Abstract
Tryptophan depletion resulting from indoleamine 2,3-dioxygenase (IDO) activity within the kynurenine pathway is one of the most prominent gamma interferon (IFN-γ)-inducible antimicrobial effector mechanisms in human cells. On the other hand, nitric oxide (NO) produced by the inducible isoform of NO synthase (iNOS) serves a more immunoregulatory role in human cells and thereby interacts with tryptophan depletion in a number of ways. We investigated the effects of NO on IDO gene transcription, protein synthesis, and enzyme activity as well as on IDO-mediated bacteriostasis in the human epithelial cell line RT4. IFN-γ-stimulated RT4 cells were able to inhibit the growth of Staphylococcus aureus in an IDO-mediated fashion, and this bacteriostatic effect was abolished by endogenously produced NO. These findings were supported by experiments which showed that IDO activity in extracts of IFN-γ-stimulated cells is inhibited by the chemical NO donors diethylenetriamine diazeniumdiolate, S-nitroso-l-cysteine, and S-nitroso-N-acetyl-d,l-penicillamine. Furthermore, we found that both endogenous and exogenous NO strongly reduced the level of IDO protein content in RT4 cells. This effect was not due to a decrease in IDO gene transcription or mRNA stability. By using inhibitors of proteasomal proteolytic activity, we showed that NO production led to an accelerated degradation of IDO protein in the proteasome. This is the first report, to our knowledge, that demonstrates that the IDO is degraded by the proteasome and that NO has an effect on IDO protein stability.
The indoleamine 2,3-dioxygenase (IDO) catalyzes the first and rate-limiting step in the kynurenine pathway of tryptophan degradation, the conversion of tryptophan to N-formyl-kynurenine. IDO is induced by gamma interferon (IFN-γ) in the course of an inflammatory response in many human cell types, including macrophages, astrocytes, fibroblasts, and epithelial cells. A strong IDO activity leads to a nearly complete depletion of the essential amino acid tryptophan at the site of infection, which results in growth arrest of several tryptophan-dependent microorganisms. Among these IDO-sensitive microorganisms are eukaryotic pathogens such as Toxoplasma gondii (10, 36) and bacteria such as group B streptococci (28) and enterococci (29). Furthermore, the immunoregulatory role of tryptophan depletion has recently received much attention. Mellor and colleagues found that T cells are unable to proliferate in a tryptophan-depleted environment and that in vivo IDO activity in the mouse placenta protects allogeneic concepti from being rejected by a T-cell-driven mechanism (33). It has been suggested that first-time activation of T cells in the absence of tryptophan may even result in the development of tolerance to the antigen presented (31).
The role of nitric oxide (NO) production by the inducible isoform of NO synthase (iNOS) in human cells is controversial. While having a clearly illustrated antimicrobial potential against a variety of pathogens in rodent cells (reviewed in reference 4), the impact of NO on the immune response in human cells, as well as on cell function and death, is complex and often appears to be contradictory. Expression of iNOS protein in immunologically active cells has been observed in humans during infection with Mycobacterium tuberculosis and Plasmodium falciparum, but the influence of NO on the outcome of the disease is not clear. However, iNOS protein has also been found in the absence of infections in a variety of human autoimmune diseases as well as in chronic inflammatory diseases (26).
There are known interactions between tryptophan depletion and NO production. Thomas et al. first described this interaction by showing the inhibition of IDO activity in IFN-γ-primed human mononuclear phagocytes by chemical NO donors (38). Later, Alberati-Giani et al. found another mechanism of NO-dependent IDO regulation in mouse macrophages. In these cells but not in mouse microglial cells, NO negatively modulated transcription of the IDO gene. Furthermore, lipopolysaccharide and the tryptophan metabolite picolinic acid, two costimulatory agents which upregulate iNOS in activated cells, decreased IDO gene transctiption in mouse macrophages. However, this effect seemed to be NO independent (1). We demonstrated functional consequences of the IDO-NO interaction by showing that the IDO-mediated bacteriostatic effect on Enterococcus faecalis is abolished by simultaneous NO production by the IDO-expressing cell and NO production by neighboring cells (9).
We further investigated the interaction between these mechanisms, both of which are known to have antimicrobial and immunoregulatory activity. We show that the human uroepithelial cell line RT4, which expresses both IDO activity (after stimulation with IFN-γ) and strong iNOS activity (after stimulation with IFN-γ and interleukin-1β [IL-1β] and/or tumor necrosis factor alpha [TNF-α]), is able to inhibit the growth of Staphylococcus aureus via IDO-mediated tryptophan depletion and that this inhibition is abolished by an endogenous NO production. Furthermore, we show that long-lasting NO production decreases the level of IDO protein in IFN-γ-stimulated RT4 and human lung carcinoma (A549) cells. This effect depends not on transcriptional but on posttranslational regulation resulting from accelerated proteasomal degradation of IDO. Thus, we describe a previously unknown mechanism of IDO regulation by NO.
MATERIALS AND METHODS
Media, chemicals, and cytokines.
RPMI 1640 medium (BioWhittaker) supplemented with 2 mM l-glutamine and 5% heat-inactivated fetal calf serum was used as the culture medium for all cell lines. All cytokines were obtained from R&D Systems (Wiesbaden, Germany). MG-132, proteasome inhibitor I, clasto-lactacystin β-lactone, cycloheximide, actinomycin D, NG-monomethyl-l-arginine, and 2-amino-5,6-dihydro-6-methyl-4H-1,3-thiazine (AMT) hydrochloride were obtained from Calbiochem (Bad Soden, Germany). All iNOS inhibitors used differed in their specificity to inhibit the inducible iNOS and were all used in concentrations sufficient to mediate a >90% inhibition of nitrite accumulation. Tritium-labeled arginine was obtained from Amersham Pharmacia Biotech (Freiburg, Germany). Sodium nitrite and l-cysteine were supplied by Sigma (Deisenhofen, Germany). Protease inhibitors were obtained from Roche (Mannheim, Germany).
Chemical nitric oxide donors.
Diethylenetriamine diazeniuondiolate (DETA/NO) was a kind gift from K.-D. Kröncke (Research Group Immunobiology, Heinrich Heine University, Düsseldorf, Germany). S-Nitrosoglutathione (GSNO) and S-nitroso-N-acetyl-d,l-penicillamine (SNAP) were obtained from Alexis Biochemicals (Grünberg, Germany). S-Nitroso-l-cysteine (SNOC) was prepared from sodium nitrite and l-cysteine as described by Kröncke and Kolb-Bachofen (25). All donor solutions were prepared immediately before use in experiments. As a control, degassed donor-NO solutions were prepared by incubating a donor solution for 7 days at 37°C. These solutions contained the parent donor compound as well as the stable reaction products of NO with water, nitrite, and nitrate.
Cells and bacteria.
The human uroepithelial carcinoma cell line RT4 was obtained from the American Type Culture Collection (ATCC HTB-2). The human glioblastoma cell line 86HG39 was characterized by immunocytochemical and immunohistological criteria and was a kind gift from T. Bilzer (Institute for Neuropathology, Heinrich Heine University). The human lung carcinoma cell line A549 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany. Cells were grown in culture medium in tissue culture flasks and divided weekly. The viability of the cells used in the experiments was tested with the trypan blue exclusion method; there was no difference in cell viability in differently stimulated cells.
Bacteria were isolated from clinical specimens. They were identified by colony morphology and positive coagulase reaction and in addition confirmed biochemically with a commercial system (API-20staph; bioMerieux, Lyon, France). Staphylococcus aureus was grown on brain heart infusion agar (Difco, Hamburg, Germany) containing 5% sheep blood and incubated at 37°C in 5% CO2-enriched atmosphere. For use in experiments, a 24-h-old single bacterial colony was picked and suspended in RPMI 1640 without l-tryptophan. Bacteria were serially diluted in the same medium, and the numbers of CFU in each dilution were calculated by plating two 10-μl aliquots onto agar plates.
Determination of bacterial growth in cultures of cytokine-treated cells.
RT4 cells were incubated in culture medium in 96-well, flat-bottomed culture plates at 3 × 104 cells/well and stimulated for 3 days with the cytokines indicated. Thereafter, staphylococci were added to RPMI 1640 without l-tryptophan (BioWhittaker). Bacterial growth was monitored after a further incubation of 16 h with a microplate photometer (SLT Labinstruments, Crailsheim, Germany), measuring the optical density at 600 nm.
Determination of IDO activity in cell extracts.
86HG39 cells were stimulated with 300 U of IFN-γ per ml for 24 h. Thereafter, cells were harvested, and the number of living cells was determined by trypan blue exclusion. For each sample, 2 × 106 living cells were resuspended in 200 μl of phosphate-buffered saline containing a protease inhibitor cocktail (2 μg/ml each of leupeptin, aprotinin, and pepstatin, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA) and lysed by three to five cycles of freezing in liquid nitrogen and thawing. After centrifugation (10, 000 × g, 10 min, 4°C), supernatants were used as cell extracts; 200 μl of cell extract was mixed with 200 μl of 2× IDO reaction buffer (100 mM potassium phosphate buffer [pH 6.5], 40 mM ascorbate, 20 μM methylene blue, 800 μM l-tryptophan, 200 μg of catalase per ml) as described by Feng and Taylor (14) and incubated for 30 min at 37°C. Freshly prepared nitric oxide donors were added to the reaction buffer immediately before addition of the cell extract. After incubation at 37°C, 30 μl of 30% trichloroacetic acid was added, followed by further incubation for 30 min at 50°C (hydrolysis of N-formylkynurenine to l-kynurenine). After centrifugation (10, 000 × g, 10 min), the resulting supernatant was used for determination of IDO activity by colorimetric measurement of the l-kynurenine content as previously described (12).
Quantification of IDO mRNA.
RT4 cells were stimulated for 8 h with the indicated combinations of cytokines in 25-cm2 culture flasks. After stimulation, total RNA was prepared according to standard protocols by extraction with guanidinium thiocyanate; 1 μg of total RNA was used for reverse transcription with random hexanucleotide primers and the Advantage reverse transcription-PCR kit (BD Biosciences) following the manufacturer's instructions. The cDNA was used for quantification of IDO mRNA by real-time PCR (TaqMan technique) in 96-well optical plates (Eurogentec, Seraing, Belgium); 5 μl of each cDNA sample was mixed with 25 μl of qPCR Mastermix (Eurogentec), 0.3 μM each primer, 2 μM fluorescently labeled probe, and bidistilled water (total volume, 50 μl). A solution with known molecule numbers of a construct consisting of the full-length IDO cDNA cloned into plasmid pMEP4 was used as a standard. The amplification reaction was carried out in an ABI Prism 5700 sequence detector (Applied Biosystems) with the following cycling program: 50°C for 2 min, 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min. The last two steps represented melting (95°C) and annealing-synthesis (60°C) and were repeated 40 times. For amplification, the following oligonucleotides were used as primers and probe: IDO forward, 5′-CGC CTT GCA CGT CTA GTT CTG; IDO reverse, 5′-CGG ACA TCT CCA TGA CCT TTG; and IDO probe, 5′-(FAM) ATG CAT CAC CAT GGC ATA TGT GTG GG (TAMRA). The primer pair creates an amplicon comprising bases 868 to 938 of IDO mRNA (NCBI accession no. M34455). As an internal standard for each sample, the number of molecules of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was determined with the GAPDH TaqMan endogenous control kit (Eurogentec) according to the instructions of the manufacturer. For presentation, the copy numbers of IDO mRNA were normalized to the copy number of GAPDH mRNA in the corresponding sample.
Determination of iNOS activity in cell extracts.
RT4 cells were stimulated with different combinations of cytokines for various times in 25-cm2 culture flasks. After stimulation, cell extracts were prepared, and the iNOS activity in these extracts was determined with the NOSdetect assay kit (Stratagene) according to the manufacturer's instructions, with minor modifications. Briefly, after harvesting in phosphate-buffered saline and centrifugation, the cells were resuspended in 100 μl of homogenization buffer containing a protease inhibitor cocktail and lysed by cycles. After a further centrifugation (13,000 × g, 5 min, 4°C), the protein concentration in the cell extracts was determined by the Bradford method, and 100 μg of protein was used for the assay. Samples were incubated with reaction buffer and [3H]arginine for 20 min at 37°C. After separation on a column matrix, the content of [3H]citrulline in each sample was measured in a liquid scintillation counter (Beckman LS3801).
IDO degradation experiments.
In experiments testing IDO degradation by endogenous nitric oxide, RT4 cells were stimulated with the cytokines indicated for 8, 24, and 48 h in the presence and absence of the selective iNOS inhibitor AMT hydrochloride. Cell extracts were then prepared by lysing the cells in bidest water containing a protease inhibitor cocktail by freezing and thawing. After centrifugation, the total protein concentration in the supernatant was determined by the Bradford method, and 10 μg was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, proteins were blotted on nitrocellulose membranes by semidry transfer, and IDO and GAPDH proteins were detected with monoclonal antibodies. The IDO monoclonal antibody was a kind gift from O. Takikawa (Department of Pharmacology, Hokkaido University), and the GAPDH monoclonal antibody was from HyTest (Turku, Finland). Chemiluminescent detection was performed with the ECL kit (Apbiotech, Freiburg, Germany) as a substrate, followed by membrane exposure to Kodak X-Omat film.
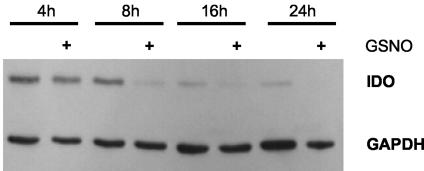
To show the influence of exogenous nitric oxide on IDO protein stability, A549 cells were stimulated with 50 U of IFN-γ per ml for 8 to 16 h. Thereafter the cells were washed with phosphate-buffered saline, and active substances (cycloheximide and proteasome inhibitors) were added to fresh culture medium. One hour later, the NO donor GSNO was added to a final concentration of 750 μM, and the cells were incubated for another 4 to 24 h. Cell extracts were then prepared, and IDO protein was detected as described above.
Computational analysis.
Densitometric analysis was performed with ScionImage software. Statistical analysis was performed with the unpaired t test and GraphPad Prism software.
RESULTS
IDO induction as an antimicrobial effector mechanism.
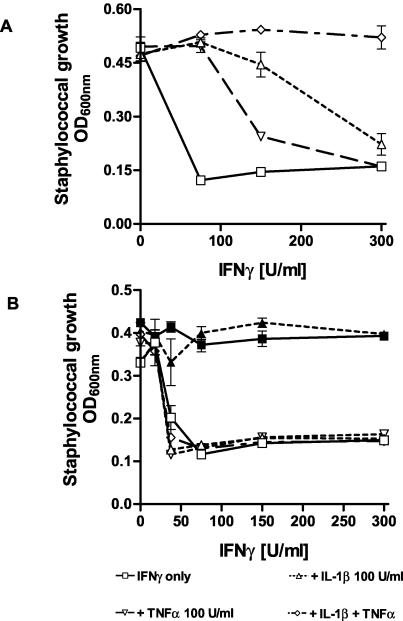
We and others have shown that IDO-mediated tryptophan depletion acts as an antimicrobial effector mechanism against a number of eukaryotic and prokaryotic pathogens. In this paper we analyzed the capacity of IFN-γ-stimulated, IDO-expressing human cells to restrict the growth of S. aureus. S. aureus causes a variety of infections ranging from localized abscesses and faruncles to severe sepsis and toxic shock syndrome. Since it was not known if S. aureus is susceptible to growth inhibition by tryptophan depletion, we tested the ability of the human uroepithelial cell line RT4 to restrict the growth of these bacteria after IFN-γ stimulation. As shown in Fig. 1A, IFN-γ-stimulated RT4 cells were able to inhibit the growth of S. aureus in an in vitro cell culture system. The bacteriostasis was IDO dependent, as shown by the antagonistic effect of l-tryptophan. In contrast, the antibacterial effect was abolished if the RT4 cells were stimulated with IFN-γ and IL-1β and/or TNF-α. The cell line RT4 has been shown to express measurable iNOS activity after stimulation with IFN-γ, IL-1β, and TNF-α, and it was therefore likely that the reduced growth inhibition under these conditions was due to NO inhibition of IDO activity. To confirm this, the same experiment was performed with cells stimulated in the presence of the NOS inhibitor NG-monomethyl-l-arginine. As shown in Fig. 1B, staphylococcal growth was inhibited in all experiments regardless of the cytokine combination used.
FIG. 1.
Inhibitory effect of IL-1β and TNF-α on IFN-γ-induced bacteriostasis in RT4 cells. We stimulated 3 × 104 RT4 cells per well with IFN-γ alone or together with IL-1β and/or TNF-α. After 3 days of incubation, staphylococci were added (50 CFU/well). Bacterial growth was determined 16 h later by measuring the optical density at 600 nm (A). The same experimental procedure was performed in the presence of 100 μg of NG-monomethyl-l-arginine per ml. Data are given as mean optical density (OD) ± standard deviation for triplicate cultures (B). Addition of tryptophan (50 μg/ml) completely blocked IFN-γ induced bacteriostasis (B, solid symbols).
NO inhibits IDO activity in vitro.
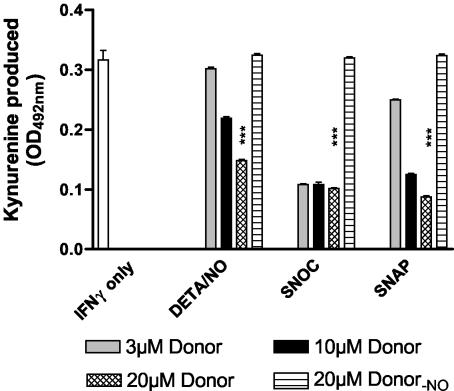
To verify that NO is able to inhibit the activity of IDO enzyme directly, we used a cell-free enzyme assay system to measure the influence of chemical NO donors on the IDO activity in cell extracts of IFN-γ-stimulated 86HG39 cells. This human glioblastoma cell line is unable to produce NO, and thus the possibility of stable NO-mediated inhibitory actions against IDO during stimulation can be excluded. Figure 2 shows that different NO donors were able to inhibit IDO activity in cell extracts, while the degassed forms of the donors had no effect. Thus, the monitored effect was due to the NO produced during test incubation, while the chemical parent compound, which is inevitably formed in the course of donor decomposition, did not influence IDO activity. The inhibitory effect did not depend on the substance class of the NO donor (DETA/NO is a diazenium diolate, and SNOC and SNAP are S-nitroso compounds) but on the concentration and the half-life of the donor. DETA/NO was the most stable NO donor used here (t1/2 of ca. 8 h at 37°C; K.-D. Kröncke, personal communication) and exerted the weakest effect on IDO activity. The unstable compound SNOC (t1/2 of ca. 25 min) exerted the maximum inhibitory effect even at the lowest concentration used, while SNAP (t1/2 of ca. 1 h) was more effective than DETA/NO but less effective than SNOC. Together, these data clearly show that NO is able to inhibit IDO activity in vitro and the level of inhibition depends on the steady-state level of NO.
FIG. 2.
NO inhibits IDO activity in extracts of IFN-γ-stimulated cells. 86HG39 cells were stimulated with 300 U of IFN-γ per ml for 24 h. Per sample, 2 × 106 cells were lysed, and the prepared cell extracts were assayed for IDO activity as described in Materials and Methods. The protein content of each sample was 0.312 μg. The NO donors DETA/NO, SNOC, and SNAP were added to the reaction buffer immediately before mixing with the cell extracts. Donor-NO represents the degassed (non-NO-producing) form of the donor. Results indicate the means ± standard error of the mean for three independent experiments done in duplicate. ***, P < 0.001, unpaired t test.
NO-dependent decrease in IDO protein level.
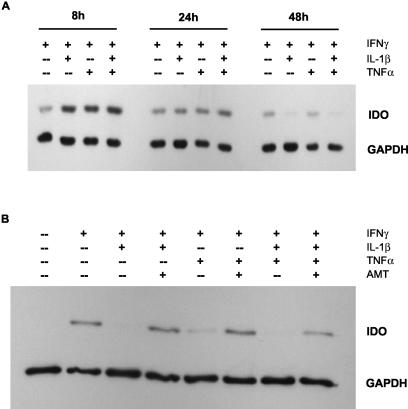
We investigated if NO has an additional effect on IDO protein besides inhibition of enzyme activity. For this purpose, we stimulated RT4 cells for various times with IFN-γ alone (non-NO-producing conditions) and with combinations of IFN-γ and IL-1β and/or TNF-α (NO-producing conditions). Figure 3A shows the results of such an experiment. After 8 h of stimulation, more IDO protein was detected in cells stimulated with cytokine combinations than in cells stimulated with IFN-γ alone. During ongoing stimulation, this effect was reversed; after 24 h of stimulation, the expression levels in cells stimulated with cytokine combinations was reduced to the level in cells stimulated with IFN-γ alone, and after 48 h, IDO protein was almost undetectable under NO-producing conditions. In contrast, the expression level in cells stimulated with IFN-γ alone was more or less constant for 48 h.
FIG. 3.
IDO protein content in RT4 cells decreases in a time- and NO-dependent manner. RT4 cells were stimulated for 8, 24, or 48 h with the indicated cytokines (each at 100 U/ml). Thereafter, cell extracts were prepared, 10 μg of total protein was separated by SDS-PAGE, and IDO protein was detected by Western blotting. As a quantity control, GAPDH was detected simultaneously (A). RT4 cells were stimulated for 48 h with the indicated cytokines (each at 100 U/ml) in the presence or absence of the selective iNOS inhibitor AMT (100 μg/ml). Thereafter, cells were lysed, and 10 μg of total protein was subjected to SDS-PAGE and Western blotting (B).
To test if the decrease in IDO protein was dependent on NO production, we stimulated RT4 cells for 48 h with the same cytokine combinations in the presence and absence of the selective iNOS inhibitor AMT. As shown in Fig. 3B, no IDO protein was detectable in cells stimulated with IFN-γ plus IL-1β and/or TNF-α, while the reduction in detectable protein after costimulation with TNF-α alone was somewhat less, resulting in a weak signal. When the cells were stimulated in the presence of AMT, IDO protein was present in all experiments regardless of the cytokine combination used for stimulation. These data demonstrate that the decrease in IDO protein level was due to the production of NO. We confirmed these results with the chemical NO donors DETA/NO and GSNO, which decreased the level of IDO protein in RT4, 86HG39, and A549 cells within 8 to 16 h of addition (data not shown).
NO does not influence IDO gene transcription in RT4 cells.
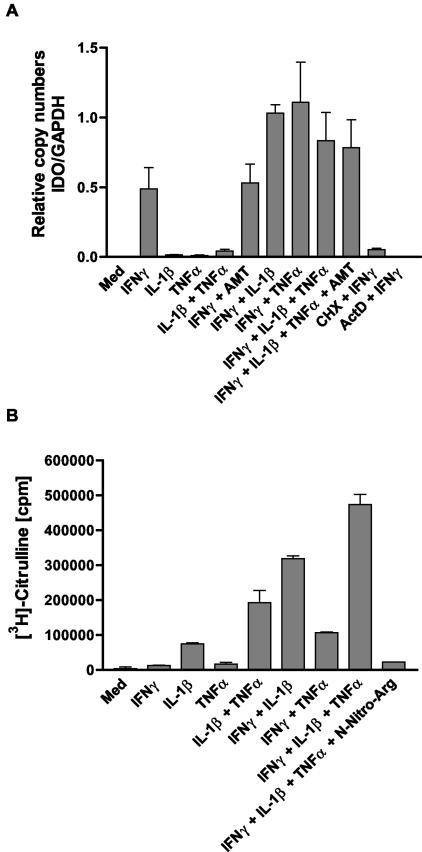
Next we were interested in elucidating the cause of the NO-dependent decrease in intracellular IDO protein. First we measured IDO mRNA in RT4 cells under different stimulation conditions by real-time PCR. Figure 4A shows the results for cells after 8 h of stimulation. We found that TNF-α and IL-1β did not decrease the IDO mRNA level. In contrast, both cytokines slightly increased steady-state IDO mRNA levels compared with the level reached after stimulation with IFN-γ alone. Furthermore, neither TNF-α nor IL-1 was able to induce IDO gene transcription in RT4 cells alone or in combination. Most important, there was no negative regulation of IDO mRNA synthesis under stimulation conditions that induced NO production in RT4 cells.
FIG. 4.
Quantification of IDO mRNA in RT4 cells under different stimulation conditions by real-time PCR. RT4 cells were stimulated with the indicated cytokines (each at 100 U/ml) for 8 h. Thereafter, RNA was extracted, and 1 μg of total RNA was used for reverse transcription. The resulting cDNA was amplified by real-time PCR as described in Materials and Methods. Standard solutions of a plasmid carrying the full-length IDO cDNA were used for quantification. Unstimulated cells and cells treated with cycloheximide (CHX, 10 μg/ml) or actinomycin D (ActD, 5 μg/ml) 1 h before addition of IFN-γ served as controls. The copy numbers of IDO mRNA were normalized to the number of GAPDH mRNA copies in the same probe. The values are the means ± standard error of the mean for the results of three independent experiments (A). There was no significant inhibition of IDO mRNA in cells treated with IFN-γ in the presence of IL-1 and/or TNF-α (P < 0.05, unpaired t test). iNOS activity in cell extracts of differently stimulated RT4 cells stimulated for 8 h with the indicated cytokines (each 100 U/ml) was determined. Cell extracts were prepared, and 100 μg of total protein per sample was assayed for iNOS activity with the NOSdetect assay kit as described in Materials and Methods. Cell extracts from unstimulated cells and samples containing the NOS inhibitor N-nitro-l-arginine (N-Nitro-Arg) served as internal controls as described in Materials and Methods. Data are given as means ± standard error of the mean for two independent experiments done in duplicate (B).
To serve as controls for a negative regulation cells were treated with cycloheximide (CHX) and actinomycin D 1 h before stimulation with IFN-γ. IDO gene transcription is known to depend on de novo protein synthesis, a fact that is normally ascribed to the important role of the transcription factor interferon regulatory factor 1 on IDO gene transcription, which is produced de novo after IFN-γ stimulation. The findings of these experiments correlate well with the Western blot experiments (Fig. 3A), which showed an increase in IDO protein levels after 8 h of stimulation with IFN-γ and/or TNF-α. As shown in Fig. 4B, RT4 cells produced relatively large amounts of NO after 8 h of stimulation with IFN-γ plus IL-1β and/or TNF-α the combination of all three being most potent and IFN-γ plus TNF-α being least effective. Again, these results are consistent with the Western blot studies (Fig. 3A), where the combinations IFN-γ plus IL-1β and IFN-γ plus IL-1β plus TNF-α were more effective in decreasing IDO protein than the combination IFN-γ plus TNF-α. Thus, we may conclude that the degree of decrease in IDO protein correlates quantitatively with NO production.
Although the cells produced large amounts of NO, the amount of IDO mRNA clearly increased under these stimulation conditions compared with stimulation with only IFN-γ. Furthermore, when cells were stimulated with all three cytokines, the IDO mRNA steady-state level did not increase in the presence of the NOS inhibitor AMT. Moreover, Northern blot analysis revealed the same IDO mRNA kinetics over 24 h in cells stimulated with IFN-γ and cells stimulated with IFN-γ plus TNF-α, with the expression level being higher in cells treated with IFN-γ plus TNF-α at each time point examined and the IDO mRNA levels in RT4 cells stimulated with IFN-γ alone or with IFN-γ plus DETA/NO did not differ over 36 h of incubation (data not shown). Together, these data strongly argue against a negative regulation of IDO gene transcription and IDO mRNA stability by NO, as any of these effects should result in a decreased steady-state level of IDO mRNA in NO-producing cells.
NO effect on IDO protein does not depend on de novo protein biosynthesis.
To find out more about the effect of NO on IDO, we tested if the decrease in IDO protein levels required de novo synthesis of protein factors. Previous experiments showed that the decrease in cellular IDO protein can be induced by chemical NO donors at concentrations of 0.5 to 1 mM (data not shown). If de novo synthesis of proteins is required to produce this, incubation of cells with cycloheximide should abolish this effect. For these experiments, we used the human lung carcinoma cell line A549, because RT4 cells exhibited extensive cell death after treatment with cycloheximide. In Fig. 5 A549 cells were stimulated with IFN-γ and then incubated with 10 μg of cycloheximide per ml, a concentration which proved to reduce protein biosynthesis in these cells by 90% without causing cell death (data not shown). One hour later, the NO donor GSNO was added to a final concentration of 750 μM, and the cells were incubated for another 4 to 24 h. Under these conditions, the level of IDO protein decreased much sooner in cells treated with GSNO than in cells treated with cycloheximide alone. The difference in IDO protein levels was most prominent after 8 h of incubation. From these data, we conclude that the decrease in cellular IDO protein content is strictly NO dependent, as it could be induced by endogenously produced (Fig. 3) and exogenously administered NO, and that de novo protein synthesis is not required for this effect. Thus, NO seems to promote the degradation of IDO protein rather than inhibit its synthesis.
FIG. 5.
Effect of NO on IDO protein is not cycloheximide sensitive. A549 cells were stimulated with 50 U of IFN-γ per ml for 16 h. Thereafter, the cells were washed, and 10 μg of cycloheximide per ml was added in fresh culture medium. One hour later, the NO donor GSNO was added to a final concentration of 750 μM to some cells, and the cells were incubated for a further 4 to 24 h. Thereafter, cell extracts were prepared, and 10 μg of total protein was separated by SDS-PAGE. After blotting, IDO and GAPDH protein levels were determined.
NO induces accelerated degradation of IDO in the proteasome.
Though the mRNA experiments showed that NO did not influence the transcription of the IDO gene, we further analyzed the possibility that an NO-dependent effect takes place at the posttranscriptional level. A scenario that could explain the decrease in IDO protein was accelerated degradation of the protein; 80 to 90% of all proteins in mammalian cells are degraded by the proteasome, a multicatalytic protease complex. However, to date there have been no studies showing that IDO is a substrate for the proteasome. Therefore, we investigated the influence of proteasome inhibitors on NO-dependent and -independent IDO degradation. Once again, A549 cells were used for this set of experiments because prolonged exposure to proteasome inhibitors proved to be toxic for RT4 cells.
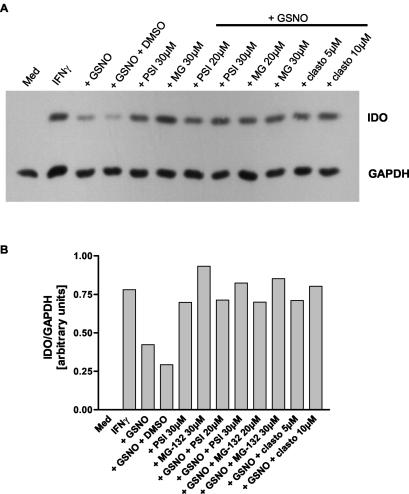
Figure 6A shows a representative experiment in which A549 cells were stimulated with IFN-γ and then incubated with the NO donor GSNO and the proteasome inhibitors MG-132, proteasome inhibitor I, and clasto-lactacystin β-lactone. Figure 6B shows the densitometric analysis of the signals seen in Fig. 6A. All proteasome inhibitors used were able to abolish the decrease in IDO protein level induced by GSNO. In the presence of the proteasome inhibitors, the IDO protein level reached about 90% (proteasome inhibitor I, 30 μM) to 109% (MG-132, 30 μM) of that in cells stimulated with IFN-γ alone. Since all inhibitors were dissolved in dimethyl sulfoxide, a sample in which cells were incubated with GSNO and dimethyl sulfoxide (0.5% final concentration) was used as a solvent control and showed that dimethyl sulfoxide itself did not have an effect on IDO protein level (lane 4). Moreover, in cells that were incubated (after stimulation with IFN-γ) with the proteasome inhibitor MG-132 in the absence of GSNO (lane 6), the IDO protein content increased to a level slightly higher than the level reached after incubation with IFN-γ alone (lane 2). These data suggest that IDO protein is degraded by the proteasome under “normal” and NO-producing conditions and that the NO-dependent effect on IDO protein can be ascribed to an increased rate of degradation. Further experiments revealed that the total protein content in RT4 cells was not decreased by NO (data not shown), and thus the monitored effect is specific for IDO and a group of proteins that include IDO.
FIG. 6.
Proteasome inhibitors abolish NO-dependent IDO degradation. A549 cells were stimulated with 50 U of IFN-γ per ml for 9 h. Thereafter, the cells were washed, and the proteasome inhibitors MG-132, proteasome inhibitor I (PSI), and clasto-lactacystin β-lactone (clasto) were added at the indicated final concentrations to fresh culture medium. The addition of dimethyl sulfoxide to a final concentration of 0.5% served as a solvent control (GSNO plus DMSO, lane 4), and unstimulated cells as a negative control (Med, lane 1). One hour later, the NO donor GSNO was added to a final concentration of 750 μM, and the cells were further incubated for 16 h. IDO and GAPDH protein levels were determined by Western blotting (A). The signal intensity of the IDO bands was measured by densitometry and normalized to the intensity of the corresponding GAPDH signal. The results shown are representative of one of three independent experiments (B).
DISCUSSION
IFN-γ-induced tryptophan depletion has been described as an antimicrobial effector mechanism in different in vitro culture systems. No in vivo models to analyze IDO-mediated effects in vivo are currently available. However, in the case of infection with S. aureus, several facts suggest that IDO might play a role in the defense against this pathogen. First, many cells found in the tissue surrounding a staphylococcal abscess are capable of expressing IDO activity after stimulation with IFN-γ (36). Second, many strains of S. aureus are capable of producing superantigens, which induce strong T-cell activation, and activated T cells produce large amounts of IFN-γ (15). In addition, several in vivo studies have shown that after treatment of humans with interferons and during infections known to induce strong IFN-γ production, the concentration of tryptophan is reduced and the kynurenine content is increased in several body fluids, e.g., serum and cerebrospinal fluid (5, 21). Furthermore, the influence of IDO activation in vivo on T-cell stimulation in animal models has been well described (33). Therefore, we suggest that the in vitro IDO-mediated antimicrobial effects described in this report might also be active in vivo.
Tryptophan depletion and NO production are both known to have an antimicrobial effect. For a long time it was thought that these two mechanisms were species specific, the microbicidal NO restricted to rodent cells and IDO-mediated tryptophan depletion occurring only in humans. Recently, it has become clear that the situation is far more complex in that both mechanisms interact in various ways.
The first experiments showing the antimicrobial potential of IFN-γ-induced tryptophan degradation were performed with the obligate intracellular parasite Toxoplasma gondii (36) and the intracellular bacterium Chlamydia psittaci (6). Later, we and others demonstrated that this effector mechanism is also able to inhibit the growth of extracellular bacteria. We showed that the human uroepithelial cell line RT4 restricted the growth of enterococci in an IDO-dependent manner and that this bacteriostatic effect was abolished by NO. In contrast, NO-producing murine macrophages (RAW 264.7 cells) were not able to inhibit enterococcal growth in a cell culture system (9). In this paper, we show growth inhibition experiments with S. aureus as the infectious agent. The bacteriostatic effect was abolished under conditions (IFN-γ plus IL-1β) that caused the RT4 cells to induce both IDO and iNOS and was reversed by an NOS inhibitor. These data led us to the conclusion that NO inhibits the IDO activity in RT4 cells, resulting in uninhibited staphylococcal growth.
NO reacts readily with heme moieties, and this reaction may result in activation or inhibition of the affected enzyme (reviewed in reference 8). The first findings demonstrating that NO is able to inhibit the IDO came from Thomas et al., who found that incubation of cell extracts or intact cells of IFN-γ-stimulated human monocyte-derived macrophages with authentic NO gas or chemical NO donors resulted in a decrease in IDO activity and that this effect could be reversed by addition of oxygenated hemoglobin, which reacts with NO to form Fe3+-hemoglobin and nitrate at diffusion-controlled rates (38). Moreover, if murine peritoneal macrophages were stimulated with IFN-γ and lipopolysaccharide in the presence of an NOS inhibitor, these cells expressed measurable IDO activity.
With regard to these findings, we were interested to confirm our own results gained with intact cells in culture systems by testing the effect of NO donors on IDO activity in extracts of IFN-γ-stimulated cells. Three NO donors belonging to different classes were all able to inhibit IDO activity in an ascorbate-methylene blue assay. The degree of inhibition correlated with the steady-state concentration of NO, which is dependent on both the half-life and the concentration of the NO donor. Additionally, we have previously shown that the IDO activity in 86HG39 cells could be inhibited in the presence of IFN-γ-stimulated murine macrophages (9), and so, taken together, the data obtained by us and others clearly demonstrate that human (and murine) IDO is inhibited by endogenous and exogenous NO. Thomas et al. suggested that the binding of NO to the heme moiety of IDO is responsible for enzyme inhibition. Though there are no reports showing direct binding of NO to IDO, we also believe that this is the most likely scenario.
Nevertheless, complete inhibition of IDO activity could not be achieved in any of the experiments with the ascorbate-methylene blue assay, and we therefore looked at the effect of NO on IDO expression. The results presented showed that IDO gene transcription is increased under conditions in which iNOS activity in RT4 cells is stimulated, but IDO protein content is dramatically decreased. All the evidence points to increased IDO protein degradation in the proteasome in an NO-dependent manner.
These results are in contrast to those reported by Alberati-Giani et al., who demonstrated that NO and picolinic acid, a tryptophan metabolite that increases IFN-γ-induced NO production, decreased IDO mRNA levels in an immortalized murine thymic macrophage cell line (1). Further research will be needed to clarify if this is a cell type- or species-specific effect. Although NO is known to modulate gene transcription events by interfering with signal transduction (3) and inhibition of transcription factors (24, 30), human IDO gene expression does not seem to be regulated by NO. Nevertheless, IDO gene expression is under tight immunological control through cytokines, with IFN-γ being the main inducer and IL-1β and lipopolysaccharide (22), TNF-α (11), transforming growth factor beta (39), IL-4 (34), and IL-13 (7) all modulating the rate of gene transcription.
Our finding that NO accelerates the proteasomal degradation of IDO marks a new feature of the IDO-NO interaction and further promotes recent reports that investigated the relatively new field of cellular NO action. It is becoming clear that NO modulates the stability of several proteins in human and rodent cells. The first report came from Kim and Ponka, who investigated iron metabolism in murine macrophages and showed that the iron regulatory protein 2 was degraded after stimulation of the cells with IFN-γ/lipopolysaccharide and incubation with the NO donor sodium nitroprusside (23). In 2002, Franzen et al. found NO-induced degradation of neutral ceramidase in rat mesangial cells (16), and Bebok et al. showed a decrease in cystic fibrosis transmembrane conductance regulator protein in human airway epithelial cells after exposure to NO (2). Thus, to the best of our knowledge, IDO is the fourth protein that is destabilized by NO to be described.
To date, the mechanism by which NO influences protein stability is not known. Clearly, the affected proteins must somehow be marked for faster degradation. NO activates soluble guanylate cyclase (reviewed in reference 13) and Ras protein (27), so it would appear that a signal transduction process is required in the NO-mediated enhancement of IDO degradation. NO might also act directly on IDO (or different proteins which subsequently induce IDO degradation) via S-nitrosylation of reduced cysteine residues, but any thoughts regarding this problem must remain speculative because at present there are no data concerning the interactions of IDO with other cellular proteins, e.g., protein kinases and ubiquitin protein ligases. It would be tempting to speculate that S-nitrosylation works as a degradation signal. The IDO protein contains eight cysteine residues (NCBI accession no. AAA36081), but according to the primary sequence, none of them matches the criteria that could be used to predict an S-nitrosylation event, i.e., the direct vicinity of an acid and a basic residue (reviewed in reference 20). Nevertheless these motifs may result from protein folding, and therefore S-nitrosylation, as a prerequisite for accelerated degradation, cannot be excluded. Further research will be required to shed light on these problems.
Taken together, our data clearly show that NO exerts at least two effects on the IDO protein in human epithelial cells, direct inhibition occurring immediately after exposure of the enzyme to NO, and accelerated induction of degradation by the proteasome, a process which requires longer incubation of cells with NO. Given that both effects also occur in murine cells (which produce much more NO in the course of an inflammation reaction), these results provide further insight into species-specific differences in cellular reactivity to cytokine stimulation between mice and humans. Nevertheless, both tryptophan depletion and NO production contribute to pathogen clearance in both species. For example, IDO activity seems to be at least partly responsible for the reduction of pathogen load in the mouse lung in the case of infections with T. gondii (18) and Mycobacterium avium (19). On the other hand, human pulmonary macrophages are only able to kill Mycobacterium bovis if they express iNOS (35). Another common feature of tryptophan depletion and NO production besides microbial killing is the ability to inhibit T-cell proliferation (3, 32). In this respect, NO appears to interfere with IL-2 signal transduction, and the IDO-dependent mechanism seems to be conferred through tryptophan metabolites (17, 37). Thus, examination of the fine tuning of the regulation and interaction of these antimicrobial and immunoregulatory mechanisms in vivo will be an interesting task for future research.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm Infektionen des Endothels).
We thank K.-D. Kröncke (Research Group Immunobiology, Heinrich Heine University) for providing the DETA/NO.
Editor: F. C. Fang
REFERENCES
- 1.Alberati-Giani, D., P. Malherbe, P. Ricciardi-Castagnoli, C. Köhler, S. Denis-Donini, and A. M. Cesura. 1997. Differential regulation of indoleamine-2,3-dioxygenase expression by nitric oxide and inflammatory mediators in IFN-γ-activated murine macrophages and microglial cells. J. Immunol. 159:419-426. [PubMed] [Google Scholar]
- 2.Bebok, Z., K. Varga, J. K. Hicks, C. J. Venglarik, T. Kovacs, L. Chen, K. M. Hardiman, J. F. Collawn, E. J. Sorscher, and S. Matalon. 2002. Reactive oxygen nitrogen species decrease cystic fibrosis transmembrane conductance regulator expression and cAMP-mediated Cl− secretion in airway epithelia. J. Biol. Chem. 277:43041-43049. [DOI] [PubMed] [Google Scholar]
- 3.Bingisser, R. M., T. A. Tilbrook, P. G. Holt, and U. R. Kees. 1998. Macrophage-derived nitric oxide regulates T cell activation via reversible disruption of the Jak3/STAT5 signaling pathway. J. Immunol. 160:5729-5734. [PubMed] [Google Scholar]
- 4.Bogdan, C., M. Röllinghoff, and A. Diefenbach. 2000. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr. Opin. Immunol. 12:64-76. [DOI] [PubMed] [Google Scholar]
- 5.Byrne, G. I., L. K. Lehmann, J. G. Kirschbaum, E. C. Borden, C. M. Lee, and R. R. Brown. 1986. Induction of tryptophan degradation in vitro and in vivo: a g-Interferon-stimulated activity. J. Interferon Res. 6:389-396. [DOI] [PubMed] [Google Scholar]
- 6.Byrne, G. I., L. K. Lehmann, and G. J. Landry. 1986. Induction of tryptophan catabolism is the mechanism for gamma interferon mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect. Immun. 53:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaves, A. C. L., I. P. Ceravolo, J. A. S. Gomes, C. L. Zani, A. J. Romanha, and R. T. Gazzinelli. 2001. IL-4 and IL-13 regulate the induction of indoleamine 2,3-dioxygenase activity and the control of Toxoplasma gondii replication in human fibroblasts activated with IFN-γ. Eur. J. Immunol. 31:333-344. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, C. E. 1999. Nitric oxide and iron proteins. Biochim. Biophys. Acta. 1411:290-309. [DOI] [PubMed] [Google Scholar]
- 9.Däubener, W., C. Hucke, K. Seidel, U. Hadding, and C. R. MacKenzie. 1999. Interleukin-1 inhibits gamma interferon-induced bacteriostasis in human uroepithelial cells. Infect. Immun. 67:5615-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Däubener, W., K. Pilz, P. Seghrouchi, T. Bilzer, H. G. Fischer, and U. Hadding. 1993. Induction of toxoplasmostasis in a human glioblastoma by interferon gamma. J. Neuroimmunol. 43:32-38. [DOI] [PubMed] [Google Scholar]
- 11. Däubener, W., C. Remscheid, S. Nockemann, K. Pilz, P. Seghrouchi, C. R. MacKenzie, and U. Hadding. 1996. Anti-parasitic effector mechanisms in human brain tumor cells: role of interferon-γ and tumor necrosis factor-α. Eur. J. Immunol. 26:487-492. [DOI] [PubMed] [Google Scholar]
- 12.Däubener, W., N. Wanagat, K. Pilz, S. Seghrouchni, H. G. Fischer, and U. Hadding. 1994. A new, simple bioassay for human IFN-γ. J. Immunol. Methods 168:39-47. [DOI] [PubMed] [Google Scholar]
- 13.Denninger, J. W., and M. A. Marletta. 1999. Guanylate cyclase and the NO/cGMP signaling pathway. Biochim. Biophys. Acta 1411:334-350. [DOI] [PubMed] [Google Scholar]
- 14.Feng, G. S., and M. W. Taylor. 1989. Interferon gamma-resistant mutants are defective in the induction of indoleamine 2,3-dioxygenase. Proc. Natl. Acad. Sci. USA 86:7144-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleischer, B. amd H. Schrezenmeier. 1988. T cell stimulation by staphylococcal enterotoxins. Clonally variable response and requirement for major histocompatibility complex class II molecules on accessory or target cells. J. Exp. Med. 167:1697-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franzen, R., D. Fabbro, A. Aschrafi, J. Pfeilschifter, and A. Huwiler. 2002. Nitric oxide induces degradation of the neutral ceramidase in rat renal mesangial cells and is counter regulated by protein kinase C. J. Biol. Chem. 277:46184-46190. [DOI] [PubMed] [Google Scholar]
- 17.Frumento, G., R. Rotondo, M. Tonetti, G. Damonte, U. Benatti, and G. B. Ferrara. 2002. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J. Exp. Med. 196:459-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujigaki, S., K. Saito, M. Takemura, N. Maekawa, Y. Yamada, H. Wada, and M. Seishima. 2002. l-Tryptophan-l-kynurenine pathway metabolism accelerated by Toxoplasma gondii infection is abolished in gamma interferon-gene-deficient mice: cross-regulation between inducible nitric oxide synthase and indoleamine 2,3-dioxygenase. Infect. Immun. 70:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi, T., S. P. Rao, K. Takabayashi, J. H. Van Uden, R. S. Kornbluth, S. M. Baird, M. W. Taylor, D. A. Carson, A. Catanzaro, and E. Raz. 2001. Enhancement of innate immunity against Mycobacterium avium infection by immunostimulatory DNA is mediated by indoleamine 2,3-dioxygenase. Infect. Immun. 69:6156-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess, D. T., A. Matsumoto, R. Nudelman, and J. S. Stamler. 2001. S-nitrosylation: spectrum and specifity. Nat. Cell Biol. 3:E1-E3. [DOI] [PubMed] [Google Scholar]
- 21.Heyes, M. P., B. J. Brew, K. Saito, B. J. Quearry, R. W. Price, K. Lee, R. B. Bhalla, M. Der, and S. P. Markey. 1992. Inter-relationships between quinolonic acid, neuroactive kynurenines, neopterin and β2-microglobulin in cerebrospinal fluid and serum of HIV-1-infected patients. J. Neuroimmunol. 40:71-80. [DOI] [PubMed] [Google Scholar]
- 22.Hissong, B. D., and J. M. Carlin. 1997. Potentiation of interferon-induced indoleamine 2,3-dioxygenase mRNA in human mononuclear phagocytes by lipopolysaccharide and interleukin-1. J. Interferon Cytokine Res. 17:387-393. [DOI] [PubMed] [Google Scholar]
- 23.Kim, S., and P. Ponka. 1999. Control of transferrin receptor expression via nitric oxide-mediated modulation of iron-regulatory protein 2. J. Biol. Chem. 274:33035-33042. [DOI] [PubMed] [Google Scholar]
- 24.Kröncke, K.-D., and C. Carlberg. 2000. Inactivation of zinc finger transcription factors provides a mechanism for a gene-regulatory role of nitric oxide. FASEB J. 13:166-173. [DOI] [PubMed] [Google Scholar]
- 25.Kröncke, K.-D., and V. Kolb-Bachofen. 1996. Detection of nitric oxide interaction with zinc finger proteins. Methods Enzymol. 269:279-284. [DOI] [PubMed] [Google Scholar]
- 26.Kröncke, K.-D., C. V. Suschek, and V. Kolb-Bachofen. 2000. Implications of inducible nitric oxide synthase expression and enzyme activity. Antiox. Redox Signal. 2:585-605. [DOI] [PubMed] [Google Scholar]
- 27.Lander, H. M., J. S. Ogiste, S. F. A. Pearce, R. Levi, and A. Novogrodsky. 1995. Nitric oxide-stimulated guanine nucleotide exchange on p21ras. J. Biol. Chem. 270:7017-7020. [DOI] [PubMed] [Google Scholar]
- 28.MacKenzie, C. R., U. Hadding, and W. Däubener. 1998. Interferon-γ-induced activation of indoleamine 2,3-dioxygenase in cord blood monocyte-derived macrophages inhibits the growth of group B streptococci. J. Infect. Dis. 178:875-878. [DOI] [PubMed] [Google Scholar]
- 29.MacKenzie, C. R., C. Hucke, D. Müller, K. Seidel, O. Takikawa, and W. Däubener. 1999. Growth inhibition of multiresistant enterococci by interferon-γ-activated human uro-epithelial cells. J. Med. Microbiol. 48:935-941. [DOI] [PubMed] [Google Scholar]
- 30.Marshall, H. E., and J. S. Stamler. 2001. Inhibition of NF-κB by S-nitrosylation. Biochemistry 40:1688-1693. [DOI] [PubMed] [Google Scholar]
- 31.Mellor, A. L., D. B. Keskin, T. Johnson, P. Chandler, and D. H. Munn. 2002. Cells expressing indoleamine 2,3-dioxygenase inhibit T cell responses. J. Immunol. 168:3771-3776. [DOI] [PubMed] [Google Scholar]
- 32.Munn, D. H., E. Shafizadeh, J. T. Attwood, I. Bondarev, A. Pashine, and A. L. Mellor. 1999. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 189:1363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munn, D. H., M. Zhou, J. T. Attwood, I. Bondarev, S. J. Conway, B. Marshall, C. Brown, and A. L. Mellor. 1998. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 281:1191-1193. [DOI] [PubMed] [Google Scholar]
- 34.Musso, T., G. L. Gusella, A. Brooks, D. L. Longo, and L. Varesio. 1994. Interleukin-4 inhibits indoleamine 2,3-dioxygenase expression in human monocytes. Blood 83:1408-1411. [PubMed] [Google Scholar]
- 35.Nozaki, Y., Y. Hasegawa, S. Ichiyama, I. Nakashima, and K. Shimokata. 1997. Mechanism of nitric oxide-dependent killing of Mycobacterium bovis BCG in human alveolar macrophages. Infect. Immun. 65:3644-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfefferkorn, E. R. 1984. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc. Natl. Acad. Sci. USA 81:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terness, P., T. M. Bauer, L. Röse, C. Dufter, A. Watzlik, H. Simon, and G. Opelz. 2002. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. J. Exp. Med. 196:447-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas, S. R., D. Mohr, and R. Stocker. 1994. Nitric oxide inhibits indoleamine 2,3-dioxygenase activity in Interferon-γ-primed mononuclear phagocytes. J. Biol. Chem. 269:14457-14464. [PubMed] [Google Scholar]
- 39.Yuan, W., A. Collado-Hidalgo, T. Yufit, M. Taylor, and J. Varga. 1998. Modulation of cellular tryptophan metabolism in human fibroblasts by transforming growth factor-β: selective inhibition of indoleamine 2,3-dioxygenase and tryptophanyl-tRNA synthetase gene expression. J. Cell Physiol. 177:174-186. [DOI] [PubMed] [Google Scholar]