Abstract
Here we investigate bacterial tryptophan dimer (TD) biosynthesis by probing environmental DNA (eDNA) libraries for chromopyrrolic acid (CPA) synthase genes. Functional and bioinformatics analyses of TD clusters indicate that CPA synthase gene sequences diverge in concert with the functional output of their respective clusters, making this gene a powerful tool for guiding the discovery of novel TDs from the environment. Twelve unprecedented TD biosynthetic gene clusters that can be arranged into five groups (A–E) based on their ability to generate distinct TD core substructures were recovered from eDNA libraries. Four of these groups contain clusters from both cultured and culture independent studies, while the remaining group consists entirely of eDNA-derived clusters. The complete synthetic refactoring of a representative gene cluster from the latter eDNA specific group led to the characterization of the erdasporines, cytotoxins with a novel carboxy-indolocarbazole TD substructure. Analysis of CPA synthase genes in crude eDNA suggests the presence of additional TD gene clusters in soil environments.
INTRODUCTION
The oxidation and subsequent dimerization of tryptophan are the initial steps in the biosynthesis of a structurally diverse collection of bacterial natural products (Fig. 1).1 High frequency of association of tryptophan dimers (TDs) with biological activity2 suggests that the TD motif may be a privileged natural substructure, making its biosynthesis an appealing target for sequence-guided bioactive natural product screening. Many previously identified TDs are produced by soil dwelling bacteria.3 While thousands of unique bacterial species may be found in a single gram of soil, the TD biosynthetic potential encoded within the genomes of these organisms remains largely unexplored due to the difficulties associated with culturing the majority of environmental microbes.4 Here we used a culture independent approach to explore TD biosynthesis diversity in soil environments and to guide the discovery of novel bioactive TDs. Screening of three environmental DNA (eDNA) libraries led to the identification of 12 TD clusters that are predicted to be distinct in gene content from any previously sequenced gene clusters. Complete synthetic refactoring of an eDNA specific family of TD clusters in E. coli led to the identification of the erdasporines (1–3), cytotoxins with a novel carboxy-indolocarbazole core (Fig. 2).
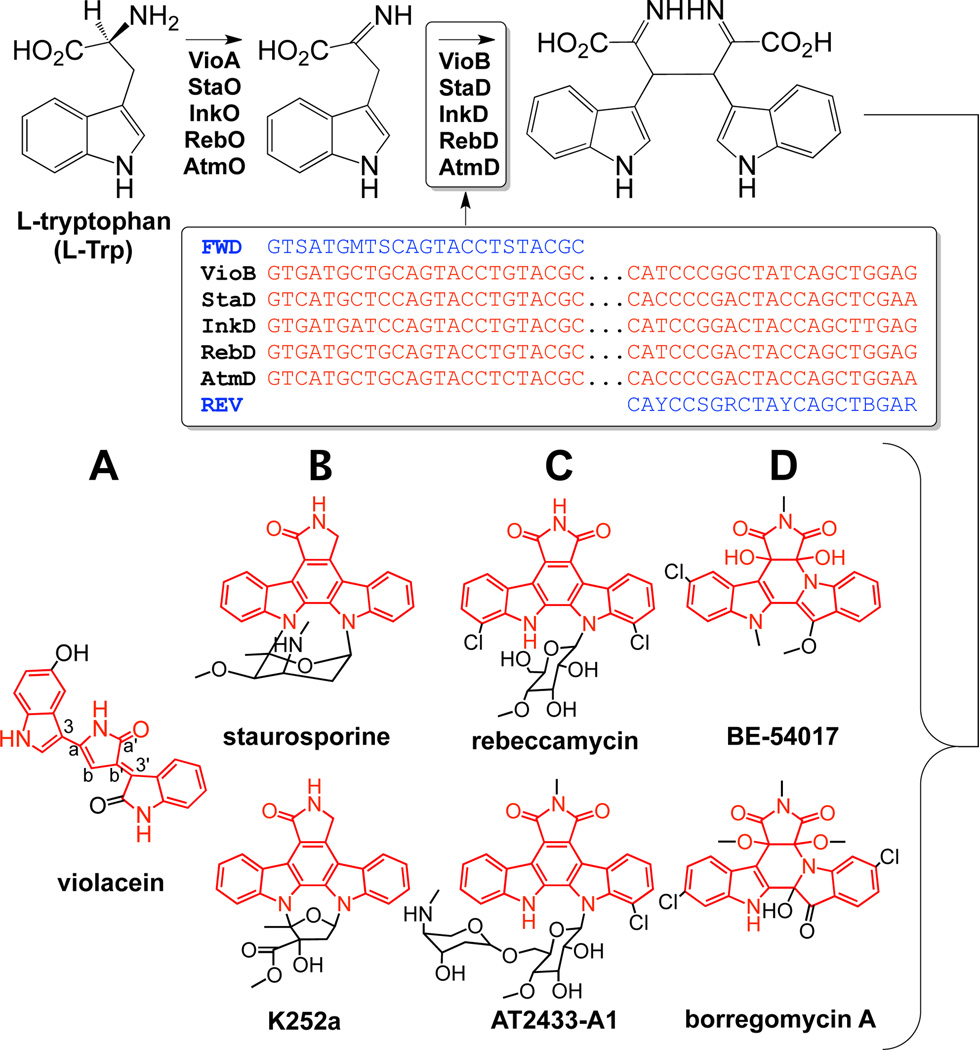
Figure 1.
Conserved biosynthetic steps by IPA-imine synthase and CPA synthase that initiate the production of known bacterial TDs with varied substructures (A–D). Degenerate PCR primers were designed based on conserved regions of CPA synthase genes.
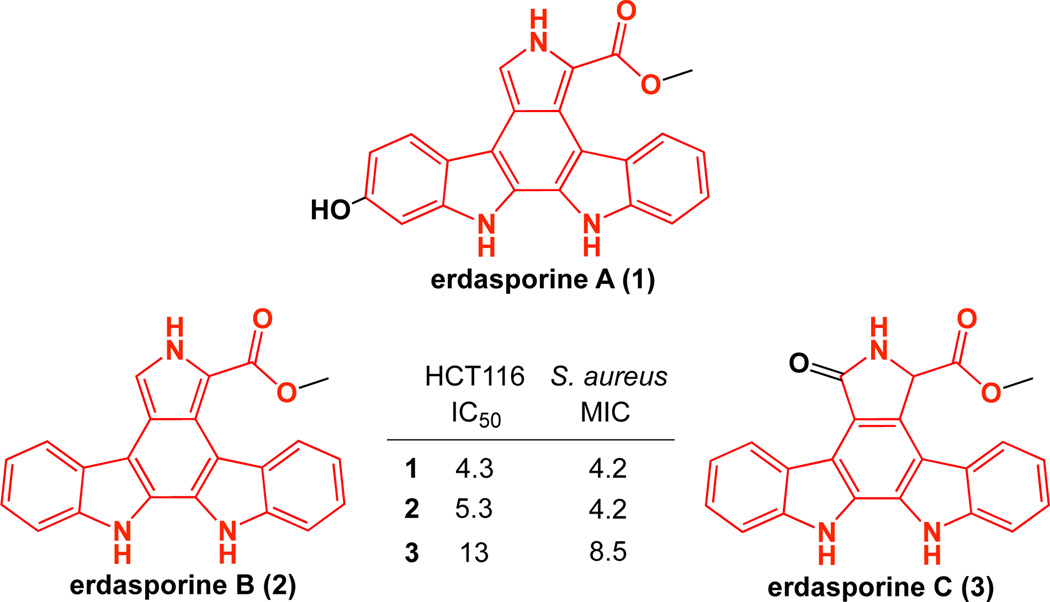
Figure 2.
Chemical structure and cytotoxicity data of erdasporine A–C (1–3) encoded by the esp cluster. The carboxy-indolocarbazole core that is representative of Group E is colored in red. Cytotoxicity (µM) against human HCT116 cells and Staphylococcus aureus is shown.
RESULTS AND DISCUSSION
Prior to our culture independent discovery efforts, clusters for five bacterial TDs (violacein, staurosporine, re beccamycin, K252a, AT2433-A1) had been sequenced and functionally characterized in culture-based studies (Fig. 1).5 The biosynthesis of each of these TDs shares two initial steps: i) the oxidation of tryptophan by an indole-3-pyruvic acid imine (IPA imine) synthase (e.g. StaO) to yield IPA imine and ii) the dimerization of IPA imine by a chromopyrrolic acid (CPA) synthase (e.g. StaD) to give CPA. TD biosynthetic pathways subsequently diverge, resulting in the production of distinct TD substructures (Fig. 1).
While IPA imine synthase genes show limited sequence similarity, CPA synthase gene sequences are highly conserved across known bacterial TD clusters. To survey the TD cluster diversity present in the environment, we used CPA synthase-specific degenerate PCR primers (Fig. 1) to amplify CPA gene homologs from three previously arrayed eDNA libraries.6 Although the majority of bacteria present in soil environments are not readily cultured, clusters from the collective metagenome can be examined by extracting DNA directly from environmental samples and cloning this DNA into model cultured bacterial hosts. The three libraries used in this study were constructed with DNA isolated from soils collected in the Anza-Borrego desert of California (AB), the Sonoran desert of Arizona (AR) and the Chi-huahuan desert of New Mexico (NM). Each library contains >10,000,000 unique eDNA cosmid clones.
In total, 16 unique CPA synthase-like amplicons were identified from PCR screens of these libraries. Cosmid clones associated with each unique CPA synthase-like sequence were recovered from the libraries, sequenced and annotated (9, 4 and 3 clones from the AB, AR and NM libraries, respectively). All 16 clones were found to contain putative TD clusters, in that each possessed a CPA synthase-like sequence along with a set of genes that is similar to those found in previously annotated TD clusters (Fig. 3). eDNA clusters AB1350 and AR1455 are identical in gene content to characterized staurosporine and rebeccamycin clusters, respectively.5 Two eDNA-derived clusters, AB1091 and AB1650, were characterized by us previously and found to encode indolotryptoline structures.7 The remaining 12 clusters are different in gene content from any previously sequenced TD clusters. When these clusters are organized according to CPA synthase gene phylogeny, the clusters with similar gene content group together. CPA synthase genes, and in turn the TD clusters from which they arise, form 5 distinct clades (Fig. 3, Groups A–E). Based on the TD chemical structures encoded by functionally characterized gene clusters, the groupings appear to correlate with the production of distinct TD substructures (Fig. 1 and 3).
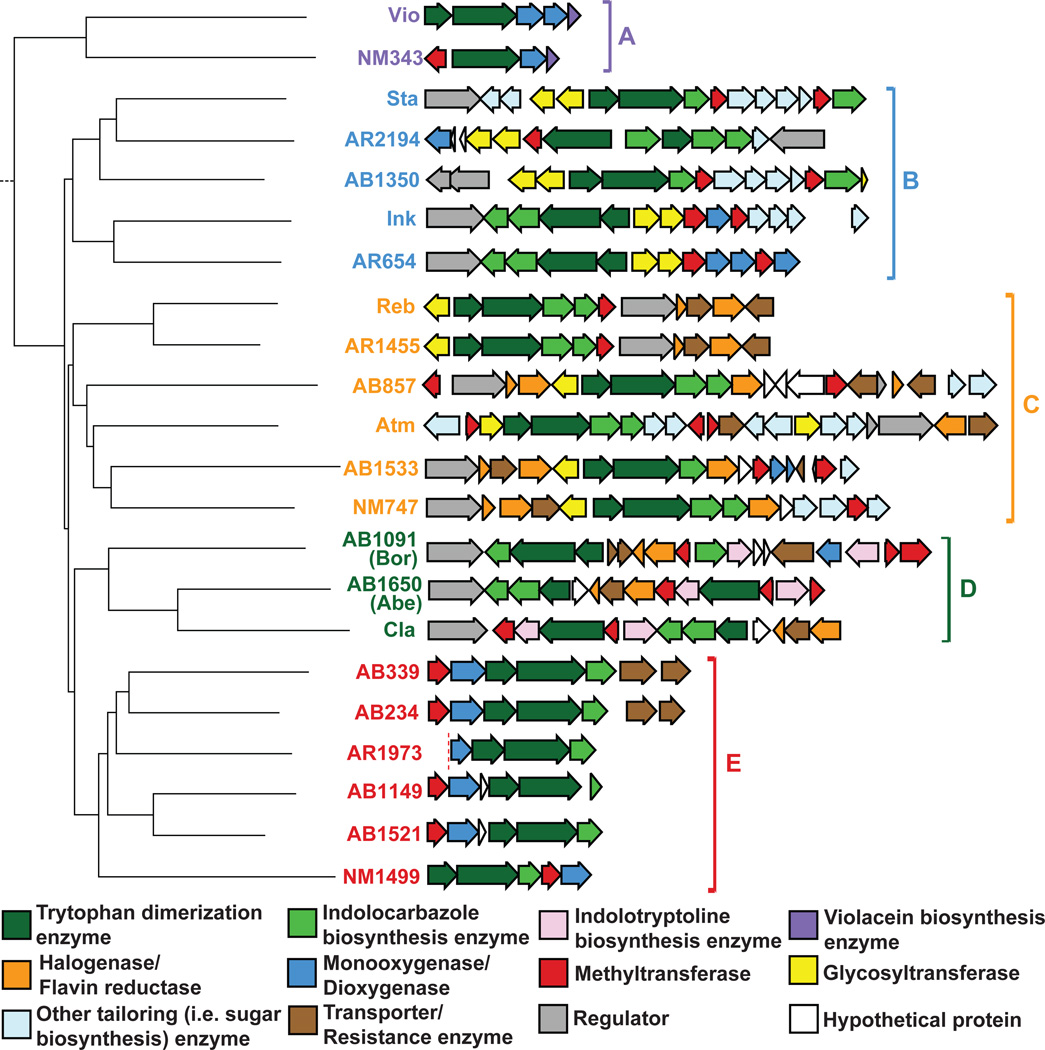
Figure 3.
ClustalW-based phylogenetic tree based on culture-derived (Vio, Sta, Ink, Reb, Atm, Cla) and eDNA-derived (AB#, AR#, or NM#) CPA synthase genes. TD gene clusters are shown next to each CPA synthase gene. Five functionally distinct groups of TD clusters (A–E) are predicted based on the clustering of the tree. AR1973 is truncated by the vector.
Group A contains violecein-like clusters, characterized by the presence of vioE homologs, which are responsible for introducing the unusual C3-Cβ to C3-Cα carbon connectivity in violacein.8 Groups B and C contain clusters that encode functionally and bioinformatically distinct FAD-binding monooxygenases (StaC/RebC) that produce mono and dioxygenated indolocarbazole cores, respectively (Fig. S1).9 Group D contains functionally characterized, indolotryptoline encoding clusters found in both cultured and culture independent studies. These clusters are characterized by the presence of a pair of oxidoreductase genes responsible for the oxidative rearrangement of an indolocarbazole into an indolotryptoline.7,10 Group E contains no functionally characterized relatives, suggesting that clusters in this group could encode a new TD motif.
More than 100 TDs have been described from culture-based studies.3 As the majority of these are not associated with a sequenced cluster, predicting whether a newly discovered TD cluster might encode a novel metabolite is often challenging. Most known TDs are monooxygenated indolocarbazole-based (Group B) compounds, making it particularly difficult to determine whether eDNA-derived Group B clusters (AB2194, AB1350, AR654) encode for novel metabolites. In contrast, only a handful of dioxygenated indolocarbazole (Group C) metabolites are known. Group C eDNA-derived clusters (AB857, AB1533, TX747) all contain collections of genes that are predicted to allow them to encode for novel dioxygenated indolocarbazole-based TDs (e.g. additional halogenases and sugar tailoring enzymes).
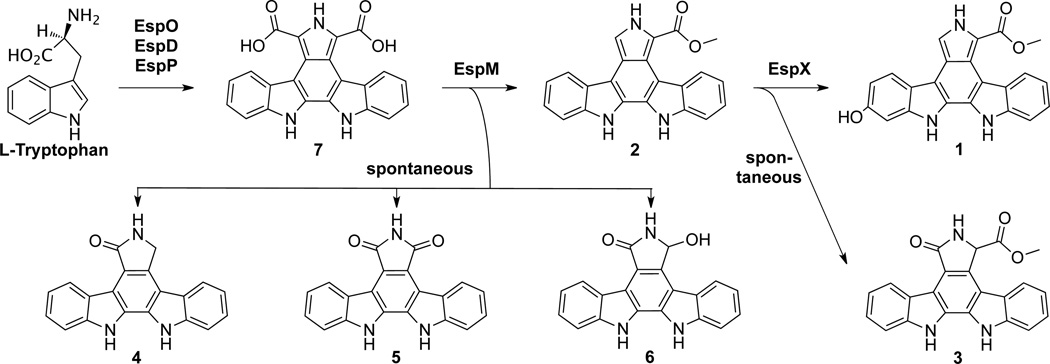
Group E clusters are comprised of single operons containing three conserved indolocarbazole biosynthesis genes (espO, D, P), a predicted methyltransferase (espM) and a FAD-binding monooxygenase (espX) (Fig. 3). Although Group E clusters were recovered from all three metagenomic libraries (AB234, AB339, AB1149, AB1521, AR1973, TX1499), no clusters with the same gene content were found in the NCBI database of sequenced bacterial genomes. Since Group E clusters are unprecedented in sequenced bacterial genomes and have not been previously associated with any known TD core substructure, we elected to investigate the biosynthetic output of these clusters through heterologous expression. The TD cluster from clone AB339 (Fig. S2), which we have called the esp cluster, was selected as a representative member of Group E for expression studies.
Initial heterologous expression efforts, including the introduction of the esp cluster into model bacterial hosts (e.g. E. coli, Streptomyces spp., Burkholderia spp.) and induced expression of the esp operon under a T7 promoter in E. coli did not yield any detectable clone specific small molecules. Previous work with TD gene clusters identified in culture-based studies has shown the potential for accessing TDs through induced expression of partial, complete and mixed gene clusters.5,8–9,11 Presuming that a similar method could be used to induce expression of otherwise silent eDNA-derived TD gene clusters, we chose to synthetically refactor the esp cluster by individually cloning each esp gene in front of a T7 promoter and inducing the expression of the esp genes in E. coli.
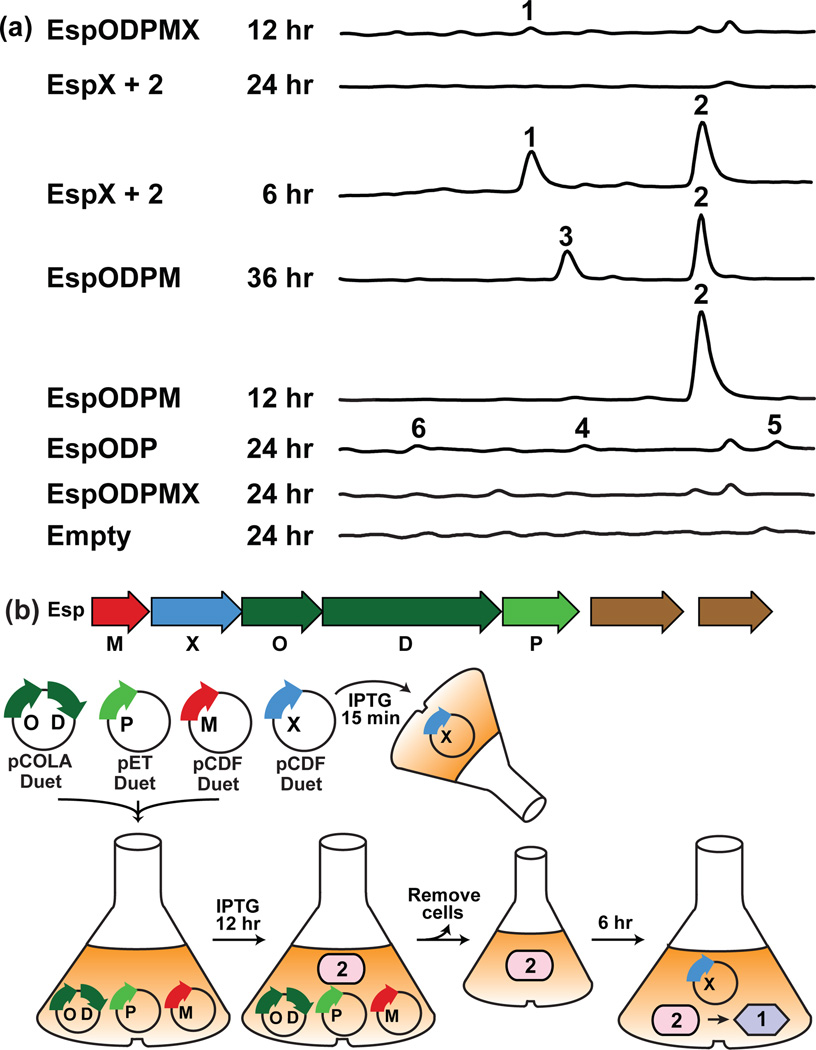
As homologs of espO, D and P are seen in a number of well-studied TD indolocarbazole biosynthetic gene clusters, their functions are predictable.12 Co-expression of these three genes in E. coli resulted in the low level production of the expected indolocarbazole-based intermediates (4–6), indicating that esp is, in fact, a TD gene cluster (Fig. 4a, espODP 24 hr). Addition of espM, a predicted methyltransferase gene, to espODP led to the appearance of compound 2 (espODPM 12 hr). After extended incubation periods, either in culture broth or as a purified compound in DMSO (Fig. S3), 2 spontaneously oxidized to 3 (espODPM 36 hr). Interestingly, when espX, a predicted FAD-binding monooxygenase gene, was co-expressed with espODPM, no clone specific metabolites were detected in the culture broth extract (es-pODPMX 24 hr), mimicking the result from the expression of the native esp operon in E. coli.
Figure 4.
(a) HPLC-UV traces of organic extracts from E. coli cultures expressing various combinations of esp genes. “+ 2” refers to the addition of compound 2 in the form of spent medium from EspODPM cultures. (b) Schematic of the method used for the complete refactoring of the esp cluster.
Suspecting that the product of the entire esp operon might be rapidly degraded, we investigated the EspX-catalyzed transformation reaction by feeding compound 2 in the form of spent culture broth from cultures of E. coli expressing espODPM to EspX-expressing E. coli cultures (Fig. 4b). Within a narrow time window (<6–8 hr) we observed the accumulation of a new compound (1) in concert with the disappearance of 2 (Fig. 4a, EspX + 2, 6 hr). After longer incubations, neither 1 nor 2 could be detected in the culture broth. Retrospectively, we reexamined the espODPMX monoculture at shorter time points and were able to detect very small qualities of 1 in these E. coli cultures (Fig. 4a, EspODPMX 12hr). Ultimately, the temporal control over individual esp gene expression that was possible in our refactoring study permitted the isolation of a natural product that would otherwise have been too transiently present to identify.
Compound 1 was purified from the culture broth of espX expressing cultures fed with spent “espODPM” culture broth, while compounds 2 and 3 were isolated from E. coli cultures expressing espODPM. Based on extensive 1-D and 2-D NMR analysis, compounds 1–3 were determined to be novel methylcarboxylated indolocarbazoles and were named erdasporine A–C, respectively (Fig. 2, Fig. S4–S6). As seen with many known TDs,2,3 the erdasporines are potent cytotoxins with low µM activity against bacteria and human cell lines (Fig. 2).
Based on our heterologous expression studies and previous studies on indolocarbazole biosynthesis with well-characterized transient indolocarbazole intermediates,12–13 erdasporine biosynthesis is proposed to initially proceed like many other indolocarbazole pathways, with EspO, D, and P giving rise to dicarboxy-indolocarbazole (7) from two tryptophans (Fig. 5). In the absence of additional enzymes, this intermediate is known to spontaneously decarboxylate and oxidize to form three indolocarbazole products (4–6). In many previously characterized indolocarbazole pathways, an FAD-binding monooxygenase (e.g. StaC/RebC) directs the formation of a single indolocarbazole product (monooxygenated 4 for Group B clusters and dioxygenated 5 for Group C clusters). In the esp cluster, methylation by EspM appears to preclude these spontaneous oxidative decarboxylation events, resulting in the formation of 2 and, with time, 3. In the presence of EspX, compound 2 undergoes oxidation to form 1 as the product of the esp cluster.
Figure 5.
Proposed biosynthetic scheme for the erdasporines. Key steps include methylation of the dicarboxy-indolocarbazole (7) by EspM to generate 2 and the hydroxylation of 2 by EspX to give 1.
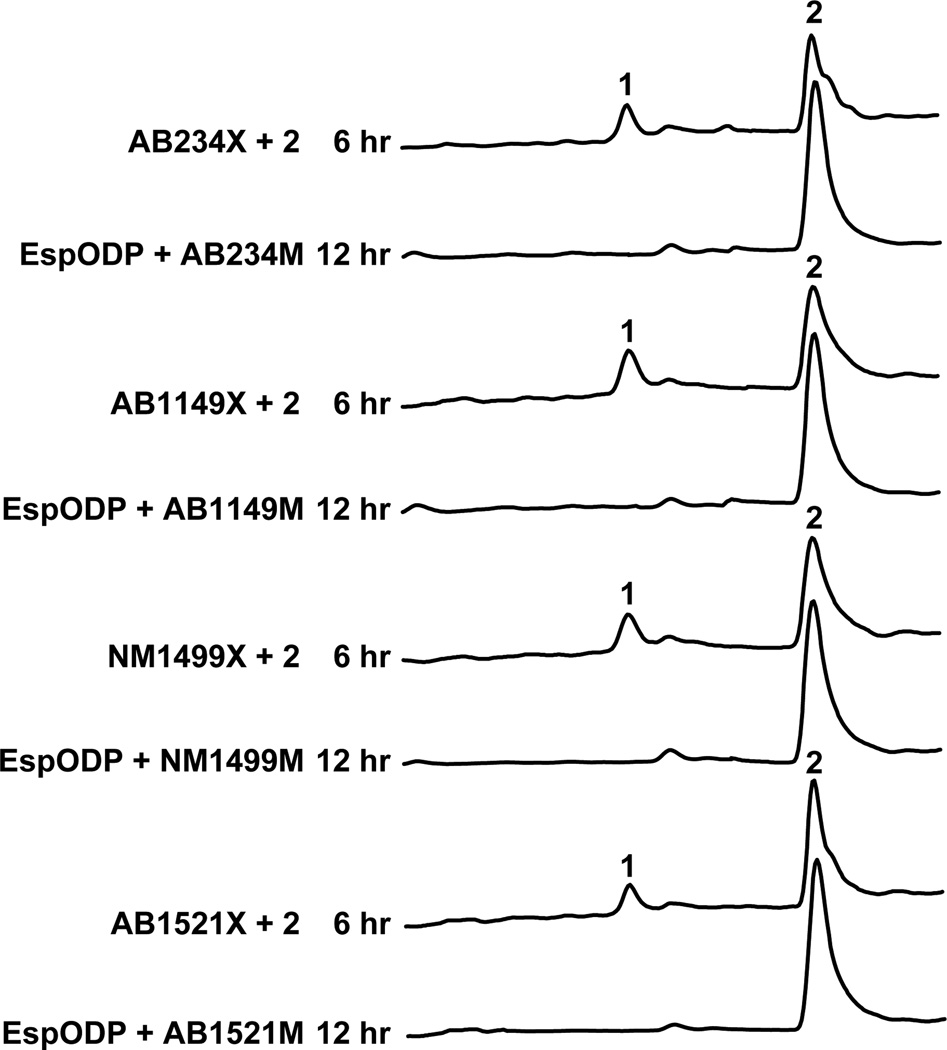
The remaining eDNA-derived Group E clusters are predicted to contain the same five genes as the esp cluster. While EspODP homologs are functionally equivalent across all characterized TD clusters, the exact role of the EspM-like methyltransferases and the EspX-like monooxygenases in each Group E gene cluster was not certain. The remaining four complete Group E clusters were therefore characterized by expressing each unique EspM-like methyltransferase (AB234M, AB1149M, NM1499M, AB1521M) and unique EspX-like monooxygenase (AB234M, AB1149M, NM1499M, AB1521M) gene in the EspODP expression system that was used to characterize the original AB339 esp gene cluster. In each case, the expression of the pathway-specific methyltransferase resulted in the accumulation of 2 and the subsequent expression of the monooxygenase led to the production of 1 (Fig. 6). Based on these studies, all Group E clusters were determined to be functionally equivalent to the esp cluster.
Figure 6.
HPLC-UV traces of organic extracts from E. coli cultures expressing the indicated EspM-like methyltransferase and EspX-like monooxygenase in the EspODP background. “+ 2” refers to the addition of compound 2 in the form of spent medium from EspODP culture coexpressing each pathway-specific methyltransferase.
The functional characterization of the esp cluster defines a new TD group (Fig. 3, Group E) that is based on a carboxy-indolocarbazole core substructure (Fig. 2). Bio-informatics analysis of the genes surrounding eDNA-derived Group E clusters suggests that they likely originate from diverse species within the phylum actinobacteria (Fig. S2). Although esp-like clusters were found in all three soil eDNA libraries examined, neither sequence nor functional analyses of cultured bacteria have identified this family of TDs.
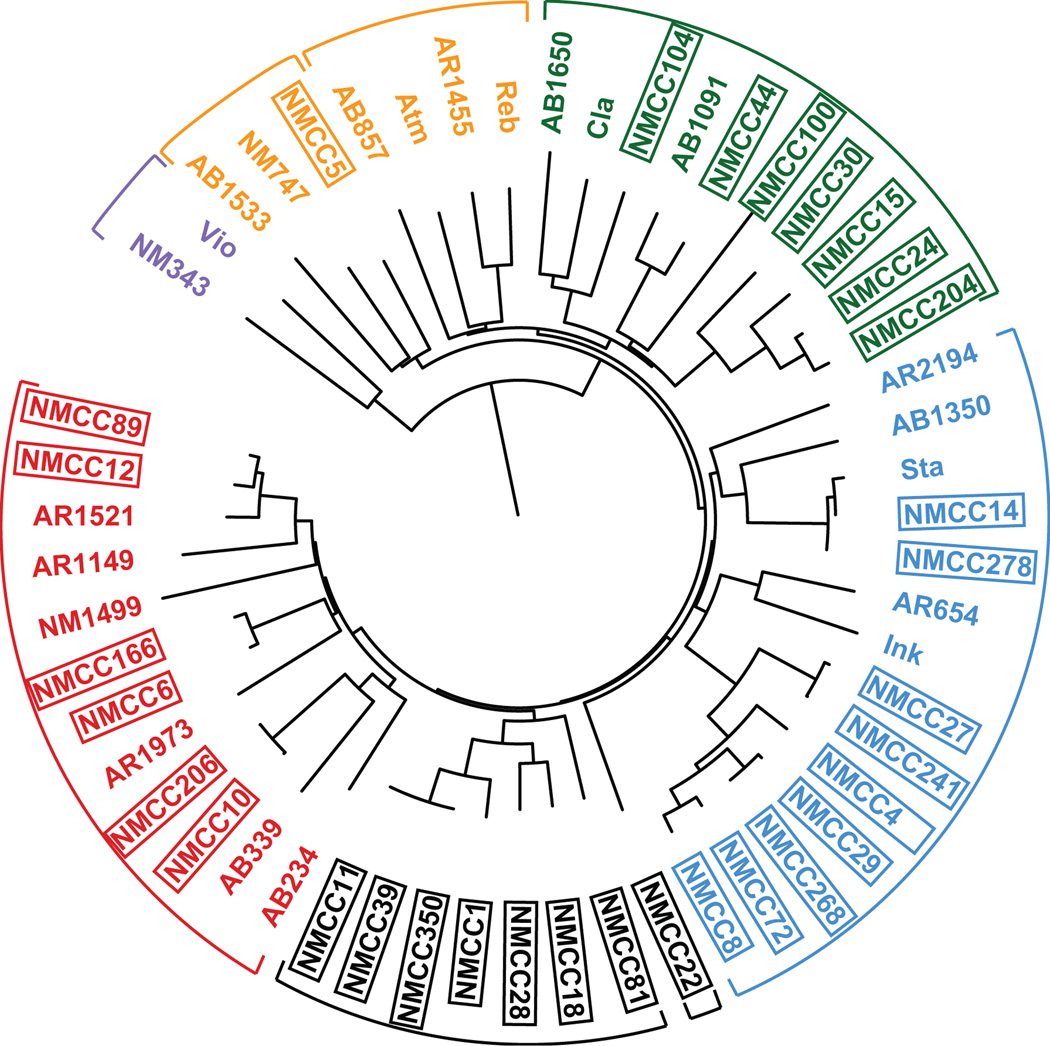
Our functional and bioinformatics analyses of TD clusters suggest that CPA synthase gene sequences diverge in concert with the functional outputs of their respective TD clusters, making CPA gene sequences alone a good marker to guide the discovery of novel TDs. To further explore TD diversity in the environment, DNA extracted directly from 20 soil samples from geographically distinct sites in New Mexico was screened by PCR using our CPA synthase degenerate primers, and the resulting PCR amplicons were sequenced (Table S1). A phylogenetic analysis of these sequences shows that they form new clades both within (e.g. NMCC27) and outside (e.g. NMCC11) the characterized TD groups, suggesting these CPA synthase homologs may appear in gene clusters that encode novel TDs with known substructures as well as novel TD substructures (Fig. 7).
Figure 7.
ClustalW-based phylogenetic tree of trimmed CPA synthase gene amplicons. The eDNA-derived sequences (boxed) fall into known TD groups A–E (colored), and form new clades (black) that encode potentially novel TD core substructures.
CONCLUSIONS
Here we have shown the utility of CPA synthase gene screening for guiding the discovery of novel bioactive TDs. While functional characterization of cryptic clusters remains a significant hurdle in sequence guided natural product discovery programs, we show that complete synthetic gene cluster refactoring in simple to use hosts like E. coli can be used to generate novel biologically active metabolites from eDNA-derived TD gene clusters. The recovery and functional analysis of gene clusters associated with additional novel CPA clades should provide a means of identifying structurally diverse natural bioactive TDs from the environment.
EXPERIMENTAL PROCEDURES
Soil environmental DNA (eDNA) library construction
The three eDNA megalibraries, each consisting of over 10,000,000 unique cosmid clones, were constructed previously from the soil samples collected from the Anza-Borrego desert of California (AB), the Sonoran desert of Arizona (AR) and the Chihuahuan desert of New Mexico (NM), using published methods.14 Briefly, soil was resuspended in lysis buffer (100 mM Tris-HCl, 100 mM EDTA, 1.5 M NaCl, 1% (w/v) CTAB, 2% (w/v) SDS, pH 8.0) and heated for 2 hr at 70 °C. Soil particulates were removed by centrifugation (30 min, 4000 X g, 4 °C). Crude eDNA was precipitated from the resulting supernatant through the addition of 0.7 vol of isopropanol, pelleted (30 min, 4000 X g, 4 °C), washed with 70% ethanol, and pelleted once more (10 min, 4000 X g, 4 °C) to yield crude eDNA.
High molecular weight (HMW; ≥25 kb) eDNA was purified from crude eDNA by agarose gel electrophoresis (1% agarose gel, 16 hr, 20 V). The electroeluted (2 hr, 100V) HMW eDNA was concentrated (100 KDa molecular weight cut off), blunt-ended (End-It), ligated into cosmid vector, packed into λ phage (MaxPlax), and transfected into E. coli (EC100, Epicentre). The eDNA libraries were archived as unique sublibraries, each containing 4000–5000 clones. Matching DNA miniprep and glycerol stock pairs were generated for each sublibrary. DNA minipreps were arrayed such that sets of 8 sublibraries were combined to generate unique “row pools.”
eDNA library homology guided screening of chromopyrrolic acid (CPA) synthase gene
A degenerate primer set was designed based on conserved regions of known CPA synthase genes from culture-based studies (accession no.: vioB AF172851.1, staD AB088119.1, rebD AJ414559.1, inkD DQ399653.1, atmD DQ297453.1.) Primers: StaDVF: GTS ATG MTS CAG TAC CTS TAC GC, StaDVR: YTC VAG CTG RTA GYC SGG RTG. The eDNA libraries were screened by performing PCR on miniprep DNA from each of the unique “row pools”. Each 20 µl reaction consisted of 8.3 µl of water, 10 µl of FailSafe PCR Buffer G (Epicentre), 0.5 µl each of StaDVF and StaDVR primers (final concentration of 2.5 µM each), 0.5 µl of template “row pool” eDNA (100 ng), and 0.2 µl Taq DNA polymerase (New England Biolabs). PCR cycling conditions were as follows: 1 cycle of 95°C for 5 min; 7 cycles of 95°C for 30 sec, 65°C for 30 sec with 1°C decrement per cycle to 59°C, 72°C for 40 sec; 30 cycles of 95°C for 30 sec, 58°C for 30 sec, 72°C for 40 sec; 1 cycle of 72°C for 7 min; 4°C hold. Amplicons of the correct size (561 base pairs) were gel purified, reamplified and sequenced using the same degenerate primers. Amplicons that were confirmed to be CPA synthase gene sequences based on BLASTX homology searches (NCBI) were used to guide the recovery of the corresponding cosmid clones from within our eDNA megalibraries.
Recovery of cosmid clones harboring tryptophan dimer (TD) gene clusters
Cosmid clones containing CPA synthase genes were recovered from the archived eDNA libraries using a serial dilution approach. For each amplicon of interest, a specific PCR primer set was designed to recognize the sequence of that particular amplicon. These primers were used to identify, from a given “row pool,” the corresponding sublibrary that contains the clone of interest. The sublibrary glycerol stock was resuspended into LB to an OD600 of 0.5, diluted 2 × 105 fold and arrayed as 60 µl aliquots (about 25 cells) into 4 sterile 96 well plates. Upon overnight growth, the well containing the clone of interest was identified by whole cell PCR. The culture broth from this well was then spread onto LB plates and single colonies were screened by colony PCR to identify the specific clone harboring the targeted CPA synthase gene. Cosmid clones were de novo sequenced at the Sloan Kettering Institute DNA Sequencing Core Facility using 454 pyrosequencing technology (Roche). Clone assemblies were annotated using FGENESB (Softberry) or CloVR15 for gene prediction and BLASTP (NCBI) for protein homology relationsips. The gene clusters reported in this paper have been deposited in the GenBank database under the accession numbers: KF551861 (NM1499), KF551862 (NM747), KF551863 (NM343), KF551864 (AB234), KF551865 (AB339), KF551866 (AB857), KF551867 (AB1149), KF551868 (AB1350), KF551869 (AB1521), KF551870 (AB1533), KF551871 (AR654), KF551872 (AR1455), KF551873 (AR1973), KF551874 (AR2194).
Phylogenetic tree construction of CPA synthase genes
The ClustalW alignment was performed on the sequences of culture-derived and eDNA-derived CPA synthase genes using MacVector version 12.0.3 (Open Gap Penalty: 10.0; Extend Gap Penalty: 5.0; Pairwise Alignment Mode: Slow). The corresponding phylogenetic tree was constructed from the alignment using the non-TD pathway related CPA synthase-like hypothetical gene Riv7116_4841 from Rivularia sp. PCC 7116 (accession no.: CP003549.1) as an outgroup for rooting (Best Tree Mode; Tree Building Method: Neighbor Joining; Distance: Absolute).
Synthetic refactoring of the esp gene cluster
The biosynthetic genes from the esp gene cluster in cosmid clone AB339 were amplified using the manufacturer’s recommended Phusion Hot Start Flex DNA polymerase reaction conditions (New England Biolabs). PCR primers are listed in the supplementary material in Table S2. PCR cycling conditions were as follows: 1 cycle of 95°C for 5 min; 30 cycles of 95°C for 10 sec, 62°C for 30 sec, 72°C for 30 sec/kb; 1 cycle of 72°C for 7 min; 4°C hold. The resulting amplicons were digested and cloned into the following Duet vectors: EspM NdeI/MfeI site of pCDFDuet-1; EspX NcoI/HindIII site of pCDFDuet-1; EspO NcoI/HindIII site of pCOLADuet-1; EspD NdeI/MfeI site of pCOLADuet-1; EspP NcoI/HindIII site of pETDuet-1.
Induced expression analysis of refactored esp gene cluster
Electrocompetent E. coli BL21 cells were transformed with constructs containing various combinations of esp genes, grown in LB medium in the presence of the appropriate antibiotics (spectinomycin 100 µg/ml; kanamycin 30 µg/ml; ampicillin 100 µg/ml) and induced at OD600 of 0.5 by the addition of IPTG to a final concentration of 0.1 mM. After growth for between 6 and 36 h (200 rpm, 25 °C), the culture was extracted with ethyl acetate and dried in vacuo. Upon resuspension in methanol, the samples were subjected to reversed phase LC/MS analysis (150 × 4.6 mm, 5 µm XBridge C18: linear gradient of 80:20 water:methanol to 0:100 water:methanol). Analytical LC/MS was obtained using Micromass ZQ mass spectrometer (Waters).
Isolation and purification of the erdasporines
For compound 1, 2 liters of EspODPM expressing E. coli BL21 culture was grown for 12 hr (200 rpm, 25 °C) after IPTG induction. The culture was pelleted by centrifugation (10 min, 4000 X g, 25 °C) and the resulting supernatant was added to 2 liters of EspX expressing E. coli BL21 culture that had been grown for 15 minutes after IPTG induction. The combined culture was then grown for 6 hr (200 rpm, 25 °C) before extraction with 2 volumes of ethyl acetate.
For compounds 2 and 3, 2 liters of EspODPM expressing E. coli BL21 culture grown for 12 hr (compound 2) or 36 hr (compound 3) after IPTG induction was extracted with 2 volumes of ethyl acetate.
Organic extracts were separated by silica gel RediSep flash chromatography (RediSepRf 12 gram silica flash column: 3 min 100% chloroform, 27 min linear gradient from 100% chloroform to 90:10 chlorofom:methanol). Compound 2 eluted with 99:1, 3 eluted with 96:4 and 1 eluted with 95:5. Compounds 1–3 were purified from these fractions using isocratic reserved phase HPLC (150 × 10 mm, 5 µm XBridge C18). 2 (2.3 mg) was purified using 64:38 water:acetonitrile. 3 (1.1 mg) was purified using 68:32 water:acetonitrile. Compound 1 was first fractionated using 70:30 water:acetonitrile and subsequently purified (0.4 mg) using silica gel flash chromatography (0.5 gram of Silica Gel 60 packed in a glass pipette) with an isocratic 70:30 hexane:ethyl acetate mobile phase. HRMS data was obtained using a LTQ-Orbitrap mass spectrometer (Thermo Scientific). NMR data was acquired using a 600-MHz spectrometer (Bruker). Specific rotation was measured using a P-1020 Polarimeter (Jasco).
Induced expression analysis of eDNA-derived Group E clusters
The pathway-specific methyltransferase and monooxygenase genes from the Group E clusters found in cosmid clones AB234, AB1149, NM1499 and AB1521 were amplified using the same PCR reaction and cycling conditions that was done for clone AB339 amplification reactions. PCR primers are listed in the supplementary material in Table S2. The methyltransferase and the monooxygenase amplicons were cloned into the NdeI/MfeI or NcoI/HindIII sites of pCDFDuet-1, respectively.
The heterologous expression was conducted in a similar manner to that used to study the esp gene cluster. Briefly, 25 mL of each methyltransferase/EspODP co-expressing E. coli BL21 culture was grown for 12 hr after IPTG induction. This culture was pelleted by centrifugation and the resulting supernatant was added to 25 mL of a monooxygenase expressing E. coli BL21 culture that had been grown for 15 minutes after IPTG induction. The cultures were then grown for an additional 6 hr and then extracted with ethyl acetate. Extracts were subjected to reversed phase LC/MS analysis as described above.
Bioactivity assays
For the antibacterial assay, an overnight culture of Staphylococcous aureus 6538P grown in LB medium was diluted 106 fold and distributed as 100 µl aliquots into a sterile 96 well microtiter plate. Compounds 1–3, along with an ampicillin positive control and a compound-free negative control, all resuspended in DMSO, were added to the first well at the initial concentration of 50 µg/ml and were serially diluted 2-fold across the plate such that the final concentrations of the wells were 50, 25, 13, 6.3, 3.1, 1.6, 0.78, 0.39, 0.20, 0.098, 0.049, 0.024 µg/ml. The culture was grown for 18 hr (500 rpm, 30 °C), and the lowest concentration with no observable growth (OD600 < 0.05) was reported as the minimum inhibitory concentration (MIC) of each compound.
For the human cell line assay, human colon cancer cells HCT116 (ATCC: CCL-247) grown in McCoy's 5A Media (modified, Invitrogen) supplemented with 10% fetal bovine serum and 1% (w/v) Penicillin/Streptomycin were seeded as 100 µl aliquots into a sterile 96 well microtiter plate at a titer of approximately 1,000 cells per plate and incubated (24 hr, 37 °C, 5% CO2). Compounds 1–3 resuspended in DMSO and a compound-free DMSO control were diluted in fresh medium and added to the appropriate wells at final concentrations of 50, 25, 13, 6.3, 3.1, 1.6, 0.78, 0.39, 0.20, 0.098, 0.049, 0.024 µg/ml. These plates were then cultured for an additional 72 hr. The cell density in each well was determined via the crystal violet assay.16 Briefly, the cells were washed with phosphate-buffered-saline (PBS), fixed with 4% formaldehyde in PBS (10 min, room temp), washed again with PBS and stained with 0.1 % (w/v) filtered crystal violet solution (30 min, room temp). The stained cells were washed three times with water, air dried and 100 ml of 10% acetic acid was then added to extract the dye. The absorbance of the dye extract was measured using a microplate reader (Epoch Microplate Spectrophotometer; BioTek) at 590 nm. The normalized absorbance values were plotted and the resulting curve fitted, using Graphpad Prism, to determine the half maximal inhibitory concentrations (IC50).
TD diversity analysis from crude eDNA
Topsoil was collected from 20 distinct sites in New Mexico. Crude eDNA was extracted from each sample using the same initial protocol as described for eDNA library construction. However, instead of purifying HMW eDNA by gel electrophoresis, crude eDNA was cleaned with two rounds of column based purification (PowerClean DNA Clean-Up Kit, MO-BIO).
For crude eDNA screening, the forward primer of the degenerate primer (StaDVF) was modified at the 5’ end to permit the direct 454 sequencing (Roche) of amplicons from all 20 samples simultaneously. Forward primers each contained a 454 sequencing adapter tag (CGTATCGCCTCCCTCGCGCCATCAG), followed by a unique 8 base pair barcode for each soil sample and then the StaDVF degenerate sequence. A unique StaDVF/StaDVR pair was then used to amplify CPA synthase gene fragments from each soil sample. Each 20 µl PCR reaction consisted of 8.3 µl of water, 10 µl of Fail-Safe PCR Buffer D (Epicentre), 0.5 µl each of modified StaDVF and StaDVR primers (final concentration of 2.5 µM each), 0.5 µl of template crude eDNA (100 ng) and 0.2 µl Taq DNA polymerase (New England Biolabs). PCR cycling conditions were as follows: 1 cycle of 95°C for 5 min; 30 cycles of 95°C for 30 sec, 59°C for 30 sec, 72°C for 40 sec; 1 cycle of 72°C for 7 min; 4°C hold. Amplicons of the correct size (561 base pairs) were gel purified and processed for single-end read sequencing using the 454 GS-GLX Titanium platform at the Sloan Kettering Institute DNA Sequencing Core Facility.
The raw reads were initially processed using the Qiime software suite (version 1.6)17 which utilizes size cutoff, quality cutoff, insertion/deletion removal and chimera removal filters to retain only high quality reads. Only reads with lengths >500 base pairs were retained and they were subsequently trimmed to 450 base pairs (from the 3’ end where sequencing errors occur most frequently). Reads were then clustered at 95% identity and only the amplicon sequences that were populated with more than 30 reads from a single soil sample were retained for phylogenetic analysis. Consensus amplicons that were either unrelated to CPA synthase genes or were potentially chimeric based on BLASTX homology searches (NCBI) were also removed, leaving 31 sequences for analysis (Table S1).
A ClustalW alignment was performed on these 31 amplicon sequences plus all known full-length CPA synthase genes using MacVector version 12.0.3 (Open Gap Penalty: 10.0; Extend Gap Penalty: 5.0; Pairwise Alignment Mode: Slow). Using this alignment as a guide, full-length CPA synthase genes were trimmed to match the 450 bp amplicon sequences and a final phylogenetic tree was constructed using the hypothetical gene Riv7116_4841 as an outgroup for rooting (Best Tree Mode; Tree Building Method: Neighbor Joining; Distance: Tajima-Nei). This tree was reformatted to a circular display using iTOL.18
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH GM077516. SFB is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
Experimental details and additional data. This material is available free of charge via the Internet at http://pubs.acs.org.
No competing financial interests have been declared.
REFERENCES
- 1.Ryan KS, Drennan CL. Chem Biol. 2009;16:351. doi: 10.1016/j.chembiol.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakano H, Omura S. J Antibiot (Tokyo) 2009;62:17. doi: 10.1038/ja.2008.4. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez C, Mendez C, Salas JA. Nat Prod Rep. 2006;23:1007. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- 4.Torsvik V, Goksoyr J, Daae FL. Appl Environ Microbiol. 1990;56:782. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Pemberton JM, Vincent KM, Penfold RJ. Current Microbiology. 1991;22:355. [Google Scholar]; (b) Onaka H, Taniguchi S, Igarashi Y, Furumai T. Journal of Antibiotics. 2002;55:1063. doi: 10.7164/antibiotics.55.1063. [DOI] [PubMed] [Google Scholar]; (c) Sanchez C, Butovich IA, Brana AF, Rohr J, Mendez C, Salas JA. Chem Biol. 2002;9:519. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]; (d) Chiu HT, Chen YL, Chen CY, Jin C, Lee MN, Lin YC. Mol Biosyst. 2009;5:1180. doi: 10.1039/b905293c. [DOI] [PubMed] [Google Scholar]; (e) Gao Q, Zhang C, Blanchard S, Thorson JS. Chem Biol. 2006;13:733. doi: 10.1016/j.chembiol.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 6.(a) Reddy BV, Kallifidas D, Kim JH, Charlop-Powers Z, Feng Z, Brady SF. Appl Environ Microbiol. 2012;78:3744. doi: 10.1128/AEM.00102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Owen JG, Reddy BVB, Ternei MA, Charlop-Powers Z, Calle PY, Kim JH, Brady SF. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11797. doi: 10.1073/pnas.1222159110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Chang FY, Brady SF. J Am Chem Soc. 2011;133:9996. doi: 10.1021/ja2022653. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chang FY, Brady SF. Proc Natl Acad Sci U S A. 2013;110:2478. doi: 10.1073/pnas.1218073110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Sanchez C, Brana AF, Mendez C, Salas JA. Chembiochem. 2006;7:1231. doi: 10.1002/cbic.200600029. [DOI] [PubMed] [Google Scholar]; (b) Balibar CJ, Walsh CT. Biochemistry. 2006;45:15444. doi: 10.1021/bi061998z. [DOI] [PubMed] [Google Scholar]
- 9.(a) Groom K, Bhattacharya A, Zechel DL. Chembiochem. 2011;12:396. doi: 10.1002/cbic.201000580. [DOI] [PubMed] [Google Scholar]; (b) Goldman PJ, Ryan KS, Hamill MJ, Howard-Jones AR, Walsh CT, Elliott SJ, Drennan CL. Chem Biol. 2012;19:855. doi: 10.1016/j.chembiol.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan KS. PLoS One. 2011;6:e23694. doi: 10.1371/journal.pone.0023694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Hyun CG, Bililign T, Liao J, Thorson JS. Chembiochem. 2003;4:114. doi: 10.1002/cbic.200390004. [DOI] [PubMed] [Google Scholar]; (b) Zhang C, Albermann C, Fu X, Peters NR, Chisholm JD, Zhang G, Gilbert EJ, Wang PG, Van Vranken DL, Thorson JS. Chembiochem. 2006;7:795. doi: 10.1002/cbic.200500504. [DOI] [PubMed] [Google Scholar]; (c) Sanchez C, Zhu L, Brana AF, Salas AP, Rohr J, Mendez C, Salas JA. Proc Natl Acad Sci U S A. 2005;102:461. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Salas AP, Zhu L, Sanchez C, Brana AF, Rohr J, Mendez C, Salas JA. Mol Microbiol. 2005;58:17. doi: 10.1111/j.1365-2958.2005.04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard-Jones AR, Walsh CT. J Am Chem Soc. 2006;128:12289. doi: 10.1021/ja063898m. [DOI] [PubMed] [Google Scholar]; (b) Howard-Jones AR, Walsh CT. J Am Chem Soc. 2007;129:11016. doi: 10.1021/ja0743801. [DOI] [PubMed] [Google Scholar]
- 13.Asamizu S, Hirano S, Onaka H, Koshino H, Shiro Y, Nagano S. Chembiochem. 2012;13:2495. doi: 10.1002/cbic.201200535. [DOI] [PubMed] [Google Scholar]
- 14.Brady SF. Nat Protoc. 2007;2:1297. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- 15.Angiuoli SV, Matalka M, Gussman A, Galens K, Vangala M, Riley DR, Arze C, White JR, White O, Fricke WF. BMC Bioinformatics. 2011;12:356. doi: 10.1186/1471-2105-12-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zivadinovic D, Watson CS. Breast Cancer Res. 2005;7:R130. doi: 10.1186/bcr959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. Nat Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letunic I, Bork P. Bioinformatics. 2007;23:127. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.