Abstract
Escherichia coli strain LF82 recovered from a chronic lesion of a patient with Crohn's disease (CD) is able to adhere to and invade cultured intestinal epithelial cells and to replicate within macrophages. One mutant selected for its impaired ability to invade epithelial cells had an insertion of a Tn phoA transposon within the nlpI gene encoding the lipoprotein NlpI. A NlpI-negative isogenic mutant showed a 35-fold decrease in its ability to adhere to and a 45-fold decrease in its ability to invade Intestine-407 cells, but its ability to survive and to replicate within macrophages was similar to that of wild-type strain LF82. In addition, this mutant did not express flagella and synthesized very small amounts of type 1 pili. Downregulation of type 1 pili in the NlpI-negative mutant resulted from a preferential switch toward the OFF position of the invertible DNA element located upstream of the fim operon. The FimB and FimE recombinases act in concert to control the switch, and a large decrease in fimB and fimE mRNA levels was observed. The absence of flagellar structures correlated with a drastic 19-fold decrease in the fliC mRNA level, regardless of the FlhD2C2 transcriptional regulator and of the σ28 transcription factor. The key role of NlpI in virulence is independent of type 1 pili and motility, since induced type 1 pilus expression and/or forced contact between bacteria and intestinal epithelial cells did not restore the ability of the NlpI mutant to adhere to and to invade intestinal epithelial cells.
Crohn's disease (CD) is an inflammatory bowel disease characterized by chronic transmural, segmental, and granulomatous inflammation of the intestine in humans (10). CD has features that might be the result of a microbial process in the gut. Various studies have addressed the hypothesis that pathogenic bacteria contribute to the pathogenesis of inflammatory bowel disease (4, 22, 23, 33, 36). Bacterial adhesion to intestinal epithelial cells represents the first step in the pathogenicity of many organisms involved in infectious bowel diseases since it allows the bacteria to colonize the gut mucosa and resist mechanical removal from the intestine. The ileal mucosa of patients with CD is abnormally colonized with adherent Escherichia coli strains, which suggests that these bacteria are involved in the initiation of the inflammatory process (7).
Colonization of host tissues is usually mediated by adhesins on the surface of the bacteria, responsible for binding to specific receptor moieties of the host cell. Enterobacteria elaborate either adhesive filamentous appendages from their surface, termed fimbriae or pili (such as type 1 pili, pap pili, or type IV pili), or nonfimbrial adhesins (for a review, see references 34 and 39). We previously reported that type 1 pili play an important role in the interaction between E. coli isolated from patients with CD and intestinal epithelial cells (2). However, other structures expressed on the bacterial surface could play a direct or indirect role in the adhesion process. For example, flagella allow some microorganisms to adhere to epithelial cells via active motility, in particular enteropathogenic E. coli (12) and Salmonella enterica (35). In addition, outer membrane proteins, such as OmpA, promote E. coli K1 adhesion to and invasion of brain microvascular endothelial cells (29). Finally, bacterial lipoproteins are also involved in adhesion to and invasion of epithelial cells (42). For example, the surface-exposed lipoprotein JlpA specific to Campylobacter jejuni mediates adherence to HEp-2 epithelial cells and Lsp, which belongs to the LraI lipoprotein family, is involved in Streptococcus pyogenes adhesion to and invasion of A549 cells (11, 16).
E. coli strain LF82, isolated from a chronic ileal lesion of a patient with CD, belongs to a new pathogenic group of invasive E. coli strains mainly associated with CD and designated AIEC (for “adherent invasive E. coli”) (3). It is a true invasive pathogen since its uptake by HEp-2 epithelial cells and Intestine-407 intestinal epithelial cells is dependent on actin microfilaments and microtubules, it survives intracellularly, and it replicates in the host cell cytoplasm after lysis of endocytic vacuoles (3). However, strain LF82 has none of the invasive determinants of Salmonella, Shigella, and invasive E. coli strains, which are known to be involved in gastrointestinal infections. In addition, E. coli strain LF82 is able to survive and/or replicate intracellularly within J774-A1 macrophage cells, murine peritoneal macrophages, and human monocyte-derived macrophages (13). Type 1 pilus-mediated adherence plays an essential part in the invasive ability of strain LF82 by inducing membrane extensions, which surround the bacteria at the sites of contact between the entering bacteria and the epithelial cells (2). However, type 1 pili have to be expressed in the genetic background of strain LF82 to promote bacterial uptake since their expression in E. coli K-12 is not sufficient to confer invasiveness. We recently showed that flagella also play a direct role in the adhesion process via active motility and an indirect role in the interaction between bacteria and epithelial cells by downregulating the expression of type 1 pili (1). Finally, a study of the ability of the LF82-ΔfliC isogenic mutant to adhere to and invade intestinal epithelial cells showed that another factor, in addition to flagella and type 1 pili, is involved in adherence and invasion of strain LF82 (1).
To identify new genetic determinants involved in the ability of strain LF82 to adhere and/or invade, we screened 5,329 Tn5 phoA mutants for a decreased ability to interact with HEp-2 epithelial cells. Sixteen mutants were identified that showed a decreased invasion and/or adhesion to HEp-2 cells, and analysis of transposon insertion sites revealed mutated genes associated with type 1 pilus biogenesis, genes encoding a putative outer membrane protein and a putative serine-threonine protein kinase, and the nlpI gene encoding the NlpI lipoprotein (2). The present study focuses on the NlpI lipoprotein. We showed that NlpI, for which no biological role has been described, is a key component of AIEC strain LF82 virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cell lines.
Strain LF82 was isolated from a chronic ileal lesion of a patient with CD and belongs to E. coli serotype O83:H1. It is sensitive to most antibiotics but not to amoxicillin. It adhered to and invaded HEp-2, Intestine-407, and Caco-2 cells (3). E. coli strain SM10 (λpir) was used for Tn phoA mutagenesis as a source of the mini-Tn5 phoA donor plasmid pUTKml (9). E. coli strains JM109 and C600 were used as host strains for cloning experiments. The bacterial strains, plasmids, and cosmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and cosmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| LF82 | E. coli isolated from an ileal biopsy specimen from a CD patient | 7 |
| LF82-ΔnlpI | LF82 isogenic mutant with nlpI deleted | This study |
| LF82-ΔfliC | LF82 isogenic mutant with fliC deleted | 1 |
| Plasmids and cosmids | ||
| pUC18 | E. coli cloning vector, oriColE1, ampicillin resistant | Biolabs |
| pHSG575 | E. coli cloning vector, chloramphenicol resistant | Biolabs |
| pKOBEG | pBAD cloning vector harboring λ phage redγβα operon | 5 |
| pHC79 | E. coli cloning cosmid, ampicillin resistant | 6 |
| pPBI01 | pHSG575 harboring the 11.2-kb SalI fragment with the entire fim operon of E. coli K-12 strain J96 | 2 |
| pPBI02 | pUC18 containing the 1.3-kb PstI fragment with the complete nlpI gene of LF82 | This study |
| pPBI03 | pHSG575 containing the 1.3-kb PstI fragment with the complete nlpI gene of LF82 | This study |
| Cosmid 14C11 | Cosmid pHC79 harboring the entire nlpI gene of LF82 | This study |
Plasmid vectors pUC18 and pHSG575 and cosmid vector pHC79 (6) were used in cloning procedures. Bacteria were grown routinely in Luria-Bertani (LB) broth or on LB agar plates (Institut Pasteur Production) overnight at 37°C. Antibiotics were added to the media at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 25 μg/ml.
Intestine-407 cells (derived from human intestinal embryonic jejunum and ileum) were purchased from Flow Laboratories Inc., McLean, Va. Cultured cells were maintained in an atmosphere containing 5% CO2 at 37°C in modified Eagle medium (Seromed; Biochrom KG, Berlin, Germany) supplemented with 10% (vol/vol) fetal calf serum (FCS) (Seromed), 1% nonessential amino acids (Life Technologies, Cergy-Pontoise, France), 1% l-glutamine (Life Technologies), 200 U of penicillin, 50 mg of streptomycin, 0.25 mg of amphoterocin B per liter, and 1% minimal essential medium (MEM) vitamin mix X-100 (Life Technologies).
The murine macrophage-like cell line J774-A1 (American Type Culture Collection no. TIB67) was maintained in an atmosphere containing 5% CO2 at 37°C in RPMI 1640 medium (Seromed) supplemented with 10% (vol/vol) FCS, 1% nonessential amino acids, 1% l-glutamine, and 1% MEM vitamin mix X-100.
Assays of adhesion to and invasion of Intestine-407 cells and bacterial survival and replication within macrophages.
The assays of bacterial invasion of Intestine-407 epithelial cells and bacterial survival and replication within macrophages were performed using the gentamicin protection assay as described previously (3, 13). Briefly, monolayers were seeded in 24-well tissue culture plates (Polylabo, Strasbourg, France) with 4 × 105 cells per well for Intestine-407 cells and 2 × 105 cells per well for J774-A1 macrophages and incubated for 20 h. Monolayers were then infected in 1 ml of the cell culture medium without antibiotics and with heat-inactivated FCS at a multiplicity of infection of 10 bacteria per cell for adhesion and invasion assays and 100 bacteria per cell for bacterial uptake, survival, and replication within macrophages.
For assay of adhesion to Intestine-407 cells, after a 3-h incubation period at 37°C, monolayers were washed five times in phosphate-buffered saline (PBS; pH 7.2). The epithelial cells were then lysed with 1% Triton X-100 (Sigma Chemical Co., St. Louis, Mo.) in deionized water. Samples were diluted and plated onto Muller-Hinton agar plates to determine the number of CFU corresponding to the total number of cell-associated bacteria (adherent and intracellular bacteria). To determine the number of intracellular bacteria, fresh cell culture medium containing 100 μg of gentamicin per ml (Sigma) was added for 1 h to kill extracellular bacteria. The monolayers were then lysed with 1% Triton X-100, and the bacteria were quantified as described above. When needed, adhesion and invasion assays were performed after centrifugation for 8 min at 1,000 × g.
For bacterial survival and replication within macrophages, infected monolayers were centrifuged at 1,000 × g for 10 min at 20°C and then incubated for 10 min at 37°C. The monolayers were washed twice in PBS, and the number of intracellular bacteria were determined after a 1-h and a 24-h incubation period at 37°C in fresh cell culture medium containing 20 μg of gentamicin per ml.
Colony and Western immunoblotting.
The rabbit antiserum raised against purified type 1 pilus preparations was a generous gift from Karen Krogfelt (18). Bacteria were grown overnight at 37°C in LB broth without agitation. A 1-ml volume of culture was centrifuged, and the pellet of bacteria was suspended in 100 μl of PBS. A 5-μl sample was spotted onto nitrocellulose membranes (Amersham International). The membranes were blocked with 2% (wt/vol) bovine serum albumin (Sigma) in Tris-buffered saline-0.05% Tween (TBST) at room temperature for 2 h. The membranes were incubated with the type 1 pilus antiserum diluted in 1% (wt/vol.) bovine serum albumin in TBST at room temperature for 2 h. Immunoreactants were then detected using a secondary anti-rabbit antibody conjugated with alkaline phosphatase. A substrate composed of 5-bromo-4-chloro-3-indolyl phosphate and 4-nitroblue tetraolium chloride (Sigma) in a detection buffer (100 mM Tris, 100 mM NaCl, 5 mM MgCl2 [pH 9.5]) was used to visualize reaction products.
Western immunoblotting using E. coli antiserum H1 (a generous gift from the World Health Organization International Escherichia coli Centre) was performed by the procedure of Towbin et al. (40) with minor modifications. Bacterial in stationary growth were harvested, suspended in sodium dodecyl sulfate sample buffer, and heated for 5 min. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 12% polyacrylamide gels (21) and electroblotted onto nitrocellulose membranes. The membranes were then dried and processed as described above.
Motility assays.
Motility assays were used to analyze the motility function of AIEC strain LF82 and different mutants. Bacterial strains were grown overnight at 37°C in LB broth. A 5-μl volume of this culture was inoculated onto 0.3% agar plates. These plates were incubated at 37°C for 16 h, and motility was assessed qualitatively by examining the circular swim formed by the growing motile bacterial cells.
Construction of the LF82-ΔnlpI isogenic mutant.
The LF82-ΔnlpI isogenic mutant with the nlpI gene deleted was generated with a PCR product, using the method described by Datsenko and Wanner (8) and modified by Chaveroche et al. (5). Briefly, the basic strategy was to replace a chromosomal sequence with a selectable antibiotic resistance gene (kanamycin) generated by PCR. This PCR product was generated by using primers with 50-nucleotide extensions that are homologous to regions adjacent to the nlpI and template E. coli strain harboring the kanamycin resistance gene (Table 2). In addition, strain LF82 was transformed with plasmid pKOBEG, a plasmid that encoded Red proteins from phage λ, which protect linear DNA in bacteria, expressed under the control of an inductible promoter in the presence of 1 mM l-arabinose. This plasmid was maintained in bacteria at 30°C with 25 μg of chloramphenicol per ml and was killed at 37°C.
TABLE 2.
Oligonucleotides used and PCR product sizes
| Primer | Oligonucleotide sequence (5′-3′) | PCR product size (bp) | Use |
|---|---|---|---|
| MI nlpI-1 | AGCAACCGGGAACAGGACGTTCATTCAACCGTGGTCTTCGGGAGTGGGAAAAAGCCACGTTGTGTCTCAA | 1,057 | LF82-ΔnlpI isogenic |
| MI nlpI-2 | CGTTAAGGTGATGGCAATCAAAAAAGATTACGGGCTGATGTGTACGTCAGTTAGAAAAACTCATCGAGCA | mutant construction | |
| nlpI-1 | AACTCGACAATGTTCCATCCCC | 244 | nlpI probe |
| nlpI-2 | CTTGATCCAACTTACAACTACC | ||
| nlpI-3 | AACCGCCAGTTTGAACAGTGCC | 735 | Allelic replacement of |
| nlpI-4 | AGGATGCAGTAATACTTCCTGGC | nIpI in LF82-ΔnIpI | |
| nlpI-5 | AACGCGTTGAGAAAGTGACCG | 1,348 | Allelic replacement of |
| nlpI-6 | CAGCCATGTAGTACGTGTGCC | nIpI in LF82-ΔnIpI | |
| A2-GBL-3 | AAAGCCACGTTGTGTCTCAA | 957 | Kanamycin resistance |
| B2-GBLnp-5 | TTAGAAAAACTCATCGAGCA | cassette amplification | |
| FIME | GCAGGCGGTTTGTTACGGGG | 750 | OFF-oriented invertible |
| INV | GAGGTGATGTGAAATTAATTTAC | element | |
| FIMA | GATGCGGTACGAACCTGTCC | 450 | ON-oriented invertible |
| INV | GAGGTGATGTGAAATTAATTTAC | element | |
| RNA 16S 1 | ATGACCAGCCACACTGGAAC | 157 | RT-PCR |
| RNA 16S 2 | CTTCCTCCCCGCTGAAAGTA | ||
| flhDC1 | CACCAATGTCCAGGCACGGG | 211 | RT-PCR |
| flhDC2 | GATGCTGCCATTCTCAACCG | ||
| fliA-F | CGCCGTGCTCTTCGCGCC | 202 | RT-PCR |
| fliA-R | CGTGCGACGCAACGCGCG | ||
| motB-F | TGAAAGCCCAATTCCCGGCG | 509 | RT-PCR |
| motB-R | CTTAAGCGCATCGTTGCCGC | ||
| fliC-5 | TGGTGCTGCAACTGCTAACGC | 212 | RT-PCR |
| fliC-6 | TTATCGGCATATTTTGCGCTAGC | ||
| fimB-F | CCTGACCCATAGTGAAATCG | 159 | RT-PCR |
| fimB-R | CTTTGCCTTAAGATCAATATC | ||
| fimE-F | GGCATATCGGCATGGGATGC | 170 | RT-PCR |
| fimE-R | CGTTCCTGGGTCCAGCGTTC |
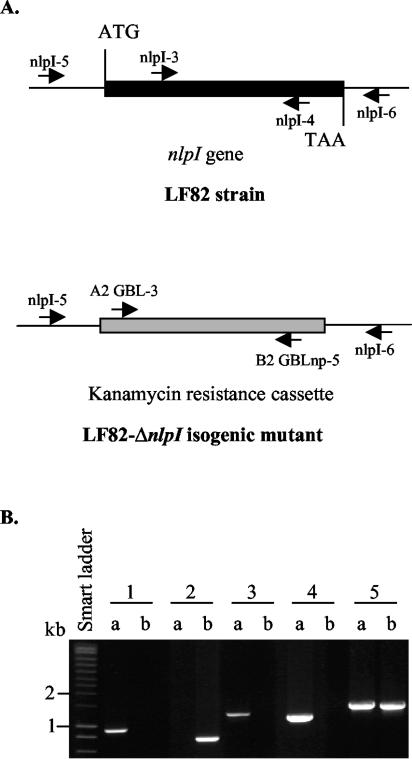
Strain LF82/pKOBEG was cultivated at 30°C with 1 mM l-arabinose to induce Red expression. When the optical density at 620 nm reached 0.5, the bacterial culture was incubated for 10 min at 42°C to kill the plasmid. The bacteria were washed three times with 10% glycerol, and PCR products were electroporated. The LF82-ΔnlpI isogenic mutant was selected onto LB agar containing 50 μg of kanamycin per ml. Replacement of the nlpI gene by the kanamycin resistance cassette in the LF82-ΔnlpI isogenic mutant was confirmed by PCR (Fig. 1).
FIG. 1.
Allelic replacement of the nlpI gene by the kanamycin resistance cassette in the LF82-ΔnlpI isogenic mutant. (A) Schematic representation of the locations of different primers on the DNA of strain LF82 and the LF82-ΔnlpI isogenic mutant. (B) PCR amplification product analysis. Amplification products were generated by using specific primers for kanamycin resistance cassette sequence (A2- GBL-3 and B2-GBLnp-5) (lanes 1), for the intragenic region of the nlpI gene (nlpI-3 and nlpI-4) (lanes 2), for location of the kanamycin resistance cassette (nlpI-5 and B2-GBLnp-5) (lanes 3) and (nlpI-6 and A2-GBL-3) (lanes 4), and to amplify the extragenic region of nlpI gene (nlpI-5 and nlpI-6) (lanes 5). DNA to be amplified was released by boiling from LF82-ΔnlpI bacteria (lanes a) and LF82 bacteria (lanes b).
Cosmid cloning and transcomplementation assays.
A cosmid library of 2,300 clones of total genomic DNA from strain LF82 was generated in cosmid vector pHC79 (2). Cosmids harboring the nlpI gene were detected by hybridization with a 244-base nlpI probe generated by PCR (Table 2). Cosmid 14C11 harboring the nlpI gene was extracted and digested with PstI (Boehringer, Mannheim, Germany). A 1.3-kb fragment containing the entire nlpI gene was cloned into the pUC18 and pHSG575 vectors, and the products were named pPBI02 and pPBI03, respectively. The pPBI03 construct was used to transform the LF82-ΔnlpI isogenic mutant.
DNA sequencing and sequence analysis.
Plasmid DNA templates were prepared using the Qiagen plasmid minikit. Single-stranded DNA was sequenced by Euro Sequence Gene Service (Cybergene) by the dideoxynucleotide chain termination method. DNA sequences were translated and analyzed using GENE JOCKEY II software. DNA and amino acid sequence comparisons were performed with BLAST programs available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nhi.gov).
DNA manipulations, hybridization, and PCR experiments.
Standard DNA procedures and hybridization were performed as described elsewhere (32). PCR conditions and all PCR primer sequences are listed in Table 2.
RNA manipulations, RT, and real-time RT-PCR.
Total RNA was extracted from bacteria and treated with DNase (Roche Diagnostics, Mannheim, Germany) to remove any contaminating genomic DNA. Total RNA was reverse transcribed and amplified using primers specific to fimB, fimE, flhDC, fliA, motB, and fliC mRNAs or 16S rRNA (Table 2). Amplification of a single expected PCR product was confirmed by electrophoresis on 2% agarose gels and ethidium bromide staining. Real-time reverse transcription-PCR (RT-PCR) was performed using a light cycler (Roche Diagnostics). Quantification of mRNA levels or 16S rRNA (as controls) was done using RNA master SYBR Green I (Roche Diagnostics) with 0.5 μg of total RNA.
Statistical analysis.
For analysis of the significance of differences in adhesion and invasion levels, and uptake and survival within J774-A1 macrophages, Student's t-test was used for comparison of two groups of data. All experiments were repeated at least three times. A P-value less than or equal to 0.05 was considered statistically significant.
RESULTS
Identification of the Tn phoA insertional site in mutant 2D2.
Nucleotide sequence analysis of the Tn phoA insertion region of mutant 2D2 revealed that this transposon was inserted in the putative yhbM open reading frame (2), which is annotated as coding for a putative regulator in E. coli K-12 strain MG1655 (GenBank accession no. AE000397). This open reading frame was later reported to encode a lipoprotein and has been renamed gene nlpI, for “new lipoprotein I” (27). The nucleotide sequence of the LF82 nlpI gene demonstrated 99% identity to that of E. coli K-12. Alignment of deduced amino acid sequences revealed 100% identity between NlpI of strain LF82 and that of E. coli K-12. To study the role of the nlpI gene in AIEC strain LF82, an isogenic mutant with the nlpI gene deleted was constructed. PCR controls confirmed deletion of the nlpI gene (Fig. 1).
Growth of the LF82-ΔnlpI isogenic mutant in vitro.
A previously published study of the nlpI mutant WU62 of E. coli K-12 bearing an insertion of a chloramphenicol cassette within the nlpI gene showed that the NlpI lipoprotein may be involved in an undefined step in the overall process of cell division (27). In contrast to results reported by Ohara et al. for the nlpI insertion mutant WU62 (27), microscopic examination of the LF82-ΔnlpI isogenic mutant showed that the bacteria were not elongated (data not shown). Therefore, we checked whether deletion of the nlpI gene in strain LF82 could interfere with the bacterial growth rate. Growth of wild-type strain LF82 and the LF82-ΔnlpI isogenic mutant took place at 37°C without shaking in LB broth (data not shown) and in the bacterium-cell incubation medium used for adhesion and invasion experiments (Eagle minimal essential medium cell culture medium supplemented with 10% heat-inactivated FCS). The growth curves for wild-type strain LF82 and the LF82-ΔnlpI isogenic mutant were similar all time points (from 1 to 6 h) in both media. With an initial inoculum of 3 × 106 CFU/ml and a 6-h incubation in Eagle minimal essential medium, the number of bacteria reached 1.06 × 108 ± 0.06 × 108 and 1.21 × 108 ± 0.10 × 108 for the wild-type strain LF82 and the LF82-ΔnlpI isogenic mutant, respectively. Thus, nlpI gene deletion had no effect on the ability of the LF82-ΔnlpI isogenic mutant to grow in vitro. Differences in the number of cell-associated bacteria (adherent and invasive bacteria) between wild-type strain LF82 and isogenic mutant LF82-ΔnlpI observed in subsequent experiments were not due to growth deficiency of the isogenic mutant.
Phenotype of the LF82-ΔnlpI isogenic mutant.
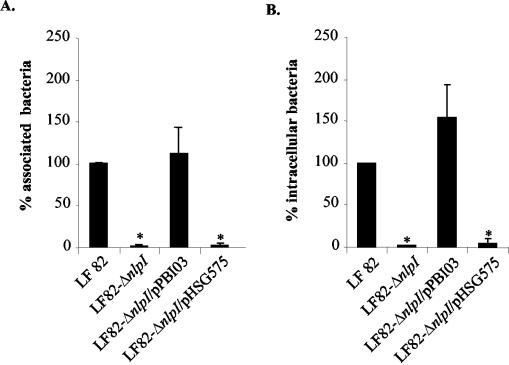
Quantitative adhesion assays showed that the LF82-ΔnlpI isogenic mutant was consistently reduced in its ability to adhere to Intestine-407 epithelial cells, having a 2.8% ± 0.8% residual adhesion level compared to wild-type strain LF82, taken as 100% (Fig. 2A). In addition, the LF82-ΔnlpI isogenic mutant was unable to invade Intestine-407 epithelial cells, with 2.2% ± 0.3% residual invasion level compared to wild-type strain LF82 (Fig. 2B). Transcomplementation experiments were performed with the nlpI gene cloned into plasmid vector pHSG575, forming pPBI03. The adhesion level of the LF82-ΔnlpI isogenic mutant transcomplemented with the cloned nlpI gene (pPBI03) was 111.7% of that of strain LF82, while the adhesion level of the LF82-ΔnlpI isogenic mutant transcomplemented with the vector alone was 3.8%. The invasion level of the LF82-ΔnlpI isogenic mutant transcomplemented with the cloned nlpI gene was 153.7% of that of strain LF82, while the invasion level of the LF82-ΔnlpI isogenic mutant transcomplemented with the vector alone was 4.4% (Fig. 2). Hence, the nlpI gene plays a crucial role in the ability of AIEC strain LF82 to adhere to and invade intestinal epithelial cells. Based on similar residual adhesion and invasion levels of the LF82-ΔnlpI isogenic mutant, the role of the nlpI gene in strain LF82 invasion could be an indirect consequence of the loss of adhesion.
FIG. 2.
Transcomplementation of the adhesion (A) and invasion (B) defects of the LF82-ΔnlpI isogenic mutant with plasmid pPBI03 harboring the entire LF82 nlpI gene. Cell-associated bacteria were quantified after a 3-h infection. Invasion was determined after gentamicin treatment for an additional 1 h. The results are expressed as cell-associated (adherent plus intracellular) or intracellular bacteria relative to those obtained for wild-type strain LF82, taken as 100%. Each value is the mean of at least five separate experiments. Error bars indicate standard errors of the means for five separate experiments. *, P < 0.001.
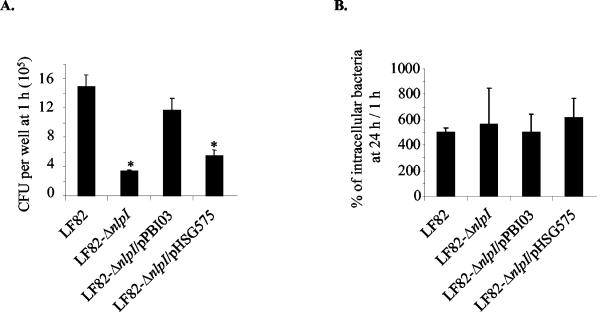
AIEC strains, in addition to being able to adhere to and invade epithelial cells, are able to survive and replicate intracellularly within J774-A1 macrophages. We therefore investigated the role of nlpI gene in such abilities. Bacterial uptake was determined after 1 h of gentamicin treatment, and the number of intracellular bacteria surviving the macrophage killing was determined after 24 h of gentamicin treatment. The level of intracellular bacteria at 24 h postinfection was expressed as a mean percentage of the number of bacteria recovered after 1 h of gentamicin treatment, defined as 100%. As shown in Fig. 3A, the uptake of the LF82-ΔnlpI isogenic mutant by J774-A1 macrophages was fourfold less efficient than that of strain LF82. The number of internalized bacteria was 3.35 × 105 ± 0.79 × 105 CFU per well for the LF82-ΔnlpI isogenic mutant and 14.9 × 105 ± 1.70 × 105 CFU per well for wild-type strain LF82. Transcomplementation with the cloned nlpI gene restored the uptake of the isogenic mutant to a level not significantly different of that of strain LF82. The LF82-ΔnlpI isogenic mutant survived within macrophages at the same levels as wild-type strain LF82, suggesting that the nlpI gene was not involved in resistance to macrophage killing (Fig. 3B). Similar rates of replication of intracellular bacteria were observed for wild-type strain LF82 and the LF82-ΔnlpI isogenic mutant, indicating that nlpI gene deletion had no effect on the ability of the bacteria to grow intracellularly.
FIG. 3.
Phenotype of the LF82-ΔnlpI isogenic mutant within J774-A1 macrophages cells. (A) Uptake of LF82, the LF82-ΔnlpI isogenic mutant, the LF82-ΔnlpI isogenic mutant transformed with the cloned nlpI gene (pPBI03), and the LF82-ΔnlpI isogenic mutant harboring the vector alone (pHSG575). Results were expressed in CFU per well after 1 h of gentamicin treatment. (B) Bacterial survival and replication after 24 h of gentamicin treatment. Results are expressed as the number of intracellular bacteria at 24 h relative to that obtained at 1 h after gentamicin exposure, taken as 100%. Data are means ± standard errors of the means for five separate experiments. *, P < 0.05.
Loss of type 1 pilus expression in LF82-ΔnlpI isogenic mutant.
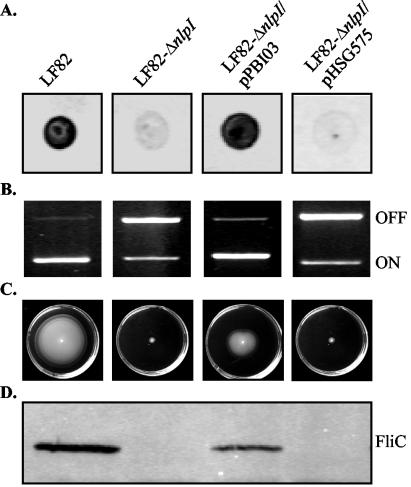
A decreased ability to adhere to and invade epithelial cells has already been observed and is characteristic of type 1 pilus negative mutants of strain LF82 (2). Type 1 pilus expression of the LF82-ΔnlpI isogenic mutant was assessed by colony immunobloting using polyclonal antibodies raised against type 1 pili and by determining the ability of the bacteria to aggregate yeast cells via binding to d-mannose residues. As shown in Fig. 4A, this mutant was unable to synthesize type 1 pili. It was also unable to aggregate yeast cells (Table 3). Type 1 pilus expression is mediated by a process called phase variation, in which the bacteria can switch between piliated and nonpiliated states under the control of a switch invertible element, located upstream of the type 1 pilus-encoding operon (fim operon). PCR experiments using two sets of primers specific to the phase-ON and phase-OFF orientations of the invertible element (37) clearly demonstrated that the decrease in type 1 pilus expression in the LF82-ΔnlpI isogenic mutant correlated with a switch of the invertible element to the phase-OFF orientation (Fig. 4B). Transcomplementation of the LF82-ΔnlpI isogenic mutant with the cloned nlpI gene (pPBI03) restored type 1 pili expression, while transformation with the vector alone (pHSG575) did not restore the wild-type phenotype (Fig. 4A and B; Table 3). Since the orientation of the invertible element is controlled by site-specific DNA inversion catalyzed by the two fim recombinases FimB and FimE, we performed RT-PCR experiments to quantify fimB and fimE mRNA levels. The levels of both fimB and fimE mRNAs in the LF82-ΔnlpI isogenic mutant were decreased (12- and 9-fold, respectively) compared to those in wild-type strain LF82 (Table 4). However, only the decrease in the fimE mRNA level was statistically significant (P < 0.05). The fimB and fimE mRNA levels in the LF82-ΔnlpI isogenic mutant transcomplemented with the cloned nlpI gene (pPBI03) were similar to those observed in the wild-type strain LF82.
FIG. 4.
Type 1 pilus and flagellum expression and/or regulation. Experiments were performed with strain LF82, the LF82-ΔnlpI isogenic mutant, the LF82-ΔnlpI isogenic mutant transcomplemented with the cloned nlpI gene (pPBI03), and the LF82-ΔnlpI isogenic mutant transformed with the vector alone (pHSG575). (A) Colony immunoblotting using polyclonal antibodies raised against purified type I pili. (B) Determination of the invertible element orientation of the fim operon. The orientation was determined by PCR analysis, as described in Materials and Methods. A 450-bp product revealed the ON-orientation and a 750-bp product revealed the OFF-orientation of the invertible element. (C) Motility on a 0.3% agar plate after 16 h at 37°C. Motility was visualized as a halo of radial diffusion of bacteria around the primary inoculum. (D) Western immunoblot analysis of FliC synthesis, using polyclonal antibodies raised against purified flagellin H1.
TABLE 3.
Expression of pili in wild-type strain LF82 compared to that in nlpI mutants
| Strain | Yeast aggregationa | % Positive for pilib |
|---|---|---|
| LF82 | 1/8 | 96 |
| LF82-ΔnlpI | 1 | 9 |
| LF82-ΔnlpI/pPBI03 | 1/8 | 88 |
| LF82-ΔnlpI/pPBI01 | 1/8 | NDc |
Aggregation was monitored visually, and the titer was recorded as the last dilution giving a positive aggregation reaction.
The percentage of bacteria expressing pili was monitored by electron microscopy examination of negatively stained bacteria.
ND, not done.
TABLE 4.
Quantification of fimB, fimE, flhDC, fliA, motB, and fliC expression by RT-PCR in wild-type strain LF82 and mutant strains
| Mutant | Fold decrease in mRNA levels relative to those of wild-type strain LF82a
|
|||||
|---|---|---|---|---|---|---|
| fimB | fimE | flhDC | fliA | motB | fliC | |
| LF82-ΔnlpI | 12.54 ± 9.16 | 9.45 ± 3.98b | 0.95 ± 0.05 | 0.99 ± 0.05 | 0.98 ± 0.18 | 19.13 ± 3.60b |
| LF82-ΔnlpI/pPBI03 | 1.55 ± 0.77 | 3.63 ± 0.92 | 2.22 ± 0.89 | 1.07 ± 0.02 | 0.89 ± 0.13 | 3.63 ± 0.92 |
Fold decrease in mRNA levels relative to that of wild-type strain LF82 using real time RT-PCR. As controls, 16S rRNA levels were measured. Only experiments showing the same levels of 16S rRNA for each sample were taken in to account. Data are mean ± SEM of at least three separate experiments.
P < 0.05.
Biogenesis of type 1 pili in the LF82-ΔnlpI isogenic mutant.
Since some lipoproteins play a role in the biogenesis of fimbriae (30, 31, 41), we investigated whether the LF82-ΔnlpI isogenic mutant could express functional type 1 pili on the bacterial surface. The LF82-ΔnlpI isogenic mutant was transformed with plasmid pPBI01 harboring the entire fim operon to force the bacteria to express these pili. As a consequence of induced type 1 pilus expression, the LF82-ΔnlpI/pPBI01 bacteria aggregated yeast cells (Table 3) and showed a similar amount of type 1 pili to that of strain LF82 by colony immunoblotting assays with antibodies raised against type 1 pili (data not shown). Thus, the absence of biogenesis of type 1 pili in the LF82-ΔnlpI isogenic mutant was not directly related to the absence of NlpI in the outer membrane.
Loss of flagellum expression in LF82-ΔnlpI isogenic mutant.
We recently observed that the lack of expression of flagella in the LF82-ΔfliC isogenic mutant induced the switch of the type 1 pilus invertible element to the phase-OFF orientation (1); we therefore decided to analyze whether the decreased type 1 pilus expression was due to the absence of flagella in a NlpI-negative mutant. The LF82-ΔnlpI isogenic mutant was nonmotile onto 0.3% agar plates (Fig. 4C). In addition, electron microscope examination of negatively stained bacteria indicated that no bacteria synthesized flagella in the LF82-ΔnlpI isogenic mutant whereas 78% of the bacteria were flagellated in the wild-type strain LF82. The lack of flagella was confirmed by Western immunobloting using polyclonal antibodies raised against E. coli flagellin H1 (Fig. 4D). Transcomplementation of the LF82-ΔnlpI isogenic mutant with the cloned nlpI gene (pPBI03) restored motility and synthesis of the FliC flagellin, while transformation with the vector alone (pHSG575) did not (Fig. 4C and D).
Molecular analysis of flagellar downregulation.
Since three levels of transcriptional regulation of flagellar biogenesis exist, we measured the mRNA levels of the flhDC operon encoding the class I master transcriptional regulator FlhD2C2, the fliA gene encoding the class II σ28 sigma factor, and the class III fliC and motB genes encoding the FliC flagellin and the MotB protein (necessary for motor rotation), respectively. The levels of the flhDC and fliA mRNAs in the LF82-ΔnlpI isogenic mutant were similar to those measured in wild-type strain LF82 (Table 4). In contrast, the mRNA level of fliC was drastically decreased (19-fold) in the LF82-ΔnlpI isogenic mutant. Surprisingly, the mRNA level of motB, which, like fliC, belongs to the class III genes of the flagellar regulon and whose transcription is also controlled by the σ28 sigma factor, was not modified. These results showed that the absence of the NlpI lipoprotein induces a decrease in fliC transcription or fliC mRNA stability.
Involvement of NlpI in adhesion and invasion of strain LF82.
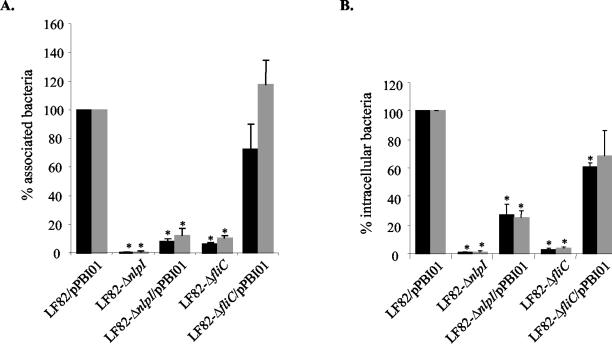
We analyzed the involvement of NlpI in adhesion to and invasion of Intestine-407 epithelial cells with the LF82-ΔnlpI isogenic mutant transformed with the cloned fim operon (pPBI01) in order to force the bacteria to express type 1 pili. The adhesion and invasion levels of the transformed mutant were compared to those of strain LF82 also transformed with the multicopy plasmid pPBI01 harboring the fim operon, since we previously reported that the adhesion and invasion levels of strain LF82/pPBI01 were 1.6- and 6.9-fold increased, respectively, compared to the levels of wild-type strain LF82 (1). The adhesion and invasion levels of the LF82-ΔnlpI isogenic mutant transformed with pPBI01 were 8.3 and 27.1%, respectively, of those of strain LF82/pPBI01 (Fig. 5). Thus, the large decreases in adhesion and invasion levels observed in the LF82-ΔnlpI isogenic mutant were not related only to the absence of type 1 pilus expression.
FIG. 5.
Adhesion (A) and invasion (B) abilities of the isogenic mutant LF82-ΔnlpI with induced type 1 pilus expression and forced contact between bacteria and Intestine-407 epithelial cells. Induced type 1 pilus expression was determined by transformation with pPBI01 harboring the entire fim operon. Experiments were performed without (black) or with (grey) centifugation. See the legend to Fig. 2. Data are means ± standard errors of the means for five separate experiments. *, P < 0.05.
Since the decreases in adhesion and invasion of this NlpI-negative mutant may be related to bacterial motility, we added a centrifugation step to establish a close contact between bacteria and epithelial cells. As controls, we performed experiments with the LF82-ΔfliC isogenic mutant with induced type 1 pilus expression. As already reported (1), centrifugation fully restored the adhesion level of LF82-ΔfliC/pPBI01. In contrast, after centrifugation, the adhesion level of LF82-ΔnlpI/pPBI01 was not significantly increased in comparison with that in assays performed without centrifugation (Fig. 5A). In addition, there was no significant increase in the invasion level of LF82-ΔnlpI/pPBI01 (Fig. 5B). Thus, the lack of flagella and/or motility, and consequently the lack of type 1 pili, cannot explain the decreased adhesion and invasion abilities of LF82-ΔnlpI isogenic mutant.
DISCUSSION
A Tn phoA insertion within the gene encoding the NlpI lipoprotein induced a loss of the ability of mutant 2D2 to invade intestinal epithelial cells (2). For this study, an isogenic mutant with the nlpI gene deleted was constructed. The LF82-ΔnlpI isogenic mutant showed a 35-fold decrease in its ability to adhere to and a 45-fold decrease in its ability to invade Intestine-407 epithelial cells. Phagocytosis by J774-A1 macrophages of the LF82-ΔnlpI isogenic mutant showed a fourfold decrease, but the mutant was still able to survive and replicate intracellularly at the same level as that of wild-type strain LF82. In addition, the LF82-ΔnlpI isogenic mutant was nonmotile and did not express flagella. It synthesized very small amounts of type 1 pili, as confirmed by immunoblotting and electron microscope examination of negatively stained bacteria. The impaired functions were specifically linked to the lack of NlpI since transcomplementation of the LF82-ΔnlpI isogenic mutant with the cloned nlpI gene restored the wild-type phenotype.
Surface-exposed lipoproteins can play a role as adhesins. Such a role has been reported for the surface-exposed lipoprotein JlpA expressed by Campylobacter jejuni (16). NlpI is supposed to be targeted to the outer membrane in the E. coli cell envelope (M. Ohara, personal communication). This was confirmed by the protein sequence analysis indicating that the second amino acid residue of the NlpI protein is not an aspartate, a characteristic of inner membrane-anchored proteins (25, 43). In contrast to a jlpA isogenic mutant of C. jejuni, which lost the ability to adhere to HEp-2 cells but was still invasive, the LF82-ΔnlpI isogenic mutant was affected in various virulence phenotypes (adhesion, invasion, uptake by macrophages, and flagellar and type 1 pilus biogenesis). However, the effect of the absence of NlpI on the decreased invasion of the LF82-ΔnlpI isogenic mutant could be an indirect consequence of the loss of adhesion, since similar residual adhesion and invasion levels were observed. NlpI might function directly to promote strain LF82 adhesion to intestinal epithelial cells, but transformation of strain LF82 with a multicopy recombinant plasmid harboring the cloned nlpI gene did not lead to any significant increased adhesion levels (data not shown).
Some lipoproteins are described as playing a role in the biogenesis of fimbriae. For instance, the outer membrane lipoprotein BfpB is required for the biogenesis of type IV bundle-forming pili in enteropathogenic E. coli (30), lipoprotein HifD takes part in fimbria biogenesis in Haemophilus influenzae type b (41), and lipoprotein VirB7 is associated with the Agrobacterium tumefaciens T-pilus assembly (31). The very low level of type 1 pili observed in the LF82-ΔnlpI isogenic mutant could therefore be a direct consequence of the absence of lipoprotein NlpI in the outer membrane. We investigated whether the LF82-ΔnlpI isogenic mutant could express functional type 1 pili on the bacterial surface. The transformation of the LF82-ΔnlpI isogenic mutant with pPBI01 harboring the entire fim operon fully restored the expression of type 1 pili on the surface of bacteria, as shown by yeast cell aggregation assays. Thus, the NlpI lipoprotein is not essential for type 1 pilus biogenesis on the bacterial surface.
The decreased expression of type 1 pili in the LF82-ΔnlpI isogenic mutant correlated with a switch of the invertible element located upstream of the type 1 pilus-encoding fim operon to the preferential phase-OFF orientation. A similar result was recently shown for LF82 mutants deficient for flagellar biogenesis or motility, for which very small amounts of type 1 pili are expressed on the bacterial surface (1). Since a deletion of the nlpI gene in AIEC strain LF82 yielded bacteria unable to produce both type 1 pili and flagella, a genetic link probably exists between the expression of type 1 pili and that of flagella.
The orientation of the invertible element is determinated by the influence of two upstream, trans-acting gene products, FimB and FimE (17). FimB recombinase mediates both ON-to-OFF and OFF-to-ON inversion, whereas FimE recombinase mediates mainly ON-to-OFF inversion (26). A possible explanation for the specific ON-to-OFF phase transition in the LF82-ΔnlpI isogenic mutant could have been an increase of the FimE activity. However, the fimE mRNA levels were decreased 12-fold in the LF82-ΔnlpI isogenic mutant compared to the wild-type strain. An undetectable FimE recombinase activity and a marked decrease in levels of fimE mRNA when the fim switch is in the OFF orientation have already been reported (19, 38). Since it has been shown that switch OFF-specific fimE mRNAs are more susceptible to exoribonuclease digestion (38), the decrease in the levels of fimE and fimB mRNA observed in the LF82-ΔnlpI isogenic mutant compared to the wild-type strain could result from mRNA stability.
The absence of flagellar structures in the LF82-ΔnlpI isogenic mutant correlated with a lack of flagellin synthesis. The flagellar operons are organized as a hierarchy within the regulon, with the expression of operons at a given level affecting the expression of operons at lower levels (20, 24). The flhDC operon lies on the top of the hierarchy as the sole class I operon, with both of its gene products being absolutely required for the expression of all other genes of the flagellar regulon (15), i.e., those belonging to the class II and III operons. The σ28 sigma factor (a class II product) is required for the expression of the fliC gene and the motABcheAW operon, which belongs to the class III operons. We previously reported that the flhDC transcript level was downregulated in a fliC LF82 isogenic mutant (1). We thus investigated whether the lack of flagella in the LF82-ΔnlpI isogenic mutant correlated with a decreased level of flhDC mRNAs. This level was similar in the nonflagellated LF82-ΔnlpI isogenic mutant and the wild-type strain, LF82. Moreover, the level of mRNAs encoding the σ28 sigma factor was identical to that for the wild-type strain, confirming that the absence of flagella was not dependent on transcriptional regulation via the FlhD2C2 complex. Since the transcription of both fliC gene and motABcheAW operon is known to be under the same control, similar levels of fliC and motB mRNA were expected. Surprisingly, the level of fliC mRNAs was drastically decreased (19-fold) in the LF82-ΔnlpI isogenic mutant in comparison with the wild-type strain LF82, while the motB mRNA level was not modified. That the levels of specific class III transcripts differ in the LF82-ΔnlpI isogenic mutant is intriguing, and this difference has not been reported previously. Since we have shown that FlhD2C2 and σ28 sigma factor are expressed and active in the absence of NlpI, the decrease in the fliC mRNA level is not dependent on the known flagellar transcriptional hierarchy. It can be speculated that the link between the lipoprotein NlpI and the fliC transcript level depends either on an unknown regulator involved in a transcriptional control or on mRNA stability.
The biological role of outer membrane lipoproteins in bacterial virulence has been previously reported (14, 28). Lipoproteins can be involved in two-component signal transduction pathways. As an example, the NlpE lipoprotein in E. coli K-12 is involved in the Cpx two-component signal transduction pathway, which plays a key role in the regulation of induced adhesion to hydrophobic surfaces (28). The loss in the NlpI negative mutant of a broad spectrum of virulence factors (lack of flagella and type 1 pili, reduced bacterial uptake by J774-A1 macrophages, and drastic decreases in the abilities to adhere to and to invade Intestine-407 epithelial cells) could indicate that a regulatory system is also involved in NlpI-mediated full virulence in strain LF82. Two-component signal transduction pathways in which NlpI could be involved are under investigation.
Acknowledgments
This study was supported by a grant from the Ministère de la Recherche et de la Technologie (PRFMMIP 2000) and by grants from the Association F. Aupetit (AFA) and Institut de Recherche des Maladies de l'Appareil Digestif (IRMAD; Laboratoire Astra France). N.B. was supported by a grant from AFA.
We thank Karen Krogfelt and the WHO International Escherichia coli Centre (Copenhagen Denmark) for providing E. coli type 1 pili and flagellin H1 antisera, and we thank Jean-Marc Ghigo (Institut Pasteur, Paris, France) for providing plasmid pKOBEG.
Editor: V. J. DiRita
REFERENCES
- 1.Barnich, N., J. Boudeau, L. Claret, and A. Darfeuille-Michaud. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol. Microbiol. 48:781-794. [DOI] [PubMed] [Google Scholar]
- 2.Boudeau, J., N. Barnich, and A. Darfeuille-Michaud. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39:1272-1284. [DOI] [PubMed] [Google Scholar]
- 3.Boudeau, J., A. L. Glasser, E. Masseret, B. Joly, and A. Darfeuille-Michaud. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect. Immun. 67:4499-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke, D. A., and A. T. Axon. 1988. Adhesive Escherichia coli in inflammatory bowel disease and infective diarrhoea. Br. Med. J. 297:102-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, J. 1979. Escherichia coli plasmids packageable in vitro in lambda bacteriophage particles. Methods Enzymol. 68:309-326. [DOI] [PubMed] [Google Scholar]
- 7.Darfeuille-Michaud, A., C. Neut, N. Barnich, E. Lederman, P. Di Martino, P. Desreumaux, L. Gambiez, B. Joly, A. Cortot, and J. F. Colombel. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115:1405-1413. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duchmann, R., and M. Zeitz. 1999. Crohn's disease, p. 1055-1080. In P. L. Ogra et al. (ed.), Mucosal immunology, 2nd ed. Academic Press, San Diego, Calif.
- 11.Elsner, A., B. Kreikemeyer, A. Braun-Kiewnick, B. Spellerberg, B. A. Buttaro, and A. Podbielski. 2002. Involvement of Lsp, a member of the LraI-lipoprotein family in Streptococcus pyogenes, in eukaryotic cell adhesion and internalization. Infect. Immun. 70:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 13.Glasser, A. L., J. Boudeau, N. Barnich, M. H. Perruchot, J. F. Colombel, and A. Darfeuille-Michaud. 2001. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 69:5529-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagiwara, D., M. Sugiura, T. Oshima, H. Mori, H. Aiba, T. Yamashino, and T. Mizuno. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmann, J. D., and M. J. Chamberlin. 1987. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc. Natl. Acad. Sci. USA 84:6422-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin, S., A. Joe, J. Lynett, E. K. Hani, P. Sherman, and V. L. Chan. 2001. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol. Microbiol. 39:1225-1236. [DOI] [PubMed] [Google Scholar]
- 17.Klemm, P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 5:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogfelt, K. A., and P. Klemm. 1988. Investigation of minor components of Escherichia coli type 1 fimbriae: protein chemical and immunological aspects. Microb. Pathog. 4:231-238. [DOI] [PubMed] [Google Scholar]
- 19.Kulasekara, H. D., and I. C. Blomfield. 1999. The molecular basis for the specificity of fimE in the phase variation of type 1 fimbriae of Escherichia coli K-12. Mol. Microbiol. 31:1171-1181. [DOI] [PubMed] [Google Scholar]
- 20.Kutsukake, K., and T. Iino. 1994. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol. 176:3598-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lamps, L. W., K. T. Madhusudhan, J. M. Havens, J. K. Greenson, M. P. Bronner, M. C. Chiles, P. J. Dean, and M. A. Scott. 2003. Pathogenic yersinia DNA is detected in bowel and mesenteric lymph nodes from patients with Crohn's disease. Am. J. Surg. Pathol. 27:220-227. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Y., H. J. van Kruiningen, A. B. West, R. W. Cartun, A. Cortot, and J. F. Colombel. 1995. Immunocytochemical evidence of Listeria, Escherichia coli, and Streptococcus antigens in Crohn's disease. Gastroenterology 108:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macnab, R. M. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26:131-158. [DOI] [PubMed] [Google Scholar]
- 25.Masuda, K., S. Matsuyama, and H. Tokuda. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. USA 99:7390-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClain, M. S., I. C. Blomfield, and B. I. Eisenstein. 1991. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 173:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohara, M., H. C. Wu, K. Sankaran, and P. D. Rick. 1999. Identification and characterization of a new lipoprotein, NlpI, in Escherichia coli K-12. J. Bacteriol. 181:4318-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasadarao, N. V., C. A. Wass, M. F. Stins, H. Shimada, and K. S. Kim. 1999. Outer membrane protein A-promoted actin condensation of brain microvascular endothelial cells is required for Escherichia coli invasion. Infect. Immun. 67:5775-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramer, S. W., D. Bieber, and G. K. Schoolnik. 1996. BfpB, an outer membrane lipoprotein required for the biogenesis of bundle-forming pili in enteropathogenic Escherichia coli. J. Bacteriol. 178:6555-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Sartor, R. B., H. C. Rath, S. N. Lichtman, and E. A. van Tol. 1996. Animal models of intestinal and joint inflammation. Baillieres Clin. Rheumatol. 10:55-76. [DOI] [PubMed] [Google Scholar]
- 34.Sauer, F. G., M. A. Mulvey, J. D. Schilling, J. J. Martinez, and S. J. Hultgren. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 3:65-72. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt, C. K., J. S. Ikeda, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, and A. D. O'Brien. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultsz, C., M. Moussa, R. van Ketel, G. N. Tytgat, and J. Dankert. 1997. Frequency of pathogenic and enteroadherent Escherichia coli in patients with inflammatory bowel disease and controls. J. Clin. Pathol. 50:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwan, W. R., H. S. Seifert, and J. L. Duncan. 1992. Growth conditions mediate differential transcription of fim genes involved in phase variation of type 1 pili. J. Bacteriol. 174:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sohanpal, B. K., H. D. Kulasekara, A. Bonnen, and I. C. Blomfield. 2001. Orientational control of fimE expression in Escherichia coli. Mol. Microbiol. 42:483-494. [DOI] [PubMed] [Google Scholar]
- 39.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Ham, S. M., L. van Alphen, F. R. Mooi, and J. P. van Putten. 1994. The fimbrial gene cluster of Haemophilus influenzae type b. Mol. Microbiol. 13:673-684. [DOI] [PubMed] [Google Scholar]
- 42.Wu, H. C. 1996. Biosynthesis of lipoproteins, p. 1005-1014. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 43.Yamaguchi, K., F. Yu, and M. Inouye. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53:423-432. [DOI] [PubMed] [Google Scholar]