008 (il-12a); 0.03 (tnfα). n, number of independent embryos analyzed; dpf, day postfertilization. Error bars show s.e.m.; *, p < 0.05; **, p<0.01, ***, p < 0.001, all p-values are two-tailed.
Because macrophage activation is a prominent component of the inflammatory immune response after infection or injury (Mosser and Edwards, 2008), we examined whether the macrophage abnormalities in nlrc3-like mutants were linked to inappropriate inflammation. Using quantitative PCR, we found a highly significant increase in all pro-inflammatory cytokines tested (il-1β, il-8, il-12a, tnfα)(Rock et al., 2010), but no detectable change in expression of the β-actin control or the anti-inflammatory cytokine il-10 (Moore et al., 2001) in the mutants relative to wildtype siblings (Fig. 3G and data not shown). The highly elevated expression of pro-inflammatory cytokines indicated that macrophages in the mutants were activated in the context of an inflammatory environment. These inappropriately activated embryonic macrophages (microglia precursor cells) do not migrate properly and undergo premature cell death, thus accounting for the lack of microglia in nlrc3-like mutants.
Systemic inflammation in nlrc3-like mutants
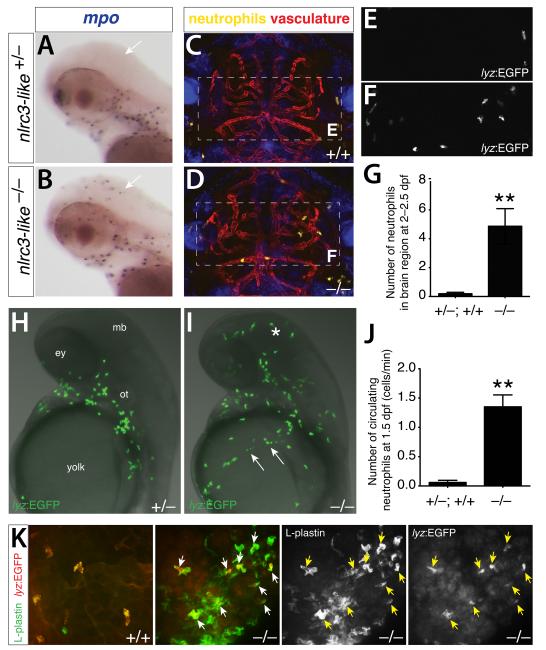
The abnormal activation of macrophages in nlrc3-like mutants raised the possibility of a broader dysregulation of the immune system. To investigate whether other immune cells are disrupted in nlrc3-like mutants, we examined neutrophils, the only other functional leukocyte present during embryonic stages, before the adaptive immune system becomes activate (Lam et al., 2004). Whereas neutrophils were excluded from the brain of wildtype embryos, they aberrantly infiltrated the brains of nlrc3-like mutants (Fig. 4), as demonstrated by examination of the neutrophil markers myeloperoxidase (mpo) and lysozyme C (lyz)(Meijer and Spaink, 2011). In vivo time-lapse imaging of the neutrophil reporter transgene lyz:EGFP (Hall et al., 2007) (Fig. 4A-F, Movies S5 and S6) revealed neutrophils freely roaming in the brain of nlrc3-like mutants. Quantitation of lyz:EGFP+ neutrophil numbers revealed a significant number of brain neutrophils in nlrc3-like mutants, compared with essentially none in wildtype siblings (Fig. 4G). Transverse sections confirmed the mislocalization of mutant neutrophils in the brain parenchyma and around vasculature (Fig. S3). In addition to brain infiltration, live imaging in nlrc3-like mutants also revealed a highly significant increase of neutrophils in circulation (Fig. 4H-J, Movies S7 and S8), starting as early as 1.5 dpf, when primitive macrophages begin to enter the brain in wildtype embryos. Using markers to distinguish neutrophils (lyz:EGFP+ and pan-leukocyte marker L-plastin+) and macrophages (L-plastin+ only), we also found that neutrophils joined the aberrant aggregates of macrophages described above (Fig. 2B-C; Fig. 4K). The increase of neutrophils in circulation, infiltration of neutrophils into the brain, intermixing of neutrophils and macrophages in abnormal aggregates, and extremely high levels of pro-inflammatory cytokines collectively indicate systemic inflammation in nlrc3-like mutants.
Figure 4. Systemic inflammation in nlrc3-like−/− mutants in the absence of infection or injury.
Neutrophil-specific marker mpo shows no brain neutrophils in wildtype siblings (arrow, A) but abundant infiltration in mutants (arrow, B). Lateral views, anterior to the left. (C,D) Representative frames from time-lapse imaging in the intact 2.5 dpf embryos (see Movies S5 and S6) show wandering neutrophils (lyz:EGFP) in the brain of mutants (D) but none in wildtype (C), with reference to vasculature (kdrl:mCherry-CAAX). Dorsal views, anterior to the top. (E,F) Higher magnification showing GFP channel alone (dotted box region in C and D, respectively). (G) Plot shows that high numbers of neutrophil infiltrate the brain in mutants (n=15) but nearly zero in wildtype (n=16, p = 0.0018). (H,I) Representative images from live imaging of neutrophils at 1.5 dpf show a large number of neutrophils in brain (asterisk) and circulation (arrows) in mutants (I) (see Movies S7 and S8). Lateral views, anterior to the top. (J) Plot shows significantly higher numbers of circulating neutrophils in mutants (n=4) compared with wildtype (n=5; p = 0.0083). (K) Neutrophils (lyz:EGFP as pseudo-colored in red) are closely intermixed with the aberrantly coalescing macrophages (L-plastin expression only as shown in green) in the mutants, whereas wildtype neutrophils are dispersed and not clustered with macrophages. n, number of embryos analyzed. Error bars show s.e.m.; **, p < 0.01; all p-values are two-tailed; ey, eye; mb, midbrain; ot, otic vesicle. See also Figure S3.
nlrc3-like acts autonomously in macrophages to mediate normal microglia development
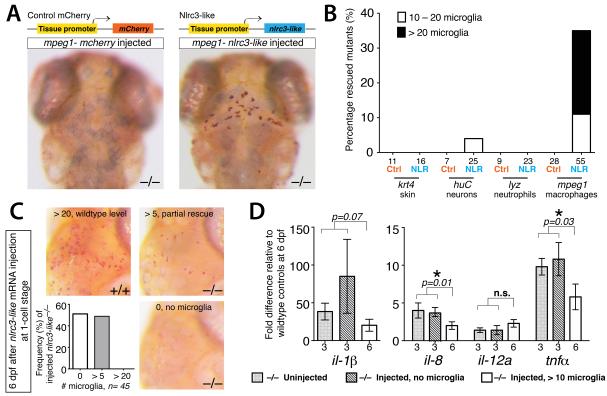
In light of such systemic inflammation, we sought to determine whether NLRC3-like acts within macrophages or in other cell types. Using transient Tol2-mediated transgenesis in nlrc3-like mutants, we introduced constructs that expressed either the wildtype nlrc3-like or control mCherry coding sequence in skin, neurons, neutrophils, or macrophages using cell-type specific regulatory elements (Fig. 5A,B and Fig. S4). Expression of the wildtype nlrc3-like gene in macrophages rescued microglia (>20 microglia/embryo; 24% of the mutants, n=55), but expression in the other cell types did not (Fig. 5). Partial rescue (10 – 20 microglia/embryo) was also observed in 11% of mutants injected with the macrophage expression construct (Fig. 5B), and in one case with the neuronal nlrc3-like expression construct (one mutant with partial rescue among 25 examined), perhaps due to leaky promoter expression (Fig. 5B). No rescue or partial rescue was obtained with any of the control mCherry expression constructs (Fig. 5B). Driving nlrc3-like expression using the myeloid lineage specific pu.1/spi1-GAL4 line also rescued microglia in the mutants, providing further evidence that nlrc3-like is required in macrophages (Fig. S4).
Figure 5. nlrc3-like has an autonomous and continuous function in macrophages that become microglia.
(A) Top, diagram of transgenic constructs driving either control mCherry or nlrc3-like expression by tissue-specific gene promoters in skin (keratin 4/krt4), neurons (elavl3/huC), neutrophils (lyz), or macrophages (mpeg1). Bottom, representative images showing rescue of neutral red+ microglia in nlrc3-like−/− mutants when expression of wildtype nlrc3-like is restored in macrophages (right) but not by control mCherry expression (left). (B) Plot quantifying the percentage of microglia rescue using different tissue-specific expression vectors. Ctrl, control mCherry injected; NLR, nlrc3-like construct injected. (C) Images and plot showing microglia by neutral red staining at 6 dpf, after nlrc3-like mRNA injection at the 1-cell stage. Injected nlrc3-like−/− mutants show no microglia or partial rescue (> 5) at 6 dpf, but no mutants are rescued to the wildtype level. In contrast, RNA injection does fully rescue microglia in some mutants at earlier stages (Fig. S1). (D) Graph showing relative pro-inflammatory transcript levels after transient rescue of nlrc3-like−/− mutants by injection of wildtype nlrc3-like mRNA at 1-cell stage. Injected mutants with no microglia at 6 dpf have elevated levels of pro-inflammatory cytokine transcripts that are similar to uninjected mutants. In contrast, mutants with partial rescue of microglia at 6 dpf have lower levels of il-8 and tnf-α expression, indicating that the number of microglia correlates inversely to the extent of inflammatory cytokine expression. Individual plots are normalized to their corresponding wildtype siblings, either uninjected or injected. Numbers below bar graphs represent n, number of embryos analyzed. Error bars show s.e.m. *, p < 0.05, one-tailed Student’s t-test; n.s., not significant. See also Figures S4 and S5.
nlrc3-like continues to be required after microglial progenitors enter the brain and differentiate
To test whether nlrc3-like is required after macrophages have enter the brain and differentiate as microglia, we transiently restored wildtype nlrc3-like expression by synthetic mRNA injection at the 1-cell stage. Injection of nlrc3-like mRNA into mutants can restore microglia at 5 dpf and earlier stages (Fig. S1 and data not shown). By contrast, at 6 dpf no mutants injected with nlrc3-like mRNA exhibited wildtype numbers of microglia (n= 45), and more than half of the injected mutants had no microglia detectable by neutral red staining (Fig. 5C). Consistent with a continuous requirement in microglia development, nlrc3-like mRNA is expressed at every stage we examined in wildtype embryos, from embryogenesis through the early larval period (1-9 dpf; Fig. S5). These data indicate that nlrc3-like continues to be required to maintain the proper number of microglia at later stages.
To assess inflammation in nlrc3-like mutants at later stages, we measured cytokine expression by qPCR. As at 3 dpf (Fig. 3G), transcripts for il-1β, il-8, and tnfα were significantly elevated in nlrc3-like mutants at 6 dpf, but il-12a expression was similar to wildtype siblings (Fig. 5D). To determine whether early expression of wildtype nlrc3-like is sufficient to suppress inflammation at later stages, we also examined cytokine levels in mutants injected with nlrc3-like mRNA. Pro-inflammatory cytokine expression at 6 dpf was elevated relative to wildtype controls, and the extent of the increase correlated with the number of microglia in the transiently rescued mutants (Fig. 5D). In these transient rescue experiments, mutants with >10 detectable microglia had significantly lower levels of il-8 and tnfα (p=0.01 and 0.03, respectively) than mutants with no microglia. These data indicate that nlrc3-like continues to be required to suppress inflammation and allow the development of the full population of microglia, even after microglia progenitors have entered the brain and differentiated.
Macrophages are required for increased inflammatory signaling in nlrc3-like mutants
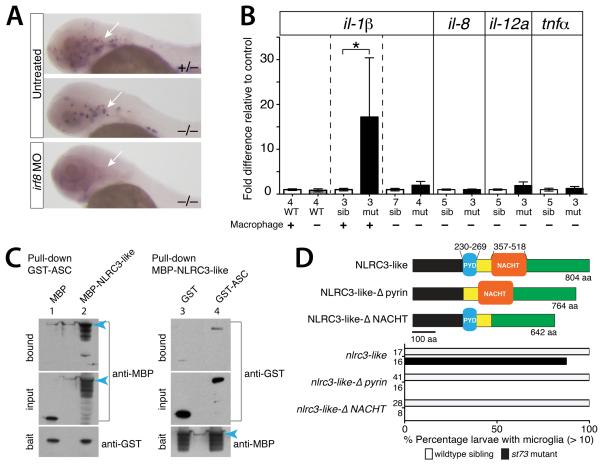
The autonomous action of nlrc3-like in the macrophage lineage and the integral roles of macrophages in mediating and responding to inflammation (Rock et al., 2010, Mosser and Edwards, 2008) prompted us to ask whether macrophages contributed to the systemic inflammation in nlrc3-like mutants. We thus examined pro-inflammatory cytokine expression after ablating macrophages in nlrc3-like mutants with a morpholino (MO) against interferon regulatory factor-8 (irf8)(Li et al., 2011) (Fig. 6A). Macrophage ablation reduced il-1β expression in nlrc3-like mutants to wildtype levels (Fig. 6B). In control assays, macrophage ablation in wildtype embryos did not alter il-1β levels, while il-1β expression remained elevated in unablated mutants (Fig. 6B), consistent with our previous analysis (Fig. 3G). All other pro-inflammatory cytokines (il-8, il12a, and tnfα) examined were also comparable to wildtype levels in macrophage-ablated mutants (Fig. 6B). Taken together, these results demonstrate that macrophages are required for the increased inflammatory signaling in nlrc3-like mutants.
Figure 6. NLRC3-like negatively regulates inflammatory signaling through macrophages, and may interact with the inflammasome component ASC and other pyrin and NACHT containing proteins.
(A) Macrophage mfap4 expression showing complete macrophage ablation by irf8 MO injection at 2 dpf (arrow). Injected wildtype embryos show complete loss of neutral red+ microglia (n=29/29, data not shown). (B) Graph showing relative il-1β mRNA levels, with pair-wise comparisons indicated by the dotted lines. Depletion of macrophages in nlrc3-like mutants reduces il-1β and other pro-inflammatory cytokine levels similar to wildtype levels. (C) 4–12% SDS-PAGE analyses of reciprocal pull-down assays show binding of zebrafish ASC with NLRC3-like (lanes 2 and 4), and minimal binding to tag-alone controls (lanes 1 and 3). MBP-tagged full-length NLRC3-like runs at ~160kDa (blue arrowhead) with several smaller processed forms; GST-ASC at ~50kDa; MBP alone from pMAL-c2X vector at ~50kDa; GST alone at ~30kDa. Blot shows 0.4% of the input prey protein per lane. (D) Top, schematic of the wildtype NLRC3-like protein and deletion versions: NLRC3-like-Δpyrin with deletion at amino acids (aa) 230–269 and NLRC3-like-ΔNACHT with deletion at aa 357–518. Bottom, bar graph showing large percentage of rescue of st73 mutants (87.5%, n=16) by mRNA injection encoding wildtype NLRC3-like but none using the mutant constructs. Numbers at left edge of bar graphs represent n, number of embryos analyzed. Error bars show s.e.m. *, p < 0.05, **, p < 0.01, ***, p < 0.001; n.s., not significant; MO, morpholino; WT, wildtype; sib, siblings; mut, mutants.
NLRC3-like requires both the pyrin and NACHT domains and may interact with the inflammasome component ASC
The inflammatory phenotypes of nlrc3-like mutants suggested that this unusual NLR family member might regulate the production of pro-inflammatory cytokines and induction of pyroptosis by inhibiting the inflammasome (Davis et al., 2011). Previous in vitro studies indicate that cellular and viral proteins with similar domain organizations, containing only the PYD or PYD combined with NACHT, can inhibit the inflammasome through homotypic interactions with other inflammasome components containing these domains (Dorfleutner et al., 2007a, Imamura et al., 2010, Dorfleutner et al., 2007b, Stehlik et al., 2003). To examine the possibility that NLRC3-like interacts with components of the inflammasome, we asked whether NLRC3-like protein can bind to the PYD-containing adaptor protein of the inflammasome, ASC (apoptosis-associated speck-like protein)(Davis et al., 2011). Reciprocal pull-down assays showed that GST-tagged zebrafish ASC and MBP-tagged NLRC3-like can bind (Fig. 6C). The unusual pyrin-NACHT structure of NLRC3-like, the systemic inflammation in nlrc3-like mutants, and the potential interaction between the NLRC3-like and ASC proteins suggest that NLRC3-like may negatively regulate inflammasome activity or other pathways mediating inflammatory signaling that involve ASC (Sarkar et al., 2006, Kolly et al., 2009).
To determine which domains may be essential for nlrc3-like function, we performed a structure-function analysis. We designed deletion constructs that express mRNAs encoding NLRC3-like proteins that lack either the NACHT domain (Nlrc3-like-ΔNACHT), or the pyrin domain (Nlrc3-like-Δpyrin), which are the two recognizable conserved domains in NLRC3-like. Wildtype nlrc3-like mRNA injection can fully rescue microglia in nlrc3-like mutants at 3-5 dpf (Fig. 6D and S1), and we assayed the ability of mRNAs encoding the deletion mutants to rescue nlrc3-like mutants (Fig. 6D). Both domains are required to rescue nlrc3-like mutants (Fig. 6D), providing evidence that the pyrin and NACHT domains are both essential for NLRC3-like function. The requirement for the pyrin domain supports the possibility that NLRC3-like may functionally interact with other pyrin-containing proteins such as ASC (Dorfleutner et al., 2007a, Vajjhala et al., 2012, Liepinsh et al., 2003), and the requirement for the NACHT domain indicates that NLRC3-like may also interact with itself or other NLR proteins that regulate inflammatory signaling.
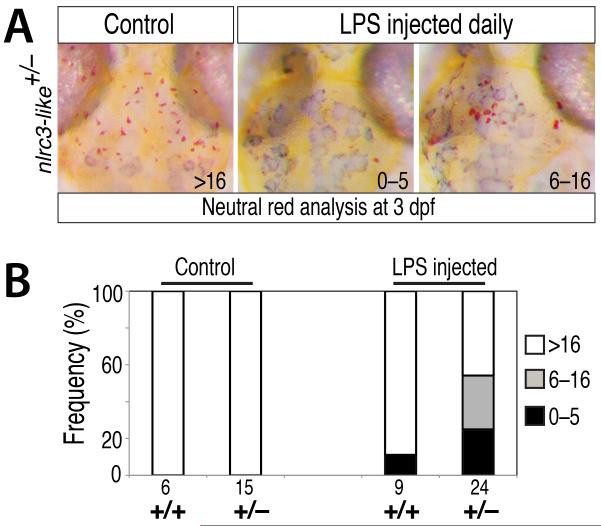
LPS challenge compromises microglia development in nlrc3-like heterozygous embryos
To further test the relationship between systemic immune activation and nlrc3-like function, we injected LPS into the bloodstream of embryos from nlrc3-like+/− heterozygous intercrosses at 1 dpf and 2 dpf, and analyzed microglial development at 3 dpf. LPS injection reduced microglia formation in many of the injected nlrc3-like+/− heterozygous embryos (54%, n=24) and affected one of nine injected wildtype siblings (11%)(Fig. 7). These data indicate that nlrc3-like+/− heterozygotes are more susceptible to disruptions of microglia development when exposed to the LPS inflammatory stimulus than wildtype siblings. These results are consistent with the autonomous function of nlrc3-like in the macrophage lineage (Fig. 5A and 5B), but also show that extrinsic inflammatory stimuli can disrupt the development of microglia when nlrc3-like function is partially reduced.
Figure 7. Exposure of nlrc3-like heterozygous embryos to systemic LPS challenge disrupts microglia development.
(A) Neutral staining at 3 dpf showing normal formation of microglia in control uninjected nlrc3-like heterozygous embryos. In contrast, many nlrc3-like heterozygous embryos injected with LPS at 1 dpf and 2 dpf have strongly reduced (6–16 microglia) or eliminated (0–5 microglia) microglia. (B) Bar graph showing frequency of loss of microglia in nlrc3-like heterozygous fish injected with LPS, compared with little effect on the wildtype siblings.
DISCUSSION
Macrophages utilize an array of pattern recognition receptors, including the Toll-like receptors (TLRs) and NLRs, to recognize and respond rapidly to infection, injury, and cellular stress (Chen et al., 2009, Davis et al., 2011, Mosser and Edwards, 2008). The signaling activities of these receptors must be regulated to maintain homeostasis and prevent inappropriate or chronic inflammation. Previous studies show that TLR signaling is attenuated through multiple mechanisms, including production of decoy receptors and inhibitors of downstream signaling complexes (Anwar et al., 2013, Coll and O’ Neill, 2010). Much less is known about the negative regulation of NLRs in vivo, despite their far-reaching roles in regulating innate immunity (Chen et al., 2009, Davis et al., 2011, Coll and O’Neill, 2010). Our results demonstrate that nlrc3-like is an essential negative regulator of macrophage activation and inflammation. Loss of NLRC3-like causes an inflammatory state in macrophages that profoundly alters their normal course of development. Instead of seeding the brain to form microglia, embryonic macrophages in the mutant actively migrate to form aberrant cellular aggregations in the yolk, fail to enter the brain, and eventually undergo inflammatory cell death. Activity of nlrc3-like continues to be required even after microglial progenitors enter the brain and differentiate, because transient expression of the wildtype gene is not sufficient for long-term rescue of microglia or suppression of inflammation. These results highlight the importance of regulating the activation of embryonic macrophages to allow normal microglia development, in addition to the more widely recognized need to control activation of adult microglia to prevent chronic or acute inflammation that is common in many neurodegenerative and autoimmune diseases such as Alzheimer’s disease, Parkinson’s disease, and multiple sclerosis (Cunningham, 2013, Amor et al., 2010).
Our data support a model in which NLRC3-like competes with pro-inflammatory NLRs for ASC binding to set a threshold level of activated canonical NLRs required to trigger inflammation. According to this view, NLRC3-like would prevent widespread inflammation in response to limited or transient stimuli, such as exposure to non-pathogenic microbes, but still allow rapid initiation of inflammation when NLRs are activated by a robust stimulus such as an injury or infection. This model is also consistent with the proposed mechanism of cellular and viral proteins containing only the pyrin domain, only the NACHT domain, or a pyrin and NACHT, which inhibit the inflammasome through homotypic binding (Dorfleutner et al., 2007a, Imamura et al., 2010, Dorfleutner et al., 2007b, Stehlik et al., 2003). Furthermore, similar to these inflammasome inhibitory proteins, NLRC3-like has a distinctive pyrin-NACHT structure that lacks the canonical LRRs thought to be important for self-inhibition (Rosenstiel et al., 2007, Chen et al., 2009), suggesting that it may be a constitutively active inhibitor. Our structure-function analysis indicates that both the pyrin and NACHT domains are required for NLRC3-like function, suggesting that NLRC3-like may interact with other proteins containing one or both of these domains, including the adaptor ASC.
The zebrafish genome contains a large number of NLR genes homologous to mammalian NLRC3 (Laing et al., 2008), and our analysis provides functional insights into the first of this group of zebrafish NLRC3-like genes. Although its NACHT domain is most similar to the mammalian NLRC3 (Laing et al., 2008, Hughes, 2006), the central NACHT domain is the only major region conserved between the zebrafish NLRC3-like and mammalian NLRC3 proteins. Moreover, the zebrafish NLRC3-like protein has only pyrin and NACHT domains, whereas the mammalian NLRC3 has C-terminal LRRs but lacks an N-terminal effector motif. Our biochemical studies suggest that zebrafish NLRC3-like may suppress inflammation by interfering with interactions between ASC and other inflammasome components. The mammalian NLRC3 protein can also suppress inflammation. A recent study shows that NLRC3 reduces TLR signaling and NF-kB activity after LPS exposure through its interaction with TRAF6 (Schneider et al., 2012). It is also possible that zebrafish NLRC3-like regulates NF-kB activation in an inflammasome-independent manner, because highly elevated levels of mRNAs for pro-inflammatory cytokines were detected in nlrc3-like deficient embryos. Consistent with this possibility, zebrafish NLRC3-like has three predicted TRAF interaction motifs (following the consensus Pro/Ser/Ala/Thr-X-Gln/Glu-Glu). Thus NLRC3-like may repress NFkB activity by interference with TRAF adaptor proteins in the recruitment of NF-kB, similar to the anti-inflammatory action of NLRC3 and NLRP12 after immune stimulation (Schneider et al., 2012, Allen et al., 2012, Zaki et al., 2011). Alternatively, inappropriate activation of the inflammasome may secondarily induce cytokine expression in nlrc3-like mutants.
Another important and intriguing difference between zebrafish nlrc3-like and the mouse NLR genes known to suppress inflammation, including NLRC3 and NLRP12 (Schneider et al., 2012, Allen et al., 2012, Zaki et al., 2011)), is that the mouse hyperinflammatory mutant phenotypes are evident only after LPS injection or infection, whereas systemic inflammation is found in nlrc3-like mutant zebrafish in the absence of immune perturbation. Under standard rearing conditions, wildtype and heterozygous siblings show no signs of inflammation, but nlrc3-like mutants display severe, systemic dysregulation of the immune system. This difference reveals that nlrc3-like is an essential suppressor of inflammation at steady state, highlighting the importance of the regulatory mechanism that controls immune activation not only in response to overt immune challenges, but also during normal development.
In summary, we identify an essential role for an NLR that acts as a checkpoint on inflammatory processes and permits the development of microglia in zebrafish. Our analysis of nlrc3-like mutants highlights the deleterious effect that inflammatory activation has on embryonic macrophages and their subsequent development into microglia. The negative regulatory function of NLRC3-like may provide important insights into the maintenance of homeostasis and the etiology of autoimmune and inflammatory disorders.
EXPERIMENTAL PROCEDURES
Zebrafish lines and embryos
Embryos from wildtype (TL, AB/TU, and WIK), transgenic (Tg(lyz:EGFP)(Hall et al., 2007), Tg(mpeg1:EGFP)(Ellett et al., 2011), Tg(kdrl:mCherry-CAAX)(Fujita et al., 2011), and Tg(pu.1:Gal4-UAS-EGFP)(Peri and Nusslein-Volhard, 2008)), and nlrc3-likest73/+ heterozygous intercross backgrounds were raised at 28.5°C, and staged as described (Kimmel et al., 1995). Embryos were treated with 0.003% 1-phenyl-2-thiourea (PTU) in methylene blue embryo water to inhibit pigmentation.
ENU mutagenesis, microglia screen, and neutral red assay
Founder P0 wildtype males were mutagenized with N-ethyl-N-nitrosourea (ENU) and subsequently outcrossed to raise F1 and F2 families for screening as described (Pogoda et al., 2006). A pilot F3 genetic screen was conducted to identify putative mutants with specific defects in microglia (e.g. loss of microglia) but no apparent anatomical morphological defects at 5 dpf. A total of 74 F2 families were screened, which amounted to analysis of 50.7 mutagenized genomes. Two putative microglia mutants were identified, but only st73 was recovered. Microglia were scored in the live larvae by a neutral red vital dye staining assay (Herbomel et al., 2001) with modifications. For phenotypic analyses, neutral red assay was used at 3 dpf and later stages by incubating larvae in embryo water containing 2.5 μg/ml neutral red at 28.5°C for 2-3 hours, followed by 1-2 water changes, and then analyzed 0.5-24 hours later using a dissecting microscope.
Genetic mapping
st73 mutant embryos were phenotypically sorted from wildtype siblings at 4–5 dpf by a lack of neutral red positive microglia staining in the head. Bulk segregant analysis with 480 simple sequence length polymorphisms (SSLPs)(Talbot and Schier, 1999) identified markers on LG15 flanking the st73 mutation. High-resolution mapping was conducted using additional SSLPs and single nucleotide polymorphisms linked to the mutation, which were found from sequencing PCR fragments amplified from genomic DNA of mutants and wildtype siblings. Sequencing segments of genes in the critical interval by PCR from genomic DNA templates identified the st73 lesion in the nlrc3-like gene (LOC100538217). Genotyping the st73 lesion is based on a PCR/restriction digest assay using the following forward (5′-CAACAATTTCATCAAAATCTTCAA) and reverse (5′-CAGACATATTCTGGAAGCAAACA) PCR primers and MseI digest.
Whole mount RNA in situ hybridization
In situ hybridization on whole zebrafish embryos and larvae from 20 hpf to 5 dpf was performed using standard methods as described (Pogoda et al., 2006). Antisense riboprobes were transcribed from the following gene coding sequences cloned in pCRII-Topo vector: lyz (518 bp), mfap4 (627 bp using primer sequences as described (Zakrzewska et al., 2010)), l-plastin (1541 bp of lcp1), and apoe (505 bp of apoeb). Other probes were made from vectors encoding mpo (full-length, Open Biosystems clone 6960294) and kdrl (flk1)(Thisse, 2008).
Whole mount immunostaining and TUNEL assay
TUNEL assay (In situ Cell Death Detection Kit, TMR Red, Roche) was performed as previously described (Chen et al., 2010). Subsequent immunostaining on whole zebrafish embryos or larvae was performed using the anti-L-plastin antibody (Redd et al., 2006) at 1:250-500 dilution and anti-GFP antibody (GT859,GeneTex) at 1:500 dilution, followed by DAPI staining.
Time-lapse and fluorescent imaging
For live imaging, embryos were embedded in 1.5% low melting point agarose on glass slides. For cyrosectioning, post-fixed embryos were equilibrated to 30% sucrose, embedded in O.C.T. medium, snap frozen in dry ice-ethanol bath, sectioned at 10–12 μm, and subsequently imaged. All time-lapse and fluorescent images, except images for analysis of vacuolation and apoptosis, were taken on a Zeiss LSM 5 Pascal confocal microscope using the 10× (NA 0.30) and 20× (NA 0.75) objectives, and 488 nm and 543 nm laser lines with bright-field. Images of yolksac macrophages for assessing cellular vacuolation and cell death were taken on an upright Zeiss Axio Imager.M2 microscope using the 20× (NA 0.8) and 63× (NA 1.4) plan-apochromatic objectives. See also the Extended Experimental Procedures for additional information.
Expression constructs
Full-length nlrc3-like coding sequence (XM_003200091.1) was cloned from a 2.5 dpf embryonic cDNA pool, and directionally inserted into the pCS2+ plasmid at XhoI/XbaI sites to make pCS2-FL-nlrc3-like for mRNA transcription. Truncated forms (pCS2-nlrc3-like-Δpyrin and pCS2-nlrc3-like-ΔNACHT) were made using fusion PCR and also directionally cloned into the pCS2+ plasmid at XhoI/XbaI sites. All transgenic expression constructs contained the Tol2-transposon sequences for genomic integration. Primers used in cloning are listed in Table S1. See also Extended Experimental Procedures for further details.
mRNA, plasmid, and morpholino injections
pCS2-FL-nlrc3-like and truncated forms (pCS2-nlrc3-like-Δpyrin and pCS2-nlrc3-like-ΔNACHT) were transcribed using the Sp6 mMessage machine (Ambion) to produce 5′ capped mRNAs with a poly-A tail. A range of 50–200 pg of nlrc3-like mRNA was injected into 1–4 cell stage embryos; all amounts of RNA tested in this range can rescue microglia in nlrc3-like mutants. Structure-function analysis was conducted using injections of 150-200 pg of mRNA in 1–2 cell stage embryos. Transgenes were transiently expressed by co-injecting ~12–25 pg of Tol2-plasmid as described above and ~50-100 pg of Tol2 transposase mRNA at 1-cell stage. A splice-blocking morpholino against irf8 was synthesized by Gene Tools, LLC, and was injected at 2.5–5 ng into 1-cell stage embryos as previously described (Li et al., 2011). As negative controls, embryos were injected with water or not injected.
RNA extraction, RT-PCR, and quantitative PCR (qPCR)
Total RNA was extracted from individual embryos using the RNAqueous-Micro RNA Isolation Kit (Ambion). Embryos were pre-sorted by neutral red phenotype prior to lysis, and genotyped using a PCR-restriction digest assay as described above. cDNA was made using oligo dT primer and SuperScript II or III reverse transcriptase (Invitrogen). qPCR was performed on the ABI 7300 Real-Time PCR System or the Bio-Rad CFX384 Real-Time PCR Detection System with SYBR GreenER qPCR reagent (Invitrogen). Additional information is provided in the Extended Experimental Procedures.
Protein interaction pull down assay
DNA constructs were made using Gateway cloning to express control gluthatione S-transferase (GST) or GST-ASC fusion protein containing the full-length zebrafish ASC sequence (NM_131495.2) from modified pGEX-4T-1 vector (Meireles et al., 2009) with Gateway cassettes, and control maltose-binding protein (MBP) or full-length Nlrc3-like protein (XP_003200139.1) fused to MBP from modified pMAL-c2X (Meireles et al., 2009). All proteins were expressed in Escherichia coli BL-21 cells (Invitrogen) using IPTG induction. See also the Extended Experimental Procedures for more details.
LPS microinjection
3-5 nl of lipopolysaccharides (LPS) from Pseudomonas aeroginosa 10 (Sigma) at 5 mg/ml in PBS buffer with rhodamine dextran (10,000 MW at 1:5 dilution) was injected into the circulation valley (duct of Cuvier) as described (Milligan-Myhre et al., 2011). Embryos were injected at 22–24 hpf and at 48 hpf; detection of the fluorescent dextran throughout vasculature was used to select for successful injections for later analysis. Injected embryos and control uninjected siblings were analyzed at 3 dpf using the neutral red assay for microglia.
Supplementary Material
HIGHLIGHTS.
Genetic screen identifies NLRC3-like as a key regulator of microglia development
NLRC3-like acts cell-autonomously in the macrophage lineage
NLRC3-like prevents unwarranted macrophage activation and systemic inflammation
Suppression of macrophage activation is essential for microglia development
ACKNOWLEDGEMENTS
We are grateful to G. J. Lieschke, A.T. Look, F. Marlow, S. Mercurio, H. Ohkura, F. Peri, M. Redd, B. Weinstein, and G. Wright for fish strains and plasmids; A. Meireles for cloning reagents; and M. Barna, M. Fuller, and S. Kim for sharing equipment. Special thanks to M. Krasnow, B. Barres, and Talbot lab members for critical comments on the manuscript and T. Reyes and C. Hill for fish care. C.E.S. recovered and mapped the st73 mutation, and conducted all experiments. K.R.M, W.J., and W.S.T. performed the genetic screen. C.E.S. and W.S.T. analyzed the data and wrote the manuscript. C.E.S. was supported by NIH NRSA postdoctoral fellowship 5F32NS067754, and K.R.M. by NMSS fellowship FG 1719-A-1. This work was supported by NIH grant R01 NS065787 to W.S.T.
Footnotes
Supplemental Information includes five figures, one table, eight movies, Extended Experimental Procedures, and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- AGUZZI A, BARRES BA, BENNETT ML. Microglia: scapegoat, saboteur, or something else? Science. 2013;339:156–61. doi: 10.1126/science.1227901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALLEN IC, WILSON JE, SCHNEIDER M, LICH JD, ROBERTS RA, ARTHUR JC, WOODFORD RM, DAVIS BK, URONIS JM, HERFARTH HH, JOBIN C, ROGERS AB, TING JP. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity. 2012;36:742–54. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMOR S, PUENTES F, BAKER D, VAN DER VALK P. Inflammation in neurodegenerative diseases. Immunology. 2010;129:154–69. doi: 10.1111/j.1365-2567.2009.03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANWAR MA, BASITH S, CHOI S. Negative regulatory approaches to the attenuation of Toll-like receptor signaling. Exp Mol Med. 2013;45:e11. doi: 10.1038/emm.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN G, SHAW MH, KIM YG, NUNEZ G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009;4:365–98. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- CHEN HL, YUH CH, WU KK. Nestin is essential for zebrafish brain and eye development through control of progenitor cell apoptosis. PLoS One. 2010;5:e9318. doi: 10.1371/journal.pone.0009318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLL RC, O’NEILL LA. New insights into the regulation of signalling by toll-like receptors and nod-like receptors. J Innate Immun. 2010;2:406–21. doi: 10.1159/000315469. [DOI] [PubMed] [Google Scholar]
- CUNNINGHAM C. Microglia and neurodegeneration: The role of systemic inflammation. Glia. 2013;61:71–90. doi: 10.1002/glia.22350. [DOI] [PubMed] [Google Scholar]
- DAVIS BK, WEN H, TING JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–35. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DORFLEUTNER A, BRYAN NB, TALBOTT SJ, FUNYA KN, RELLICK SL, REED JC, SHI X, ROJANASAKUL Y, FLYNN DC, STEHLIK C. Cellular pyrin domain-only protein 2 is a candidate regulator of inflammasome activation. Infect Immun. 2007a;75:1484–92. doi: 10.1128/IAI.01315-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DORFLEUTNER A, TALBOTT SJ, BRYAN NB, FUNYA KN, RELLICK SL, REED JC, SHI X, ROJANASAKUL Y, FLYNN DC, STEHLIK C. A Shope Fibroma virus PYRIN-only protein modulates the host immune response. Virus Genes. 2007b;35:685–94. doi: 10.1007/s11262-007-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLETT F, PASE L, HAYMAN JW, ANDRIANOPOULOS A, LIESCHKE GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117:e49–56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUJITA M, CHA YR, PHAM VN, SAKURAI A, ROMAN BL, GUTKIND JS, WEINSTEIN BM. Assembly and patterning of the vascular network of the vertebrate hindbrain. Development. 2011;138:1705–15. doi: 10.1242/dev.058776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GINHOUX F, GRETER M, LEBOEUF M, NANDI S, SEE P, GOKHAN S, MEHLER MF, CONWAY SJ, NG LG, STANLEY ER, SAMOKHVALOV IM, MERAD M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–5. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALL C, FLORES MV, STORM T, CROSIER K, CROSIER P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERBOMEL P, THISSE B, THISSE C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–45. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- HERBOMEL P, THISSE B, THISSE C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238:274–88. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- HUGHES AL. Evolutionary relationships of vertebrate NACHT domain-containing proteins. Immunogenetics. 2006;58:785–91. doi: 10.1007/s00251-006-0148-8. [DOI] [PubMed] [Google Scholar]
- IMAMURA R, WANG Y, KINOSHITA T, SUZUKI M, NODA T, SAGARA J, TANIGUCHI S, OKAMOTO H, SUDA T. Anti-inflammatory activity of PYNOD and its mechanism in humans and mice. J Immunol. 2010;184:5874–84. doi: 10.4049/jimmunol.0900779. [DOI] [PubMed] [Google Scholar]
- KIMMEL CB, BALLARD WW, KIMMEL SR, ULLMANN B, SCHILLING TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- KOLLY L, KARABABA M, JOOSTEN LA, NARAYAN S, SALVI R, PETRILLI V, TSCHOPP J, VAN DEN BERG WB, SO AK, BUSSO N. Inflammatory role of ASC in antigen-induced arthritis is independent of caspase-1, NALP-3, and IPAF. J Immunol. 2009;183:4003–12. doi: 10.4049/jimmunol.0802173. [DOI] [PubMed] [Google Scholar]
- LAGE SL, AMARANTE-MENDES GP, BORTOLUCI KR. Evaluation of pyroptosis in macrophages using cytosolic delivery of purified flagellin. Methods. 2013 doi: 10.1016/j.ymeth.2013.02.010. [DOI] [PubMed] [Google Scholar]
- LAING KJ, PURCELL MK, WINTON JR, HANSEN JD. A genomic view of the NOD-like receptor family in teleost fish: identification of a novel NLR subfamily in zebrafish. BMC Evol Biol. 2008;8:42. doi: 10.1186/1471-2148-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAM SH, CHUA HL, GONG Z, LAM TJ, SIN YM. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev Comp Immunol. 2004;28:9–28. doi: 10.1016/s0145-305x(03)00103-4. [DOI] [PubMed] [Google Scholar]
- LAMKANFI M, DIXIT VM. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe. 2010;8:44–54. doi: 10.1016/j.chom.2010.06.007. [DOI] [PubMed] [Google Scholar]
- LI L, JIN H, XU J, SHI Y, WEN Z. Irf8 regulates macrophage versus neutrophil fate during zebrafish primitive myelopoiesis. Blood. 2011;117:1359–69. doi: 10.1182/blood-2010-06-290700. [DOI] [PubMed] [Google Scholar]
- LIEPINSH E, BARBALS R, DAHL E, SHARIPO A, STAUB E, OTTING G. The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol. 2003;332:1155–63. doi: 10.1016/j.jmb.2003.07.007. [DOI] [PubMed] [Google Scholar]
- MASON DR, BECK PL, MURUVE DA. Nucleotide-binding oligomerization domain-like receptors and inflammasomes in the pathogenesis of non-microbial inflammation and diseases. J Innate Immun. 2012;4:16–30. doi: 10.1159/000334247. [DOI] [PubMed] [Google Scholar]
- MEIJER AH, SPAINK HP. Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets. 2011;12:1000–17. doi: 10.2174/138945011795677809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEIRELES AM, FISHER KH, COLOMBIE N, WAKEFIELD JG, OHKURA H. Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol. 2009;184:777–84. doi: 10.1083/jcb.200811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLIGAN-MYHRE K, CHARETTE JR, PHENNICIE RT, STEPHENS WZ, RAWLS JF, GUILLEMIN K, KIM CH. Study of host-microbe interactions in zebrafish. Methods Cell Biol. 2011;105:87–116. doi: 10.1016/B978-0-12-381320-6.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOORE KW, DE WAAL MALEFYT R, COFFMAN RL, O’GARRA A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- MOSSER DM, EDWARDS JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUJAWAR Z, ROSE H, MORROW MP, PUSHKARSKY T, DUBROVSKY L, MUKHAMEDOVA N, FU Y, DART A, ORENSTEIN JM, BOBRYSHEV YV, BUKRINSKY M, SVIRIDOV D. Human immunodeficiency virus impairs reverse cholesterol transport from macrophages. PLoS Biol. 2006;4:e365. doi: 10.1371/journal.pbio.0040365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAOLICELLI RC, BOLASCO G, PAGANI F, MAGGI L, SCIANNI M, PANZANELLI P, GIUSTETTO M, FERREIRA TA, GUIDUCCI E, DUMAS L, RAGOZZINO D, GROSS CT. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–8. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- PERI F, NUSSLEIN-VOLHARD C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–27. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- PERRY VH, NICOLL JA, HOLMES C. Microglia in neurodegenerative disease. Nat Rev Neurol. 2010;6:193–201. doi: 10.1038/nrneurol.2010.17. [DOI] [PubMed] [Google Scholar]
- POGODA HM, STERNHEIM N, LYONS DA, DIAMOND B, HAWKINS TA, WOODS IG, BHATT DH, FRANZINI-ARMSTRONG C, DOMINGUEZ C, ARANA N, JACOBS J, NIX R, FETCHO JR, TALBOT WS. A genetic screen identifies genes essential for development of myelinated axons in zebrafish. Dev Biol. 2006;298:118–31. doi: 10.1016/j.ydbio.2006.06.021. [DOI] [PubMed] [Google Scholar]
- RANSOHOFF RM, CARDONA AE. The myeloid cells of the central nervous system parenchyma. Nature. 2010;468:253–62. doi: 10.1038/nature09615. [DOI] [PubMed] [Google Scholar]
- RANSOHOFF RM, PERRY VH. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–45. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- REDD MJ, KELLY G, DUNN G, WAY M, MARTIN P. Imaging macrophage chemotaxis in vivo: studies of microtubule function in zebrafish wound inflammation. Cell Motil Cytoskeleton. 2006;63:415–22. doi: 10.1002/cm.20133. [DOI] [PubMed] [Google Scholar]
- RENSHAW SA, TREDE NS. A model 450 million years in the making: zebrafish and vertebrate immunity. Dis Model Mech. 2012;5:38–47. doi: 10.1242/dmm.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROCK KL, LATZ E, ONTIVEROS F, KONO H. The sterile inflammatory response. Annu Rev Immunol. 2010;28:321–42. doi: 10.1146/annurev-immunol-030409-101311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSENSTIEL P, TILL A, SCHREIBER S. NOD-like receptors and human diseases. Microbes Infect. 2007;9:648–57. doi: 10.1016/j.micinf.2007.01.015. [DOI] [PubMed] [Google Scholar]
- SARKAR A, DUNCAN M, HART J, HERTLEIN E, GUTTRIDGE DC, WEWERS MD. ASC directs NF-kappaB activation by regulating receptor interacting protein-2 (RIP2) caspase-1 interactions. J Immunol. 2006;176:4979–86. doi: 10.4049/jimmunol.176.8.4979. [DOI] [PubMed] [Google Scholar]
- SCHAFER DP, LEHRMAN EK, KAUTZMAN AG, KOYAMA R, MARDINLY AR, YAMASAKI R, RANSOHOFF RM, GREENBERG ME, BARRES BA, STEVENS B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHNEIDER M, ZIMMERMANN AG, ROBERTS RA, ZHANG L, SWANSON KV, WEN H, DAVIS BK, ALLEN IC, HOLL EK, YE Z, RAHMAN AH, CONTI BJ, EITAS TK, KOLLER BH, TING JP. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol. 2012;13:823–31. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIRACUSA MC, REECE JJ, URBAN JF, JR., SCOTT AL. Dynamics of lung macrophage activation in response to helminth infection. J Leukoc Biol. 2008;84:1422–33. doi: 10.1189/jlb.0308199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEHLIK C, KRAJEWSKA M, WELSH K, KRAJEWSKI S, GODZIK A, REED JC. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-kappa B and pro-caspase-1 regulation. Biochem J. 2003;373:101–13. doi: 10.1042/BJ20030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TALBOT WS, SCHIER AF. Positional cloning of mutated zebrafish genes. Methods Cell Biol. 1999;60:259–86. doi: 10.1016/s0091-679x(08)61905-6. [DOI] [PubMed] [Google Scholar]
- THISSE B, WRIGHT GJ, THISSE C. Embryonic and Larval Expression Patterns from a Large Scale Screening for Novel Low Affinity Extracellular Protein Interactions. ZFIN Direct Data Submission. 2008 doi: 10.1101/gr.7187808. http://zfin.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAJJHALA PR, MIRAMS RE, HILL JM. Multiple binding sites on the pyrin domain of ASC protein allow self-association and interaction with NLRP3 protein. J Biol Chem. 2012;287:41732–43. doi: 10.1074/jbc.M112.381228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAKI MH, VOGEL P, MALIREDDI RK, BODY-MALAPEL M, ANAND PK, BERTIN J, GREEN DR, LAMKANFI M, KANNEGANTI TD. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20:649–60. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAKRZEWSKA A, CUI C, STOCKHAMMER OW, BENARD EL, SPAINK HP, MEIJER AH. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116:e1–11. doi: 10.1182/blood-2010-01-262873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.