Abstract
Neisseria meningitidis expresses a two-component TonB-dependent receptor, HpuAB, which mediates heme-iron (Hm-Fe) acquisition from hemoglobin and hemoglobin-haptoglobin complexes. Due to genetic polymorphisms in the human haptoglobin gene, haptoglobin (and hemoglobin-haptoglobin) exists as three structurally distinct phenotypes. In this study, we examined the influence of the haptoglobin phenotype on the interactions of HpuAB with apo-haptoglobin and hemoglobin-haptoglobin. Growth assays confirmed that HpuAB utilizes hemoglobin-haptoglobin more efficiently than hemoglobin as an Fe source and revealed a preference for human-specific, polymeric 2-2 or 2-1 hemoglobin-haptoglobin complexes. We developed a flow cytometry-based assay to measure the binding kinetics of fluorescein-labeled ligands to HpuAB on live, intact meningococci. The binding affinity of HpuAB for hemoglobin-haptoglobin phenotypes correlated well with the ability of each ligand to support Neisseria meningitidis growth, with higher affinities exhibited for types 2-2 and 2-1 hemoglobin-haptoglobin. Saturable binding of Hb and apo-haptoglobin suggested that HpuAB-mediated utilization of hemoglobin-haptoglobin involves specific interactions with both components. In contrast to previous studies, we detected binding of HpuB expressed alone to hemoglobin, apo-haptoglobin, and hemoglobon-haptoglobin of all three phenotypes. However, in the absence of HpuA, the binding capacity and/or affinity of the receptor was reduced and the dissociation of hemoglobin was impaired. We did not detect binding of HpuA alone to hemoglobin, apo-haptoglobin, or hemoglobin-haptoglobin; however, the lipoprotein is crucial for optimal recognition and use of ligands by the receptor. Finally, this study confirmed the integral role of TonB and the proton motive force in the binding and dissociation of Hb and hemoglobin-haptoglobin from HpuAB.
The human body is protected by specific and nonspecific defense mechanisms and presents a hostile environment to invading microorganisms. A significant challenge faced by potential pathogens is the scarcity of essential nutrients such as iron (Fe). Virtually all free Fe in the human body is sequestered by carrier proteins, including lactoferrin (Lf) in exocrine secretions or transferrin (Tf), hemoglobin (Hb), and hemoglobin-haptoglobin (HbHp) in the bloodstream. To thrive in this environment, a pathogen must be able to overcome this host defense, known as nutritional immunity (85). Neisseria meningitidis is the leading cause of bacterial meningitis in the United States (9, 74) as well as of substantial morbidity and mortality worldwide (57, 66, 75). The vital role of Fe assimilation in meningococcal pathogenesis has been established in vitro and in vivo by using animal models of infection (5, 35-37, 70, 78, 81). This obligate human pathogen has evolved mechanisms to exploit multiple host Fe sources including ferric and ferrous salts, heme (Hm), siderophores, Tf, Lf, Hb, and HbHp (68, 73), making it an excellent model for the study of bacterial strategies for Fe acquisition.
The best-characterized group of neisserial Fe transporters consists of two-component receptors for Tf, Lf, and Hb/HbHp that contain an accessory lipoprotein and a TonB-dependent gated porin (20, 47, 50, 55, 56, 71, 72). The conserved interaction of a family of high-affinity outer membrane (OM) transporters with the TonB complex suggests that many, if not all, gram-negative bacteria have adopted similar strategies to transport Fe across the OM. TonB is anchored in the inner membrane in association with ExbBD and spans the periplasm, presumably to transduce the energy of the proton motive force (PMF) to OM receptors (4, 34, 38, 40). Although TonB physically interacts with OM receptors (1, 8, 32, 45, 83), the way in which TonB might convey PMF energy to these receptors to drive solute uptake remains unclear (10, 46). Homologues of tonB, exbB, and exbD have been identified and characterized in N. meningitidis (80). Mutations in the N. meningitidis TonB-ExbB-ExbD operon resulted in pleiotropic defects in Fe uptake from Tf, Lf, Hb, and HbHp, indicating a central role for TonB in Fe transport across the OM in the meningococcus (80). Five TonB-dependent Fe transporters have been confirmed in N. meningitidis, and an additional six putative members of this family have been identified in published meningococcal genome sequences (63, 82, 84). Unlike TonB-dependent siderophore receptors, which internalize the entire ligand, neisserial two-component receptors extract Fe (or Hm-Fe) directly from large host Fe carrier proteins by an unknown mechanism.
This study was focused on the meningococcal two-component receptor HpuAB, previously shown to bind Hb, apo-Hp, and HbHp (47, 49). Based on its homology to the TonB-dependent receptor family, we proposed that the 85-kDa HpuB interacts with TonB and forms a beta-barrel transport channel across the OM that is gated by an N-terminal periplasmic “plug” domain (49, 68). This model was based on the highly conserved three-dimensional X-ray crystallographic structures of three E. coli TonB-dependent siderophore receptors: FepA, FhuA, and FecA (6, 23-25, 52). The 35-kDa HpuA accessory lipoprotein is peripherally associated with the OM via an N-terminally linked fatty acid tether (49, 69). No known homologues of HpuA exist in public databases, and its role in HpuAB function remains to be determined. We have reported data suggesting that HpuA and HpuB physically interact to form a functional receptor complex (51, 69). Characterization of isogenic single mutants of HpuA and HpuB suggested that both receptor components are absolutely required for the utilization of Hb or HbHp (48, 69).
To begin to define the biochemical mechanisms involved in HpuAB Hm-Fe acquisition, we have characterized the binding interactions of this unique receptor with its multiple ligands. Numerous investigators have relied heavily on solid-phase dot blot assays to estimate receptor-ligand binding kinetics. However, Cornelissen and Sparling reported that solid-phase assays did not accurately represent the binding of ligand to live cells, perhaps because receptor conformations important for ligand binding were disrupted by the drying process (15). Therefore, we had previously developed a liquid-phase 125I-Hb binding assay to measure the equilibrium binding kinetics of Hb to HpuAB on live meningococci (69). In those experiments, wild-type HpuAB saturably bound 125I-Hb with a moderate affinity approximately 50-fold lower than that of the interaction of Tf with the TbpAB receptor (15, 69). We did not detect significant Hb binding to mutant strains lacking either HpuA, HpuB, or both receptor components by using that assay or a solid-phase dot blot analysis (69). Based on those observations, we concluded that both HpuA and HpuB were required to bind and to utilize Hb. Competitive binding assays established that Hb recognition by HpuAB was not species specific and that Hm did not effectively compete with intact Hb for binding to HpuAB. We identified at least one role of TonB and an intact PMF for HpuAB function by demonstrating that mutation of TonB or protonophore treatment severely impaired the dissociation of Hb from HpuAB (69).
Although Hb is efficiently utilized by N. meningitidis via HpuAB, Hb is probably not directly available to invading meningococci. On release into the bloodstream by erythrocyte lysis, Hb α2β2 tetramers dissociate into α1β1 dimers that are rapidly and irreversibly bound by Hp (18, 33, 59), an α2-sialoglycoprotein present at significant concentrations (∼1 to 2 mg/ml) in the sera of all mammals. Thus, HbHp complexes are the most physiologically relevant source of Hm-Fe encountered by the meningococcus during invasive disease. Hp is an acute-phase protein induced by cytokines during conditions such as infection, neoplasia, pregnancy, trauma, and acute myocardial infarction (44). Hp exhibits broad-spectrum activities ranging from anti-inflammatory properties (inhibition of prostaglandin synthesis) and angiogenesis to protection against free radicals (44). The most important biological function of Hp is Hb binding, which serves to prevent Fe loss and kidney damage by targeting the clearance of Hb through the liver. A single molecule of Hp is an α2β2 tetramer composed of two α1β1 dimers linked via a disulfide bond between the α-subunits, and it can bind to two Hb α1β1 dimers. In humans, there are three Hp phenotypes, types 1-1 and 2-2 (homozygous) and type 2-1 (heterozygous), that result from molecular heterogeneity of the α-chain gene (44). All Hp types have identical 40-kDa β-chains, whereas the Hp phenotype is determined by the α-chain present. The Hp1 allele encodes an 8.9-kDa α1-chain, whereas the Hp2 allele produces a 16-kDa α2-chain. The larger Hp2 allele results from the fusion of two α1 alleles and is found only in humans (53). Type 1-1 Hp exists as a “monomeric” α2β2 molecule as described above, whereas human-specific type 2-1 and 2-2 Hp can form very large disulfide-linked polymers (77) due to the presence of a duplicated cysteine residue within the Hp2 allele. Each Hp β-chain is capable of binding a single α1β1 Hb dimer; thus, type 1-1 Hp has two Hb binding sites while polymeric 2-2 and 2-1 Hp have a correspondingly larger Hb binding capacity per molecule.
Polymorphism of the multifunctional human Hp protein has numerous proven as well as potential clinical consequences. Epidemiological studies have shown that specific Hp phenotypes are linked to various inflammatory diseases, atherosclerosis, and autoimmune disorders (16, 17, 44). In addition, the gene frequencies of Hp alleles show markedly unequal geographic distributions, with the Hpα1 allele more prevalent in Africa and South America but relatively rare in Southeast Asia (44). The Hpα2 allele is thought to have originated in India about 2 million years ago and subsequently spread under genetic pressure, displacing the Hpα1 allele. Similar to the influence of malaria on the persistence of the sickle cell gene in Africa, infectious diseases may also be influenced by Hp polymorphisms. If a pathogen that utilizes HbHp in the bloodstream displays a preference for a particular Hp structure, one could hypothesize that an individual's Hp phenotype could contribute to disease susceptibility.
Extending our recent analysis of HpuAB-Hb interactions (69), we have determined the role of Hp in HpuAB-mediated utilization of HbHp as an Fe source. To do this, we developed a flow cytometry assay to characterize the binding kinetics of all three phenotypes of apo-Hp and HbHp complexes to HpuAB. In control experiments, we also conducted flow cytometric analysis of Hb binding to HpuAB and compared the results with previous data from equilibrium binding assays using radiolabeled Hb (69). We found that the flow cytometry binding assay provided a more reproducible and more sensitive method for examining receptor-ligand interactions that allowed us to measure previously undetectable ligand binding activity of HpuB expressed alone. Finally, we compared the effect of TonB inactivation and PMF dissipation on the interaction of HpuAB with Hb versus HbHp. Taken together, our findings significantly advance our understanding of the complex, energy-dependent mechanisms by which HpuAB recognizes and utilizes its seven structurally diverse ligands.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The construction of all N. meningitidis strains used in this study has been described previously (69) and is summarized in Table 1. N. meningitidis strains used in flow cytometry assays were cultured on Fe-deplete Chelex-treated defined medium (CDM0) plates overnight (18 h), resuspended in CDM0 broth, and grown to mid-log phase (optical density at 600 nm [OD600] = 0.3) in acid-washed sidearm flasks at 37°C with shaking. Conditions used to assay growth dependent on Fe acquisition from Hb and HbHp have been described previously (47, 69). Briefly, Hb and HbHp were added to CDM0 broth cultures to a final Fe concentration of 4 μM. Note that compared to monomeric-type 1-1 HbHp complexes, a smaller molar amount of the polymeric HbHp complexes containing type 2-2 or 2-1 Hp is required to provide 4 μM Fe because more Hm-Fe moieties are carried per complex. Lyophilized human ferrous Hb (type A0; Sigma) was dissolved in 10 mM HEPES buffer (pH 7.4) as a 100 μM stock solution. The purification of types 1-1, 2-2, and 2-1 Hp and preparation of the corresponding HbHp complexes is detailed below. Desferal (Df; Ciba-Geigy) was added at 10 μM to all Hb- and HbHp-supplemented cultures to remove free Fe. In addition, 16 μM human serum albumin (SA) was added to bind and sequester trace free Hm. Because N. meningitidis is unable to use Fe or Hm-Fe bound to Df or SA, respectively, these controls ensured that any growth observed was not the result of HpuAB-independent uptake of free Fe or Hm released into the medium due to denaturation of Hb during the assay (20).
TABLE 1.
Meningococcal strains used in this study
Native PAGE, SDS-PAGE, and Western blotting.
Using the discontinuous buffer system of Laemmli (41), protein samples were separated by either native polyacrylamide gel electrophoresis (PAGE) or sodium dodecyl sulfate (SDS)-PAGE. To denature Hp and HbHp, proteins were solubilized by boiling in SDS sample buffer (54) and electrophoresed on 15% acrylamide gels (10 by 12mm) at 25 mA per gel for 2 h. To visualize intact Hp and HbHp complexes, samples were prepared in sample buffer free of SDS and reducing agents and run on 7.5% native acrylamide gels without boiling. The gels were then either stained with Coomassie brilliant blue R-250 (Fisher) or transferred to nitrocellulose (Nitrobind; GE Osmonics, Fisher) at 0.98 A for 30 min. Western blots were blocked for 1 h at room temperature with blocking buffer containing 1% skim milk (Difco) in 1× TSB (50 mM Tris-HCl, 150 mM NaCl [pH 7.5]). Hp and HbHp were detected by probing blots for 1 h at room temperature with rabbit anti-human haptoglobin immunoglobulin G (IgG) (Sigma) diluted 1:10,000 in blocking buffer. The blots were then washed three times (10 min each) with 1× TSB before being incubated for 1 h at room temperature with goat anti-rabbit Ig heavy plus light chains conjugated with horseradish peroxidase (HRP; Southern Biotechnology Associates) diluted 1:2,000 in blocking buffer. After being washed as described above, the blots were developed using the 4-chloro-1-naphthol HRP color development reagent as specified by the manufacturer (Bio-Rad).
Solid-phase binding assays (dot blots).
The binding of ligands to immobilized meningococci was assessed essentially as described previously (47). Briefly, Fe-starved meningococcal cultures were grown in CDM0 to an OD600 of 0.45. Using a 96-well vacuum manifold (Stratagene), 100 μl of cells per well (dot) was filtered onto nitrocellulose. Dot blots were blocked with 1% skim milk in 1× TKB (100mM Tris-HCl, 150 mM KCl [pH 7.2]) for 1h at room temperature. Hp and HbHp (3 μg/ml each) were added to separate strips of immobilized meningococci and allowed to bind for 1 h at room temperature. Unbound ligand was removed by washing three times (15 min each) with 1× TKB containing 0.02% Tween 20. Ligands specifically bound to meningococcal cells were detected using a rabbit anti-human Hp primary antibody and goat anti-rabbit IgG-HRP conjugate as detailed above. Conjugate controls in which no ligand was added were used to account for nonspecific binding of antibodies to the cell surface or nitrocellulose membrane.
Hp screening and purification of Hp and HbHp.
To avoid the prohibitive cost and unreliable availability of commercial purified Hp, we purified Hp directly from human serum. Blood samples from anonymous donors were kindly provided by the Oklahoma Blood Institute (Oklahoma City, Okla.). Immediately following collection in nonheparinized tubes, blood samples were incubated on ice for 1 h to allow clot formation and retraction. Following packing of clotted blood samples by centrifugation in a clinical swinging-bucket centrifuge, the serum fraction was removed, aliquoted, and stored at −20°C. Prior to Hp purification, the Hp phenotype of each donor was determined by SDS-PAGE and Western blotting of 2 μl of serum. All Hp samples exhibited the invariant 40-kDa β-chain common to all Hp types, type 1-1 samples had only a ∼9-kDa α1S or α1F chain, type 2-2 samples showed only a 16-kDa α2 band, and heterozygous type 2-1 Hp samples had both the smaller α1 chain and larger α2 subunit (data not shown) (44). The characteristic monomeric (type 1-1) and polymeric (types 2-2 and 2-1) quaternary structures of each Hp phenotype were also observed as expected by native PAGE (data not shown).
Hp was purified using an affinity matrix consisting of polyclonal rabbit anti-human Hp IgG (Sigma) bound to recombinant protein A immobilized on 6% Fast Flow beaded agarose (Sigma). This matrix was made by incubation of 32 mg of Hp-specific antibody with 8 ml of protein A-agarose suspension (4 ml of resin) for 1 h at room temperature. The resin was washed twice with 50 ml of 0.2 M sodium borate (pH 8.0) and then suspended in 50 ml of borate buffer. Dimethyl pimelimidate (Pierce) was added to a final concentration of 20 mM for 30 min at room temperature to irreversibly cross-link anti-Hp IgG via its Fc region to immobilized protein A. The cross-linking reaction was stopped by adding 50 ml of 0.2 M ethanolamine (pH 8.0) (Sigma). The resin was collected by centrifugation and suspended in an additional 50 ml of 0.2 M ethanolamine (pH 8.0) with gentle shaking for 2 h to remove un-cross-linked antibody. Finally, the affinity resin was washed with 50 ml of phosphate-buffered saline (PBS; pH 7.4)-0.01% merthiolate, resuspended in 5 ml of PBS, and packed into a gravity-flow drip column.
Serum samples were passed over the affinity column five times to allow adsorption of Hp, and the column was washed with 200 ml of ice-cold wash buffer (50 mM Tris-HCl, 0.5 M NaCl, 0.1% Triton X-100 [pH 8.0]) at 4°C. The column was equilibrated with 100 ml of PBS, which served to remove residual salt and detergent. Affinity-purified Hp was eluted with 80 ml of cold 100 mM glycine (pH 2.5) and immediately neutralized with 1/10 volume of 1 M Tris (pH 8.0). Pooled eluate fractions were concentrated and equilibrated into Q-Sepharose buffer A (10 mM HEPES, 1 mM EDTA [pH 8.0]) (see below), using Centricon Plus-20 centrifugal filters (Millipore). SDS-PAGE revealed that this preparation was contaminated with two serum proteins whose molecular weights corresponded to the serum proteins albumin and IgG. To remove these contaminants, this material was loaded onto a Fast Flow Q-Sepharose (Sigma) high-performance liquid chromatography (HPLC) column previously equilibrated in buffer A, and 5 ml of affinity-purified Hp was loaded onto the column. The column was washed at a constant flow rate of 2 ml/min for 30 min with buffer A, and Hp was eluted using a 120-min linear gradient from 0 to 30% buffer B (10 mM HEPES, 1 mM EDTA, 1 M NaCl [pH 8.0]). To regenerate the Q-Sepharose, the column was stripped by a 25-min washing step with 100% buffer B and then equilibrated for 25 min with 100% buffer A. The purity, phenotype, structural integrity, and functionality of purified apo-Hp (ability to bind Hb and to bind to HpuAB) were established by native PAGE, SDS-PAGE, Western blot analysis, and dot blot analysis (data not shown). The absence of Hb-specific 412-nm Soret peaks in spectroscopic wavelength scans of all apo-Hp ligands confirmed the absence of contaminating Hb or HbHp that could also bind HpuAB.
HbHp complexes were formed by mixing Hp with at least a twofold excess Hb (for 30 min at room temperature with gentle agitation) to ensure that all Hp was saturated with Hb. Taking advantage of the disparate isoelectric points of free Hb (pI ≈ 7.0) and HbHp (pI = 5.5), HbHp complexes were separated from excess free Hb by using Q-Sepharose anion-exchange HPLC as described above. At pH 8.0, the strong anion HbHp eluted as a distinct peak from Hb, requiring significantly higher salt concentrations for elution than did the weak anion Hb. Thus, HbHp complexes used in binding assays were purified following formation and did not contain free Hb or Hp. Following the separation of excess free Hb from HbHp complexes by Q-Sepharose chromatography, purified 1-1 HbHp appeared as a single band on native gels migrating slower than Hb or Hp alone (data not shown). Complexes of Hb with polymeric Hps could not be verified due to their large size and laddering of bands on native gels. However, the reactivity with specific antibodies, the HPLC elution profile, and the red color of the samples were consistent with the presence of both Hb and Hp in type 2 Hp-containing complexes. In addition, dot blot and growth assays verified that purified HbHp complexes could bind to and support the growth of HpuAB-expressing meningococcal strain DNM140 (data not shown).
Fluorescent labeling of ligands.
To detect the binding of Hb, Hp, and HbHp to meningococci by flow cytometry, proteins were fluorescently labeled with the amine-reactive dye fluorescein-5-EX succinimidyl ester (FEX). FEX was prepared fresh as a 5-mg/ml stock in dimethyl sulfoxide and was reacted with target proteins at a 25:1 dye-to-protein ratio in 0.2 M sodium bicarbonate (pH 8.3) for 1 h at room temperature. Unreacted dye was removed by passage over a PD-10 desalting column (Amersham-Pharmacia Biotech) and elution with 0.2 M sodium bicarbonate (pH 8.3). The concentration of FEX-labeled protein conjugates was determined using the bicinchoninic acid assay (Pierce). To determine the degree of labeling (DOL), wavelength scans (240 to 600 nm) of FEX conjugates were conducted using a Beckman DU650 spectrophotometer. Based on the absorbance maximum (Amax) at 494 nm, the DOL was calculated using the following equation: DOL = (Amax × mw)/([protein] × ɛdye) where Amax is absorbance at 494 nm, mw is the molecular weight of the labeled protein, [protein] is the concentration of protein, and ɛdye is the extinction coefficient of dye at the Amax of 494 nm. The molar extinction coefficient for FEX determined at 494 nm is 76,000 cm−1 M−1 (31). Based on the ability to bind specifically to HpuA+B+ strain DNM140 as well as unmodified proteins in dot blot assays, no significant deleterious effects were induced in FEX-labeled ligands by fluorophore conjugation.
Flow cytometric binding kinetics assay.
Meningococci were grown in CDM0 to mid-log phase (OD600 = 0.3), and 100 μl of cells per reaction was blocked with 1% filter-sterilized skim milk. Bovine serum albumin (BSA) was not used as a blocking agent because 1% BSA inhibited the binding of Hb and HbHp to HpuAB as detected by flow cytometry. FEX-labeled Hb, Hp, or HbHp ligands were added to the indicated final concentrations and equilibrated for 15 min at room temperature. Cells were centrifuged at 5,000 × g for 1.5 min and washed with 0.5 ml of Hanks' balanced salts solution (pH 7.6) (HBSS; Sigma) to remove unbound ligand. A single wash step was sufficient to remove nonspecifically bound ligand proteins from negative control strain DNM68 (HpuA−B−). Bacteria with bound ligand were resuspended in 100 μl of cold HBSS-1% formalin and incubated on ice for 15 min to fix. Control experiments measuring the binding of FEX-labeled ligands to live, unfixed cells confirmed that formalin fixation had no effect on the binding or the ability of the Alexa conjugate (see below) to recognize ligands bound to the bacterial surface. Following washing with HBSS as above, samples were stained for 30 min on ice with 2 μg of anti-fluorescein/Oregon Green rabbit IgG fraction conjugated with Alexa Fluor 488 (Molecular Probes) in 100 μl of cold HBSS. Stained cells were washed as above before finally being suspended in 0.5 ml of HBSS. The Alexa 488-labeled secondary detection reagent was used to amplify the low but detectable levels of fluorescence resulting from the binding of fluorescein-tagged ligands to meningococci. Background levels of fluorescence detected in the presence of excess unlabeled ligand (routinely only 1 to 2 arbitrary fluorescein units [AU]) were subtracted from total ligand binding fluorescence values to calculate specific binding. Conjugate control experiments in which only anti-fluorescein conjugate (no ligand) was added indicated that nonspecific binding to cells was minimal. Stained samples were stored at 4°C and analyzed within 24 h by flow cytometry using a FACSCaliber equipped with a 488-nm laser (Flow and Image Cytometry Laboratory, Warren Medical Research Institute, University of Oklahoma Health Sciences Center, Oklahoma City, Okla.) to collect 10,000 events per sample. Storage of samples for this period did not affect the results of binding assays (data not shown). To determine the role of the PMF in HpuAB-ligand interactions, meningococci were incubated with the protonophore carbonylcyanide-m-chlorophenylhydrazone (CCCP) at 10μM (prepared fresh in dimethyl sulfoxide) for 10 min prior to ligand binding analysis. Conditions for CCCP treatment were optimized in an earlier study (69) such that energy-dependent utilization of Tf and Hb was maximally inhibited without deleterious effects on cell viability.
Competitive-binding flow cytometry assays were carried out as described above with the following modifications. Cells were mixed with 150 nM FEX-labeled type 1-1 HbHp and unlabeled competitors at concentrations ranging from 1.5 nM to 7.5 μM. The fluorescence of control cells to which no competitor was added served as the measure of 100% binding.
To measure the dissociation kinetics of Hb and type 1-1 HbHp from the Hpu receptor, multiple samples of cells were incubated with 200 nM FEX-Hb or FEX-HbHp for 15 min. Then a 20-fold excess (4 μM) unlabeled Hb or HbHp was added to individual tubes at various time points to “chase” dissociated FEX-labeled ligand. The cells were then washed, fixed, and stained as described previously. The 100% binding level (t = 0) was defined as the fluorescence values from control samples to which no competitor was added.
In flow cytometry assays involving type 2-1 or 2-2 Hp and HbHp, ligands were added to yield mass/volume concentrations equal to the concentration of type 1-1 Hp or HbHp used in the same experiment. The polymeric structure of type 2 Hp-containing proteins prohibits the calculation of the exact molar amounts of these proteins added to the binding-assay mixture. Therefore, at the same protein concentration, the molar amount of type 2-1 and 2-2 Hp or HbHp is approximately 2- to 10-fold lower than that of the corresponding type 1-1 Hp ligand. Thus, binding affinities determined for type 2-1 and 2-2 ligands are estimates and are effectively ∼2- to 10-fold higher than actually measured.
Analysis of binding kinetics data.
For a single flow cytometry binding assay, binding at each ligand concentration was measured in duplicate. The data presented in this study represent the mean fluorescence values from at least two independent experiments (minimum of four total data sets). The software programs CellQuest and FlowJo were used to analyze the flow cytometric data and prepare composite histograms. The geometric mean fluorescence resulting from the binding of FEX ligands to meningococci (minus the fluorescence of conjugate controls) was used to quantify the average level of ligand bound to the sample population of bacteria (10,000 cells/sample) at a given ligand concentration. GraphPad PRISM version 2.0 software was used to perform nonlinear regression curve-fitting analyses of binding data to estimate dissociation constants (Kd), competitive inhibition constants (Ki) and dissociation half-lives (t1/2).
RESULTS
Effect of Hp phenotype on the growth of N. meningitidis with HbHp Fe sources.
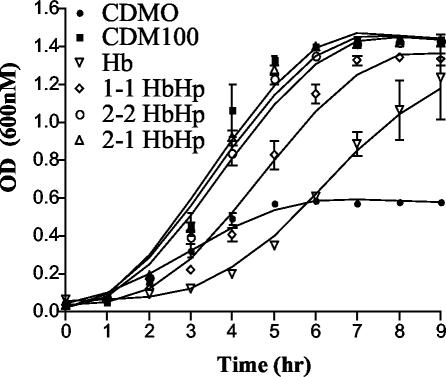
Our laboratory has previously demonstrated that, based on growth kinetics assays, HpuAB appears to utilize HbHp more efficiently than Hb alone (47). These experiments utilized commercial Hp (Sigma) from pooled human plasma that corresponds to heterozygous 2-1 Hp. In this study, we examined whether the Hp phenotype affected the ability of meningococci to use HbHp as an Fe source (Fig. 1). When provided with Hb as the sole Fe source, strain DNM140 exhibited a noticeable lag time but grew to a higher final OD (doubling time, ∼2.25 h) than did the CDM0 Fe-depleted control (Fig. 1). Cultures supplemented with HbHp grew at higher rates (doubling times, ∼1 h) and to higher final densities, regardless of Hp phenotype. However, the Hp phenotype influenced Fe acquisition from HbHp by HpuAB. HbHp complexes containing either polymeric Hp phenotype (types 2-2 and 2-1) supported growth kinetics equivalent to that of the CDM100 Fe-replete control containing 100 μM ferric nitrate (Fig. 1). Meningococci utilized monomeric type 1-1 HbHp to grow slightly slower than type 2-2 and 2-1 HbHp-supplemented cultures, but with a notable lag phase. These data indicated that the human-specific, polymeric type 2-2 and 2-1 HbHp complexes were the preferred ligand and Fe source for HpuAB.
FIG. 1.
Growth kinetics of HpuA+B+ DNM140 with Hb and HbHp complexes. Meningococci expressing wild-type HpuAB were grown overnight on Fe-depleted CDM0 plates before being used to inoculate CDM0 broth cultures supplemented with Hb, type 1-1 HbHp, type 2-2 HbHp, or type 2-1 HbHp. Control cultures grown in Fe-replete (CDM100) and Fe-depleted (CDM0) media were included. The data presented are the averaged results of two independent experiments.
Flow cytometric analysis of HpuAB-Hb binding kinetics.
Previously, we reported the binding kinetics of 125I-Hb to HpuAB determined by using a liquid-phase assay (69). In attempting to extend those results using 125I-labeled HbHp complexes, we found that the 125I-labeled ligand gave erratic and irreproducible binding isotherms. We speculate that the oxidative conditions required for 125I labeling damaged the HbHp and compromised its ability to interact properly with HpuAB. We also noted unacceptably high levels of nonspecific binding to numerous filter matrices used to separate free ligand from cell-bound ligand (data not shown). We could not remedy these problems by optimizing the labeling conditions. Previously, iodination of tyrosine residues of Hp was shown to dramatically reduce its biological activity and induce Hp polymer formation, which may account for our results (19). An alternative approach, suggested by El Ghmati et al., employed flow cytometry to assay fluorescein-labeled type 1-1 Hp binding to monocytes and lymphocytes (22). Gauduchon et al. also have measured the binding kinetics of a staphylococcal leucocidin to human neutrophils by using a flow cytometric assay (27).
Thus, we developed a nonradioactive flow cytometric assay to measure the equilibrium binding kinetics of ligands to HpuAB on live meningococcal cells. The fluorescent dye FEX was chosen for this application for several reasons. Labeling of protein ligands with the amine-reactive dye FEX is less likely to compromise their structural integrity than is the highly oxidative iodination reaction. Fluorescein derivatives exhibit good water solubility and excellent fluorescence yield and have an excitation maximum (494 nm) close to the wavelength of 488-nm lasers used in flow cytometers. Finally, FEX has a seven-atom hydrophilic spacer between the fluorophore and reactive group to decrease quenching after conjugation to target proteins and increase the accessibility of the fluorophore to secondary detection reagents (31). Although the specific binding of FEX-labeled Hb and HbHp to HpuAB was detectable, the fluorescence intensities were only two- to threefold above those of background and DNM68 negative controls (data not shown). The low signal intensity was perhaps due to strong quenching of dye fluorescence by proximity to the Fe atoms of Hb or to the protein itself. To exploit FEX as a hapten and amplify the signal strength, we used an Alexa Fluor 488 antifluorescein conjugate to stain cells with FEX ligands bound to their surface. The cell fluorescence, resulting from binding of Hb or HbHp to HpuAB, was increased by approximately 10-fold. This could be due to the increased distance between conjugate-linked dyes and Fe quenchers, the enhanced fluorophore properties of Alexa Fluor 488 compared to fluorescein (increased photostability, quantum yield, decreased pH sensitivity, and identical absorption spectra), and/or the higher density of dye molecules associated with each ligand molecule (31).
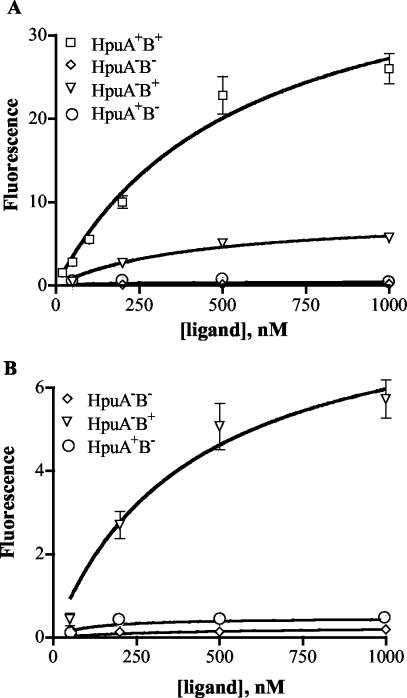
Using this assay, we first compared the kinetics of the Hb-HpuAB interaction to previous 125I-Hb binding data to validate the results of the flow cytometric binding analysis (69). The saturable binding of FEX-labeled Hb to HpuA+B+ strain DNM140 is shown in Fig. 2A. Nonlinear regression analysis yielded an apparent Kd of 555 nM and a predicted Bmax of 42 AU, indicative of the maximum amount of Hb bound at equilibrium (Table 2). As noted previously, Hb dissociates into dimers after release from erythrocytes and dilution into the serum (7, 30, 67), and only α1β1 Hb can form complexes with Hp (44, 60). Thus, calculations of Hb molar concentrations in this study are based on the molecular weight of α1β1 Hb dimers to accurately reflect the ligand as N. meningitidis encounters it in vitro and in vivo. Our previously reported affinity of HpuAB for 125I-Hb (Kd = 149.2 nM) was calculated based on the molecular weight of tetrameric Hb and should be adjusted by a factor of 2 (Kd = 298.4 nM) (69). Analysis of Hb-HpuAB binding kinetics by flow cytometry indicated a moderate-affinity interaction comparable to that measured by the radioligand binding assay. The 1.8-fold higher apparent Kd observed by flow cytometry might be due to differences between the two assays such as the number of cells used or ligand alterations induced by different labeling methods.
FIG. 2.
(A) Binding kinetics of Hb. Binding of fluorescein-labeled Hb to N. meningitidis strains DNM140 (HpuA+B+), DNM68 (HpuA−B−), DNM69 (HpuA−B+), and DNM143 (HpuA+B−) was detected using an Alexa Fluor 488 anti-fluorescein conjugate and measured by flow cytometry. The geometric mean fluorescence (AU) is shown as a function of fluorescent-ligand concentration. Binding at each ligand concentration was assessed in duplicate, and the data points shown represent the mean of at least two independent experiments. Error bars indicate the standard error of the mean for each combined data set. (B) Hb binding data for DNM68, DNM69, and DNM143 from panel A were plotted on a smaller y-axis range to illustrate the low-capacity binding to DNM69 for comparison with DNM140 (A). Error bars for DNM68 and DNM143 are too small to be seen due to the scale of the graph.
TABLE 2.
Summary of HpuAB binding kinetics of Hb, apo-Hp, and HbHpa
| Ligand | DNM140 (wild type)
|
DNM69 (HpuA−B+)
|
||
|---|---|---|---|---|
| Kd (nM) | Bmax (AU) | Kd (nM) | Bmaxb (AU) | |
| Hb | 555 | 42 | 409 | 9 |
| 1-1 Hp | 4,249 | 29 | >5,000 | ++ |
| 2-2 Hp | 808 | 26 | >5,000 | ++ |
| 2-1 Hp | 486 | 24 | >5,000 | ++ |
| 1-1 HbHp | 119 | 23 | >5,000 | ++ |
| 2-2 HbHp | 84 | 21 | >5,000 | ++ |
| 2-1 HbHp | 39 | 20 | >5,000 | + |
No binding to DNM68 was detected, and only very low binding of types 2-2 and 1-1 HbHp to DNM143 was detected.
+ and ++ indicate relative binding (nonsaturable) of ligands to DNM69.
The extremely low level of background staining seen in this assay is evident in the lack of fluorescence of the HpuA−B− control strain, DNM68 (Fig. 2), even when incubated with high concentrations of FEX-labeled Hb. Consistent with our earlier data, HpuA expressed in the absence of HpuB by DNM143 was unable to bind detectable levels of Hb (Fig. 2). Similar analysis of DNM69 (HpuA−B+), however, revealed low but detectable saturable binding of Hb to HpuB alone (Fig. 2). In fact, HpuB bound Hb almost as avidly as did the intact receptor (DNM69 Kd = 409 nM versus DNM140 Kd = 555nM) (Table 2). However, in the absence of HpuA, HpuB bound ca- fivefold less Hb than did wild-type HpuAB at saturating ligand concentrations (DNM69 Bmax = 9 AU, DNM140 Bmax = 42 AU [Table 2]). The decreased Hb binding capacity of DNM69 was not attributed to decreased levels of receptor protein expression, since DNM69 produced amounts of HpuB comparable to those found in wild-type cells (69). Even though HpuB alone was capable of binding Hb with a moderate affinity, HpuA was required for wild-type binding capacity and subsequent uptake of Hm-Fe from Hb.
Binding of HbHp and apo-Hp to HpuAB.
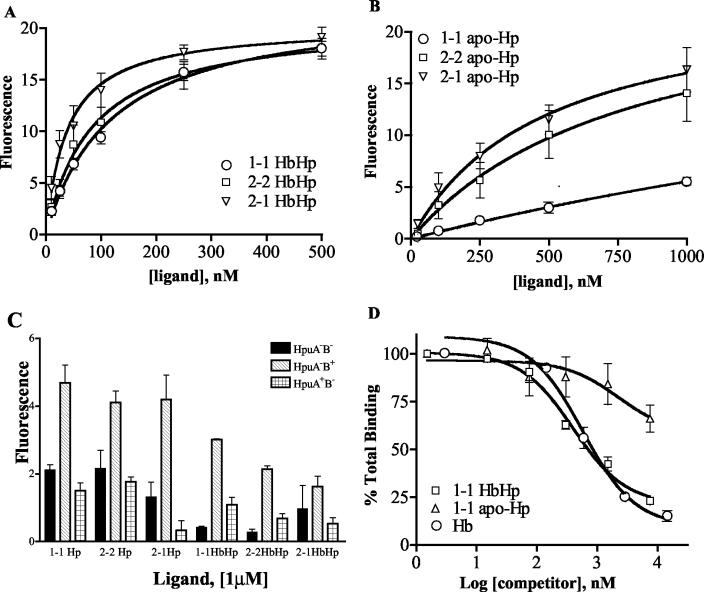
We observed saturable binding of type 1-1, 2-2, and 2-1 HbHp complexes to wild-type HpuAB expressed by strain DNM140, illustrated by the binding isotherms shown in Fig. 3A. HpuAB exhibited 4.7-fold (type 1-1), 6.6-fold (type 2-2), and 14-fold (type 2-1) higher affinities for these HbHp phenotypes than for Hb (Table 2). The enhanced growth kinetics of N. meningitidis with HbHp correlated well with the higher affinity of HpuAB for all types of HbHp complexes relative to Hb. However, the Hp phenotype did influence interactions with HpuAB. Polymeric type 2-2 and 2-1 HbHp complexes bound with higher affinity than type 1-1 HbHp and, as shown in Fig. 1, served as better Fe sources for meningococci. As noted in Materials and Methods, type 2-2 and 2-1 HbHp were added to equivalent mass/volume concentrations as type 1-1 HbHp. Therefore, at a given concentration there are fewer molar equivalents of the larger, polymeric complexes (types 2-2 and 2-1) than the monomeric complexes (type 1-1). As a result, the higher affinity of HpuAB for human-specific type 2-2 and 2-1 HbHp compared to type 1-1 complexes is likely to be even more pronounced than the reported Kd values indicate. The lower affinity of HpuAB for type 1-1 HbHp was paralleled by slower growth kinetics of N. meningitidis with that Fe source (Fig. 1). Comparison of the Bmax values (maximum number of binding sites) of Hb and type 1-1 HbHp with HpuAB (Table 2) suggests that HpuA+B+ N. meningitidis contains more binding sites for Hb than for HbHp. Correcting for differences in the DOL between the two ligands (∼7 dye/Hb molecule versus 12 dye/1-1 HbHp molecule), HpuAB bound approximately two- to threefold more molar equivalents of Hb than HbHp complexes (compare the plateau levels of the curves in Fig. 2A with those in Fig. 3A). The possible implications of this observation for the stoichiometry of HpuAB-ligand interactions are addressed in Discussion.
FIG. 3.
(A) Kinetics of binding of types 1-1, 2-2, and 2-1 HbHp to strain DNM140 (HpuA+B+). The data are presented as described in the legend to Fig. 2. The data for each ligand represent the mean of at least three independent experiments conducted in duplicate (total of six data sets per data point on the graph). (B) Kinetics of binding of types 1-1, 2-2, and 2-1 apo-Hp to strain DNM140. As above, data presented are the mean of three separate assays in which each sample was assayed in duplicate. (C) Binding of all three apo-Hp and HbHp phenotypes to DNM68 (HpuA−B−), DNM69 (HpuA−B+), and DNM143 (HpuA+B−). The height of each bar represents the amount bound on incubation of cells with 1 μM each fluorescein-labeled ligand. (D) Ability of Hb and apo-Hp to compete with HbHp for binding to strain DNM140. Cells were mixed with 150 nM FEX-labeled type 1-1 HbHp and 1.5 nM to 7.5 μM unlabeled competitors (type 1-1 HbHp, type 1-1 apo-Hp, and Hb). Results are expressed as the percentage of total binding, with 100% binding determined from control samples to which no competitor was added. Each data point represents the mean of two independent assays, with each sample assayed in duplicate.
Consistent with dot blot binding analysis (data not shown), HpuAB was able to bind specifically to all three phenotypes of apo-Hp. Figure 3B shows the saturable binding of polymeric types 2-2 (Kd = 808 nM) and 2-1 (Kd = 486 nM) apo-Hp and the much weaker binding of type 1-1 apo-Hp (Kd = 4.2 μM). As noted above, the apparent Kd values for types 2-2 and 2-1 Hp underestimate the actual affinity of HpuAB for these ligands. The relative affinity of HpuAB for the various apo-Hp types (Hp 2-1 > Hp 2-2 > Hp 1-1) also reflects the relative affinity of HpuAB for the corresponding HbHp complex (Table 2). Thus, Hp contributes directly to the physical interaction of HbHp with HpuAB, and the Hp phenotype influences the affinity of that interaction.
We then characterized the ability of N. meningitidis mutants lacking one or both HpuAB components to bind the six apo-Hp and HbHp ligands (Fig. 3C). We found that in addition to interacting with Hb, HpuB (in the absence of HpuA) bound with a low affinity to all three phenotypes of apo-Hp and HbHp complexes (hatched bars in Fig. 3C). However, we did not observe saturable binding of ligands to DNM69 at ligand concentrations up to 1 μM, which precluded calculation of the apparent dissociation constants. The binding of Hp and HbHp to DNM143 (HpuA+B−) was not significantly greater than the levels of nonspecific binding to the negative control strain DNM68 (HpuA−B−) (Fig. 3C). Hence, despite the absolute requirement for HpuA in HpuAB-mediated Fe acquisition (48, 69), in the absence of HpuB we did not detect specific involvement of this lipoprotein in binding to any of the seven ligands recognized by HpuAB.
To confirm these data, we performed competitive binding assays. Both unlabeled type 1-1 HbHp (Ki = 179 nM) and Hb (Ki = 258 nM) competed effectively with 1-1 FEX-HbHp, reducing the binding of the labeled ligand by more than 75% (Fig. 3D). Unlabeled type 2-2 and 2-1 HbHp complexes yielded very similar competition curves, indicating that these ligands also specifically compete with type 1-1 FEX-HbHp for binding to HpuAB (data not shown). However, types 1-1, 2-2, and 2-1 apo-Hp only partially competed with FEX-HbHp, reducing labeled ligand binding by ∼25 to 30% of total binding even at saturating concentrations (50-fold molar excess of competitor) (Fig. 3D and data not shown).
HpuAB ligand interactions are TonB and PMF dependent.
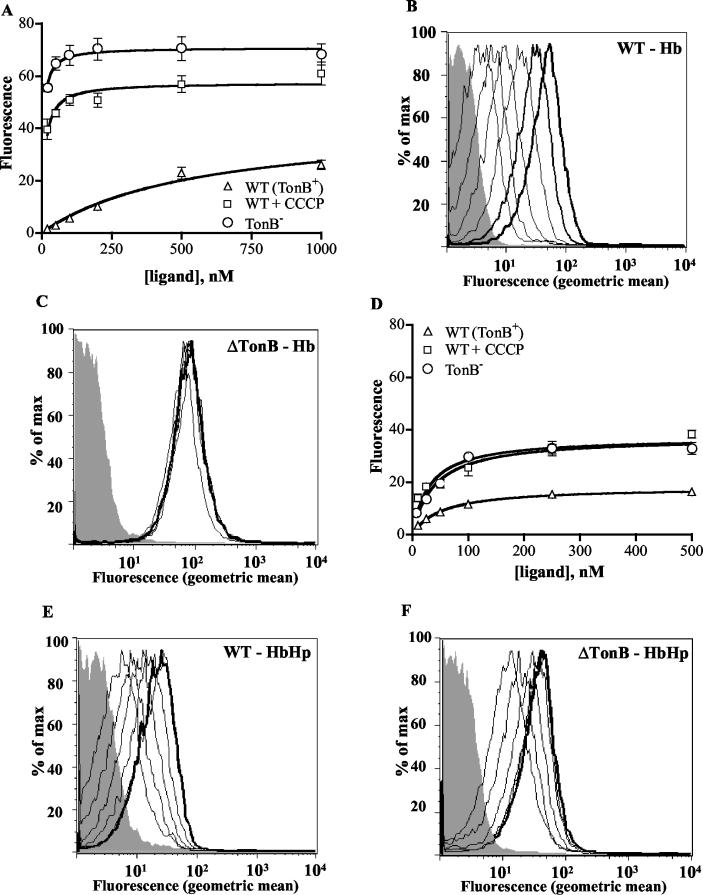
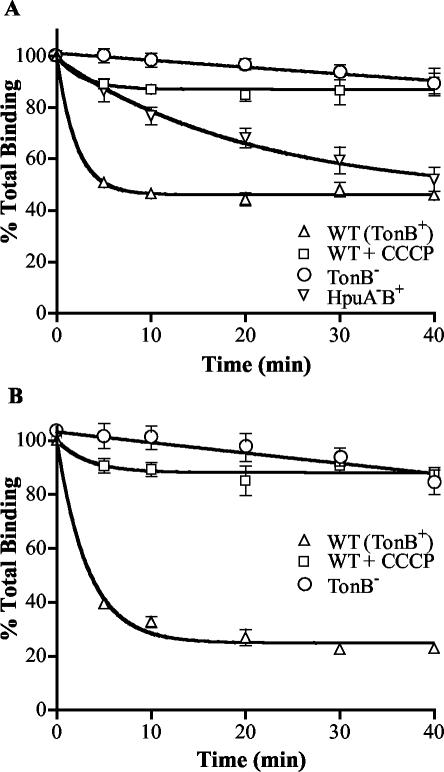
Previous studies have shown that the binding and dissociation of ligands from TonB-dependent receptors is energy dependent, requiring both the presence of TonB and an intact PMF (2, 3, 14, 29, 69). To characterize the interaction of deenergized HpuAB receptors with Hb and HbHp, we studied the binding of these ligands to the TonB− mutant DNM146 and to wild-type strain DNM140 treated with the protonophore CCCP. Figure 4 illustrates the dramatic effect of the energy state of the receptor on ligand binding kinetics. The absence of TonB in DNM146 resulted in a ∼100-fold increase in the apparent affinity of HpuAB for Hb, a phenotype closely mimicked by treatment with CCCP (Fig. 4A; Table 3). Similarly, the Hb binding capacity of deenergized meningococci was significantly elevated compared to that of wild-type cells (Table 3). This is illustrated dramatically by the flow cytometric histograms in Fig. 4B and C, which show the data used to generate the binding isotherms in Fig. 4A. Incubation of TonB− cells with even the lowest FEX-Hb concentration (1.5 nM) resulted in almost maximal fluorescence (compare Fig. 4B and C).
FIG. 4.
Hb and HbHp binding to deenergized HpuAB. The data are presented as described in the legend to Fig. 2. Each data point represents the mean of two independent experiments conducted in duplicate. (A) Isotherm of Hb binding to wild-type (DNM140) and deenergized (DNM146) HpuAB. (B) Flow cytometry histogram depicting FEX-Hb binding to TonB+ DNM140. From left to right, peaks correspond to unstained control (grey) and 20, 50, 100, 200, 500, and 1,000 nM Hb. (C) Flow cytometry histogram of FEX-Hb binding to TonB− DNM146. The shaded grey peak is the fluorescence of the unstained negative control. Ligand was added at same concentrations as used for the experiment panel B. (D) Isotherm of type 1-1 HbHp binding to wild-type and deenergized HpuAB. (E) Histogram of flow cytometry analysis of HbHp binding to DNM140. Left to right, fluorescence peaks of unstained control (grey) and 10, 25, 50, 100, 250, and 500 nM ligand. Note that the 250 and 500 nM (bold) peaks are superimposed. (F) Histogram of type 1-1 HbHp binding to DNM146. Ligand was added at same concentrations as used for the experiment in panel E. Fluorescence peaks of the 100, 250, and 500 nM (bold) samples are superimposed.
TABLE 3.
Role of TonB and an intact PMF in binding and dissociation kinetics
| Ligand | DNM140 (wild type)
|
DNM146 (TonB−)
|
DNM140 + CCCP
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Kd (nM) | Bmax (AU) | t1/2 (min) | Kd (nM) | Bmax (AU) | t1/2 (min) | Kd (nM) | Bmax (AU) | t1/2 (min) | |
| Hb | 555 | 42 | 1.4 | 5 | 71 | 186 | 11 | 57 | 1.8 |
| 1-1 HbHp | 119 | 23 | 2.3 | 39 | 37 | 442 | 31 | 37 | 2.3 |
The interaction of HpuAB with HbHp complexes was also found to be sensitive to the presence of TonB and an intact PMF (Fig. 4D to F). DNM146 (TonB−) and CCCP-treated DNM140 cells showed a modest (1.6-fold) increase in their HbHp binding capacity, comparable to the effect on the Bmax of Hb binding (Fig. 4D; Table 3). However, the apparent affinity of deenergized HpuAB for HbHp increased by only 3- to 4-fold (Table 3), compared with the 100-fold-enhanced affinity of HpuAB for Hb noted above. This discrepancy in the degree of energy dependence between Hb and HbHp binding can be visualized by comparing the histograms in Fig. 4B and C (Hb) with those in Fig. 4E and F (HbHp). The ability of HpuAB to bind more Hb than HbHp is again evident from the level of fluorescence at the plateau of the binding curves shown in Fig. 4A and D.
Dissociation of HbHp from HpuAB requires TonB and an intact PMF.
In a previous study, we showed that TonB and the PMF were crucial for the dissociation of 125I-Hb from HpuAB (69). For comparison to that earlier data, we determined the rate of dissociation of FEX-Hb from DNM140 (wild type), DNM146 (TonB−), and CCCP-treated DNM140. Approximately 50% of bound FEX-Hb dissociated very rapidly from energized HpuAB within the first 5 min, with no further dissociation detected over the 40-min assay (Fig. 5A), which closely mimicked dissociation of 125I-Hb (69). The half-life (t1/2) of Hb was estimated to be 1.4 min (Table 3) based on nonlinear regression analysis fitting these data to a one-phase exponential decay. Figure 5A clearly shows that Hb dissociation from both deenergized cell types is impaired. However, the t1/2 of Hb dissociation from TonB− cells was increased by >100-fold whereas treatment with CCCP had little or no effect on the half-life of the ligand (Table 3). Although CCCP treatment did not change the half-life of Hb, only ∼10% of the bound ligand was released from receptors on cells in which the PMF was dissipated. We also investigated whether a functional two-component receptor is necessary for ligand utilization and release by analyzing Hb dissociation from DNM69 (HpuA−B+). Over the 40-min course of the assay, ∼50% of the bound FEX-Hb was able to dissociate from HpuB, but at a rate 10 times lower than that of dissociation from wild-type HpuAB (DNM69 t1/2 = 14.4 min, DNM140 t1/2 = 1.4 min) (Fig. 5A). These data suggest that HpuA is important for some aspect of Hb utilization that is a prerequisite for release of the ligand from the receptor.
FIG. 5.
Energy-dependent dissociation of Hb and HbHp from HpuAB. DNM140, DNM146, and CCCP-treated DNM140 cells were equilibrated with 200 nM FEX-labeled Hb (A) or type 1-1 HbHp (B). Then, 20-fold excess unlabeled competitor was added at various time points and the amount of labeled ligand remaining bound to cells over time was determined. Control samples to which no competitor was added defined 100% binding. Each strain was tested twice in duplicate for each ligand (total of four data sets).
Finally, we characterized the effect of the energy state of the receptor on the dissociation kinetics of type 1-1 HbHp from HpuAB. We did not examine type 2-2 and 2-1 complexes because of the polymeric nature of these ligands. Like Hb, HbHp dissociated very rapidly from wild-type energized HpuAB, with a half-life of 2.3 min. Within the initial 5 min, ∼60% of the bound HbHp had dissociated from the cells, with an additional 10 to 15% decrease in the level of bound HbHp over the next 10 min (Fig. 5B). Note the discrepancy between Hb and HbHp in the maximal amount of ligand that can be dislodged from the receptor by unlabeled competitor (compare Fig. 5A and B). HbHp incubated with TonB− DNM146 became virtually irreversibly bound, having an estimated half-life of >7 h (Table 3). As we observed with Hb, DNM140 treated with CCCP released only 10% of bound HbHp, but that released fraction had a half-life equivalent to that of HbHp dissociating from wild-type HpuAB (Table 3).
DISCUSSION
The binding of Hb by Hp exerts a bacteriostatic effect on many organisms like Escherichia coli, which are unable to acquire Fe from the HbHp complex (21). However, in addition to N. meningitidis, there are several microbial species able to bind and use HbHp as an Fe source. Nonetheless, the bacterial mechanisms involved in binding and scavenging Fe from HbHp remain poorly characterized. Even less is known about the influence of the Hp phenotype on receptor-ligand interactions. Bacteroides fragilis can obtain Hm from Hb and HbHp, with Hb supporting growth better than HbHp (62). Lammler et al. have demonstrated that gram-positive group A streptococci bind to all three types of apo-Hp and HbHp (42, 43). In both cases, a specific receptor for HbHp or alternative mechanism for acquiring Hm-Fe from HbHp remains to be characterized. Vibrio vulnificus is able to acquire Hm-Fe from types 1-1 and 2-2 HbHp but not from type 2-1 HbHp (88). This pathogen relies on a Hm-inducible Hb protease, not a surface receptor, to acquire Hm-Fe from HbHp (61). To satisfy its absolute requirement for exogenous Hm, Haemophilus influenzae type b expresses at least three distinct single component TonB-dependent HbHp receptors designated HgpA, HgpB, and HgpC (39, 58, 64, 65). Thus far, quantitative biochemical analyses of the interactions of the phase-variable Hgp receptors with HbHp of any phenotype have not been reported. Finally, a homologue of N. meningitidis HpuAB has been identified in Neisseria gonorrhoeae that also binds Hb, apo-Hp, and HbHp (13). Studies of the gonococcal receptor have focused on its interaction with Hb, whereas the more complex HbHp ligands have not yet been examined (11-13). To our knowledge, this study is the first detailed biochemical analysis of the kinetics of binding of all Hp and HbHp phenotypes to a bacterial receptor.
We developed a flow cytometric method for analyzing HbHp binding by N. meningitidis. This assay has several important advantages to previously described techniques using radiolabeled ligands (69). Like the radioactive assay format, flow cytometry monitors the binding of ligand to live cells in the liquid phase. This is important to accurately reflect the kinetics of the dynamic, energy-dependent interaction of ligands at equilibrium with the receptor. This method allowed us to analyze the same number of cells in each sample and provided a unique ability to observe binding to cell populations instead of just capturing the average number of cell-associated counts. This method allowed us to exclude potential binding to membrane blebs and look only at cell-associated binding events. The nonoxidative labeling of Hp and HbHp with fluorescein (FEX) proved less damaging than 125I modification of the structural integrity of ligand proteins. Also, FEX-labeled ligands would not be subjected to radiolysis during storage. Finally, the reproducibility and sensitivity of flow cytometry coupled with low levels of nonspecific fluorescence allowed us to measure previously undetected binding of ligands with HpuB.
We reexamined the kinetics of binding of Hb to HpuAB in order to validate the flow cytometric binding assay (69). However, one unavoidable caveat of the flow cytometric assay for direct quantiation of ligand binding was the use of the Alexa conjugate amplification step. The modest apparent affinity of wild-type HpuAB for FEX-labeled Hb estimated by flow cytometry (555 nM) was less than twofold different from the affinity we determined for 125I-Hb dimers (298 nM). This suggested that the two techniques yield comparable results and that the indirect detection method did not adversely affect our results. The minor differences between the Kd values may have arisen from effects of the distinct labeling methods on Hb, batch-to-batch variation in Hb samples, or other assay-specific factors. We observed very low levels of nonspecific Hb binding to our negative control strain DNM68 (HpuA−B−). In agreement with dot blot analyses and radioligand binding data (69), the HpuA lipoprotein was unable to bind detectable levels of Hb when expressed alone by strain DNM143 (HpuA+B−). In contrast to earlier results, we found that HpuA−B+ strain DNM69 was able to specifically and saturably bind Hb (69). This observation is consistent with the ability of Hb to protect HpuB expressed by DNM69 from digestion with trypsin (69). In addition, Chen et al. have reported the ability to affinity purify gonococcal HpuB from an hpuA mutant by using a Hb matrix (11). In the absence of HpuA, HpuB exhibited a near-wild-type affinity but a four- to fivefold-reduced binding capacity for Hb. HpuB expressed in the absence of HpuA also bound all three phenotypes of Hp and HbHp, although with a much lower affinity than that of the wild-type HpuAB receptor. Our data suggest that the lipoprotein is important for optimal interactions of HpuAB with Hb, apo-Hp, and HbHp, even though we did not detect direct binding of HpuA to any of these ligands. It is possible that HpuA does participate directly in ligand binding but is dependent on HpuB to fold into a binding-competent conformation. The ability of Hb or HbHp bound to wild-type HpuAB to protect HpuA from protease digestion is consistent with the participation of HpuA in ligand binding (69). The absence of HpuA more severely impaired interactions with Hp and HbHp, perhaps indicating that HpuA is involved in making contacts with Hp. Consistent with that hypothesis, the N. meningitidis single-component TonB-dependent receptor HmbR lacks an accessory lipoprotein and is able to bind Hb but not Hp or HbHp (78, 79). Alternatively, HpuA may act indirectly and serve as a sort of chaperone that promotes the proper folding of HpuB into a binding competent conformation. The observation that only a fraction of HpuB expressed alone is capable of binding Hb, but does so with wild-type affinity, may reflect this type of contribution to ligand binding from HpuA. Although its specific role in receptor function is unclear, our data indicate that HpuA is essential for Hb utilization and is required for optimal ligand binding.
In growth assays, we demonstrated that HpuAB preferred HbHp to Hb as an Fe source, regardless of Hp phenotype. Interestingly, Francis et al. have reported that Staphylococcus aureus and Streptococcus pyogenes exhibit a similar ligand preference and were able to extract more 59Fe-Hm from HbHp complexes than from free Hb (26). Although HpuAB is able to utilize all three phenotypes of HbHp complexes, human-specific, polymeric types 2-2 and 2-1 HbHp enhanced N. meningitidis growth kinetics to a greater extent than did monomeric type 1-1 HbHp complexes. There are several possible reasons for the preference of HpuAB for HbHp over Hb. The formation of a complex with Hp may render the Hm-Fe moiety of Hb easier for HpuAB to extract and internalize. In this regard, Smith and Beck (76) reported increased exposure of Hm after Hp binding to Hb due to conformational changes of the globin chains. Circular dichroism spectra of the Soret region of HbHp and Hb and increased sensitivity of HbHp-associated Hm to dithionite also indicate a significant difference in the Hm environment between free Hb and HbHp (28). Furthermore, HpuAB bound apo-Hp, indicating that there are binding sites on the receptor for both Hb and Hp. Thus, HpuAB binds HbHp more avidly than it binds free Hb. This study also revealed that ligand recognition by HpuAB is sensitive to Hp structure, as indicated by higher affinities for human-specific polymeric type 2-1 and 2-2 HbHp complexes than for monomeric type 1-1 HbHp. Similarly, HpuAB bound directly to Hp with a low affinity (compared to HbHp) in a phenotype-dependent manner that paralleled the recognition of HbHp by HpuAB.
The Hp phenotype dependence of HpuAB ligand binding exhibits the following order: type 2-1 > type 2-2 > type 1-1. It is perhaps not surprising that HpuAB is able to distinguish between Hp phenotypes. The primary sequence and molecular weight of each Hp type are different due to the Hpα gene duplication that gave rise to the Hp2 allele. The much larger type 2-2 and 2-1 Hp ligands may engage the receptor with a much higher avidity than the monomeric type 1-1 does. Circular dichroism examination of Hp structures by Hamaguchi revealed significant conformational differences between type 1-1 Hp and types 2-2 and 2-1 Hp (28). High-resolution scanning transmission electron microscopy of Hp by Wejman et al. showed that 2-1 Hp exists as linear polymers of various lengths terminated by type 1 molecules lacking an extra cysteine residue (86, 87). Type 2-2 Hp polymers, on the other hand, form closed circular molecules (86, 87). The higher affinity of HpuAB for type 2-1 Hp and HbHp may reflect a steric hindrance or reduced avidity for the more structurally constrained type 2-2 cyclic polymers.
Despite the importance of Hp in the interaction of HbHp with the receptor, HpuAB probably interacts more extensively with the Hb portion of the complex. This would seem logical, given that HpuAB must extract Hm-Fe from Hb, not from Hp. In addition, HpuAB bound two to three times more molar equivalents of Hb than of HbHp complexes. Although the significance of this observation is unclear, it is tempting to speculate that each functional HpuAB receptor may have two binding sites for Hb dimers. Recall that a molecule of type 1-1 HbHp is composed of an α2β2 Hp tetramer with an α1β1 Hb dimer tightly bound to each Hp β chain. HbHp could bind to HpuAB such that the Hb dimers occupy the two putative Hb binding sites, with the more distal Hp chains involved in important but secondary receptor contacts. Competitive binding assays also supported the conclusion that the major contacts between HpuAB and HbHp complexes involve Hb. Hb was able to compete almost as well as HbHp with fluorescein-labeled HbHp complexes, whereas a 50-fold molar excess of Hp (type 1-1, 2-2, or 2-1) inhibited FEX-HbHp binding by only 50%.
The apparent affinity of HpuAB for Hb and HbHp was dramatically increased when the receptor was deenergized by the absence of TonB. Similar results were obtained following treatment of wild-type meningococci with the protonophore CCCP, confirming the close link between TonB activity and an intact PMF gradient. Our observations are consistent with the dramatic increase in the apparent affinity of the gonococcal TbpAB receptor for Tf when TonB is absent (14). The receptor energy state had a larger impact on the observed affinity of HpuAB for Hb than for HbHp (compare Fig. 4B and C with Fig. 4E and F). The reason for this disparity in the magnitude of the effect of TonB on the two ligands is not known. It should be noted that estimated Kd values for deenergized HpuAB might not accurately reflect equilibrium dissociation constants for several reasons. The extremely high level of binding at even the lowest ligand concentrations indicates that ligand depletion is probably disrupting equilibrium conditions. In addition, Hb and HbHp binding to deenergized HpuAB does not follow the law of mass action used to describe receptor-ligand equilibria, which assumes that binding is a reversible event.
Deenergized HpuAB also exhibited an increased binding capacity for Hb and HbHp that reflects a very high-affinity, irreversible interaction (see below) in which virtually all receptors are occupied, in contrast to the lower capacity observed for energized receptors at equilibrium. In contrast, previously reported radioligand binding assay data showed that deenergized HpuAB bound less, not more, Hb than did the wild type (69). The oxidative radiolabeling of Hb with 125I may have damaged the ligand structure such that energy-dependent interactions with HpuAB were altered. Alternatively, the apparently low Hb binding capacity might be the result of high levels of background seen in radioligand assays of deenergized HpuAB. The amount of specific 125I-Hb binding was determined by subtracting the amount of nonspecific binding (binding detected in the presence of excess unlabeled Hb) from the total binding. The extremely high-affinity and high-capacity binding of Hb to deenergized HpuAB may have allowed a significant amount of 125I-Hb to bind even in the presence of cold competitor. In contrast, the flow cytometric data closely parallel the description by Cornelissen et al. of energy-dependent TbpAB-Tf interactions (14).
In examining dissociation kinetics, we found that 50% of Hb that was bound to wild-type HpuAB dissociated very rapidly within 5 min after the addition of excess unlabeled Hb, with no further Hb dissociation over the course of the experiment. This was consistent with previous observations (69). Although the dissociation rate of HbHp was comparable to that of Hb, a notably larger fraction (almost 80%) of bound HbHp dissociated from HpuAB. It is unclear how the distinct dissociation kinetics of Hb and HbHp are related to the mechanism of HpuAB Fe acquisition from these ligands. If a substantial fraction of HpuAB receptors are occupied by Hb but unable to release the ligand and reset for another cycle of Fe transport, this could contribute to the reduced ability of Hb to serve as an Fe source. As noted previously (69), binding of Hb and HbHp to HpuAB in the absence of TonB was, in effect, irreversible. Although this phenotype was closely mimicked by CCCP-treated wild-type cells, about 10% of bound Hb and HbHp dissociated from HpuAB with normal dissociation rates despite dissipation of the PMF. This could indicate that CCCP did not completely abolish the PMF in all cells or that a weakened proton gradient was able to energize Hb release from a few receptors.
These data clearly demonstrated that TonB and an intact PMF energy gradient are required for the dissociation of both Hb and HbHp from HpuAB. TonB might play a direct role in triggering ligand release by inducing conformational changes in the receptor that disrupts ligand interactions. On the other hand, one could speculate that a step that requires the input of energy from TonB, such as the extraction of Hm-Fe from Hb, must occur prior to the release of Hb or HbHp. This second hypothesis is substantiated by the partially impaired dissociation of Hb from DNM69, which expresses HpuB in the absence of HpuA. Like DNM146 (TonB−), DNM69 is unable to grow using Hb or HbHp as an Fe source, presumably because it cannot extract Hm-Fe from Hb (69). The extraction of Hm-Fe from Hb might induce conformational changes in the ligand structure that lower its affinity for HpuAB and promote its release. Whereas the presence of HpuA appears to facilitate ligand release, TonB-derived energy is absolutely required for the dissociation of Hb or HbHp from HpuAB.
Acknowledgments
We thank Arlene Wilson (Oklahoma Blood Institute) for her help procuring serum samples for haptoglobin purification and Jim Henthorn (OUHSC Flow and Image Cytometry Laboratory) for flow cytometry training and suggestions. We also thank Allison Gillaspy and Thomas Ducey for critical reading of the manuscript.
Editor: D. L. Burns
REFERENCES
- 1.Bell, P. E., C. D. Nau, J. T. Brown, J. Konisky, and R. J. Kadner. 1990. Genetic suppression demonstrates interaction of TonB protein with outer membrane transport proteins in Escherichia coli. J. Bacteriol. 172:3826-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradbeer, C. 1993. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 175:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradbeer, C., and M. L. Woodrow. 1976. Transport of vitamin B12 in Escherichia coli: energy dependence. J. Bacteriol. 128:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V., S. Gaisser, C. Herrmann, K. Kampfenkel, H. Killmann, and I. Traub. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brener, D., I. W. De Voe, and B. E. Holbein. 1981. Increased virulence of Neisseria meningitidis after in vitro iron-limited growth at low pH. Infect. Immun. 33:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 7.Bünn, H. F. 1987. Subunit assembly of hemoglobin: an important determinant of hematologic phenotype. Blood 69:1-6. [PubMed] [Google Scholar]
- 8.Cadieux, N., C. Bradbeer, and R. J. Kadner. 2000. Sequence changes in the ton box region of BtuB affect its transport activities and interaction with TonB protein. J. Bacteriol. 182:5954-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2000. Prevention and control of meningococcal disease. Recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. 49:1-10. [Google Scholar]
- 10.Chang, C., A. Mooser, A. Pluckthun, and A. Wlodawer. 2001. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276:27535-27540. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. J., C. Elkins, and P. F. Sparling. 1998. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect. Immun. 66:987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C. J., D. McLean, C. E. Thomas, J. E. Anderson, and P. F. Sparling. 2002. Point mutations in HpuB enable gonococcal HpuA deletion mutants to grow on hemoglobin. J. Bacteriol. 184:420-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, C. J., P. F. Sparling, L. A. Lewis, D. W. Dyer, and C. Elkins. 1996. Identification and purification of a hemoglobin-binding outer membrane protein from Neisseria gonorrhoeae. Infect. Immun. 64:5008-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornelissen, C. N., J. E. Anderson, and P. F. Sparling. 1997. Energy-dependent changes in the gonococcal transferrin receptor. Mol. Microbiol. 26:25-35. [DOI] [PubMed] [Google Scholar]
- 15.Cornelissen, C. N., and P. F. Sparling. 1996. Binding and surface exposure characteristics of the gonococcal transferrin receptor are dependent on both transferrin-binding proteins. J. Bacteriol. 178:1437-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahlqvist, S. R., G. Beckman, and L. Beckman. 1988. Serum protein markers in systemic lupus erythematosus. Hum. Hered. 38:44-47. [DOI] [PubMed] [Google Scholar]
- 17.Dahlqvist, S. R., and N. Frohlander. 1985. Haptoglobin groups and rheumatoid arthritis Hum. Hered. 35:207-211. [DOI] [PubMed] [Google Scholar]
- 18.Dobryszycka, W. 1997. Biological functions of haptoglobin—new pieces to an old puzzle. Eur. J. Clin. Chem. Clin. Biochem. 35:647-654. [PubMed] [Google Scholar]
- 19.Dobryszycka, W., and I. Bec-Katnik. 1975. Effect of modification on physico-chemical and biological properties of haptoglobin. Acetylation, iodination and nitration. Acta Biochim. Pol. 22:143-153. [PubMed] [Google Scholar]
- 20.Dyer, D. W., E. P. West, and P. F. Sparling. 1987. Effects of serum carrier proteins on the growth of pathogenic neisseriae with heme-bound iron. Infect. Immun. 55:2171-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eaton, J. W., P. Brandt, J. R. Mahoney, and J. T. Lee, Jr. 1982. Haptoglobin: a natural bacteriostat. Science 215:691-693. [DOI] [PubMed] [Google Scholar]
- 22.El Ghmati, S. M., E. M. Van Hoeyveld, J. G. Van Strijp, J. L. Ceuppens, and E. A. Stevens. 1996. Identification of haptoglobin as an alternative ligand for CD11b/CD18. J. Immunol. 156:2542-2552. [PubMed] [Google Scholar]
- 23.Ferguson, A. D., V. Braun, H. P. Fiedler, J. W. Coulton, K. Diederichs, and W. Welte. 2000. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 9:956-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 26.Francis, R. T., Jr., J. W. Booth, and R. R. Becker. 1985. Uptake of iron from hemoglobin and the haptoglobin-hemoglobin complex by hemolytic bacteria. Int. J. Biochem. 17:767-773. [DOI] [PubMed] [Google Scholar]
- 27.Gauduchon, V., S. Werner, G. Prevost, H. Monteil, and D. A. Colin. 2001. Flow cytometric determination of Panton-Valentine leucocidin S component binding. Infect. Immun. 69:2390-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamaguchi, H. 1969. Purification and some properties of the three common genetic types of haptoglobins and the hemoglobin-haptoglobin complexes. Am. J. Hum. Genet. 21:440-456. [PMC free article] [PubMed] [Google Scholar]
- 29.Hancock, R. W., and V. Braun. 1976. Nature of the energy requirement for the irreversible adsorption of bacteriophages T1 and phi80 to Escherichia coli. J. Bacteriol. 125:409-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hargrove, M. S., T. Whitaker, J. S. Olson, R. J. Vali, and A. J. Mathews. 1997. Quaternary structure regulates hemin dissociation from human hemoglobin. J. Biol. Chem. 272:17385-17389. [DOI] [PubMed] [Google Scholar]
- 31.Haugland, R. P. 2002. Handbook of fluorescent probes and research products, 9th ed. Molecular Probes, Inc., Eugene, Orag.
- 32.Heller, K. J., R. J. Kadner, and K. Gunther. 1988. Suppression of the btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene 64:147-153. [DOI] [PubMed] [Google Scholar]
- 33.Hershko, C. 1975. The fate of circulating haemoglobin. Br. J. Haematol. 29:199-204. [DOI] [PubMed] [Google Scholar]
- 34.Higgs, P. I., P. S. Myers, and K. Postle. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holbein, B. E. 1981. Enhancement of Neisseria meningitidis infection in mice by addition of iron bound to transferrin. Infect. Immun. 34:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holbein, B. E. 1980. Iron-controlled infection with Neisseria meningitidis in mice. Infect. Immun. 29:886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holbein, B. E., K. W. Jericho, and G. C. Likes. 1979. Neisseria meningitidis infection in mice: influence of iron, variations in virulence among strains, and pathology. Infect. Immun. 24:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaskula, J. C., T. E. Letain, S. K. Roof, J. T. Skare, and K. Postle. 1994. Role of the TonB amino terminus in energy transduction between membranes. J. Bacteriol. 176:2326-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin, H., Z. Ren, P. W. Whitby, D. J. Morton, and T. L. Stull. 1999. Characterization of hgpA, a gene encoding a haemoglobin/haemoglobin-haptoglobin-binding protein of Haemophilus influenzae. Microbiology 145:905-914. [DOI] [PubMed] [Google Scholar]
- 40.Karlsson, M., K. Hannavy, and C. F. Higgins. 1993. A sequence-specific function for the N-terminal signal-like sequence of the TonB protein. Mol. Microbiol. 8:379-388. [DOI] [PubMed] [Google Scholar]
- 41.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 42.Lammler, C., and H. Blobel. 1986. Binding of human haptoglobin to streptococci of serological group A. Zentbl. Bakteriol. Mikrobiol. Hyg. Ser. A 261:161-166. [DOI] [PubMed] [Google Scholar]
- 43.Lammler, C., T. Guszezynski, and W. Dobryszycka. 1988. Further characterization of haptoglobin binding to streptococci of serological group A. Zentbl. Bakteriol. Mikrobiol. Hyg. Ser. A 269:454-459. [DOI] [PubMed] [Google Scholar]
- 44.Langlois, M. R., and J. R. Delanghe 1996. Biological and clinical significance of haptoglobin polymorphism in humans. Clin. Chem. 42:1589-1600. [PubMed] [Google Scholar]
- 45.Larsen, R. A., D. Foster-Hartnett, M. A. McIntosh, and K. Postle. 1997. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J. Bacteriol. 179:3213-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letain, T. E., and K. Postle. 1997. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol. Microbiol. 24:271-283. (Erratum, 25:617, 1997.) [DOI] [PubMed] [Google Scholar]
- 47.Lewis, L. A., and D. W. Dyer. 1995. Identification of an iron-regulated outer membrane protein of Neisseria meningitidis involved in the utilization of hemoglobin complexed to haptoglobin. J. Bacteriol. 177:1299-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis, L. A., M. Gipson, K. Hartman, T. Ownbey, J. Vaughn, and D. W. Dyer. 1999. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol. Microbiol. 32:977-989. [DOI] [PubMed] [Google Scholar]
- 49.Lewis, L. A., E. Gray, Y. P. Wang, B. A. Roe, and D. W. Dyer. 1997. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol. Microbiol. 23:737-749. [DOI] [PubMed] [Google Scholar]
- 50.Lewis, L. A., K. Rohde, M. Gipson, B. Behrens, E. Gray, S. I. Toth, B. A. Roe, and D. W. Dyer. 1998. Identification and molecular analysis of lbpBA, which encodes the two-component meningococcal lactoferrin receptor. Infect. Immun. 66:3017-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis, L. A., M. H. Sung, M. Gipson, K. Hartman, and D. W. Dyer. 1998. Transport of intact porphyrin by HpuAB, the hemoglobin-haptoglobin utilization system of Neisseria meningitidis. J. Bacteriol. 180:6043-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 95:771-778. [DOI] [PubMed] [Google Scholar]
- 53.Maeda, N., F. Yang, D. R. Barnett, B. H. Bowman, and O. Smithies. 1984. Duplication within the haptoglobin Hp2 gene. Nature 309:131-135. [DOI] [PubMed] [Google Scholar]
- 54.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 55.Mickelsen, P. A., E. Blackman, and P. F. Sparling. 1982. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from lactoferrin. Infect. Immun. 35:915-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mickelsen, P. A., and P. F. Sparling. 1981. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from transferrin and iron compounds. Infect. Immun. 33:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore, P. S. 1992. Meningococcal meningitis in sub-Saharan Africa: a model for the epidemic process. Clin. Infect. Dis. 14:515-525. [DOI] [PubMed] [Google Scholar]
- 58.Morton, D. J., P. W. Whitby, H. Jin, Z. Ren, and T. L. Stull. 1999. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect. Immun. 67:2729-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller-Eberhard, U., J. Javid, H. H. Liem, A. Hanstein, and M. Hanna. 1968. Plasma concentrations of hemopexin, haptoglobin and heme in patients with various hemolytic diseases. Blood 32:811-815. [PubMed] [Google Scholar]
- 60.Nagel, R. L., and Q. H. Gibson. 1971. The binding of hemoglobin to haptoglobin and its relation to subunit dissociation of hemoglobin. J. Biol. Chem. 246:69-73. [PubMed] [Google Scholar]
- 61.Nishina, Y., S. Miyoshi, A. Nagase, and S. Shinoda. 1992. Significant role of an exocellular protease in utilization of heme by Vibrio vulnificus. Infect. Immun. 60:2128-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otto, B. R., M. Sparrius, D. J. Wors, F. K. de Graaf, and D. M. MacLaren. 1994. Utilization of haem from the haptoglobin-haemoglobin complex by Bacteroides fragilis. Microb. Pathog. 17:137-147. [DOI] [PubMed] [Google Scholar]
- 63.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 64.Ren, Z., H. Jin, D. J. Morton, and T. L. Stull. 1998. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect. Immun. 66:4733-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren, Z., H. Jin, P. W. Whitby, D. J. Morton, and T. L. Stull. 1999. Role of CCAA nucleotide repeats in regulation of hemoglobin and hemoglobin-haptoglobin binding protein genes of Haemophilus influenzae. J. Bacteriol. 181:5865-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Riedo, F. X., B. D. Plikaytis, and C. V. Broome. 1995. Epidemiology and prevention of meningococcal disease. Pediatr. Infect. Dis. J. 14:643-657. [DOI] [PubMed] [Google Scholar]
- 67.Riggs, A. F. 1998. Self-association, cooperativity and supercooperativity of oxygen binding by hemoglobins. J. Exp. Biol. 201:1073-1084. [DOI] [PubMed] [Google Scholar]
- 68.Rohde, K. H., and D. W. Dyer. 2003. Mechanisms of iron acquisition by the human pathogens Neisseria meningitidis and Neisseria gonorrhoeae Front. Biosci. 8:1186-1218. [DOI] [PubMed] [Google Scholar]
- 69.Rohde, K. H., A. F. Gillaspy, M. D. Hatfield, L. A. Lewis, and D. W. Dyer. 2002. Interactions of haemoglobin with the Neisseria meningitidis receptor HpuAB: the role of TonB and an intact proton motive force. Mol. Microbiol. 43:335-354. [DOI] [PubMed] [Google Scholar]
- 70.Schryvers, A. B., and G. C. Gonzalez. 1989. Comparison of the abilities of different protein sources of iron to enhance Neisseria meningitidis infection in mice. Infect. Immun. 57:2425-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schryvers, A. B., and L. J. Morris. 1988. Identification and characterization of the human lactoferrin-binding protein from Neisseria meningitidis. Infect. Immun. 56:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schryvers, A. B., and L. J. Morris. 1988. Identification and characterization of the transferrin receptor from Neisseria meningitidis. Mol. Microbiol. 2:281-288. [DOI] [PubMed] [Google Scholar]
- 73.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 74.Schuchat, A., K. Robinson, J. D. Wenger, L. H. Harrison, M. Farley, A. L. Reingold, L. Lefkowitz, B. A. Perkins, and the Active Surveillance Team. 1997. Bacterial meningitis in the United States in 1995. N. Engl. J. Med. 337:970-976. [DOI] [PubMed] [Google Scholar]
- 75.Schwartz, B., P. S. Moore, and C. V. Broome. 1989. Global epidemiology of meningococcal disease. Clin. Microbiol. Rev. 2(Suppl):S118-S124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith, M. J., and W. S. Beck. 1967. Peroxidase activity of hemoglobin and its subunits: effects thereupon of haptoglobin. Biochim. Biophys. Acta 147:324-333. [DOI] [PubMed] [Google Scholar]
- 77.Smithies, O. 1955. Zone electrophoresis in starch gels: group variations in the serum proteins of normal human adults. Biochem. J. 61:629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stojiljkovic, I., V. Hwa, L. de Saint Martin, P. O'Gaora, X. Nassif, F. Heffron, and M. So. 1995. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531-541. [DOI] [PubMed] [Google Scholar]
- 79.Stojiljkovic, I., J. Larson, V. Hwa, S. Anic, and M. So. 1996. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J. Bacteriol. 178:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stojiljkovic, I., and N. Srinivasan. 1997. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in neisseriae. J. Bacteriol. 179:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun, Y. H., S. Bakshi, R. Chalmers, and C. M. Tang. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 6:1269-1273. [DOI] [PubMed] [Google Scholar]
- 82.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 83.Tuckman, M., and M. S. Osburne. 1992. In vivo inhibition of TonB-dependent processes by a TonB box consensus pentapeptide. J. Bacteriol. 174:320-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turner, P. C., C. E. Thomas, I. Stojiljkovic, C. Elkins, G. Kizel, D. A. Ala'Aldeen, and P. F. Sparling. 2001. Neisserial TonB-dependent outer-membrane proteins: detection, regulation and distribution of three putative candidates identified from the genome sequences. Microbiology 147:1277-1290. [DOI] [PubMed] [Google Scholar]
- 85.Weinberg, E. D. 1975. Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA 231:39-41. [DOI] [PubMed] [Google Scholar]
- 86.Wejman, J. C., D. Hovsepian, J. S. Wall, J. F. Hainfeld, and J. Greer. 1984. Structure and assembly of haptoglobin polymers by electron microscopy. J. Mol. Biol. 174:343-368. [DOI] [PubMed] [Google Scholar]
- 87.Wejman, J. C., D. Hovsepian, J. S. Wall, J. F. Hainfeld, and J. Greer. 1984. Structure of haptoglobin and the haptoglobin-hemoglobin complex by electron microscopy. J. Mol. Biol. 174:319-341. [DOI] [PubMed] [Google Scholar]
- 88.Zakaria-Meehan, Z., G. Massad, L. M. Simpson, J. C. Travis, and J. D. Oliver. 1988. Ability of Vibrio vulnificus to obtain iron from hemoglobin-haptoglobin complexes. Infect. Immun. 56:275-277. [DOI] [PMC free article] [PubMed] [Google Scholar]