Abstract
Infection with Shiga toxin (Stx)-producing bacteria and the subsequent release of Stxs and endotoxins into the bloodstream may damage blood vessels in the colon, kidneys, and central nervous system, leading to bloody diarrhea, acute renal failure, and neurological complications. The proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β) may contribute to the pathogenesis of Stx-induced vascular lesions by up-regulating toxin receptor expression on endothelial cells. We previously showed that macrophages treated with purified Shiga toxin 1 (Stx1) or lipopolysaccharides (LPS) secrete TNF-α and IL-1β. Northern blot analysis revealed that treatment of the human monocytic cell line THP-1 with LPS induced a rapid and transient increase in steady-state TNF-α and IL-1β transcripts. In contrast, Stx1 induced slower but prolonged elevations in cytokine transcripts. The presence of both stimulants resulted in optimal cytokine mRNA induction in terms of kinetics and prolonged expression. Compared to LPS, Stx1 was a poor inducer of IL-1β protein expression, although levels of soluble IL-1β induced by all treatments continually increased over 72 h. IL-1β transcripts were not induced by Stx1 B-subunits. Using the transcriptional inhibitor actinomycin D, we determined that treatment with Stx1 or Stx1 plus LPS induced cytokine transcripts with increased stability compared to transcripts induced by LPS alone. For all treatments, IL-1β mRNA decay was slower than TNF-α. Collectively, our data suggest that Stxs affect cytokine expression, in part, at the posttranscriptional level by stabilizing mRNAs. Optimal TNF-α expression occurs when both Stxs and LPS are present.
Shigella dysenteriae serotype 1 and Shiga toxin-producing Escherichia coli (STEC) are causative agents of bacillary dysentery and hemorrhagic colitis, respectively. These bacteria produce potent cytotoxins known as Shiga toxins (Stxs). Shiga toxin produced by S. dysenteriae serotype 1 and Shiga toxin 1 (Stx1) produced by STEC are essentially identical toxins. STEC may also express one or more Stxs that are antigenically distinct from Stx1, called Stx2 and Stx2 variants (31, 34). The toxins are thought to damage blood vessels serving the colon (9). In addition to exacerbating intestinal damage associated with infection, Stxs are associated with the development of life-threatening postdiarrheal complications (21). The action of Stxs on glomerular and brain microvascular endothelial cells may activate prothrombotic and proinflammatory cascades that lead to the development of the hemolytic-uremic syndrome (HUS) and central nervous system (CNS) complications (36, 38, 46). Stxs are AB5 toxins, having a single enzymatic A-subunit in noncovalent association with five identical B-subunits (12, 47). Stx B-subunits bind to cells primarily through interaction with the membrane glycolipid receptor globotriaosylceramide (Gb3) (19, 26). Following clathrin-dependent endocytosis, the toxins undergo retrograde transport through the trans-Golgi network and the Golgi apparatus to reach the endoplasmic reticulum and nuclear membranes, where the toxins gain access to the cytosol (reviewed in reference 44). During transport, the A-subunit is cleaved and reduced, and once in the cytosol, the enzymatic A1 fragment mediates the depurination of a single adenine residue located near the 3′ end of 28S rRNA of the 60S ribosomal subunit (8, 45). The single cleavage event results in the inhibition of peptide elongation and the loss of protein synthesis (16, 35).
While the mechanism of action of Stxs and the resultant cytotoxicity are well described, the pathogenic mechanism(s) leading to the profound vascular damage seen in HUS is less well understood. Possible contributors to pathogenesis may include bacterial lipopolysaccharides (LPS) and the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β). Patients with bacillary dysentery or HUS caused by S. dysenteriae serotype 1 are frequently endotoxemic (23), and patients with hemorrhagic colitis or HUS caused by STEC frequently present with elevated antibody titers directed against STEC O antigens (20, 38). LPS are known to be potent inducers of cytokine production. TNF-α and IL-1β levels are elevated in the serum and urine of some HUS patients but are not consistently elevated, as in the case with endotoxic shock (reviewed in reference 49). Although the relative detrimental and beneficial effects of cytokines in the pathogenesis of HUS remain unclear, TNF-α and IL-1β treatment promotes the up-regulation of membrane Gb3 expression on cultured vascular endothelial cells derived from human umbilical veins, brain, and renal glomeruli, resulting in increased sensitivity to Stxs in vitro (7, 27, 28, 40, 54, 56). These data suggest that TNF-α and IL-1β may contribute to the pathogenesis of HUS by rendering blood vessels in the colon, kidneys, and CNS more susceptible to the destructive action of Stxs. Isogai et al. (18) showed that the exogenous administration of TNF-α to mice infected with STEC exacerbated the severity of CNS and glomerular pathology, while treatment with a TNF-α inhibitor ameliorated vascular damage. The source(s) of cytokines in target organs is not known, but macrophages are known to be major producers of proinflammatory cytokines when stimulated with microbes or microbial products (33, 41, 57). Human macrophages produce TNF-α and IL-1β when stimulated with purified Stxs in vitro (39, 55). Infusion of purified Stx1 into mice harboring a TNF-α promoter-chloramphenicol acetyltransferase transgene showed selective induction of chloramphenicol acetyltransferase activity in the kidneys (14). Collectively, these data suggest that tissue macrophages localized to target organs are possible sources of TNF-α and IL-1β.
Earlier studies by Sakiri et al. suggested that Stxs stimulate cytokine production through activation of transcription factors nuclear factor κB (NF-κB) and activator protein 1 (AP-1) (43). We noted, however, that stimulation of the differentiated human monocytic cell line THP-1 with Stx1 or Stx1 plus LPS resulted in the prolonged elevation of TNF-α transcripts compared to cells stimulated with LPS alone. These data suggested that Stxs may regulate cytokine expression at transcriptional and posttranscriptional levels. We hypothesized that cytokine mRNAs may be stabilized in macrophages stimulated with Stxs, explaining in part the prolonged high steady-state levels of TNF-α mRNA extracted from toxin-treated cells. Therefore, we measured the kinetics of TNF-α and IL-1β mRNA induction and determined the rates of mRNA decay through the use of the transcriptional inhibitor actinomycin D (ActD). Finally, we examined the kinetics of Stx1-induced IL-1β protein production and the requirement for the presence of the Stx1 A subunit in the activation of IL-1β production.
MATERIALS AND METHODS
Cells.
The human myelogenous leukemia cell line THP-1 (53) was purchased from American Type Culture Collection (Rockville, Md.). Cell cultures were maintained in RPMI 1640 (Gibco-BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, Utah), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in 5% CO2 in a humidified incubator.
Toxins.
Stx1 was expressed from E. coli DH5α transformed with plasmid pCKS112 containing the toxin operon under control of the T7 promoter (50). Stx1 in crude bacterial lysates was purified by sequential ion-exchange, chromatofocusing, and immunoaffinity chromatography as described previously (51). The toxin was assessed for homogeneity by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis with silver staining and Western blots. The toxin preparation was then passed through ActiClean Etox columns (Sterogene Bioseparations, Carlsbad, Calif.) to remove trace endotoxin contaminants and was determined to contain <0.1 ng of endotoxin per ml by the Limulus amebocyte lysate assay (Associates of Cape Cod, Falmouth, Maine). Purified Stx1 B-subunits were the kind gift of Cheleste Thorpe, Tufts University School of Medicine, Boston, Mass. Purified LPS derived from E. coli O111:B4 were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Macrophage differentiation and stimulation.
The mature macrophage-like state was induced by treating THP-1 cells (106 cells per ml) for 48 h with phorbol 12-myristate 13-acetate (PMA; Sigma) at a concentration of 50 ng/ml in 100-mm-diameter culture dishes. Differentiated, plastic-adherent cells were washed twice with cold Dulbecco's phosphate-buffered saline (Sigma) and incubated with fresh medium lacking PMA but containing 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). The medium was then changed every 24 h for 3 additional days. Experiments were performed on the fourth day after removal of PMA.
For Northern blot analyses, differentiated cells were treated with LPS (200 ng/ml), Stx1 (400 ng/ml), or both for various times. Ramegowda and Tesh demonstrated that these stimulant doses produced maximal cytokine protein secretion in differentiated THP-1 cells in vitro (39). The amount of Stxs necessary to cause systemic disease in humans is unknown, but the Stx1 dose used in these experiments represents approximately 4 × 105 50% cytotoxic doses for Vero cells and approximately 1 50% lethal dose for CD-1 mice (50). To determine mRNA decay rates, differentiated THP-1 cells were treated with the same concentrations of stimulants in the presence or absence of the transcriptional inhibitor ActD (Sigma) using the protocol of Harrold et al. (15). Since we have shown that the induction kinetics of TNF-α and IL-1β are slower in response to treatment with Stx1 than for treatment with LPS, actD (5.0 μg/ml) was added to the cells 1 h after LPS stimulation or 2 h after stimulation with Stx1 or Stx1 plus LPS. Two 1.0-ml aliquots of all cell supernatants were collected and stored at −20°C for use in enzyme-linked immunosorbent assays (ELISAs).
Probe synthesis.
The human TNF-α and IL-1β cDNA clones were purchased from American Type Culture Collection and grown in Luria-Bertani broth (Difco, Detroit, Mich.) containing ampicillin (50 μg/ml; Sigma) or tetracycline (20 μg/ml; Sigma), respectively. Plasmids containing the TNF-α and IL-1β cDNAs were isolated using the QIAprep spin miniprep kit (Qiagen, Valencia, Calif.) and subsequently digested with restriction endonucleases (New England Biolabs, Beverly, Mass.) AvaI and HindIII for TNF-α or PstI for IL-1β. Digestion of the TNF-α cDNA resulted in a 578-bp fragment containing 450 bp of the TNF-α coding region as well as 128 bp of the 3′ untranslated region (3′-UTR). Digestion of the IL-1β cDNA resulted in a 1,047-bp fragment containing the entire IL-1β coding region plus 5′- and 3′-UTRs. Fragments were visualized with ethidium bromide after electrophoresis of the digests into 1.2% agarose gels. The 578- and 1,047-bp DNA fragments were excised and purified using the QIAquick gel extraction kit (Qiagen). Approximately 25 ng of the TNF-α and IL-1β cDNA fragments were resuspended in sterile water for use in [α-32P]dCTP random primer labeling reactions employing the Rediprime II kit (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). A 316-bp human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA probe (Ambion, Inc., Austin, Tex.) was randomly prime labeled using the Rediprime II kit to detect GAPDH mRNA that served as a mRNA stability control as well as a loading control. Following labeling, unincorporated [α32P]dCTP was removed using TE-Midi-Select D G-50 Sephadex columns (Shelton Scientific, Shelton, Conn.). The labeled probes were ready to use after boiling for 5 min.
Northern blot analysis.
Total RNA from Stx1- and/or LPS-stimulated or purified Stx1 B-subunit-stimulated, differentiated THP-1 cells was extracted using the TRIzol reagent (Gibco Life Technologies) protocol for cell monolayers. Following extraction, total RNA samples (10 μg of RNA induced by LPS and by Stx1 plus LPS and 15 μg of RNA induced by Stx1 and Stx1 B-subunit) were electrophoresed into 1.0% agarose-formaldehyde gels at 50 V for 1.5 h and then transferred to positively charged nylon membranes using the Turboblotter apparatus (Schleicher & Schuell, Keene, N.H.) for 3 h. Transferred RNA was cross-linked to membranes using a UV cross-linker (Bio-Rad, Hercules, Calif.). Membranes were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% SDS for 30 to 60 min, after which they were prehybridized at 42°C for 3 h with salmon testes DNA (Sigma) in a prehybridization solution of 50% formamide (Sigma) and 10% SDS (Sigma). Radiolabeled probes were then added to the hybridization buffer overnight at 42°C. After hybridization, blots were rinsed with 2× SSC, washed twice with 2× SSC-1.0% SDS for 15 min at room temperature, and then washed once with 0.1× SSC-1.0% SDS for 30 min at 60°C. Blots were dried briefly prior to exposure to a PhosphorImager screen and analyzed using a PhosphorImager (Molecular Dynamics). Membranes were stripped by boiling in 0.1× SSC-0.1% SDS twice for 15 min and sequentially reprobed with IL-1β and GAPDH probes at 42°C. Quantitation of pixel intensities of the RNA bands was done using ImageQuant software (Molecular Dynamics). Data are expressed as percentage above basal level as determined by the following equation: [(intensity of stimulated cells − intensity of unstimulated cells)/intensity of unstimulated cells] × 100.
IL-1β ELISA.
IL-1β production was quantitated using the human IL-1β Quantikine sandwich ELISA kit from R&D Systems (Minneapolis, Minn.). Supernatants from treated cells were centrifuged to remove cellular debris. LPS and Stx1 plus LPS samples were diluted 1:10, while Stx1 and Stx1 B-subunit samples were left undiluted. Two hundred microliters of each sample was added to three replicate wells on the ELISA plates for detection of IL-1β. Following the manufacturer's protocol, A450 and A570 were measured (Dynatech MR5000; Dynatech Laboratories, Chantilly, Va.), and IL-1β production levels were calculated by using a standard curve. The sensitivity of the assay is 1.0 pg/ml.
Statistics.
Statistics for experiments were performed with the SAS statistics program (SAS Institute, Cary, N.C.) or the SPSS statistics program (SPSS, Inc., Chicago, Ill.). TNF-α and IL-1β mRNA kinetics were analyzed using two-way analyses of variance (ANOVAs) with the Duncan multiple-range test for post hoc comparisons. IL-1β protein kinetics were analyzed using one-way ANOVAs with the Duncan multiple-range test used for post hoc analysis. Two-way ANOVAs were used to compare differences in cytokine mRNA levels in cells treated with different stimuli in the presence of ActD with Bonferroni's correction for multiple comparisons used for post hoc comparisons. For mRNA half-lives, the data were rank ordered and subjected to one-way ANOVAs, using the Duncan multiple-range test, to compare mRNA half-lives between the different stimuli. P values of ≤0.05 were considered significant for all analyses. All data are presented as means and standard errors of the means from a compilation of at least three independent experiments.
RESULTS
Comparison of TNF-α and IL-1β mRNA kinetics in THP-1 cells stimulated with Stx1, LPS, and Stx1 plus LPS.
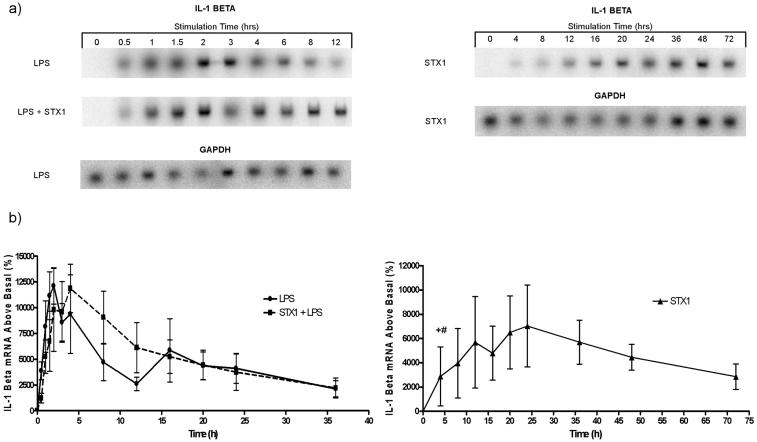
Earlier studies by Sakiri et al. demonstrated that Stx1 treatment of differentiated THP-1 cells resulted in the induction of relatively stable TNF-α mRNA transcripts (43). Therefore, we extended our initial 12-h analysis to 72 h. In this analysis, we examined TNF-α mRNA induction in differentiated THP-1 cells treated with Stx1 in the presence or absence of LPS. Treatment with LPS alone and with Stx1 plus LPS rapidly induced TNF-α transcripts with peak induction occurring at 30 to 60 min poststimulation (Fig. 1a). The peak percentage increase of TNF-α mRNA above the basal level induced by Stx1 and LPS (6,245 ± 2,050) is roughly double the peak percentage increase induced by LPS alone (3,014 ± 1,305), and the difference between the two values is statistically significant (P = 0.003). The reduction of steady-state TNF-α mRNA levels to basal values was rapid, although the reduction in mRNA levels after Stx1 plus LPS stimulation was delayed compared to treatment with LPS alone. Stx1 was not as potent an inducer of TNF-α transcripts as LPS (P = 0.04) or both stimulants (P < 0.0001), as Stx1 induced approximately 1/10 the peak percentage increase in TNF-α mRNA levels (808 ± 540) induced by Stx1 plus LPS (Fig. 1b). However, over a 4- to 12-h time frame, TNF-α transcripts induced by Stx1 appeared relatively stable, with reductions in steady-state mRNA levels appreciable only after 12 h of stimulation.
FIG. 1.
Comparison of TNF-α mRNA kinetics in THP-1 cells stimulated with Stx1, LPS, or Stx1 plus LPS. Differentiated THP-1 cells (5 × 106 cells/ml) were stimulated with Stx1 (400 ng/ml), LPS (200 ng/ml), or both for 0 to 72 h. Total RNA (10 to 15 μg) was subjected to Northern blot analysis using 32P-labeled TNF-α and GAPDH cDNA probes. Hybridization was detected and quantitated using a PhosphorImager and expressed as percentage mRNA above basal (unstimulated) expression. (a) Representative Northern blots of Stx1-, LPS-, and Stx1-plus-LPS-induced TNF-α mRNA. (b) Mean intensities of percentage mRNA above basal expression for each time point ± standard errors of the means (error bars) from three independent experiments. Values that are significantly different (P < 0.05) are indicated as follows: *, a significant difference between the values for cells treated with LPS alone and Stx1 plus LPS; #, a significant difference between the values for cells treated with Stx1 alone and Stx1 plus LPS.
Given the ability of IL-1β as well as TNF-α to increase Gb3 expression by vascular endothelial cells in vitro (54), we also determined the kinetics of IL-1β mRNA induction up to 72 h after toxin stimulation. While IL-1β mRNA induction also occurred rapidly in response to treatment with LPS or with Stx1 plus LPS (Fig. 2a), the induction kinetics were delayed compared to TNF-α, with peak values achieved at 2 h (LPS treatment) to 4 h (Stx1 plus LPS treatment). Treatment of cells with both stimulants did not significantly augment total IL-1β mRNA levels above those induced by LPS alone, in contrast to the increased production of TNF-α transcripts in Stx1-plus- LPS-treated cells. Following treatment with Stx1 alone, IL-1β mRNA was induced with slower kinetics with peak values reached at 24 h (Fig. 2b). Regardless of the stimulants used, IL-1β mRNA levels remained elevated longer compared to TNF-α transcripts. In contrast to TNF-α mRNA, IL-1β transcripts appeared to be induced to a higher level by Stx1. The peak percentage increases of IL-1β mRNA induced by treatment with LPS (12,114 ± 1,764) or Stx1 plus LPS (11,869 ± 2,350) were approximately twice that induced by Stx1 alone (P = 0.04 or P = 0.11, respectively). The prolonged elevations in steady-state transcript levels suggest that TNF-α and IL-1β expression may be regulated, in part, by Stxs at a posttranscriptional level. An important caveat in the interpretation of these data is that as the treatment times were prolonged, the number of cells detaching from the plates increased, especially with Stx1 plus LPS treatment (data not shown). Thus, at the later time points, the decline in cytokine mRNA levels may be correlated with loss of cell attachment or cell viability associated with Stx1 and/or LPS treatment.
FIG. 2.
Comparison of IL-1β mRNA kinetics in THP-1 cells stimulated with Stx1, LPS, and Stx1 plus LPS. Differentiated THP-1 cells were treated and analyzed as described in the legend to Fig. 1 except that IL-1β and GAPDH cDNA probes were used. (a) Representative Northern blots of Stx1-, LPS-, and Stx1-plus-LPS-induced IL-1β mRNA. (b) Mean intensities of percentage mRNA above basal expression for each time point ± standard errors of the means (error bars) from three independent experiments. Values that are significantly different (P ≤ 0.05) are indicated as follows: +, a significant difference between the values for cells treated with Stx1 and LPS; #, a significant difference between the values for cells treated with Stx1 alone and Stx1 plus LPS.
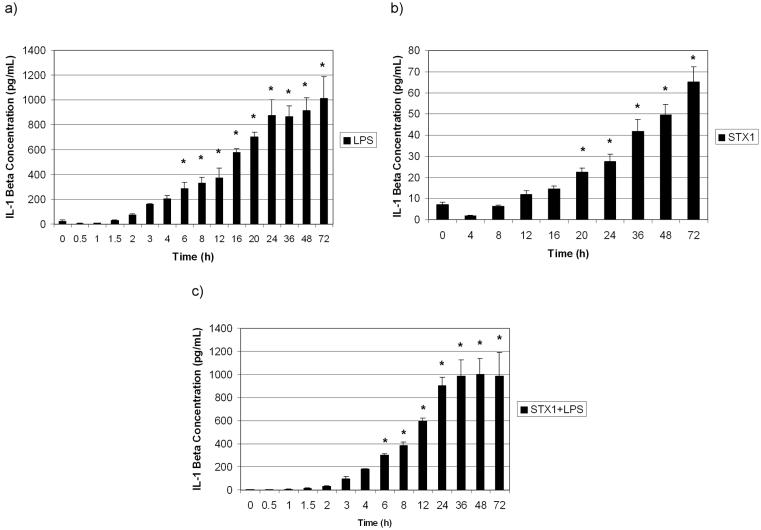
IL-1β protein production in THP-1 cells following Stx1, LPS, and Stx1 plus LPS treatment.
We previously showed that TNF-α elicited by Stx1 treatment of THP-1 cells was secreted into cell supernatants with maximal values of secreted TNF-α detected approximately 3 h after toxin stimulation (43). However, the kinetics of IL-1β protein production have not been determined in differentiated THP-1 cells treated with Stx1 in the presence or absence of LPS. To demonstrate that IL-1β transcripts were effectively translated in toxin-treated cells, we measured IL-1β protein levels in cell supernatants using a sensitive (≈1.0 pg/ml) ELISA. The results shown in Fig. 3 show that regardless of the stimulant used, IL-1β production increased over 72 h. The kinetics of IL-1β protein production correlated with IL-1β mRNA production for all the treatments, in that IL-1β mRNA production preceded IL-1β protein production. Consistent with levels of mRNA induction, Stx1 appeared to be a less potent inducer of IL-1β protein production and release than LPS alone or Stx1 plus LPS (P < 0.001 from 8 to 72 h).
FIG. 3.
IL-1β protein production by THP-1 cells treated with Stx1, LPS, and Stx1 plus LPS. Cell-free supernatants from LPS-treated (a), Stx1-treated (b), and Stx1-plus-LPS-treated (c) cells were collected and analyzed using human IL-1β-specific ELISAs. Data shown are the means ± standard errors of the means (error bars) from triplicate determinations of three independent experiments. Values that were significantly different (P ≤ 0.05) for unstimulated and treated cells are indicated (*).
IL-1β mRNA and protein production in THP-1 cells requires the Stx1 A subunit.
To determine the possible requirement of Stx1 enzymatic activity in IL-1β production, we stimulated THP-1 cells for 24 h with 400 or 800 ng of purified Stx1 B subunit per ml or with 400 ng of Stx1 per ml. Purified Stx1 B subunit did not result in a significant increase of IL-1β protein over the control value, suggesting that the presence of the A subunit may be necessary for IL-1β protein production (Fig. 4a). Furthermore, treatment of differentiated THP-1 cells with Stx1 B subunit did not result in the expression of IL-1β mRNA (Fig. 4b).
FIG. 4.
Purified Stx1 B subunits alone do not induce IL-1β protein or mRNA production in THP-1 cells. Differentiated THP-1 cells (5 × 106 cells/ml) were treated with Stx1 B subunits (400 or 800 ng/ml) for 24 h. (a) Cell-free supernatants were collected, and human IL-1β-specific ELISAs were performed to detect secreted IL-1β. Media collected from untreated cells and cells treated with Stx1 (400 ng/ml) served as negative and positive controls, respectively. Data shown are means ± standard errors of the means (error bars) from triplicate determinations of three independent experiments. Values that were significantly different (P ≤ 0.05) between unstimulated and treated cells are indicated (*). (b) Fifteen micrograms of total RNA extracted from cells treated with LPS, Stx1, Stx1 B subunits, and unstimulated cells were subjected to Northern blot analysis to detect IL-1β mRNA expression.
mRNA stability of TNF-α and IL-1β transcripts induced by Stx1, LPS, and Stx1 plus LPS.
There are multiple regulatory mechanisms that may account for the prolonged elevation of TNF-α and IL-1β mRNA levels in THP-1 cells stimulated with Stx1 or Stx1 plus LPS compared to treatment with LPS alone. It has been shown previously that Stx1 treatment of THP-1 cells results in the activation of the ribotoxic stress response and the activation of transcriptional factors NF-κB and AP-1 (10, 43). Prolonged activation of these factors may contribute to the phenomenon of elevated cytokine mRNA levels in toxin-treated cells. Cytokine expression may also be regulated at posttranscriptional levels, and one of the main mechanisms of posttranscriptional control is regulation of transcript stability (reviewed in reference 42). Thus, the toxins may disrupt mechanisms that normally facilitate cytokine mRNA decay.
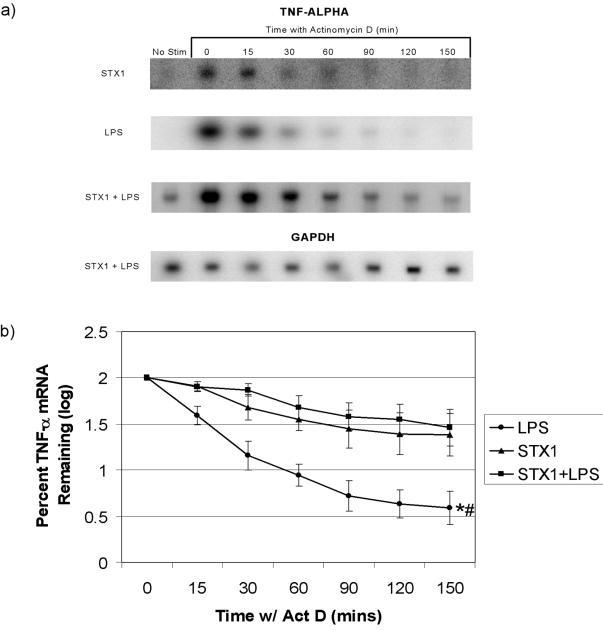
We performed Northern blot analysis using the transcriptional inhibitor ActD to compare the decay rates of TNF-α and IL-1β mRNAs in THP-1 cells stimulated with Stx1, LPS, or Stx1 plus LPS. Differentiated THP-1 cells were treated either with Stx1 or Stx1 plus LPS for 2 h or with LPS for 1 h. ActD (5.0 μg/ml) was added to each plate for 15, 30, 60, 90, 120, and 150 min. The mRNA stability and loading control, GAPDH, showed a stable transcript over the time course of all experiments conducted and allowed for normalization of TNF-α and IL-1β mRNA levels. Treatment of THP-1 cells with Stx1 or Stx1 plus LPS resulted in TNF-α transcript levels that were significantly different from LPS-induced transcript levels (P ≤ 0.001) after actD addition over time (Fig. 5).
FIG. 5.
Effects of Stx1 and/or LPS on TNF-α mRNA stability in THP-1 cells. Differentiated THP-1 cells (5 × 106 cells/ml) were treated with LPS (200 ng/ml) for 1 h, Stx1 (400 ng/ml) for 2 h, or LPS plus Stx1 for 2 h. ActD was then added to each plate at a final concentration of 5.0 μg/ml. Total RNA was isolated at each of the indicated time points after ActD addition. Northern blot analysis was performed, and TNF-α mRNA was detected and quantitated using a PhosphorImager. (a) Representative Northern blots probed with 32P-labeled TNF-α- and GAPDH-specific cDNA probes. (b) Mean log intensities of percent TNF-α mRNA remaining ± standard errors of the means (error bars) from three independent experiments. Values that were significantly different (P ≤ 0.001) are indicated as follows: *, a significant difference between the values for cells treated with Stx1 plus LPS and LPS alone; #, a significant difference between the values for cells treated with Stx1 and LPS.
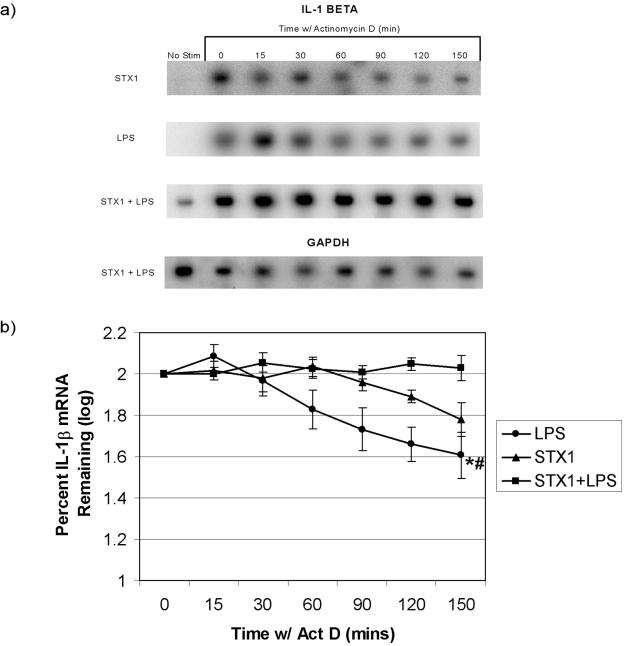
Half-lives for TNF-α transcripts induced by LPS, Stx1, and Stx1 plus LPS treatments are shown in Table 1. Statistical analysis revealed that only the half-lives of TNF-α transcripts induced by Stx1 plus LPS versus LPS alone were significantly different (P ≤ 0.05). For IL-1β transcripts in the presence of ActD, Stx1 plus LPS and Stx1 treatments resulted in significantly higher transcript levels than LPS treatment (P ≤ 0.001 and P ≤ 0.005, respectively) (Fig. 6). Half-lives for IL-1β transcripts induced by LPS, Stx1, and Stx1 plus LPS treatments are shown in Table 1. Statistical analysis did not reveal significant differences in the half-lives of IL-1β transcripts induced by LPS versus Stx1. Due to the parameters of the experiment, we cannot accurately estimate the half-life of IL-1β transcripts induced by Stx1 plus LPS (half-life of >150 min).
TABLE 1.
Estimated half-lives of TNF-α and IL-1β mRNA
| Treatment | Half-life (min)a of:
|
|
|---|---|---|
| TNF-α mRNA | IL-1β mRNA | |
| LPS | 7.28 ± 5.12*# | 114.39 ± 33.13 |
| Stx1 | 75.86 ± 44.24# | 175.60 ± 28.94 |
| Stx1 + LPS | 101.63 ± 51.49* | NDb |
Significant differences (P ≤ 0.05) between values for the different treatments with the same symbol are indicated by the * and # symbols.
ND, not determined.
FIG. 6.
Effects of Stx1 and/or LPS on IL-1β mRNA stability in THP-1 cells. Differentiated THP-1 cells were treated and analyzed as described in the legend to Fig. 5 except that IL-1β- and GAPDH-specific probes were used. (a) Representative Northern blots probed with 32P-labeled IL-1β- and GAPDH-specific cDNA probes. (b) Mean log intensities of percent IL-1β mRNA remaining ± standard errors of the means (error bars) from three independent experiments. Values that were significantly different (P ≤ 0.05) are indicated as follows: *, a significant difference between the values for cells treated with Stx1 plus LPS and LPS alone; #, a significant difference between the values for cells treated with Stx1 and LPS.
DISCUSSION
In addition to their critical roles in the innate host response and the induction of inflammation, TNF-α and IL-1β may also contribute to the pathogenesis of vascular damage that follows the systemic distribution of Stxs in the bloodstream. Our studies confirm earlier reports that LPS rapidly induce TNF-α and IL-1β transcripts in macrophages (reviewed in reference 4). Compared to LPS, Stx1 is a less robust inducer of cytokine mRNA in terms of the kinetics of induction and total amount induced, although the transcripts remained elevated for a prolonged time (Fig. 1 and 2). Stx1 induction of maximal TNF-α and IL-1β mRNA expression was delayed (12 and 24 h, respectively) compared to LPS-induced (0.5 and 2 h, respectively) or Stx1-plus-LPS-induced (1 and 4 h, respectively) maximal expression. The reason for the delay in induction of cytokine mRNAs by Stx1 is unknown but may be attributable to the necessity for toxin internalization, retrograde transport, A-subunit processing, and translocation into the cytosol for toxin enzymatic activity. In contrast, LPS rapidly initiate transmembrane signaling events in macrophages through TLR4/MyD88 after binding to the cell surface receptor CD14 (5). THP-1 cells exposed to both stimulants showed rapid mRNA induction characteristic of LPS treatment and prolonged mRNA elevation characteristic of Stx1 treatment, suggesting that the bacterial products may signal through separate pathways to elicit cytokine expression. The presence of both bacterial products in the bloodstream or inflammatory loci may optimize proinflammatory cytokine production. The kinetics data also suggest that TNF-α and IL-1β are differentially regulated. For all treatments, TNF-α mRNA reached maximal levels and returned to basal levels before IL-1β transcripts.
We previously showed that Stx1-induced TNF-α mRNA and protein expression were temporally correlated in that peak mRNA levels were reached at 2 h while peak protein levels were detected 3 h after toxin stimulation and remained elevated for up to 12 h. Compared to Stx1 stimulation, Stx1 plus LPS treatment augmented TNF-α mRNA induction, resulting in 10-fold-higher TNF-α protein levels detected in supernatants (43). We show here that the production of soluble IL-1β protein by THP-1 cells in response to Stx1 is very different. Although IL-1β mRNA was induced by Stx1 treatment, the levels of soluble IL-1β protein detected in cell supernatants (Fig. 3) were much less than predicted from the IL-1β transcript levels induced by Stx1 (Fig. 2b). IL-1β mRNA levels in THP-1 cells stimulated with Stx1 were roughly half of the IL-1β mRNA levels in cells stimulated with LPS alone or Stx1 plus LPS. IL-1β protein levels, however, were 20-fold less in THP-1 cells treated with Stx1 than in cells treated with LPS alone or Stx1 plus LPS.
The precise reason for the discrepancy between Stx1-induced IL-1β mRNA and protein levels is unclear. An obvious explanation is that Stx1 inhibits protein synthesis. However, we have previously shown that, at the concentrations used in this study, Stx1 mediated a modest (≈10%) reduction in total protein synthesis in differentiated THP-1 cells (11). An alternative explanation is that Stx1 may affect posttranslational processing and release mechanisms necessary for IL-1β expression that are not required for secretion of TNF-α.
The 26-kDa membrane-associated form of TNF-α is cleaved to a 17-kDa form by the action of the metalloproteinase disintegrin (1, 32). ProIL-1β, the 35-kDa precursor form of IL-1β, is cleaved to the active 17-kDa form by the cysteine-dependent aspartate-specific proteases caspase 1 and caspase 5 which form part of a high-molecular-weight complex called the inflammasome (30). Recently, Greenwell-Wild et al. [T. Greenwell-Wild, G. Peng, N. Vázquez, W. Jin, K. Lei, J. M. Orenstein, and S. M. Wahl, J. Leukoc. Biol. 74(Suppl.):38, abstr. 108, 2003] reported that infection of macrophages with Mycobacterium avium elicited IL-1β mRNA expression, but not IL-1β protein secretion. Failure to release soluble IL-1β was associated with reduced caspase 1 activity and reduced expression of inflammasome constituents.
Whether Stxs similarly affect inflammasome function and posttranslational processing mechanisms necessary for IL-1β secretion remains to be clarified. The presence of LPS, however, appears to restore IL-1β processing, as the levels of soluble IL-1β released in response to LPS alone versus Stx1 plus LPS are comparable. LPS signaling occurs earlier than Stx1 signaling, probably because Stx1 needs to be transported into the cell in order to elicit its effects. Thus, LPS-mediated signaling leading to the release of IL-1β may occur prior to Stx-mediated signaling events.
The contribution of Stx B subunits in various biological activities may differ. For example, purified Stx1 B-subunits were reported to trigger apoptosis of Burkitt's lymphoma B-cell lines (29), while apoptosis induction of other cell types requires holotoxin. Earlier studies have suggested that Stx enzymatic activity is essential for IL-8 and TNF-α expression (11, 52, 58). Our data support a role for Stx1 A subunit in IL-1β mRNA and protein production, since no elevations in IL-1β mRNA or protein above the unstimulated control values are detected when cells are treated with purified Stx1 B subunits (Fig. 4).
Excessive or prolonged expression of the cytokines TNF-α and IL-1β may lead to deleterious effects, such as vascular leak syndrome and hypovolemic shock, coagulopathies, and the development of autoimmune diseases. Therefore, it is not surprising that TNF-α and IL-1β expression are tightly regulated at transcriptional, posttranscriptional, and posttranslational stages. We previously demonstrated that Stx1, like other protein synthesis inhibitors acting on the 28S rRNA (17), induces the ribotoxic stress response (10). Downstream substrates of JNK and p38 mitogen-activated protein kinase cascades are known to be transcriptional activators of TNF-α and IL-1β gene expression. Nuclear lysates prepared from Stx1-treated THP-1 cells contained increased levels of proteins capable of binding oligonucleotides with NF-κB and AP-1 binding sites (43). Collectively, these data suggested that the Stxs regulate cytokine expression at a transcriptional level. The kinetic analyses in this study suggest that Stx1 has a regulatory effect on TNF-α and IL-1β mRNA stability, since steady-state cytokine transcripts from both Stx1- and Stx1-plus-LPS-treated cells remain elevated longer compared to LPS-induced transcripts.
To determine whether Stx1 regulated TNF-α and IL-1β mRNA levels at a posttranscriptional stage, we determined the decay rates of transcripts induced by Stx1, LPS, or Stx1 plus LPS (Fig. 5 and 6). The combined kinetics and mRNA stability data suggest that when Stxs and LPS are present after infection with Stx-producing bacteria, TNF-α transcript levels may be maximally increased, i.e., superinduced, via alterations in transcriptional and posttranscriptional control mechanisms. Interestingly, this phenomenon has been described for chemokine mRNAs after treatment of the human intestinal epithelial cell line HCT-8 with Stx1 (52). Our studies on the regulation of IL-1β transcript expression produced different results. Treatment of cells with LPS or Stx1 plus LPS resulted in similar levels of steady-state transcripts, i.e., IL-1β mRNA was not superinduced by Stx1 plus LPS. However, the half-life of IL-1β transcripts induced by Stx1 plus LPS appeared to be greater than that induced by LPS or Stx1 alone.
The precise mechanisms by which Stxs regulate cytokine transcript levels remain to be fully characterized, but it is known that many posttranscriptional control mechanisms map to the AU-rich elements (ARE) found within the 3′-UTRs of cytokine transcripts (2). Binding of proteins within the ARE may regulate transcript processing and translation in different ways. For example, binding of proteins within the TNF-α ARE may facilitate nuclear export of transcripts into the cytoplasm (6). Binding of the zinc finger protein tristetraprolin or butyrate response factors to TNF-α ARE leads to increased mRNA deadenylation and degradation of the body of the transcript (3, 24, 25, 48). Interaction of the RNA-binding proteins TIA-1 and TIAR with the TNF-α ARE results in translational silencing (13, 37) via a mechanism which sequesters TNF-α mRNA in cytoplasmic ribonucleoprotein complexes called stress granules (22). Experiments to further characterize the mechanism(s) of Stx-mediated cytokine mRNA stabilization are planned.
Why some patients develop uncomplicated bloody diarrhea and others progress to life-threatening sequelae is not understood. Our data suggest that the presence of both Stxs and endotoxin in the bloodstream may be a predictor of progression to systemic disease.
Acknowledgments
The expert technical assistance of Cassandra Armstrong, Gregory Foster, Todd Lasco, and Amminikutty Jeevan is gratefully acknowledged. We thank Cheleste Thorpe for the gift of purified Stx1 B-subunits, Shannon Sedberry Allen and Rajesh Miranda for assistance with statistical analyses, and David McMurray, Rajesh Miranda, and James Samuel for careful reading of the manuscript.
These studies were supported in part by United States Public Health Service grant 2RO1 AI34530 from NIAID, NIH.
Editor: A. D. O'Brien
REFERENCES
- 1.Black, R. A., C. T. Rauch, C. J. Kozlosky, J. J. Peschon, J. L. Slack, M. F. Wolfson, B. J. Castner, K. L. Stocking, P. Reddy, S. Srinivasan, N. Nelson, N. Boiani, K. A. Schooley, M. Gerhart, R. Davis, J. N. Fitzner, R. S. Johnson, R. J. Paxton, C. J. March, and D. P. Ceretti. 1997. A metalloproteinase disintegrin that releases tumour necrosis factor-α from cells. Nature 385:729-733. [DOI] [PubMed] [Google Scholar]
- 2.Caput, D., B. Beutler, K. Hartog, R. Thayer, S. Brown-Shimer, and A. Cerami. 1986. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. USA 83:1670-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 4.Dinarello, C. A. 1999. Immediate cytokine responses to endotoxin: tumor necrosis factor-α and the interleukin-1 family, p. 549-560. In H. Brade, S. M. Opal, S. N. Vogel, and D. C. Morrison (ed.), Endotoxin in health and disease. Marcel Dekker, Inc., New York, N.Y.
- 5.Dobrovolskaia, M. A., and S. N. Vogel. 2002. Toll receptors, CD14, and macrophage activation and deactivation by LPS. Microb. Infect. 4:903-914. [DOI] [PubMed] [Google Scholar]
- 6.Dumitru, C. D., J. D. Ceci, C. Tsatsanis, D. Kontoyiannis, K. Stamatakis, J. H. Lin, C. Patriotis, N. A. Jenkins, N. G. Copeland, G. Kollias, and P. N. Tsichlis. 2000. TNF-alpha induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103:1071-1083. [DOI] [PubMed] [Google Scholar]
- 7.Eisenhauer, P. B., P. Chaturvedi, R. E. Fine, A. J. Ritchie, J. S. Pober, T. G. Cleary, and D. S. Newberg. 2001. Tumor necrosis factor alpha increases human cerebral endothelial cell Gb3 and sensitivity to Shiga toxin. Infect. Immun. 69:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Endo, Y., K. Tsurugi, T. Yutsudo, Y. Takeda, T. Ogasawara, and K. Igarashi. 1988. Site of action of a verotoxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 171:45-50. [DOI] [PubMed] [Google Scholar]
- 9.Fontaine, A., J. Arondel, and P. J. Sansonetti. 1988. Role of Shiga toxin in the pathogenesis of bacillary dysentery studied by using a Tox− mutant of Shigella dysenteriae 1. Infect. Immun. 56:3099-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, G. H., and V. L. Tesh. 2002. Shiga toxin 1-induced activation of c-Jun NH2-terminal kinase and p38 in the human monocytic cell line THP-1: possible involvement in the production of TNF-α. J. Leukoc. Biol. 71:107-114. [PubMed] [Google Scholar]
- 11.Foster, G. H., C. S. Armstrong, R. Sakiri, and V. L. Tesh. 2000. Shiga toxin-induced tumor necrosis factor alpha expression: requirement for toxin enzymatic activity and monocyte protein kinase C and protein tyrosine kinases. Infect. Immun. 68:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, M. E., M. M. Chernaia, Y. V. Kozlov, and M. N. G. James. 1994. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 Å resolution. Nat. Struct. Biol. 1:59-64. [DOI] [PubMed] [Google Scholar]
- 13.Gueydan, C., L. Droogsman, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem. 274:2322-2326. [DOI] [PubMed] [Google Scholar]
- 14.Harel, Y., M. Silva, B. Giroir, A. Weinberg, T. B. Cleary, and B. Beutler. 1993. A reporter transgene indicates renal-specific induction of tumor necrosis factor (TNF) by Shiga-like toxin. Possible involvement of TNF in hemolytic uremic syndrome. J. Clin. Investig. 92:2110-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrold, S., C. Genovese, B. Kobrin, S. L. Morrison, and C. Milcarek. 1991. A comparison of apparent mRNA half-life using kinetic labeling techniques vs. decay following administration of transcriptional inhibitors. Anal. Biochem. 198:19-29. [DOI] [PubMed] [Google Scholar]
- 16.Igarashi, K., T. Ogasawara, K. Ito, T. Yutsudo, and Y. Takeda. 1987. Inhibition of elongation factor 1-dependent aminoacyl-tRNA binding to ribosomes by Shiga-like toxin I (VT1) from Escherichia coli O157:H7 and by Shiga toxin. FEMS Microbiol. Lett. 44:91-94. [DOI] [PubMed] [Google Scholar]
- 17.Iordanov, M. S., D. Pribnow, J. L. Magun, T.-H. Dinh, J. A. Pearson, S. L.-Y. Chen, and B. E. Magun. 1997. Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of peptidyl transferase reaction and by sequence-specific RNA damage to the α-sarcin/ricin loop in the 28S rRNA. Mol. Cell. Biol. 17:3373-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isogai, E., H. Isogai, K. Kimura, S. Hayashi, T. Kubota, N. Fujii, and K. Takeshi. 1998. Role of tumor necrosis factor alpha in gnotobiotic mice infected with an Escherichia coli O157:H7 strain. Infect. Immun. 66:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacewicz, M., H. Clausen, E. Nudelman, A. Donohue-Rolfe, and G. T. Keusch. 1986. Pathogenesis of Shigella diarrhea. XI. Isolation of a Shigella toxin-binding glycolipid from rabbit jejunum and HeLa cells and its identification as globotriaosylceramide. J. Exp. Med. 163:1391-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karmali, M. A. 1998. Human immune response and immunity to Shiga toxin (verocytotoxin)-producing Escherichia coli infection, p. 236-248. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 21.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koster, F., J. Levin, L. Walker, K. S. K. Tung, R. H. Gilman, M. M. Rahaman, A. Majid, S. Islam, and R. C. Williams, Jr. 1978. Hemolytic-uremic syndrome after shigellosis. Relation to endotoxemia and circulating immune complexes. N. Engl. J. Med. 298:927-933. [DOI] [PubMed] [Google Scholar]
- 24.Lai, W. S., E. Carballo, J. R. Strum, E. A. Kennington, R. S. Phillips, and P. J. Blackshear. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19:4311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 26.Lindberg, A. A., J. E. Schultz, M. Westling, J. E. Brown, S. W. Rothman, K.-A. Karlsson, and N. Stomberg. 1986. Identification of the receptor glycolipid for Shiga toxin produced by Shigella dysenteriae type 1, p. 439-446. In D. L. Lark (ed.), Protein-carbohydrate interactions. Academic Press, London, United Kingdom.
- 27.Louise, C. B., and T. G. Obrig. 1991. Shiga toxin-associated hemolytic-uremic syndrome: combined cytotoxic effects of Shiga toxin, interleukin-1β, and tumor necrosis factor alpha on human vascular endothelial cells in vitro. Infect. Immun. 59:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louise, C. B., M. C. Tran, and T. G. Obrig. 1997. Sensitization of human umbilical vein endothelial cells to Shiga toxin: involvement of protein kinase C and NF-κB. Infect. Immun. 65:3337-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangeney, M., C. A. Lingwood, S. Taga, B. Caillou, T. Tursz, and J. Wiels. 1993. Apoptosis induced in Burkitt's lymphoma cells via Gb3/CD77, a glycolipid antigen. Cancer Res. 53:5314-5319. [PubMed] [Google Scholar]
- 30.Martinon, F., K. Burns, and J. Tschopp. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-1β. Mol. Cell 10:417-426. [DOI] [PubMed] [Google Scholar]
- 31.Melton-Celsa, A. R., and A. D. O'Brien. 1998. Structure, biology and relative toxicity of Shiga toxin family members for cells and animals, p. 121-128. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 32.Moss, M. L., S.-L. C. Jin, M. E. Milla, W. Burkhart, H. L. Carter, W.-J. Chen, W. C. Clay, J. R. Didsbury, D. Hassler, C. R. Hoffman, T. A. Kost, M. H. Lambert, M. A. Leesnitzer, P. McCauley, G. McGeehan, J. Mitchell, M. Moyer, G. Pahel, W. Rocque, L. K. Overton, F. Schoenen, T. Seaton, J.-L. Su, J. Warner, D. Willard, and J. D. Becherer. 1997. Cloning of a disintegrin metalloproteinase that processes precursor tumour necrosis factor-α. Nature 385:733-736. [DOI] [PubMed] [Google Scholar]
- 33.Nau, G. J., J. F. L. Richmond, A. Schlesinger, E. G. Jennings, E. S. Lander, and R. A. Young. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl. Acad. Sci. USA 99:1503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Brien, A. D., V. L. Tesh, A. Donohue-Rolfe, M. P. Jackson, S. Olsnes, K. Sandvig, A. A. Lindberg, and G. T. Keusch. 1992. Shiga toxin: biochemistry, genetics, mode of action and role in pathogenesis. Curr. Top. Microbiol. Immunol. 180:65-94. [DOI] [PubMed] [Google Scholar]
- 35.Obrig, T. G., T. P. Moran, and J. E. Brown. 1987. The mode of action of Shiga toxin on peptide elongation of eukaryotic protein synthesis. Biochem. J. 244:287-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piecyk, M., S. Wax, A. R. P. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proulx, F., E. G. Seidman, and D. Karpman. 2001. Pathogenesis of Shiga toxin-associated hemolytic uremic syndrome. Pediatr. Res. 50:163-171. [DOI] [PubMed] [Google Scholar]
- 39.Ramegowda, B., and V. L. Tesh. 1996. Differentiation-associated toxin receptor modulation, cytokine production, and sensitivity to Shiga-like toxins in human monocytes and monocytic cell lines. Infect. Immun. 64:1173-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramegowda, B., J. E. Samuel, and V. L. Tesh. 1999. Interaction of Shiga toxins with human brain microvascular endothelial cells: cytokines as sensitizing agents. J. Infect. Dis. 180:1205-1213. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberger, C. M., M. G. Scott, M. R. Gold, R. E. W. Hancock, and B. B. Finlay. 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 164:5894-5904. [DOI] [PubMed] [Google Scholar]
- 42.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakiri, R., B. Ramegowda, and V. L. Tesh. 1998. Shiga toxin type 1 activates tumor necrosis factor-α gene transcription and nuclear translocation of the transcriptional activators nuclear factor-κB and activator protein-1. Blood 92:558-566. [PubMed] [Google Scholar]
- 44.Sandvig, K., S. Grimmer, S. U. Lauvrak, M. L. Torgersen, G. Skretting, B. van Deurs, and T. G. Iversen. 2002. Pathways followed by ricin and Shiga toxin into cells. Histochem. Cell Biol. 117:131-141. [DOI] [PubMed] [Google Scholar]
- 45.Saxena, S. K., A. D. O'Brien, and E. J. Ackerman. 1989. Shiga toxin, Shiga-like toxin II variant, and ricin are all single-site RNA N-glycosidases of 28S RNA when microinjected into Xenopus oocytes. J. Biol. Chem. 264:596-601. [PubMed] [Google Scholar]
- 46.Siegler, R. L. 1995. The hemolytic uremic syndrome. Pediatr. Clin. N. Am. 42:1505-1529. [DOI] [PubMed] [Google Scholar]
- 47.Stein, P. E., A. Boodhoo, G. J. Tyrrell, J. L. Brunton, and R. J. Read. 1992. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature 355:748-750. [DOI] [PubMed] [Google Scholar]
- 48.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tesh, V. L. 1998. Cytokine response to Shiga toxins, p. 226-235. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 50.Tesh, V. L., J. A. Burris, J. W. Owens, V. M. Gordon, E. A. Wadolkowski, A. D. O'Brien, and J. E. Samuel. 1993. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect. Immun. 61:3392-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tesh, V. L., B. Ramegowda, and J. E. Samuel. 1994. Purified Shiga-like toxins induce expression of proinflammatory cytokines from murine peritoneal macrophages. Infect. Immun. 62:5085-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorpe, C. M., W. E. Smith, B. P. Hurley, and D. W. K. Acheson. 2001. Shiga toxins induce, superinduce, and stabilize a variety of C-X-C chemokine mRNAs in intestinal epithelial cells, resulting in increased chemokine expression. Infect. Immun. 69:6140-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuchiya, S., M. Yamabe, Y. Yamaguchi, Y. Kobayashi, T. Konno, and K. Tada. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26:171-176. [DOI] [PubMed] [Google Scholar]
- 54.van de Kar, N. C. A. J., L. A. H. Monnens, M. A. Karmali, and V. W. M. van Hinsbergh. 1992. Tumor necrosis factor and interleukin 1 induce expression of the verocytotoxin receptor globotriaosylceramide on human endothelial cells: implications for the pathogenesis of the hemolytic uremic syndrome. Blood 80:2755-2764. [PubMed] [Google Scholar]
- 55.van Setten, P. A., L. A. H. Monnens, R. G. G. Verstraten, L. P. W. J. van den Heuvel, and V. W. M. van Hinsbergh. 1996. Effects of verocytotoxin-1 on nonadherent human monocytes: binding characteristics, protein synthesis and induction of cytokine release. Blood 88:174-183. [PubMed] [Google Scholar]
- 56.van Setten, P. A., V. W. M. van Hinsbergh, T. J. A. N. van der Velden, N. C. A. J. van de Kar, M. Vermeer, J. D. Mahan, K. J. M. Assman, L. P. W. J. van den Heuvel, and L. A. H. Monnens. 1997. Effects of TNF-α on verocytotoxin cytotoxicity in purified human glomerular microvascular endothelial cells. Kidney Int. 51:1245-1256. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Z.-M., C. Liu, and R. Dziarski. 2000. Chemokines are the main proinflammatory mediators in human monocytes activated by Staphylococcus aureus, peptidoglycan, and endotoxin. J. Biol. Chem. 275:20260-20267. [DOI] [PubMed] [Google Scholar]
- 58.Yamasaki, C., Y. Natori, X.-T. Zeng, M. Ohmura, S. Yamasaki, Y. Takeda, and Y. Natori. 1999. Induction of cytokines in a human colon epithelial cell line by Shiga toxin 1 (Stx1) and Stx2 but not by nontoxic mutant Stx1 which lacks N-glycosidase activity. FEBS Lett. 442:231-234. [DOI] [PubMed] [Google Scholar]