Abstract
In contrast to homotrimeric transporters of the RND (resistance-nodulation-division) superfamily, which often conduct efflux transport of a wide range of substrates by the functionally rotating mechanism, the mechanism utilized by the heterotrimeric members of this family, which also perform multidrug efflux, is unclear. We examined one heterotrimeric transporter, the MdtB2C complex of Escherichia coli, by an extensive cysteine scanning mutagenesis of residues likely involved in ligand transport. Many such mutations in MdtC strongly decreased the level of cloxacillin transport, whereas mutations of corresponding residues in MdtB did not affect transport. Furthermore, many such residues in MdtC were covalently modified by fluorescein maleimide, which acted as a substrate and presumably produced labeling of the residues in the substrate path. In contrast, few residues in MdtB were labeled. Together with the previous data showing that the inactivation of proton translocation channel in MdtC has an only modest effect on transport yet in MdtB totally inactivated the activity, these results suggest that the two subunits, MdtB and MdtC, play very different roles, MdtC likely involved in substrate binding and transport and MdtB presumably inducing the conformational change needed for transport through proton translocation. Three-dimensional models of MdtB and MdtC, based on sequence homology with the AcrB transporter, also support this interpretation.
Gram-negative bacteria usually produce multiple efflux transporters or pumps with wide specificity. Among them, those belonging to the resistance-nodulation-division (RND) family are especially noteworthy, as their collaboration with the outer membrane channel and the periplasmic accessory protein allows them to capture drugs within the periplasmic space and to export them directly into the external media.1 Most of the RND transporters exist as homotrimers, each protomer of which is known to undergo sequential changes in conformation that allow the extrusion of drugs coupled to proton influx.2-5 However, there are some RND pumps that are suggested by genetic evidence to function as heterotrimers. Previously, we showed that the heterotimeric MdtB2C complex of Escherichia coli is the functional form of the MdtB–MdtC RND family transporter,6 consistent with the genetic studies showing that MdtB and MdtC were interdependent with respect to each other for efflux activity.7 The MdtB and MdtC monomers consist of 1040 and 1025 amino acids, respectively, and their sequences are only 49% identical. It thus seems unlikely that the drug efflux in MdtB2C is mediated by a simple functionally rotating mechanism that is supposed to function in homotrimeric transporters such as AcrB.2-5 Indeed, MdtB and MdtC behave differently in relation to mutations in the presumed path of proton translocation.6 Mutations in such residues in the MdtB subunit exerted a much more severe effect on the activity of the complex than mutations in the MdtC subunit. To understand the potentially different functional roles of MdtB and MdtC subunits of B2C complex in drug efflux, we examine here whether the substrate flows through both MdtB and MdtC during the pumping activity of the B2C complex.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Growth Conditions
E. coli strains and plasmids used in this study are listed in Table 1. A protease- and recombinase-deficient E. coli B strain lacking both the AcrAB and MdtABC efflux systems, BL21KAMR,6 was used as the host strain. A giant gene encoding the linked Mdt B2C heterotrimer6 was constructed on pSPORT1 or its AatII-restriction-site-free derivative plasmid pSPORT1a, for generation of single-cysteine mutants, and the gene was expressed in BL21KAMR. pSPORT1a was constructed by using Quik-Change site-directed mutagenesis protocol (Stratagene) to facilitate the transfer of the AatII-NcoI fragment of the MdtC sequence from plasmid to plasmid. For the cloxacillin susceptibility assay, the giant mdt genes on the pSPORT1 derivative was cut out by using PstI and SphI and transferred into a pHSG576 derivative plasmid pHSGS6 containing MCS (multiple-cloning sites) derived from pSPORT1. Cells were grown in Luria-Bertani (LB) broth or on LB agar plates supplemented, when necessary, with the antibiotics.
Table 1.
E. coli Strains and Plasmids Used in This Study
| relevant genotype and characteristics | ref or source | |
|---|---|---|
| Strains | ||
| BL21 | E. coli B F− ompT hsdSB(rB− mB−) gal dcm lon | Novagen |
| BL21KA | BL21 ΔacrAB | 6 |
| BL21KM | BL21 ΔmdtABC | 6 |
| BL21KAMR | BL21 ΔacrAB ΔmdtABC Δ(srl-recA)306::Tn10(Tcr)b | 6 |
| Plasmids | ||
| pSPORT1 | pMB1 ori, medium-copy number cloning vector; lac-inducible expression; Apr | Gibco-BRL |
| pSPORT1a | pSPORT1 derivative deficient in the AatII restriction site | a |
| pHSG576 | pSC101 ori, low-copy number vector; lac-inducible expression; Cmr | 21 |
| pHSGS | pHSG576 containing pSPORT1-derived MCS sequence under lac promoter; Cmr | 6 |
| pSMA-B10 | pSPORT1 derivative harboring mdtA and mdtB with a His10 tag sequence at the 3′-end | 6 |
| pSMA-C10 | pSPORT1 derivative harboring mdtA and mdtC with an upstream engineered mdtB SD sequence and a His10 tag sequence at the 3′-end | 6 |
| pSABCHisfram | pSPORT1 derivative harboring mdtABC with a His10 tag sequence at the 3′-end of mdtC | 6 |
| pSMSacIC- linkerHindIII |
pSPORT1 derivative harboring the SacI-HindIII fragment of the second MdtC subunit and linker sequence | 6 |
| pSMHindIIIB10SphI | pSPORT1 derivative harboring the HindIII-SphI fragment containing third MdtB subunit with a His10 tag | 6 |
| pSMA-BC10 | pSPORT1 derivative harboring mdtA and an artificial gene for a linked Mdt B–C dimer with a C-terminal His10 tag | 6 |
| pSMA-BCB10 | pSPORT1 derivative harboring mdtA and an artificial gene for a linked Mdt B–C–B trimer with a C-terminal His10 tag | 6 |
| pSMA-BCLCCLBCL10 | pSPORT1 derivative harboring mdtA and an artificial gene for a cysteineless linked Mdt B–C–B trimer with a C-terminal His10 tag | a |
This study.
Abbreviations: Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Tcr, tetracycline resistant.
Sequence Analysis of MdtB and MdtC and Building of Homology Models
Homology models of MdtC were built with the Yasara program suite (http://www.yasara.org) with the binding protomers of AcrB structures of the highest resolution, Protein Data Bank entries 3NOC, 3NOG, and 2J8S, as templates. The program, using Modeler,8 built five models for each template based on slightly different alignments and improved the models by importing segments from different models. The final model was rated as “good” in terms of overall quality. The model for MdtB was built in a similar way, using the same three targeted templates. The final model is rated as “satisfactory” by the program. The models were further energy-minimized by Yasara by running a short equilibrium MD simulation routine.
Predicted Binding of Cloxacillin to MdtC and MdtB
The computer prediction was conducted using Autodock Vina,9 obtained from the Scripps Research Institute.
Assay of Fluorescein 5-Maleimide Efflux Activity in E. coli Cells
E. coli cells were inoculated into LB medium and grown to an optical density at 660 nm (OD660) of 0.5–0.8, without being shaken at 30 °C. The bacteria were harvested, washed with 10 mM Hepes-KOH (pH 7.5) buffer, and resuspended in this buffer. The cell density was adjusted to an OD660 of ~20. To energize the cells, 10 mM glucose was added, and after 5 min, the reaction was started via addition of 40 μM fluorescein 5-maleimide. During the incubation at room temperature, 200 μL portions were removed at the indicated times and the reaction was stopped with 10 mM dithiothreitol. The cells were washed twice with 0.5 mL of 10 mM Hepes-KOH (pH 7.5) buffer at 4 °C, resuspended in 100 μL of solubilization buffer [20 mM Hepes-NaOH (pH 7.5), 0.3 M NaCl, 10% (v/v) glycerol, and 2% (w/v) dodecyl maltoside (DDM)], and lysed by sonication. After 30 min on ice, the insoluble material was removed by centrifugation at 12000g for 10 min at 4 °C. The supernatant was appropriately diluted with a buffer [20 mM Hepes-NaOH (pH 7.5), 0.3 M NaCl, 10% (v/v) glycerol, and 0.02% DDM], and the fluorescence intensity was measured using an excitation wavelength of 491 nm and an emission wavelength of 520 nm. It is not clear what fraction of the fluorescence came from covalently labeled proteins, but sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) showed that there was a very large difference in the labeling of proteins between the parent and the ΔacrAB mutant (not shown).
Intact Cell Labeling of Single-Cysteine Mutants of the Linked Trimer with Fluorescein 5-Maleimide
Fresh transformants of BL21KAMR carrying a pSPORT1 derivative were inoculated into LB medium supplemented with 100 μg/mL ampicillin and 10 μM isopropyl β-D-thiogalactoside (IPTG) and grown to an OD660 of ~0.5, without being shaken at 30 °C. The bacteria were harvested, washed with 10 mM Hepes-KOH (pH 7.5) buffer, and suspended in the same buffer. One milliliter of the bacterial suspension (OD660 of ~20) was incubated in the presence of 40 μM fluorescein 5-maleimide for 30 min at room temperature. The reaction was stopped with 10 mM dithiothreitol. The cells were washed twice with 1 mL of 10 mM Hepes-KOH (pH 7.5) buffer, resuspended in solubilization buffer [20 mM Hepes-NaOH (pH 7.5), 0.3 M NaCl, 5 mM imidazole, 10% (v/v) glycerol, and 2% (w/v) DDM], and lysed by sonication in the presence of Complete, EDTA-free Protease Inhibitor Mixture (Roche). After 30 min on ice, the insoluble material was removed by ultra-centrifugation at 150000g for 35 min at 4 °C. The supernatant was transferred into a Micro Bio-Spin chromatography column (Bio-Rad), mixed with 60 μL of BD TALON cobalt chelating resin (BD Biosciences), and gently shaken for 1 h at 4 °C. The resin was washed with a buffer [20 mM Hepes-NaOH (pH 7.5), 0.3 M NaCl, 10 mM imidazole, 10% glycerol, and 0.02% DDM]. The proteins were eluted with 60 μL of elution buffer [20 mM Hepes-NaOH (pH 7.5), 0.3 M NaCl, 200 mM imidazole, 10% glycerol, and 0.02% DDM] and incubated with 15 μL of 5× sample buffer [60 mM Tris-HCl (pH 6.8), 25% glycerol, 2% SDS, 14.4 mM 2-mercaptoethanol, and 0.1% bromophenol blue] for 30 min at room temperature. The samples were subjected to SDS–PAGE (7.5%). The gel was scanned for fluorescence using the Typhoon 9400 Variable Mode Imager (Molecular Dynamics) with 488 nm excitation and 520 nm emission filters at normal sensitivity and then stained for protein with Coomassie brilliant blue R-250. The fluorescence intensity and the amount of the linked trimer protein band were analyzed using ImageJ from the National Institutes of Health.
Drug Susceptibility
BL21KAMR containing a pHSG576 derivative was used to determine susceptibility to cloxacillin by a gradient plate method, as described previously,6 except that 0.50 μg/mL cloxacillin in the lower layer was used.
Site-Directed Mutagenesis
Three pSPORT1-derived plasmids,6 pSMA-BC10, pSMSacIC-linkerHindIII, and pSMHindIIIB10SphI, were used as templates for site-directed mutagenesis of the MdtB or MdtC sequence by the QuikChange protocol (Stratagene). pSMA-BC10 harbors mdtA and an artificial gene for a covalently linked dimer of MdtB and MdtC tagged with a C-terminal His10 sequence. This plasmid provides a PstI-SacI fragment, containing mdtA and the first protomer unit of MdtB [with an upstream Shine-Dalgarno (SD) sequence and a downstream linker sequence], for the construction of pSMA-BCB10, harboring mdtA and an artificial gene for a linked Mdt B–C–BHis trimer.6 pSMSacIC-linkerHindIII and pSMHindIIIB10SphI provide the second protomer unit of MdtC and a linker sequence as a SacI-HindIII fragment and the third protomer unit of the MdtB sequence with the His10 tag as a HindIII-SphI fragment, respectively, for the construction of pSMA-BCB10.
Construction of Single-Cysteine Mutants of the Linked Mdt B–C–BHis Trimer
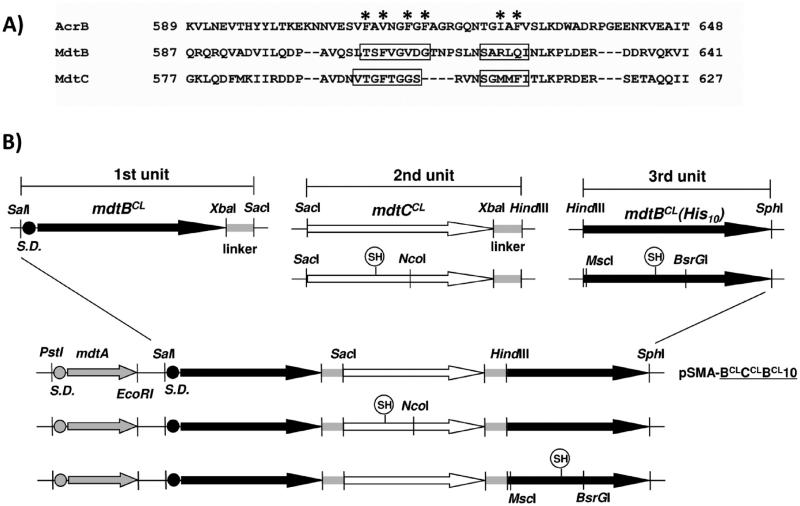
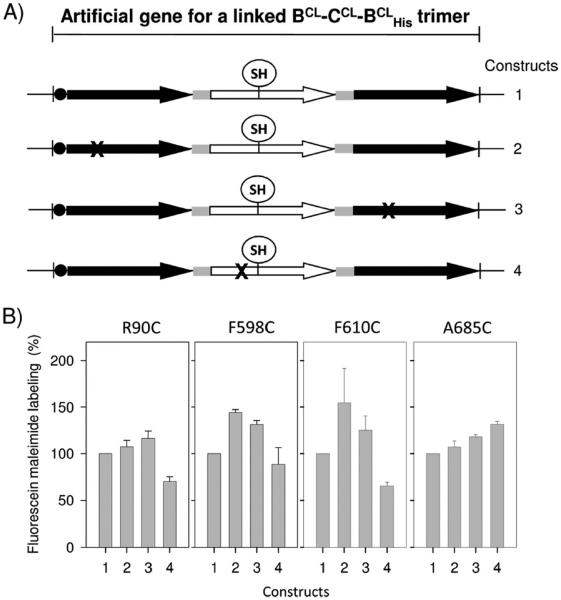
To construct the plasmid pSMA-BCLCCLBCL10 containing mdtA and an artificial gene for a covalently linked cysteine-less Mdt B–C–BHis trimer, two endogenous cysteines (C495 and C961) in the MdtB sequence of two plasmids, pSMA-BC10 and pSMHindIIIB10SphI, and four such cysteines (C24, C380, C486, and C947) in the MdtC sequence of plasmid pSMSacIC-linkerHindIII were replaced one by one with serine, resulting in plasmids pSMA-BCLC10, pSMSacICCL-linkerHindIII, and pSMHindIIIBCL10SphI (hereafter the superscript CL stands for cysteine-less). The PstI-SacI fragment from pSMA-BCLC10, the SacI-HindIII fragment from pSMSacICCL-linkerHindIII, and the HindIII-SphI fragment from pSMHindIIIBCL10SphI were cloned into pSPORT1 to produce pSMA-BCLCCLBCL10 (Figure 1B). The whole sequence for a linked Mdt BCL–CCL–BCLHis trimer was confirmed by sequencing.
Figure 1.
Construction of a pSMA-BCLCCLBCL10 derivative harboring an artificial gene for a single-cysteine mutant of a linked Mdt BCL–CCL–BCLHis trimer. (A) Sequences of MdtB and MdtC aligned with that of AcrB using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalW2). The AcrB residues below the asterisk are known to constitute the substrate-binding pocket, as determined by crystallography. To find putative binding pocket residues of MdtB and MdtC, individual residues of the MdtB or MdtC in the boxed region were replaced with cysteine in the cysteine-less background and the resulting MdtB or MdtC sequence was introduced into the second MdtC or the third MdtB subunit of the linked Mdt BCL–CCL–BCLHis trimer as in panel B. (B) Plasmid pSMA-BCLCCLBCL10 containing mdtA and a giant gene for a linked BCL–CCL–BCLHis trimer constructed as described previously.6 To introduce a single-cysteine replacement mutation into the second MdtC subunit of the linked BCL–CCL–BCLHis trimer, the SacI-NcoI fragment containing the single cysteine replaced the corresponding sequence of pSMA-BCLCCLBCL10. To introduce a single-cysteine replacement mutation into the third MdtB subunit of the linked BCL–CCL–BCLHis trimer, the MscI-BsrGI fragment containing the single-cysteine MdtB mutant sequence was prepared and replaced the corresponding sequence of the SacI-SphI fragment containing both the second MdtC and third MdtB subunits bridged by the linker. Then, the PstI-SacI fragment containing mdtA and the first MdtB protomer unit was connected.
To introduce a cysteine replacement mutation into the second, MdtCCL, subunit of this linked trimer, the intended mutation was first introduced into plasmid pSMSacICCL-linkerHindIII and confirmed by sequencing. Then the SacI-NcoI or NcoI-HindIII fragment containing the single-cysteine mutation was cut from the mutated plasmid and was used to replace the corresponding sequence in pSMA-BCLCCLBCL10. To introduce a single-cysteine replacement mutation into the third, MdtBCL, subunit of the linked trimer, the intended mutation was first introduced into plasmid pSMHindIIIBCL10SphI. The MscI-BsrGI or BsrGI-SphI fragment containing the mutation was cut from the mutated plasmid and was used to replace the corresponding sequence in pSMSacICCL-BCLSphI, with the SacI-SphI fragment containing the second MdtCCL and third MdtBCL protomer units. Then, the PstI-SacI fragment containing mdtA and the first MdtBCL protomer unit from pSMA-BCLC10 was introduced to complete the giant gene containing the three original cistrons. Likewise, we first introduced a proton translocation mutation into pSMA-BCLC10, pSMSacICCL-linkerHindIII, or pSMHindIIIBCL10SphI and then exchanged the appropriate fragments from pSMA-BCLCCLBCL10 with those containing the mutation.
RESULTS
Fluorescein 5-Maleimide Is a Substrate of AcrAB and MdtABC efflux Systems
We first investigated the possibility that a thiol-reactive fluorescent dye, fluorescein 5-maleimide, may be a substrate of the MdtABC efflux system. If it is, its specific binding to a single cysteine strategically introduced into a position along the putative substrate path within MdtB and MdtC would be enhanced. This approach was successfully applied in finding the substrate path in AcrB or monitoring conformational changes occurring around the substrate-binding pocket of AcrB, using thiol-reactive probes such as Bodipy FL maleimide and fluorescein 5-maleimide.10,11 Fluorescein 5-maleimde has a molecular weight of 427 and is somewhat hydrophobic.
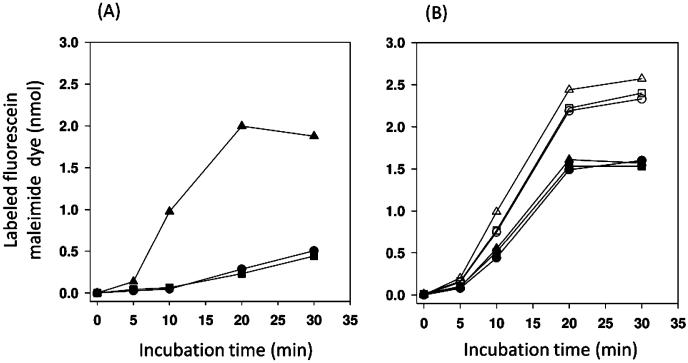
The assay is based on the idea that if fluorescein 5-maleimide is a substrate of AcrAB and MdtABC efflux systems, these systems would lower the intracellular concentration of the dye and would also lower the extent of labeling of intracellular proteins containing sulfhydryl residues. BL21 and its isogenic ΔacrAB or ΔmdtABC mutants were incubated with 40 μM fluorescein 5-maleimide in the presence of 10 mM glucose. The labeling reaction was stopped by the addition of dithiothreitol, and the labeled cells were harvested and washed to remove the extracellular dye. The cellular content was extracted into a solubilization buffer, and the concentration of the dye was estimated by fluorescence. The intracellular dye content was dramatically higher in the ΔacrAB cells than in the wild type, suggesting that AcrAB is the main molecular machinery pumping fluorescein 5-maleimide out of the cell (Figure 2A). Cells defective in the MdtABC system accumulate the dye like wild-type cells (Figure 2A), possibly because it plays a lesser role than AcrAB in dye efflux. In addition, the level of expression of MdtABC from the E. coli chromosome might have been quite low during the growth in LB broth, because it is controlled mainly by the BaeSR two-component system in response to external signals.7,12
Figure 2.
Fluorescein 5-maleimide is a substrate of AcrAB and MdtABC efflux systems. (A) Fluorescein maleimide labeling of BL21 (●) and its is ogenic ΔacrAB (▲)or ΔmdtABC (■) mutant strain. The cell suspensions, prepared as described in Materials and Methods, were incubated with 40 μM fluorescein 5-maleimide in the presence of 10 mM glucose for the indicated time, and the labeled cells were harvested. The labeled dyes were then extracted from the washed cell mass into a solubilization buffer containing 2% DDM and 10% glycerol and estimated by fluorescence measurement. The extent of labeling was represented as nanomoles of dye per cell mass harvested from 0.2 mL of a cell suspension (OD660 ~ 20). One typical example of two separate experiments is shown. (B) Fluorescein maleimide labeling of BL21KAMR cells in which Mdt proteins are expressed from a multicopy plasmid pSPORT1 derivative: pSPORT1 (○), pSMA-B10 (△), pSMA-C10 (□), pSMABCHis (●), pSMA-BCB10(▲), and pSMA-BCLCCLBCL10 (■). The transformed cells were grown in LB broth in the presence of 100 μg/mL ampicillin and labeled under the glucose-supplemented condition as described for panel A. One typical example of three separate experiments is shown.
We therefore conducted a similar experiment with E. coli BL21KAMR cells that lacked the chromosomal copies of acrAB and mdtABC, expressing MdtABC from multicopy plasmid pSABCHis under the pLac promoter. As seen in Figure 2B, the expression of MdtB and MdtC together with MdtA reduced the level of dye detectably in BL21KAMR cells. In contrast, expression of MdtB or MdtC alone (together with MdtA) from plasmid pSMA-B10 or pSMA-C10 resulted in the accumulation of the dye at a level comparable to the vector-alone control, suggesting that both MdtB and MdtC are needed for the efflux activity as expected (Figure 2B). Expression of the covalently linked Mdt B–C–BHis trimer from pSMA-BCB10 produced an effect on the accumulation of the dye in BL21KAMR cells similar to that of the expression of unlinked MdtB and MdtC from pSMABCHis. These results show that fluorescein 5-maleimide is a substrate of the MdtABC pump.
Construction of Single-Cysteine Mutants of the Linked Mdt B–C–BHis Trimer
We conducted cysteine scanning mutagenesis of MdtB and MdtC, to see if such mutational substitutions affected the efflux activity, and if any of the residues reacted strongly with fluorescein 5-maleimide presumably because they were on the substrate path (see also ref 10). Because the MdtB2C transporter is heterotrimeric,6 mutating the mdtB gene would double the dosage effect as the assembled transporter would contain two of the mutated protomers. Thus, we chose to use the covalently linked trimers3 so that we can introduce a mutation into only one of the three subunits and compare directly the effects of mutations on MdtB and MdtC subunits.
We focused here on putative residues of substrate-binding pockets of MdtB and MdtC. In AcrB, these residues are known from crystal structures2,13 and mutagenesis studies.14 The residues finally chosen (Figures 3 and 4) mostly on the basis of these AcrB residues projected onto MdtB and MdtC sequences after alignment. Because the distal substrate-binding pocket of AcrB,2 containing multiple Phe residues, was thought to play a critical role in substrate recognition and transport, we tried to cover all residues in this segment (Figure 1A). They also included residues near the bound cloxacillin, in the docked models obtained by using the three-dimensional (3D) homology models (Materials and Methods) and Autodock Vina.9 Finally, we also selected a few residues of MdtB and MdtC for cysteine replacement mutation as negative controls. These residues are presumed to be in the periplasmic loop but outside of the substrate-binding pocket. We replaced each of these residues individually with cysteine in cysteine-less MdtB or MdtC sequences, and the resulting sequences of MdtB or MdtC were introduced into the second MdtC or the (usually) third MdtB subunit of the linked Mdt BCL–CCL–BCLHis trimer (Figure 1B).
Figure 3.
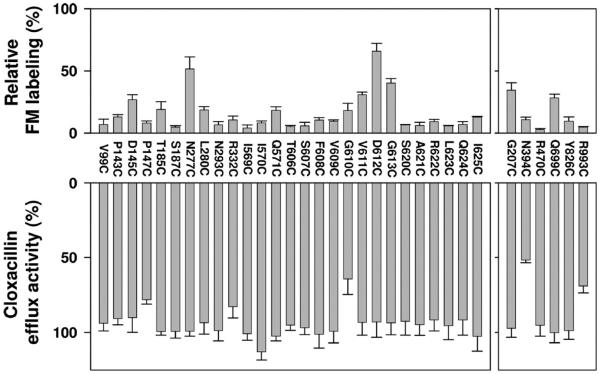
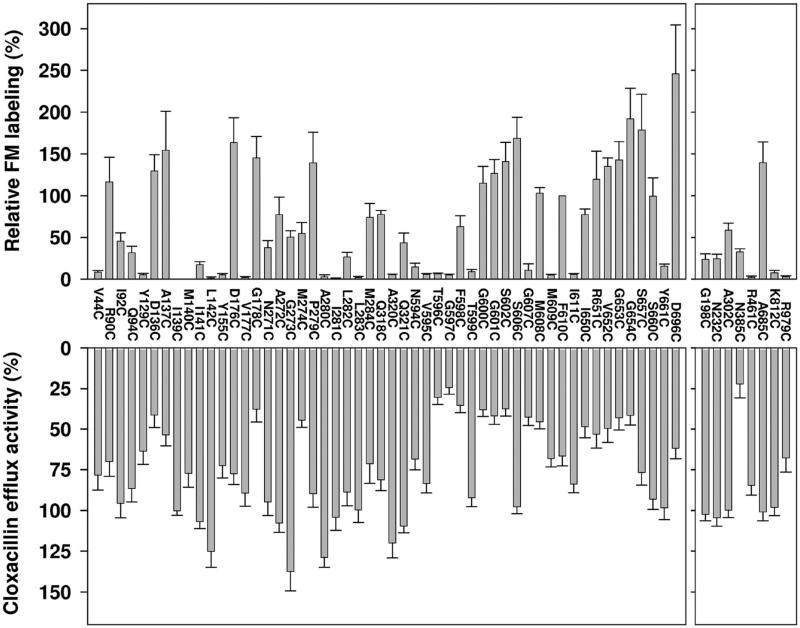
Fluorescein maleimide labeling and cloxacillin efflux activity of single-cysteine MdtB subunit mutants of the linked BCL–CCL–BCLHis trimer. A cysteine in place of a putative substrate-binding pocket residue (left) or a residue selected as a negative control (right) was introduced into the third MdtB subunit of the linked BCL–CCL–BCLHis trimer. In the top panel, the extent of fluorescein maleimide labeling of single-cysteine mutants of a linked Mdt BCL–CCL–BCLHis trimer is represented relative to the labeling of one MdtC subunit mutant (F610C) of the linked BCL–CCL–BCLHis trimer. In each experiment, each purified protein sample was loaded into two different SDS–PAGE gels and the average of two normalized fluorescence intensities was obtained. The means of three separate experiments are shown. Bars indicate the standard deviation. The bottom panel shows the activities of single-cysteine mutants of a linked BCL–CCL–BCLHis trimer in BL21KAMR. efflux activities were estimated from the levels of resistance to cloxacillin by using the gradient plate method and represented as a percentage to the activity of the linked BCL–CCL–BCLHis trimer. The means of at least two separate experiments are shown. Bars indicate the standard deviation.
Figure 4.
Fluorescein 5-maleimide labeling (top) and cloxacillin efflux activity (bottom) of single-cysteine MdtC subunit mutants of the linked BCL–CCL–BCLHis trimer. A cysteine in place of a putative substrate-binding pocket residue (left) or a residue selected as a negative control (right panel) was introduced into the second MdtC subunit of the linked BCL–CCL–BCLHis trimer. The extent of labeling and activity of the MdtC mutants were represented on a similar scale as those of MdtB mutants in Figure 3.
Cloxacillin efflux Activity of Single-Cysteine Mutants of the Linked BCL–CCL–BCLHis Trimer
We examined the effect of each cysteine substitution on the efflux activity, by measuring the ability of mutants of the linked Mdt BCL–CCL–BCLHis trimer to confer cloxacillin resistance in E. coli BL21KMAR cells. For this, pHSG576 derivative plasmids harboring mdtA and the artificial gene for the mutant linked trimer were used. In nearly all of the mutants of the putative MdtB substrate path, the efflux activity was essentially unaffected with the exception of G610C, which resulted in an activity loss of ~35% (bottom left panel of Figure 3). When the G610C mutation was introduced into both of the MdtB subunits of the covalently linked BCL–CCL–BCLHis trimer, the activity loss nearly doubled (to ~67%) (data not shown).
In contrast, in MdtC the efflux activity of cysteine mutants of putative substrate path residues was more strongly affected (bottom left panel of Figure 4). Several single-cysteine MdtC mutants such as D136C, G178C, M274C, T596C, G597C, F598C, G600C, G601C, S602C, G607C, M608C, I650C, V652C, G653C, and G654C lost more than half of the wild-type activity. Among “control” residues (right panels of Figures 3 and 4), N394C and R993C of MdtB and N385C and R979C of MdtC also showed significant activity loss. N394C of MdtB and N385C of MdtC are situated in the small periplasmic loop connecting TM3 and TM4, and R993C of MdtB and R979C of MdtC are in the small periplasmic loop connecting TM11 and TM12. The conformation of these small loops could be important for activity. Other single-cysteine mutants constructed as negative controls showed essentially unaffected activity (bottom right panels of Figures 3 and 4).
Intact Cell Labeling of Single-Cysteine Mutants of the Linked Mdt BCL–CCL–BCLHis Trimer with Fluorescein 5-Maleimide
We assume that the substrate fluorescein 5-maleimide favorably partitions into the substrate-binding pocket, if any, of each subunit of the linked Mdt BCL–CCL–BCLHis trimer, so that a single cysteine residue nearby will have more opportunity to interact with the dye than the surface-exposed cysteine situated outside of the substrate path. E. coli BL21KAMR cells, expressing a single cysteine mutation within the linked Mdt BCL–CCL–BCLHis trimer from a pSMA-BCLCCLBCL10 derivative in the presence of 10 μM IPTG, were thus incubated with 40 μM fluorescein 5-maleimide in the absence of added glucose. The linked Mdt trimer with the His10 tag was purified and subjected to SDS–PAGE analysis. Then the fluorescence of the linked trimer band was determined and was compared with the intensity of its Coomassie staining to estimate the relative amount of dye covalently bound to the single cysteine. The extent of labeling is shown relative to the labeling of MdtC F610C, which was arbitrarily set at 100% (see the top left panels of Figures 3 and 4).
Generally, cysteine residues in the last MdtB protomer in the covalently linked B–C–B trimer were much more lightly labeled than those in MdtC: only N277C and D612C of the third MdtB subunit were labeled reasonably strongly in the linked BCL–CCL–BCLHis trimer (Figure 3). This forms a striking contrast to the labeling pattern observed in AcrB, with a similar labeling reagent BODIPY FL maleimide.10 For example, we note that residues within the substrate-binding site in AcrB, such as Q89, Q176, F178, Y277, and F617, were labeled very strongly when they were mutated to cysteine,10 yet their equivalents by sequence alignment here, V99, T185, S187, L280, and G613 of MdtB, respectively, were hardly labeled at all, with the exception of G613 (Figure 3).
Because the two MdtB subunits occupy nonidentical positions in the heterotrimer, we also examined a few residues in the first MdtB subunit. Both N277C and V611C in this protomer were labeled even more weakly, at less than one-half of the intensity of these residues in the last protomer (not shown), confirming that both MdtB protomers are poorly labeled by fluorescein 5-maleimide.
In contrast to MdtB, there were many strongly labeled cysteine residues in MdtC in the putative substrate-binding pocket residues, including D136, A137, D176, G178, P279, G600, G601, S602, S606, M608, and F610, which correspond to F136, L137, Q176, F178, P285, G614, F615, T624, I626, and F628 of AcrB, respectively, on the basis of sequence alignment. Notably, an even stronger labeling was seen with some of the residues presumably at or close to the peripheral cleft entrance, such as I650, R651, V652, G653, G654, S657, S660, and D696, which corresponds to I671, V672, E673, L674, G675, T678, D681, and M712 of AcrB, respectively, which define the putative initial binding site for many substrates, based on the Bodipy FL maleimide labeling10 and earlier cocrystallization studies,15 and also on very recent cocrystallization studies.13,16 Cysteine substitutions of the MdtC residues selected as negative controls were largely weakly labeled with the exception of A685C (top right panels of Figure 4), putatively situated at the end of helix Cα3 of the PC2 domain, somewhat far from the entrance point of the external cleft.
Under the same conditions, the linked B–C–BHis trimer containing the intrinsic cysteines was very weakly labeled (4%) (data not shown), All intrinsic cysteines of MdtB or MdtC are putatively situated either in the transmembrane (TM) domain [C24 of MdtC (TM1), C380 of MdtC (TM3), and C495 of MdtB and C486 of MdtC (TM6)] or in the cytoplasmic loop connecting TM10 and TM11 (C961 of MdtB and C947 of MdtC), and their low reactivity serves as another set of negative controls in our assay. Interestingly, the cysteine-less mutant BCL–CCL–BCLHis trimer was also faintly labeled (3%), its reactivity presumably caused by the slow reaction of the maleimide group with amino groups.
Effect of Other Substrates on Fluorescein 5-Maleimide Labeling
Preincubation of cells with up to 200 μM cloxacillin for 5 min had little effect on the labeling of G600C, S602C, S606C, and F610C of MdtC or D612C of MdtB in the linked BCL–CCL–BCLHis trimer produced from a pHSG576 derivative plasmid, when 20 or 40 μM fluorescein 5-maleimide was used for the labeling reaction (data not shown). Similarly, up to 400 μM novobiocin, a substrate,7,12 or indole, an inducer of the BaeSR system,17 produced little change in the labeling of D612C of MdtB or R90C, G601C, F610C, S660C, or D696C of MdtC (data not shown). Deoxycholate (up to 2 mM), a substrate,7,12 did not significantly change the labeling of the residues of MdtC mentioned above (data not shown).
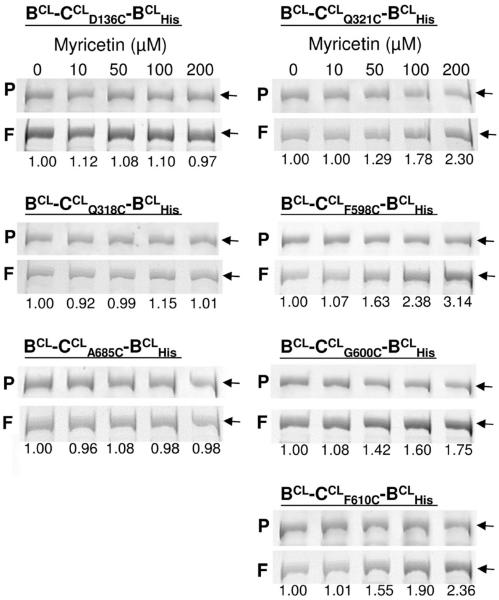
In contrast, myricetin, which may be a substrate of the MdtABC pump,18 significantly stimulated the labeling of Q321C, F598C, G600C, and F610C of MdtC at concentrations of 50–200 μM but had little effect on the labeling of D136C, Q318C, or A685C (Figure 5). It also had little effect on the labeling of V611C, D612C, or G613C of MdtB (not shown).
Figure 5.
Effect of myricetin on the labeling of some MdtC cysteine residues by fluorescein maleimide. Cells were preincubated for 5 min with the indicated concentration of myricetin before the labeling with 20 μM fluorescein maleimide was conducted for 30 min at room temperature. The linked transporter was isolated by using the His tag, and the proteins were separated via SDS–PAGE. The band containing the Mdt B–C–B linked trimer was analyzed by fluorescence (rows labeled F) and by Coomassie Blue staining for protein content (rows labeled P), and the ratio of fluorescence to the amount of protein was calculated and listed at the bottom of each lane. As seen, myricetin enhances the modification of residues 321, 598, 600, and 610 of MdtC (right) but does not affect the labeling of residues 136, 318, and 685 (left).
Conformational Change of the Substrate-Binding Pocket
The introduction of a proton translocation mutation into one of the MdtB subunits of the linked Mdt B–C–BHis trimer results in a nearly complete loss of cloxacillin efflux activity.6 On the other hand, the linked trimer having the same type of mutation in the MdtC subunit maintains some 20–80% of the activity.6 This observation suggests that MdtB may play a more important role in energy transduction in providing the energy for conformational change that would drive the substrate transport possibly through MdtC, which could be the main substrate-conducting unit in the MdtB2C complex. We therefore examined if mutations in the MdtB proton translocation pathway, thought to mimic the protonated state of one or more Asp residues,3,19 affect the accessibility of single cysteines (R90C, F598C, or F610C) within the putative substrate-binding pocket of the neighboring MdtC to fluorescein 5-maleimide. Such a mutation in either of the neighboring MdtB subunits resulted in a somewhat increased degree of labeling of MdtC (Figure 6), a result suggesting that proton translocation through MdtB affects the conformation of the substrate-binding site within MdtC. On the other hand, similar mutations within MdtC decreased the accessibility of the cysteine residues in the same MdtC subunit. The surface-exposed cysteine A685C of MdtC, far from the substrate-binding pocket, did not exhibit similar changes in the labeling pattern (Figure 6).
Figure 6.
Effect of proton translocation defect on the accessibility of cysteine substitutions of putative MdtC substrate-binding pocket residues to fluorescein 5-maleimide. A set of artificial constructs (labeled 2–4) in which one subunit of the single-cysteine MdtC mutant (R90C, F598C, F610C, and A685C) of the linked BCL–CCL–BCLHis trimer has a proton translocation mutation (D410A of the MdtB subunit or D401A of the MdtC subunit represented by X), together with a control construct containing no proton translocation mutation in any of its subunits (labeled 1), was constructed in the background of pSMA-BCLCCLBCL10. The mutant protein was expressed from each plasmid construct in BL21KAMR cells by IPTG (10 μM) induction. The cells were subjected to a labeling reaction with 40 μM fluorescein 5-maleimide, and the labeled proteins were purified. In each experiment, the protein sample was loaded into two different SDS–PAGE gels and the average of two normalized fluorescence intensities was obtained. The extent of labeling was expressed relatively as a percentage to that of the first construct shown. The means of at least three separate experiments are shown. Bars indicate the standard deviation.
Homology-Based 3D Models of MdtB and MdtC and Computer Docking of Ligands
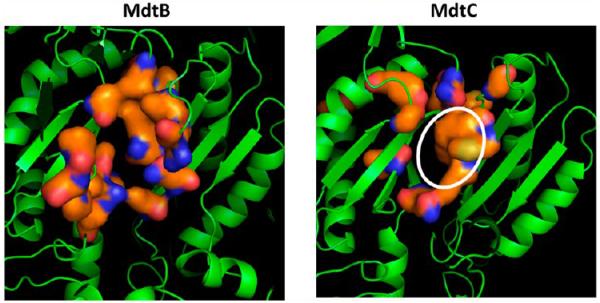
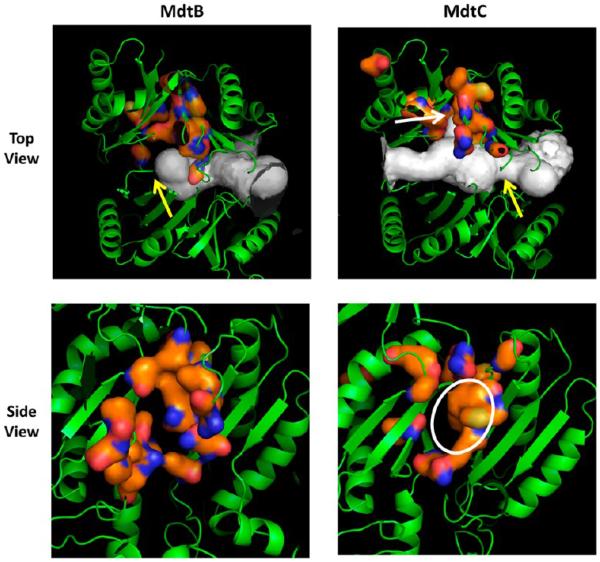
The regions corresponding to the deep substrate-binding pocket of AcrB2 were examined in the homology models. Both MdtB and MdtC models seemed to have pockets in this area at first glance (Figure 7). However, the use of Caver20 showed some differences between MdtB and MdtC models: in the MdtB model, the broad tunnel entering from the external cleft (at the right side of the protein in Figure 7) is completely terminated around the area of the binding pocket (yellow arrow in the top left panel), whereas in MdtC, the tunnel continues all the way to the interior edge of the protein. Furthermore, with MdtC, there is a significant narrowing of the tunnel before it reaches the binding pocket (yellow arrow in the top right panel), and there is a distinct protrusion of the tunnel right into the pocket (white arrow in the top right panel). The presumed binding site in MdtC seems to be a true deep pocket with a hydrophobic surface of a significant size (enclosed by a white ellipse in the bottom right panel), whereas the “pocket” appears to be less extensive and less hydrophobic in MdtB (Figure 7, bottom left panel).
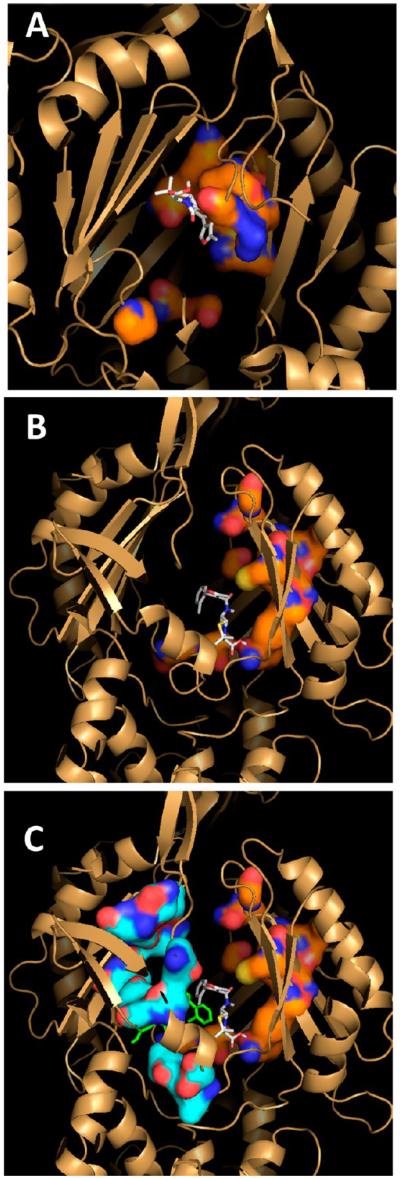
Figure 7.
Homology models of MdtB and MdtC. In the top panels, the models are shown in the orientation with the periplasmic, TolC-binding domains on top and the periplasmic cleft on the right, with the large tunnels detected by Caver colored white. The orange surface represents the putative deep binding pockets, corresponding to F136, V139, F178, I277, A278, E280, P285, Y327, F610, V612, F615, F617, I626, and F628 of AcrB.11 These are residues 146, 149, 187, 280, 282, 283, 288, 330, 606, 608, 611, 613, 622, and 624 in MdtB and residues 136, 139, 178, 271, 273, 274, 279, 321, 596, 598, 601, 608, and 610 in MdtC. In MdtB, the tunnel is closed in the middle (yellow arrow), whereas a constriction (yellow arrow) and a branch leading into the binding cavity (white arrow) are present in MdtC. In the bottom panels, the view is from the top (i.e., the TolC-binding domain). The putative substrate-binding pocket is relatively narrow in MdtB, whereas there is a substantial hydrophobic area in the wide pocket of MdtC (white ellipse). In both panels, the figures show clipped views so that the binding sites can be seen easily. They were produced by using PyMol (http://www.pymol.com).
When substrates for this pump, such as cloxacillin, were docked by Autodock Vina9 to the region centered in the deep binding pockets using a grid of 15 Å × 15 Å × 15 Å, both MdtB and MdtC were seen to bind it with a modest affinity. With MdtB, most of the substrate molecule has hardly left the broad tunnel (Figure 8A), unlike the case in AcrB where the hydrophobic parts of the substrates clearly went out of the tunnel and into the deep pocket both in crystal structures2 and in computer docking.11 However, when the grid for docking was made larger (30 Å × 30 Å × 30 Å), a more favored docking of cloxacillin was seen in MdtC near the entrance of the lateral cleft (Figure 8C), a site that was identified as being in the presumably early phase of the normal path of substrates in AcrB10,15,16. Specifically, the residues within 4 Å were D41, V44, I92, Q94, Q648, V652, S657, S660, Y663, and D696 (corresponding to A39, A42, T91, T93, P669, E673, T678, D681, L684, and N719 of AcrB, respectively) [residues 650–654, which are strongly labeled with fluorescein maleimide (Figure 4), are shown as a light blue surface in Figure 8C]. Most of these residues surround the entrance to the cleft.
Figure 8.
Computer docking of cloxacillin to the homology models of MdtB and MdtC. Docking was conducted with Autodock Vina9 with cloxacillin initially placed close to the predicted deep binding pocket. (A) MdtB with cloxacillin. The bound cloxacillin is shown as gray sticks, and the putative binding site residues (see Figure 7) are shown as an orange surface. (B) MdtC with cloxacillin, with a narrow grid of 15 Å × 15 Å × 15 Å. The putative binding site residues (see Figure 7) are shown as an orange surface. (C) MdtC with cloxacillin, with a wider (30 Å × 30 Å × 30 Å) grid. Cloxacillin is shown as green sticks, and the residues nearby (residues 83, 85, 90, 92, 660, 661, 696, 792, and 794), including those from the bottom of the cleft (residues 650, 652, and 654), are shown as a light blue surface. The cloxacillin binding pose obtained with a narrower grid (see panel B) is also shown as gray sticks for comparison.
Docking of other substrates produced similar results. For example, with myricetin, it bound most favorably to the cleft entrance to MdtC.
DISCUSSION
We examined the functional roles of the two different subunits of the MdtB2C complex by cysteine scanning mutagenesis and by the building of homology-based 3D models. When a large number of residues predicted to line the path of substrates were mutated, one by one, to cysteine, such changes in MdtB produced only minimal decreases in the pumping activity of the heterotrimeric B–C–B transporter (Figure 3, bottom). In contrast, in MdtC, a similar maneuver decreased the pumping activity by more than 25% in approximately half of the residues examined (25 of 52) (Figure 4, bottom). These results already suggested that the two essential functions of RND-type secondary transporters, energy transduction and substrate translocation, may be separately specialized in two different types of subunits in the MdtB2C complex, and that MdtC may be the main substrate-conducting unit. This assumption is also consistent with our earlier observation6 that inactivation of the putative proton translocation pathway does not abolish substrate efflux in MdtC but totally abolishes transport in MdtB.
We further tested this hypothesis by examining the modification, in intact cells, of the mutationally created cysteine residues by fluorescein maleimide. This approach is based on the earlier observation from our laboratory10 that a similarly lipophilic dye, BODIPY FL maleimide, labels only those cysteine residues on the path of the substrate, presumably because the dye also behaves as a substrate and becomes concentrated within the substrate translocation pathway. Fluorescein 5-maleimide labeled only a few residues of MdtB significantly (Figure 3, top) but again labeled approximately half of the residues (25 of 52) of MdtC strongly (Figure 4, top) if we used 50% of the level obtained with F610C of MdtC as a cutoff value. Finally, one of the presumed substrates of the MdtB2C system, a flavonoid myricetin,18 strongly enhanced the labeling of some binding site residues in MdtC (Figure 5), but not in MdtB. These studies thus strongly support the idea that substrate translocation occurs mainly through the MdtC subunit, whereas MdtB contributes to the conformational alterations of the complex through the translocation of the proton.
Some features of the homology-based 3D models are consistent with this idea. Thus, the putative deep substrate-binding site of MdtB looks more like one of the walls of the large aqueous tunnel, rather than the real substrate-binding pocket with its deep groove (as seen with the crystal structures of AcrB2,4,5,11), whereas that of MdtC appears to be a real cavity with the small branch of the main aqueous tunnel leading to it (Figure 7).
The computer docking of some known substrates also produced results that are not inconsistent with the interpretation described above, although the docking to homology models may not be quite reliable. The ligands were seen to bind to both the peripheral, more proximal site and the deep binding site of MdtC. It could be that MdtC exists in a conformation mimicking the Access protomer of the homotrimeric AcrB,2 rather than its Binding protomer, as the access to the deep binding pocket is hindered by the narrowing of the tunnel (Figure 7). Under these conditions, the substrate might first bind to the peripheral site, close to the opening of the lateral cleft, of MdtC, which happens with some substrates in the Access protomer of the homotrimeric AcrB.13,16 The existence of such a binding site had been revealed in the symmetric structure of AcrB15 and by the BODIPY FL maleimide labeling studies.10 This binding to the peripheral site may induce a conformational change to facilitate the movement of the drug molecule to the deep binding site. It is also possible that the conformational change is induced or accelerated by that in one of the neighboring MdtB subunits, possibly following proton translocation. This hypothesis also explains why the binding of myricetin, presumably to the peripheral site, enhances rather than inhibits by competition the binding of fluorescein maleimide to the deep binding pocket and the subsequent labeling of cysteine residues there. In fact, only the level of labeling of residues in the deep pocket (Q321C, F598C, G600C, and F610C) is increased, and that in the peripheral binding site (D136C, Q318C, or A685C) was not affected (Figure 5).
We can thus assume that proton translocation through the MdtB subunit(s) produces the conformational changes within MdtC needed for drug extrusion. In fact, inactivation of the proton translocation pathway in MdtB increased the level of labeling of binding site residues of MdtC (Figure 6). A similar observation was made in AcrB11 and suggested that the inactivation forces the relevant protomer and the neighboring protomer to assume the Extrusion and Binding conformations, respectively. Although MdtC is naturally adapted to bind the substrates, the initial binding presumably occurs at the peripheral site as stated above, and the opening of the tunnel and the expansion of the deep binding pocket thus may be stimulated by the MdtB protomer(s) taking the protonated conformation.
We do not know whether the proton translocation through both of MdtB protomers is needed for successful efflux. If this is the case, this may be the mechanism to facilitate the extrusion of some drugs against strong concentration gradients.
ACKNOWLEDGMENTS
We thank Attilio Vargiu for his comments on the manuscript.
Funding
This project was funded by U.S. Public Health Service Grant AI-009644-42.
ABBREVIATIONS
- DDM
dodecyl D-maltoside
- IPTG
isopropyl β-D-thiogalactoside
- RND
resistance-nodulation-division.
Footnotes
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Nikaido H. Multidrug efflux pumps of Gram-negative bacteria. J. Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 3.Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J. Bacteriol. 2009;191:1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 5.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HS, Nagore D, Nikaido H. Multidrug efflux pump MdtBC of Escherichia coli is active only as a B2C heterotrimer. J. Bacteriol. 2010;192:1377–1386. doi: 10.1128/JB.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baranova N, Nikaido H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 2002;184:4168–4176. doi: 10.1128/JB.184.15.4168-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 9.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, effcient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Husain F, Nikaido H. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol. Microbiol. 2010;78:320–330. doi: 10.1111/j.1365-2958.2010.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takatsuka Y, Chen C, Nikaido H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6559–6565. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagakubo S, Nishino K, Hirata T, Yamaguchi A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 2002;184:4161–4167. doi: 10.1128/JB.184.15.4161-4167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 14.Bohnert JA, Schuster S, Seeger MA, Fahnrich E, Pos KM, Kern WV. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J. Bacteriol. 2008;190:8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: A crystallographic and site-directed mutagenesis study. J. Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eicher T, Cha HJ, Seeger MA, Brandstatter L, El-Delik J, Bohnert JA, Kern WV, Verrey F, Grutter MG, Diederichs K, Pos KM. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raffa RG, Raivio TL. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 2002;45:1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhou L, Lei XH, Bochner BR, Wanner BL. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 2003;185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su CC, Li M, Gu R, Takatsuka Y, McDermott G, Nikaido H, Yu EW. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 2006;188:7290–7296. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damborsky J, Petrek M, Banas P, Otyepka M. Identification of tunnels in proteins, nucleic acids, inorganic materials and molecular ensembles. Biotechnol. J. 2007;2:62–67. doi: 10.1002/biot.200600208. [DOI] [PubMed] [Google Scholar]
- 21.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ α-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]