Abstract
Background
While methods for generating cardiomyocytes (CMs) from pluripotent stem cells (PSCs) have been reported, current methods produce heterogeneous mixtures of CMs and non-CM cells. Here, we report an entirely novel system in which PSC-derived CMs are purified by CM-specific molecular beacons (MBs). MBs are nano-scale probes that emit a fluorescence signal when hybridized to target mRNAs.
Method and Results
Five MBs targeting mRNAs of either cardiac troponin T or myosin heavy chain 6/7 were generated. Among five MBs, a MB targeting myosin heavy chain 6/7 mRNA (MHC1-MB) identified up to 99% of HL-1 CMs, a mouse CM cell line, but < 3% of four non-CM cell types in flow cytometry analysis, indicating that MHC1-MB is specific for identifying CMs. We delivered MHC1-MB into cardiomyogenically differentiated PSCs through nucleofection. The detection rate of CMs was similar to the percentages of cardiac troponin T (TNNT2) or cardiac troponin I (TNNI3)-positive CMs, supporting the specificity of MBs. Finally, MHC1-MB-positive cells were FACS-sorted from mouse and human PSC differentiating cultures and ~97% cells expressed TNNT2- or TNNI3 determined by flow cytometry. These MB-based sorted cells maintained their CM characteristics verified by spontaneous beating, electrophysiologic studies, and expression of cardiac proteins. When transplanted in a myocardial infarction model, MB-based purified CMs improved cardiac function and demonstrated significant engraftment for 4 weeks without forming tumors.
Conclusions
We developed a novel CM selection system that allows production of highly purified CMs. These purified CMs and this system can be valuable for cell therapy and drug discovery.
Keywords: pluripotent stem cell, cardiomyocyte, molecular beacons, cell selection, cardiac regeneration
Introduction
Human pluripotent stem cells (hPSCs), which include embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs), have emerged as a promising therapeutic option for repairing cardiac damage. While various methods for differentiating CMs from hPSCs have been developed1-3, without purification, these systems can only generate mixed populations with undifferentiated cells or non-CMs which may elicit adverse outcomes4, 5. Hence, one of the major challenges confronting the clinical use of hPSC-derived CMs is the development of efficient isolation techniques that allow enrichment of purified CMs.
Diverse approaches have been attempted to purify CMs from differentiating PSC cultures. Genetic approaches using a fluorescent reporter gene driven by a cardiac promoter such as NKX2.56, ISL17 or αMHC8 were reported to efficiently isolate CMs and/or cardiac progenitor cells, but require genetic modification and therefore are incompatible with clinical use. Another approach using a Percoll gradient capitalized on the specific density of CMs2, 9; due to its crudeness, however, the maximal purity of the cells reported was only 53% 10. Sorting cells by surface proteins for progenitor cell markers such as KDR 11 or PDGFRA 12 isolated cardiac progenitors but resulted in a mixture of CMs together with endothelial and smooth muscle cells. Fluorescence activated cell sorting (FACS) after application of the fluorescent mitochondrial dye tetramethylrhodamine methyl ester perchlorate (TMRM) allowed cells with high mitochondrial density such as mature CMs to take up the dye and become amenable for cell sorting13. However, as the dye reacts with undifferentiated PSCs and does not detect most of the immature CMs14, concerns were raised about specificity and sensitivity. Recently, studies identified two surface proteins expressed on hPSC-derived CMs, SIRPA6, 14, and VCAM-16, 15 and used them for enriching populations of CMs by cell sorting. However, these proteins are not specifically expressed in CMs. For example, SIRPA is more highly expressed in the brain and the lung than the heart and is only useful for a certain stage of CMs, raising concerns about its effectiveness for purification14.
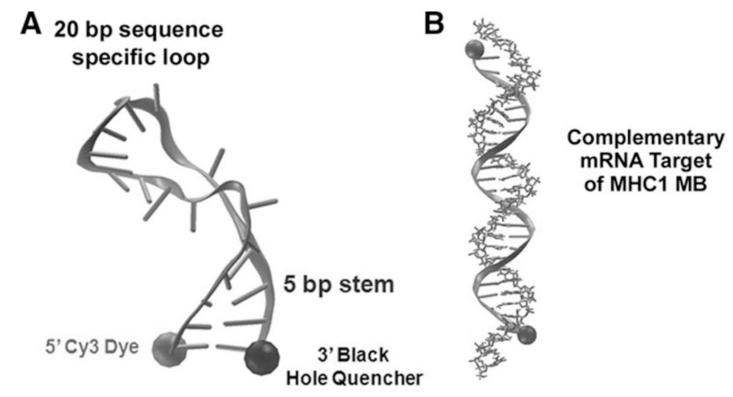
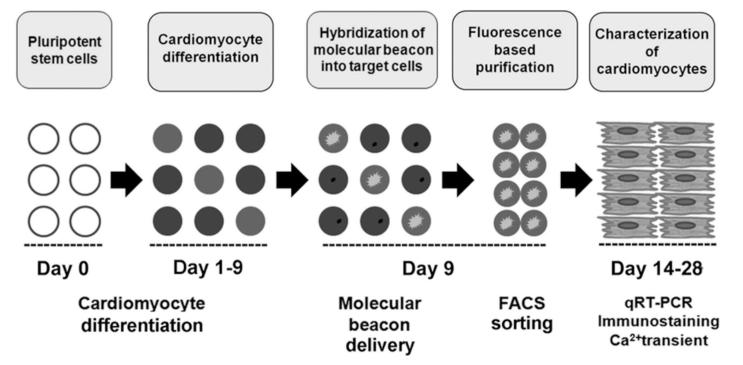
Accordingly, we aimed to develop a novel strategy to purify CMs from differentiating PSCs by targeting genes that are specifically expressed in CMs. We hypothesized that molecular beacons (MBs) hybridized to CM-specific mRNAs could enable isolation of CMs from a mixed population16. This method can be applied for both basic research and clinical applications. MBs are dual-labeled antisense oligonucleotide nano-scale probes with a fluorophore at one end and a quencher at the other end17, 18 (Figure 1A). MBs are designed to form a stem-loop (hairpin) structure in the absence of a complementary target so that fluorescence of the fluorophore is quenched. However, hybridization with the target mRNA opens the hairpin and separates the reporter from the quencher, allowing emission of a fluorescence signal (Figure 1B). MB technology has been tested in a variety of cell types to detect mRNA at various levels of expression and has been demonstrated to not alter the expression level of target genes18. We have previously demonstrated the use of MBs for detecting mESCs by simultaneous targeting of intracellular Oct4 mRNA and surface markers19. While these studies did not attempt to isolate target cells, they suggest that MB technology has unique potential for enriching target cells. In the present study, we first developed protocols to differentiate mouse and human PSCs into CMs and devised a strategy to isolate CMs by applying MBs targeting CM-specific mRNAs followed by FACS sorting (Figure 2). Our results suggest the possibility of purifying CMs at high efficiency and specificity from hPSC differentiation cultures with this innovative and clinically compatible purification system. These purified CMs are functional in vitro and in vivo.
Figure 1.
Structure of CM-specific MBs. (A) Molecular dynamics model of a MB in the closed/unbound conformation. The proximity of the 5′ Cy3 dye and the 3′ BHQ is small enough to allow FRET quenching. (B) Molecular dynamics model of a MB in the open/bound conformation. The distance between the 5′ Cy3 dye and the 3′ BHQ increases greatly to prevent dye quenching.
Figure 2.
Overall strategy to enrich hPSC-derived CMs using MBs targeting CM-specific mRNA.
Materials and Methods
The details concerning the materials and methods can be found in the online Data Supplements.
Mouse ESC culture and differentiation
mESCs (J1) were maintained as described20. To differentiate mESCs into cardiac lineage, an embryoid body (EB) method was employed with some modifications21.
Human PSC Culture and Differentiation
hESC (H1) were obtained from WiCell Research Institute (Madison, WI) and hiPSC (BJ1-iPS10) was provided by George Daley at Harvard University22. The use of hPSCs was approved by Emory University Human Embryonic Stem Cell Oversight Committee. These undifferentiated hPSCs were cultured as described previously23. To direct the differentiation of hPSCs to the cardiac lineage, we designed a staged protocol that is divided into four distinct phases
Molecular beacon synthesis and characterization
Five MBs were synthesized by MWG Operon using standard resin-based synthesis methods with HPLC purification (Table 1)18, 19, 23.
Table 1.
The list of MBs and parameters.
| Beacon Name |
Target Sequence | Beacon Sequence | Target TEM (°C) |
|---|---|---|---|
| TNT1 | CCCAAGATCCCCGATGGAGAGAG | TACCCTCTCTCCATCGGGGATCTTGGGTA | 68.1 |
| TNT2 | AGAACCGCCTGGCTGAAGAGA | CCCTCTCTTCAGCCAGGCGGTTCTGAGGG | 68.4 |
| TNT3 | GAACAGGAGGAAGGCTGAGG | ATCCTCAGCCTTCCTCCTGTTCGAGGAT | 64.8 |
| MHC1 | GTGAAGAAGAAGATGGAGG | CCTCCATCTTCTTCTTCACGGAGG | 57.5 |
| MHC2 | AAGAGCCGGGACATTGGTGCCAA | TTGGCACCAATGTCCCGGCTCTTGCCAA | 71.4 |
The in silico description of the MBs synthesized for the study. The target Tm category describes a measure of the predicted affinity for the 20 bp sequence specific loop for the indicated target sequence.
HL-1 cell culture 24
Nucleofection 25
Human heart tissue Flow cytometry and FACS-sorting 26
Immunocytochemistry and Immunohistochemistry 26
Table 2.
Primer sequences used for RT qPCR analysis.
| Primer sequences used qRT-PCR analysis (mouse) | ||
|---|---|---|
| GENE | FORWARD 5′-3′ | REVERSE 5′-3′ |
| Gapdh | TGTGATGGGTGTGAACCACGAGAA | CATGAGCCCTTCCACAATGCCAA A |
| Sox17 | ACCTACACTTACGCTCCAGTC | GCCGTAGTACAGGTGCAGAG |
| Kdr | AGGCCC ATTGAGTCCAACTACACA | ATGTCTGCATGGTCCCATACTGG T |
| Nkx2-5 | TGGGTCTCAATGCCTATGGCTACA | GACGCCAAAGTTCACGAAGTTG CT |
| Gata4 | CACGCTGTGGCGTCGTAAT | CTGGTTTGAATCCCCTCCTTC |
| Tbx5 | TGGCTGAAGTTCCACGAAGTG | TTTGGGATTAAGGCCAGTCAC |
| Tnnt2 | TCACAACCTGGAGGCTGAGAAGTT | TCATCTATTTCCAACGCCCGGTG A |
| Myh6 | AGCTGACAGGGGCCATCAT | ACATACTCGTTCCCCACCTTC |
| Myh 7 | TGCCAATGACGACCTGAAGGAGA A |
TCTTCTGGTTGATGAGGCTGGTG T |
| Myl2 | AGATGCTGACCACACAAGCAGAG A |
TCCGTGGGTAATGATGTGGACC AA |
| Myl7 | AAGGGAAGGGTCCCATCAACTTCA | AACTTGTCTGCCTGGGTCATGAG A |
| Neurod | GACGGGGTCCCAAAAAGAAAA | GCCAAGCGCAGTGTCTCTATT |
| Pecam | TCTATGACCTCGCCCTCCACAAA | GAACGGTGTCTTCAGGTTGGTAT TTCA |
| Cdh5 | TGGAGAAGTGGCATCAGTCAACAG | TCTACAATCCCTTGCAGTGTGAG |
| Gata1 | TGTCCTCACCATCAGATTCCA | TCCCTCCATACTGTTGAGCAG |
| Myod1 | CCACGACCACCTCTCAGAAC | GACAGGACAGTATGCAGTGGA |
| Ddr2 | ATGATCCCGATTCCCAGAATGC | CATATTTGGCAGCCGTGGATT |
| Acta2 | CATGTACGTCGCCATTCAAGC | TTGATGTCTCGCACAATTTCTCT |
| Primer sequences used qRT-PCR analysis (Human) | ||
|---|---|---|
| GENE | FORWARD 5′-3′ | REVERSE 5′-3′ |
| GAPDH | AGGTGGTCTCCTCTGACTTCAACA | GACAAAGTGGTCGTTGAGGGCA AT |
| OCT4 | ATGCATTCAAACTGAGGTGCCTGC | CCACCCTTTGTGTTCCCAATTCCT |
| T | TGTCCCAGGTGGCTTACAGATGAA | GGTGTGCCAAAGTTGCCAATACA C |
| KDR | ACTTTGGAAGACAGAACCAAATTAS TCTC |
TGGGCACCATTCCACCA |
| MESP1 | AGCCCAAGTGACAAGGGACAACT | AAGGAACCACTTCGAAGGTGCT GA |
| NKX2-5 | ACCTCAACAGCTCCCTGACTC | ATAATCGCCGCCACAAACTCTCC |
| TBX5 | ACAAAGTGAAGGTGACGGGC | GGGGACCACGGGATATTCTT |
| TNNT2 | TTC ACC AAA GAT CTG CTC CTC GCT |
TTATTACTGGTGTGGAGTGGGTG TGG |
| MYH6 | TCA GCT GGA GGC CAA AAG TAA AGG A |
TTCTTGAGCTCTGAGCACTCGTC T |
| MYH7 | TCGTGCCTGATGACAAACAGGAGT | ATACTCGGTCTCGGCAGTGACTT T |
| MYL2 | TGTCCCTACCTTGTCTGTTAGCCA | ATTGGAACATGGCCTCTGGATGG A |
| MYL7 | ACATCATCACCCATGGAGACGAGA | GCAACAGAGTTTATTGAGGTGCC C |
|
NEURO
D |
TCCCATGTCTTCCACGTTAAGCCT | CATCAAAGGAAGGGCTGGTGCA AT |
|
PECAM
1 |
TTCCTGACAGTGTCTTGAGTGGGT | TTTGGCTAGGCGTGGTTCTCATC T |
| ACTA2 | AATACTCTGTCTGGATCGGTGGCT | ACGAGTCAGAGCTTTGGCTAGG AA |
| MYH11 | GGTCACGGTTGGGAAAGATGA | GGGCAGGTGTTTATAGGGGTT |
| MYOD1 | CGCCATCCGCTATATCGAGG | CTGTAGTCCATCATGCCGTCG |
| DDR2 | ACCAGCCATTTGTCCTGACTCTGT | ATCACTCGTCGCCTTGTTGAAGG A |
| THY1 | ATACCAGCAGTTCACCCATCCAGT | ATTTGCTGGTGAAGTTGGTTCGG G |
| ACTA2 | AATACTCTGTCTGGATCGGTGGCT | ACGAGTCAGAGCTTTGGCTAGG AA |
Intracellular calcium (Ca2+) imaging 27
Action potential measurement 23
Induction of myocardial infarction (MI) and cell transplantation
All animal experiments were approved by Emory University IACUC. Myocardial infarction (MI) and cell implantation were performed as we described previously28. We randomly assigned the mice into three groups which received saline (PBS) (N = 10), 2 × 105 unpurified CMs differentiated from mESCs (N = 12), or 2 × 105 purified CMs with MHC1-MB from differentiated mESCs (N = 11).
Echocardiography29
Statistical analyses
The authors have full access to and take full responsibility for the integrity of the data. All authors have read and agreed to the manuscript as written.
Results
Cardiomyocyte-specific molecular beacon generation
To determine optimal candidate genes detectable by MBs, we performed quantitative reverse transcriptase PCR (qRT-PCR) analysis on known cardiac specific genes using mRNAs extracted from freshly isolated mouse adult CMs (Supplementary figure 1a), human neonatal heart tissues, and human fetal heart tissues (Supplementary figure 1b and c). The representative cardiac structural genes cardiac troponin T (TNNT2, also known as cTNT) and myosin heavy chain (MYH6/7, also known as α/βMHC) were most highly expressed in all of these samples and thus were determined targets for MBs. Based on the qRT-PCR results, we designed five MBs (Table 1) targeting unique sites in TNNT2 or MYH6/7 mRNA in both mouse and human using design rules determined by previous publications30, 31 and BLAST searches to ensure uniqueness. These MBs were synthesized with a Cy3 fluorophore or a FAM fluorophore at the 5′ end and a Black Hole Quencher 2 or a Black Hole Quencher 1 at the 3′ end as specified in Table 1. We quantified beacon fluorescence signal when hybridized to perfectly complementary and mismatched targets by incubating 500 nM MBs with targets of increasing concentrations (100 - 500 nM). MB signal was recorded using a microplate reader and normalized by the signal in wells with MB only (Supplementary figure 2). All MBs displayed a linear response to increasing concentrations of complementary target and a low response to mismatched targets.
In order to determine the most efficient transfection method to deliver MBs into living cells, we compared the following six techniques: Streptolysin O32, Lipofectamin 200033, Lullaby34, nucleofection35, electroporation36 and microinjection37. To quantify the delivery efficiency, we fabricated a non-specific MB which has a non-specific sequence with FAM dyes conjugated to both 5′ and 3′ ends, allowing the probe to fluoresce regardless of its open or closed conformation. We delivered the non-specific MB into various cell lines including an immortalized mouse CM cell line, HL-1 CMs 24, smooth muscle cells (SMCs), mouse embryonic fibroblasts (MEFs) and mouse embryonic stem cells (mESCs). Flow cytometry analysis demonstrated that regardless of the cell type, nucleofection consistently delivered MBs into > 95% of the cells, showing the highest delivery efficiency (Supplementary figure 3 and part of data not shown).
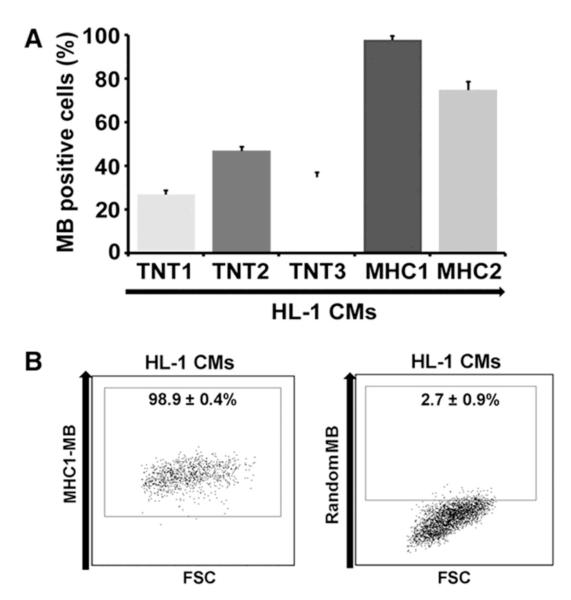
We systematically determined the sensitivity of MBs designed to target CM-specific mRNAs. As a positive control, we used HL-1 CMs. We independently confirmed the CM identity of HL-1 CMs by flow cytometry, showing that 97.2% of cells expressed Tnnt2 (Supplementary figure 4). Each of the candidate MBs targeting TNNT2 or MYH mRNA was delivered into HL-1 CMs by nucleofection and the efficacy was analyzed by flow cytometry. Among the 5 MBs (TNT1, −2, −3 and MHC1, −2) examined, the MB designated as MHC1-MB yielded the highest rate of MB-signal positive cells (98.9%) (Figure 3A).
Figure 3.
Characterization of CM-specific MBs. (A) Flow cytometry results of five different MBs designed to detect TNNT2 or MYH mRNAs on HL-1 CMs. MHC1-MB shows highest detection rate. N = 3. (B) Flow cytometric analysis of HL-1 CMs treated with random versus MHC1-MBs. The numbers represent the percentages of MB-positive cells. N = 3.
To determine the specificity of the MHC1-MB, a ‘random’-sequence MB (‘random beacon’) which has a 16-base target sequence that does not match any sequence in the mouse or human genome, was delivered as a negative control31 and displayed negligible fluorescence in HL-1 CMs (Figure 3B), ruling out the possibility that the fluorescence signal from MHC1-MB was due to nonspecific interactions and/or probe degradation by endonucleases. To further verify the specificity of the MHC1-MB, we delivered MHC1-MB into SMCs, mouse aortic endothelial cells (mECs), mouse cardiac fibroblasts (mCFs) and mESCs, which are the most likely contaminating cell types in cardiomyogenically differentiated PSC cultures (Supplementary figure 5). Flow cytometry analysis showed that < 3% of these cells displayed a detectable fluorescence signal. These results suggest high sensitivity of the MHC1-MB for CM-lineage cells.
Purification of mESC-derived CMs
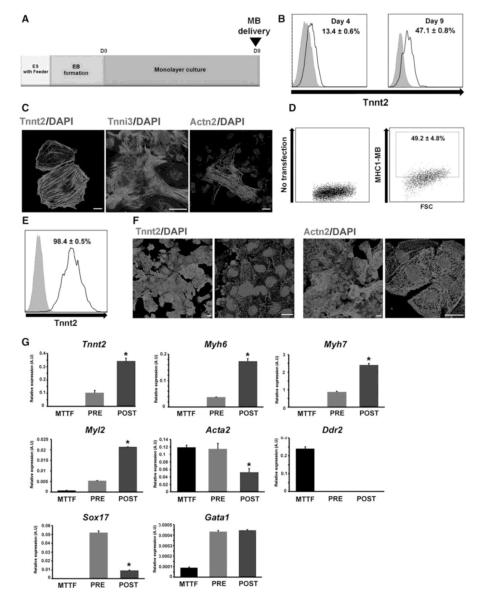
Next, we investigated whether this MB can be useful for isolating CMs from differentiating mouse PSCs. We first established an embryoid body (EB)-mediated system to efficiently differentiate mESCs into CMs (Figure 4A). In brief, undifferentiated mouse ESCs (J1) maintained on STO feeder cells were enzymatically detached to form EBs, which were cultured for 5 days and plated on a fibronectin-coated dish for CM differentiation. After 3-4 days of differentiation in the presence of ascorbic acid (50 μg/ml)20, spontaneously beating clumps began to appear (Supplementary movie 1). Flow cytometry analysis demonstrated that the percentage of Tnnt2-positive cells were 13.4% and 47.1% at days 4 and 9, respectively (Figure 4B). Immunostaining further demonstrated that cells dissociated from beating clumps displayed Tnnt2, Tnni3 and Actn2 (or α-sarcomeric actinin), confirming their CM nature (Figure 4C). The results with mouse iPSCs were similar (data not shown). After establishing the differentiation system, we attempted to isolate CMs from differentiating mESCs using MBs. The differentiating mESCs at day 9 were nucleofected with MHC1-MB and subjected to FACS-sorting. The percentage of cells positive for fluorescence signal from MHC1-MB was 49.2 ± 4.8% (Figure 4D). There was significant agreement between the detection rate of CMs using antibody-based (47.1% Tnnt2-positive cells) and MB-based (49.2% MHC1-MB-positive cells) flow cytometry results, supporting the specificity of MHC1-MB. Fluorescent microscopic imaging also confirmed these results (Supplementary figure 6). Next, cells were FACS-sorted using MHC1-MB and 98.4% of the sorted MHC1-MB-positive cells exhibited Tnnt2 in flow cytometry (Figure 4E). Immunocytochemistry verified that virtually all sorted MHC1-MB-positive cells expressed Tnnt2 and Actn2 (Figure 4F). qRT-PCR analyses demonstrated that these sorted cells expressed 2 to 6-fold higher levels of Tnnt2, Myh6, Myh7 and Myl2 compared to the pre-sorted cells (Figure 4G). Genes representing other lineages were either expressed at negligible levels (Acta2, Ddr2, Gata1 and Sox17) or were non-detectable (Pecam1, MyoD and Neuro D) in the sorted cells (Figure 4G and data not shown). Importantly, these sorted cells showed spontaneous contraction over 2 weeks suggesting that these MB-based purified CMs were functional (Supplementary movie 2). In this movie, we intentionally selected areas where individual CMs were contracting to demonstrate functionality of CMs at an individual level.
Figure 4.
Purification of cardiomyocytes from differentiating mESCs using CM-specific MBs. (A) A schematic of the protocol used for differentiating mESCs to the cardiac lineage. (B) Percent expression of Tnnt2 at day 4 and 9 during mESC differentiation into CMs. N = 3. (C) Immunocytochemistry results of mESC-derived CMs at day 9 for Tnnt2, Tnnt3 and Actn2. Scale bars, 20 μm. (D) Flow cytometry analyses of MHC1-MB signals in mESC differentiation culture at day 9. N = 6. (E) Flow cytometry analysis of Tnnt2 expression in the mESCs sorted with MHC1-MB and FACS at the differentiation day 9. N = 3. The numbers represent the percentages of MB-positive cells (D, E). (F) FACS-sorted MHC1-MB-positive cells exhibit Tnnt2 and Actn2 in immunocytochemistry. Scale bars, 20 μm. (G) qRT-PCR analyses showing difference in gene expression levels between mouse tail tip fibroblasts (MTTF), and pre-sorted and post-sorted mESCs. CM genes, Tnnt2, Myh6, Myh7 and Myl2 were significantly enriched in post-sorted cells with MHC1-MB and non-cardiac lineage genes (Acta2, Ddr2, Gata1 and Sox17) were substantially reduced compared to the pre-sorted cells. Y axis represents relative mRNA expression of target genes to GAPDH. *P < 0.05 compared to pre-sorted group. N = 3.
Purification of hESCs-derived CMs
Similarly, we investigated the utility of MB-based cell sorting for human PSC-derived CMs. We developed a novel four-step protocol for differentiating hPSCs into CMs (Figure 5A). In phase 1, undifferentiated hESCs (H1) were directly transferred onto Matrigel-coated plates and cultured as a monolayer under mTeSR® media (Stem Cell Technology) for cell expansion. Next, to induce mesodermal differentiation, several combinations of mesodermal inducers were tested and compared by qRT-PCR using mesodermal markers including T (or BRACHYURY) and KDR. We found that a combination of BMP4 (10ng/ml), Activin A (3ng/ml), and FGF2 (5ng/ml) was the most efficient for mesodermal differentiation (Supplementary figure 7a). To induce cardiac lineage differentiation in phase 3, we tested four different methods1, 3, 20, 38 and found that supplementation with conditioned media produced by mouse endodermal cell line END-2 induced the highest expression of cardiac lineage markers such as NKX2-5, TNNT2, MYH6 and −7 (Supplementary figure 7b). Finally, in phase 4, continuous treatment with a β-adrenergic receptor agonist, isoproterenol, for as short as 4 days efficiently generated spontaneously beating CMs (Supplementary movie 3).
Figure 5.
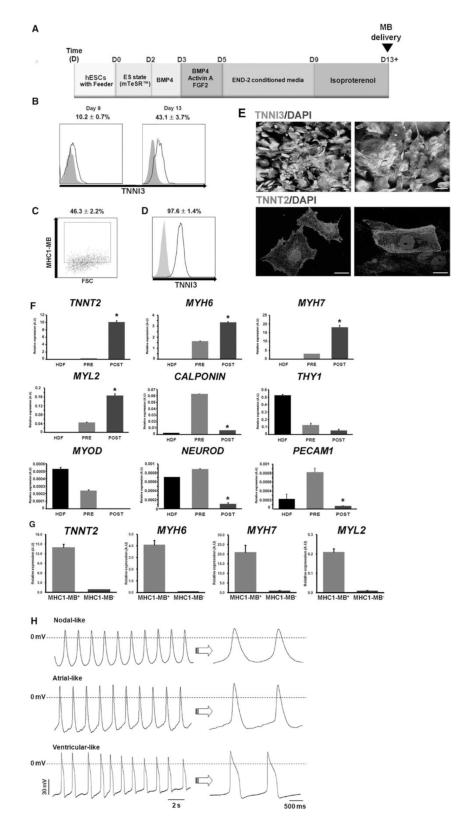
Purification of cardiomyocytes from differentiating hESCs by cell sorting via MB and FACS. (A) A schematic of the protocol to differentiate hPSCs to the cardiac lineage. (B) Percent expression of TNNI3 at day 9 and 13 determined by flow cytometry during hESC differentiation into CMs. N = 3. (C) Flow cytometry results of MHC1-MB-positive cells in hESC differentiation culture at day 13. N = 6. (D) Flow cytometry results showing TNNI3 expression of FACS-sorted hESCs at day 13 of differentiation after applying MHC1-MB. The numbers represent the percentages of MB-positive cells (C, D). N = 3. (E) Immunocytochemistry for TNNI3, and TNNT2 on MHC1-MB-positive cells sorted from hESC cultures. Scale bars, 20 μm. (F) mRNA expression levels of cardiac and non-cardiac genes measured by qRT-PCR. Comparisons were made for HDF, pre-sorted hESCs at day 13 and post-FACS sorted cells at day 13 using MHC1-MB. CM genes (TNNT2, MYH6, MYH7 and MYL2) were significantly higher in the sorted hESCs compared to the pre-sorted hESCs and HDF. Expression of the non-cardiac lineage genes (PECAM1, CALPONIN, THY1, MYOD and NEUROD) was significantly lower in the sorted cells compared to others. Y axis represents relative mRNA expression of target genes to GAPDH. *P < 0.05 compared to pre-sorted group. N = 3. (G) mRNA expression levels of cardiac genes from both MHC1-MB positive and negative cells measured by qRT-PCR. (H) Action potentials of the MHC1-MB-positive CMs. Shown are representative configurations of the nodal- (top), atrial- (middle), and ventricular (bottom) action potentials.
Flow cytometry analysis demonstrated that the percentage of TNNI3-positive cells were 10.2% and 43.1% at days 9 and 13, respectively (Figure 5B). We delivered the MHC1-MB to the cardiomyogenically differentiated hESCs at day 13 in phase 4. Flow cytometry analysis showed that the percentage of cells positive for MHC1-MB signal was 46.3% (Figure 5C). These MHC1-MB treated cells were sorted by FACS, and 97.6 ± 1.4% of the sorted MHC1-MB positive cells exhibited TNNI3 expression in flow cytometry analysis (Figure 5D). Almost all cells showed CM-like morphology and stained positive for TNNT2 and TNNI3 by immunocytochemistry (Figure 5E). qRT-PCR analyses showed a significant increase in expression of CM-specific genes (TNNT2, MYH6, MYH7, and MYL2) and decrease in expression of genes specific for SMCs (CALPONIN), fibroblast (THY1), skeletal myocyte (MYOD), neural lineage cells (NEUROD), and EC (PECAM1), suggesting enrichment of CMs and elimination of other lineage cells by cell sorting with the MB (Figure 5F-G).
Stable action potentials (APs) were recorded from CMs that were purified via MHC1-MB and cultured for 7-14 days after FACS-sorting. Three major types of APs were observed, nodal-like (6 of 46), atrial-like (11 of 46) and ventricular-like (29 of 46) APs (Figure 5H, Table 3). These results indicate that purified cells via a CM-specific MB are electrophysiologically intact, functional CMs and can maintain these characteristics in culture.
Table 3.
Action potential properties in MB-based-purified CMs derived from hESCs.
| AP types | Upstroke slope (mV/ms) |
½ AP width (ms) |
Firing frequency (Hz) |
|---|---|---|---|
| Nodal-like | 85.9 ± 5.2 | 247 ± 11.7 | 0.66 ± 0.04 |
|
| |||
| Atrial-like | 102.8 ± 6.7 | 201 ± 13.1 | 0.63 ± 0.03 |
|
| |||
| Ventricular-like | 99.7 ± 7.4 | 286 ± 13.5 | 0.55 ± 0.12 |
APs were measured in a total of 46 MHC1-MB-positive CMs: nodal-like (6 of 46), atrial-like (11 of 46) and ventricular-like (29 of 46) APs were identified.
Purification of CMs from hiPSCs
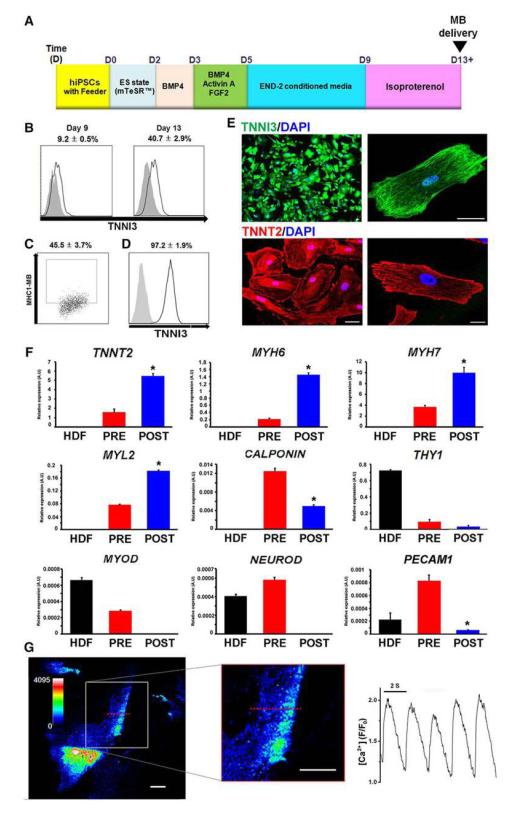
We further determined the utility of MHC1-MB for isolating CMs from differentiating hiPSCs (BJ1)22. Our differentiation system for CMs from hESCs was successfully applied to hiPSCs, yielding 40.7% of TNNI3-positive CMs at day 13 (Figure 6A, B). MHC1-MB was delivered into the differentiating hiPSCs at day 13 and the percentage of cells positive for MHC1-MB analyzed by flow cytometry was 45.5% (Figure 6C). FACS sorting based on MB signal resulted in enrichment of TNNI3-positive cells to 97.2 ± 1.9% (Figure 6D). Almost all the sorted cells stained positive for TNNI3 or TNNT2 in immunostaining (Figure 6E). qRT-PCR again showed about a 3- to 7- fold increase in CM-specific gene expression and substantial reduction or negligible levels of other lineage gene expression (Figure 6F). The sorted MHC1-MB positive CMs showed spontaneous contraction (Supplementary movie 4) and rhythmic calcium oscillations (Figure 6G and Supplementary movie 5). These data suggest that the MHC1-MB-based purified CMs are functional CMs. Taken together, our results clearly support the notion that MBs targeting CM-specific mRNA in live cells allow isolation of functional CMs from differentiating mouse and human PSCs with high specificity and efficiency.
Figure 6.
Purification of cardiomyocytes from differentiating hiPSCs by cell sorting via MB and FACS. (A) A schematic of the protocol to differentiate hiPSCs to the cardiac lineage. (B) Percent expression of TNNI3 at day 9 and 13 during hiPSC differentiation into CMs determined by flow cytometry. N = 3. (C) Flow cytometric analysis of MHC1-MB signals in hiPSC differentiation culture at day 13. N = 6. (D) Flow cytometric results showing TNNI3 expression of FACS-sorted hiPSCs at day 13 of differentiation after applying MHC1-MB. The numbers represent the percentages of MB-positive cells (C, D). N = 3. (E) Immunocytochemistry for TNNI3, TNNT2 on MHC1-MB-positive cells sorted from hiPSC cultures. Scale bars, 20 μm. (F) mRNA expression levels of cardiac and non-cardiac genes measured by qRT-PCR. Comparisons were made among human dermal fibroblast (HDF), pre-sorted hiPSCs at day 13, and MB-based FACS-sorted hiPSCs at day 13. Y axis represents relative mRNA expression of target genes to GAPDH. *P < 0.05 compared to pre-sorted group. N = 3. (G) Calcium imaging of MHC1-MB-positive CMs sorted from hiPSCs. Left panel is a confocal scan of representative cells loaded with Fluo-4 AM, with magnification of line-scanned region indicated with red dashed line (white scale bars 20μm, legend shows increasing calcium levels with blue being low calcium). Right panel is a time-course of [Ca2+]I, measured at line scan region in cell pictured and paced at 0.5hz by field stimulation. [Ca2+] is plotted as fluorescence intensity normalized to baseline (F/F0).
Engraftment and improvement of cardiac function after implantation of purified CMs
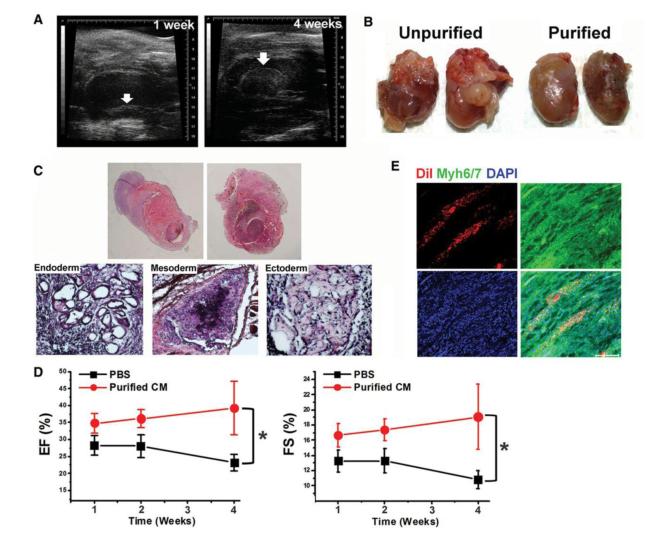
To determine the behavior and effects of MB-based purified CMs in ischemic myocardium, purified or unpurified CMs derived from mESCs or the same volume of PBS were injected into the myocardium after induction of MI in mice. Echocardiography was performed weekly to measure cardiac remodeling and function. One week later, however, in the mice receiving unpurified CMs, a distinct mass was observed in the left ventricular lumen of the hearts, which grew over 4 weeks (Figure 7A, B) Post-mortem examination at 3-4 weeks revealed tumor masses in 11 out of 12 mice. By careful gross examination, tumors invaded internally into myocardium and externally into the pericardium (Figure 7B, C). Cardiac tissues were fixed and stained with hematoxylin and eosin (H&E). Microscopic examination revealed that all tumors consisted of structures derived from all three embryonic germ layers, indicating teratomas (Figure 7C). However, we did not detect tumors in any of the mice receiving MB-based purified CMs or PBS over the same follow-up period by echocardiographic or histologic examination. Tumor formation in unpurified-CM injected mice did not allow appropriate functional comparison between mice receiving unpurified- and purified CMs. However, purified CM-injected mice showed a higher ejection fraction than PBS-injected mice (Figure 7D), indicating improved cardiac function. We next conducted immunohistochemistry for cardiac tissues injected with purified CMs. Confocal microscopic examination demonstrated that injected CMs (DiI-positive) were engrafted as clusters, survived robustly for 4 weeks and expressed representative CM proteins (Figure 7E). Taken together, these results suggest that injected MB-purified CMs are integrated into ischemic myocardium and are functional in vivo.
Figure 7.
Transplantation of unpurified and purified CMs into a mouse model of acute myocardial infarction. (A) Serial echocardiography demonstrated a growing left ventricular mass at 1 week (left) and 4 weeks (right) in the mice receiving unpurified mESC-derived CMs. (B) Images of excised hearts previously injected with unpurified or purified mESC-derived CMs. (C) H&E staining of whole cardiac sections demonstrated infiltrative growth of hypercellular tumors surrounding the heart and protruding into the cardiac lumen. H&E staining revealed cell derivatives of all three embryonic germ layers: entoderm (left), mesoderm (middle), and ectoderm (right), indicating teratomas. (D) Improvement of cardiac function of mice transplanted with MB-purified CMs derived from mESCs. Ejection fraction (EF: left) and Fractional shortening (FS: right) were significantly higher in the purified-CM-treated group compared to PBS-treated group measured by echocardiography. Repeated-measures ANOVA was used for statistical analyses. *P < 0.05. N = 5 - 10 per group. (E) Confocal microscopic images showed that the transplanted Dil-labeled MB-purified CMs (red fluorescence) express Myh6/7 (green), indicating engraftment and integration of transplanted CMs in the MI hearts. Scale bars, 100μm.
Discussion
In the present study, we have described a highly specific and efficient method for purifying CMs from differentiating mouse and human PSCs by directly targeting mRNA of CM-specific genes. By using a MB targeting the mRNA of MYH 6 and 7 followed by FACS sorting, we were able to enrich a population made up of more than 97% CMs from differentiating PSCs. Their identity as CMs was verified by a series of experiments including flow cytometry, immunocytochemistry, and qRT-PCR. Importantly, these purified CMs displayed spontaneous beating upon further culture and demonstrated stable APs and Ca2+ oscillation in electrophysiological studies. When injected into infarcted heart, MB-purified CMs were integrated and survived robustly for 4 weeks in post-MI hearts, showing improved cardiac function without forming tumors. On the other hand, mice injected with unpurified CMs developed teratoma. These results support that MB-based isolated CMs are pure and functionally intact CMs and CM purification is necessary for cell therapy.
Despite continuous improvements of current CM differentiation protocols, the resulting differentiated cell populations still contain a considerable percentage of non-CMs. Hence, one of the major challenges in recent years has been to develop stable isolation techniques that allow scalable purification of CMs. Recently, three studies employing FACS-based CM purification have attracted attention due to their sound scientific rationale, high purity and clinical applicability. Among the first, a mitochondrial dye, TMRM has been suggested to be useful for CM selection, enriching up to 99% as determined by the expression of Acnt2 ( -sarcomeric actinin) in immunocytochemistry. This approach was based on the findings that CMs have high mitochondrial content and can be purified via fluorescent dyes that label mitochondria. However, two other following studies demonstrated that TMRM not only failed to identify immature CMs early in differentiation culture14, but also detected non-cardiogenic cells or undifferentiated hESCs as these cells uptake significant amounts of TMRM6. In addition, two independent studies reported the identification of two surface marker proteins; SIRPA and VCAM-1, and suggested their utility for isolating CMs from differentiating hPSCs6, 14. To identify a candidate protein, Dubois et al screened a panel of 370 known commercially available antibodies and identified SIRPA as a specific marker for CMs differentiating from hPSCs. Subsequently, isolation via FACS using an antibody against SIRPA enriched cardiac precursors and CMs from hPSCs, yielding up to 98% TNNT2+ cells. However, Elliott et al demonstrated that SIRPA+ cells expressed significant amounts of smooth muscle marker genes such as ACTA2 and CNN1 and an endothelial cell gene, CD34, bringing the utility of SIRPA as a sole marker for CM isolation into question. This study also suggested that there are variations in the yield of cardiogenic marker expression (NKX2-5) after further culture of sorted SIRPA+ cells, reaching as low as 2.8% CMs depending on the isolation time and duration of culture. Furthermore, NKX2-5+ cells did not express SIRPA, which raised concerns about specificity and sensitivity of SIRPA. Thus Elliott et al identified another surface protein, VCAM1 through a transcriptome analysis and employed it as an additional marker for isolating CMs. However, FACS sorting with both SIRPA and VCAM1 antibodies was only able to enrich CMs ranging from 55 to 95% purity. Moreover, since both proteins are known to be expressed in other cell types, the possibility of contamination with non-CM cells even after purification with SIRPA and VCAM1 may be a critical issue14, 39.
The MB-based CM purification strategy has a number of unique advantages in comparison with previously reported CM purification methods. First of all, by directly targeting specific intracellular mRNA, there is no need to make extensive efforts to identify surface proteins for cell isolation using the corresponding antibodies 14, 40, 41. Secondly, MB-based cell purification minimizes contamination of other cells by using MBs designed to hybridize with unique sequences in the mRNAs of interest specifically expressed in the target cell type6, 14, 42, 43. While significant efforts have been made to identify SIRPA and VCAM1 as cell-surface markers for CMs, both SIRPA and VCAM1 are expressed in other organs such as brain and lung, raising questions of the utility of these surface markers in a range of differentiation systems. Another advantage of MB-based purification method is its application to any species. It allows expanding the research scope with any desired cells. Since SIRPA is not highly expressed in the mouse, its usage is limited to human cells. In contrast, the MB-based method allows isolation of virtually any cell due to unlimited accessibility of MBs to intracellular mRNA. Therefore, in addition to CMs, the MB based sorting technique described here can be broadly applied to the isolation of other cell types such as neural-lineage cells or islet cells, which are critical elements in regenerative medicine but do not have specific surface proteins identified to date.
Our cell transplantation study provides an important insight into the necessity of purified CMs for cardiac cell therapy. An unexpectedly high rate of tumors developed in mice receiving unpurified mouse ESC-derived CMs. Although unpurified, these cells were differentiated into the cardiac lineage and ~50% of cells were CMs. To date, a few studies reported syngeneic or allogeneic transplantation of unpurified mESC-derived CMs into infarct heart and all demonstrated tumor formation in hearts44, 45. Our study clearly demonstrated that purified CMs do not form tumors and this purification process is a pre-requisite for cardiac cell therapy with PSCs. Similar cardiac cell therapy studies were conducted with human PSC-derived CMs and many of them showed functional improvement with no tumor formation albeit utilizing non-purified cells 46, 47. However, these studies might have underestimated tumorigenecity because xenogeneic transplantation can induce engrafted cell death by immune reaction, particularly those non-cardiogenic cells which are not aggregated.
To ensure that MBs were safe and effective to use in target cells, we carried out several experiments to determine cell viability and functionality immediately and several days after delivery of MBs. MBs were minimally disruptive as the results did not differ significantly between hPSC-CMs and control cells in the assays performed. MBs usually degrade within an hour 16-19, 31, so cell viability or cell fate change is not a serious concern. In addition, to confirm their specificity, we carried out extensive testing to ensure that the MB signal was a robust indicator of CMs based on the binding of the probe to MYH6/7. We tested the MBs in solution with synthetic target oligonucleotides that varied from the ideal sequences by 6 bp, the closest that a BLAST search through the human transcriptome predicted that we would see. None of the MBs used in the study increased their fluorescence by more than 50% even when incubated with excess target sequence.
The purified hPSC-derived CMs sorted via MBs will be enormously useful for clinical applications as well as pre-clinical studies. A major obstacle to employing cells differentiated from hPSCs for clinical applications is potential tumorigenicity or aberrant tissue formation after cell transplantation41. By eliminating unwanted cells, this technology will advance the utilization of hPSC-derived CMs. Moreover, mechanistic studies, drug discovery, and disease modeling with non-pure PSC-derived CMs, although attempted previously, can be inaccurate due to the effects of non-cardiomyogenic cells. This purification technique in combination with CMs generated from patient-specific hiPSCs will be of great value for drug screening and disease modeling as well as cell therapy.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Emory Children’s Pediatric Research Center flow cytometry core.
Funding Sources: This work was supported in part by the NHLBI of the NIH as a Program of Excellence in Nanotechnology award (HHSN268201000043C to YSY and GB), by grants from NIDDK (DP3DK094346 to YSY), NHLBI (R01HL088488 to MBW), ACTSI (PHS grant UL1 RR025008 from the CTSA program, National Center for Research Resources, NIH to YSY) and the National Science Foundation STC award (CBET-0939511). KB is a recipient of American Heart Association postdoctoral fellowship grant.
References
- 1.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den Brink S, Hassink R, van der Heyden M, Opthof T, Pera M, de la Riviere AB, Passier R, Tertoolen L. Differentiation of human embryonic stem cells to cardiomyocytes: Role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 2.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a kdr+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 4.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, Skelton RJP, Ward-van Oostwaard D, Lim SM, Khammy O, Li X, Hawes SM, Davis RP, Goulburn AL, Passier R, Prall OWJ, Haynes JM, Pouton CW, Kaye DM, Mummery CL, Elefanty AG, Stanley EG. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 7.Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR. Human isl1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 8.Huber I, Itzhaki I, Caspi O, Arbel G, Tzukerman M, Gepstein A, Habib M, Yankelson L, Kehat I, Gepstein L. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J. 2007;21:2551–2563. doi: 10.1096/fj.05-5711com. [DOI] [PubMed] [Google Scholar]
- 9.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 10.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Growth of engineered human myocardium with mechanical loading and vascular coculture / novelty and significance. Circ Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 12.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and bmp signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Hattori F, Chen H, Yamashita H, Tohyama S, Satoh Y-s, Yuasa S, Li W, Yamakawa H, Tanaka T, Onitsuka T, Shimoji K, Ohno Y, Egashira T, Kaneda R, Murata M, Hidaka K, Morisaki T, Sasaki E, Suzuki T, Sano M, Makino S, Oikawa S, Fukuda K. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 14.Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G. Sirpa is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uosaki H, Fukushima H, Takeuchi A, Matsuoka S, Nakatsuji N, Yamanaka S, Yamashita JK. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by vcam1 surface expression. PLoS One. 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King FW, Liszewski W, Ritner C, Bernstein HS. High-throughput tracking of pluripotent human embryonic stem cells with dual fluorescence resonance energy transfer molecular beacons. Stem Cells Dev. 2011;20:475–484. doi: 10.1089/scd.2010.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyduk T, Heyduk E. Molecular beacons for detecting DNA binding proteins. Nat Biotechnol. 2002;20:171–176. doi: 10.1038/nbt0202-171. [DOI] [PubMed] [Google Scholar]
- 18.Santangelo P, Nitin N, Bao G. Nanostructured probes for rna detection in living cells. Ann Biomed Eng. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 19.Rhee WJ, Bao G. Simultaneous detection of mrna and protein stem cell markers in live cells. BMC Biotechnol. 2009;9:30. doi: 10.1186/1472-6750-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, Lee RT. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation. 2003;107:1912–1916. doi: 10.1161/01.CIR.0000064899.53876.A3. [DOI] [PubMed] [Google Scholar]
- 21.Hescheler J, Fleischmann BK, Lentini S, Maltsev VA, Rohwedel J, Wobus AM, Addicks K. Embryonic stem cells: A model to study structural and functional properties in cardiomyogenesis. Cardiovasc Res. 1997;36:149–162. doi: 10.1016/s0008-6363(97)00193-4. [DOI] [PubMed] [Google Scholar]
- 22.Park I-H, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 23.Moon S-H, Ban K, Kim C, Kim S-S, Byun J, Song M-K, Park I-H, Yu SP, Yoon Y-s. Development of a novel two-dimensional directed differentiation system for generation of cardiomyocytes from human pluripotent stem cells. Int J Cardiol. 2013 doi: 10.1016/j.ijcard.2012.09.077. doi:10.1016/j.ijcard.2012.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claycomb WC, Lanson NA, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ. Hl-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caspi O, Huber I, Kehat I, Habib M, Arbel G, Gepstein A, Yankelson L, Aronson D, Beyar R, Gepstein L. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J Am Coll Cardiol. 2007;50:1884–1893. doi: 10.1016/j.jacc.2007.07.054. [DOI] [PubMed] [Google Scholar]
- 26.Lee JY, Park C, Cho YP, Lee E, Kim H, Kim P, Yun SH, Yoon Y-s. Podoplanin-expressing cells derived from bone marrow play a crucial role in postnatal lymphatic neovascularization. Circulation. 2010;122:1413–1425. doi: 10.1161/CIRCULATIONAHA.110.941468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner MB, Wang Y, Kumar R, Tipparaju SM, Joyner RW. Calcium transients in infant human atrial myocytes. Pediatr Res. 2005;57:28–34. doi: 10.1203/01.PDR.0000148066.34743.10. [DOI] [PubMed] [Google Scholar]
- 28.Jeong J-O, Han JW, Kim J-M, Cho H-J, Park C, Lee N, Kim D-W, Yoon Y-S. Malignant tumor formation after transplantation of short-term cultured bone marrow mesenchymal stem cells in experimental myocardial infarction and diabetic neuropathy. Circ Res. 2011;108:1340–1347. doi: 10.1161/CIRCRESAHA.110.239848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho H-J, Lee N, Lee JY, Choi YJ, Ii M, Wecker A, Jeong J-O, Curry C, Qin G, Yoon Y-s. Role of host tissues for sustained humoral effects after endothelial progenitor cell transplantation into the ischemic heart. J Exp Med. 2007;204:3257–3269. doi: 10.1084/jem.20070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsourkas A, Behlke MA, Bao G. Structure-function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res. 2002;30:4208–4215. doi: 10.1093/nar/gkf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhee WJ, Santangelo PJ, Jo H, Bao G. Target accessibility and signal specificity in live-cell detection of bmp-4 mrna using molecular beacons. Nucleic Acids Res. 2008;36:e30. doi: 10.1093/nar/gkn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walev I, Bhakdi SC, Hofmann F, Djonder N, Valeva A, Aktories K, Bhakdi S. Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-o. Proc Natl Acad Sci U S A. 2001;98:3185–3190. doi: 10.1073/pnas.051429498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. Micrornas in the human heart. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 34.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. Microrna-145 regulates oct4, sox2, and klf4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 35.Terai K, Hiramoto Y, Masaki M, Sugiyama S, Kuroda T, Hori M, Kawase I, Hirota H. AMP-activated protein kinase protects cardiomyocytes against hypoxic injury through attenuation of endoplasmic reticulum stress. Mol Cell Biol. 2005;25:9554–9575. doi: 10.1128/MCB.25.21.9554-9575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klauke N, Smith G, Cooper JM. Regional electroporation of single cardiac myocytes in a focused electric field. Anal Chem. 2009;82:585–592. doi: 10.1021/ac901886j. [DOI] [PubMed] [Google Scholar]
- 37.Chen AK, Behlke MA, Tsourkas A. Efficient cytosolic delivery of molecular beacon conjugates and flow cytometric analysis of target rna. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bettiol E, Sartiani L, Chicha L, Krause KH, Cerbai E, Jaconi ME. Fetal bovine serum enables cardiac differentiation of human embryonic stem cells. Differentiation. 2007;75:669–681. doi: 10.1111/j.1432-0436.2007.00174.x. [DOI] [PubMed] [Google Scholar]
- 39.Osborn L, Hession C, Tizard R, Vassallo C, Luhowskyj S, Chi-Rosso G, Lobb R. Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell. 1989;59:1203–1211. doi: 10.1016/0092-8674(89)90775-7. [DOI] [PubMed] [Google Scholar]
- 40.Van Hoof D, Dormeyer W, Braam SR, Passier R, Monshouwer-Kloots J, Ward-van Oostwaard D, Heck AJR, Krijgsveld J, Mummery CL. Identification of cell surface proteins for antibody-based selection of human embryonic stem cell-derived cardiomyocytes. J Proteome Res. 2010;9:1610–1618. doi: 10.1021/pr901138a. [DOI] [PubMed] [Google Scholar]
- 41.Kelly OG, Chan MY, Martinson LA, Kadoya K, Ostertag TM, Ross KG, Richardson M, Carpenter MK, D’Amour KA, Kroon E, Moorman M, Baetge EE, Bang AG. Cell-surface markers for the isolation of pancreatic cell types derived from human embryonic stem cells. Nat Biotechnol. 2011;29:750–756. doi: 10.1038/nbt.1931. [DOI] [PubMed] [Google Scholar]
- 42.Kamp TJ. Recognizing heart cells in a crowd. Nat Meth. 2011;8:1013–1016. doi: 10.1038/nmeth.1780. [DOI] [PubMed] [Google Scholar]
- 43.Mummery C. Sorting cardiomyocytes: A simple solution after all? Nat Methods. 2010;7:40–42. doi: 10.1038/nmeth0110-40. [DOI] [PubMed] [Google Scholar]
- 44.Lin Q, Fu Q, Zhang Y, Wang H, Liu Z, Zhou J, Duan C, Wang Y, Wu K, Wang C. Tumourigenesis in the infarcted rat heart is eliminated through differentiation and enrichment of the transplanted embryonic stem cells. Eur J Heart Fail. 2010;12:1179–1185. doi: 10.1093/eurjhf/hfq144. [DOI] [PubMed] [Google Scholar]
- 45.Behfar A, Perez-Terzic C, Faustino RS, Arrell DK, Hodgson DM, Yamada S, Puceat M, Niederlander N, Alekseev AE, Zingman LV, Terzic A. Cardiopoietic programming of embryonic stem cells for tumor-free heart repair. J Exp Med. 2007;204:405–420. doi: 10.1084/jem.20061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Laake LW, Passier R, den Ouden K, Schreurs C, Monshouwer-Kloots J, Ward-van Oostwaard D, van Echteld CJ, Doevendans PA, Mummery CL. Improvement of mouse cardiac function by hesc-derived cardiomyocytes correlates with vascularity but not graft size. Stem Cell Res. 2009;3:106–112. doi: 10.1016/j.scr.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Martens A, Kensah G, Rojas S, Rotärmel A, Baraki H, Haverich A, Martin U, Gruh I, Kutschka I. Induced pluripotent stem cell (iPSC)-derived cardiomyocytes engraft and improve heart function in a mouse model of acute myocardial infarction. Thorac cardiovasc Surg. 2012;60:PP26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.