Highlights
-
•
DJ1 is a recessive gene involved in early onset PD.
-
•
We tested 163 Italian EOPD.
-
•
We did not find any mutation in our population.
-
•
DJ1 PD causing mutations are very rare in Italian population.
Keywords: DJ1, Early Onset Parkinson Disease, Mutation analysis
Abstract
We analyzed the DJ1 gene in a large consecutive series (N = 163) of Italian unrelated Early Onset Parkinson Disease (EOPD: onset ≤40 years of age) patients and 100 healthy controls (mean age 64 ± 7 years). No homozygous or compound heterozygous mutations with an obvious pathogenic effect were found. Several variants were identified, some of which were novels. All variants had similar frequency in patients and in controls. Our data suggest that DJ1 mutations are very rare in Italian EOPD. Other genes and risk factors for PD are still to be identified.
1. Introduction
Parkinson's disease (PD) is a common neurodegenerative disorder with a prevalence of ∼2% in persons >65 years old [17]. Mean age at onset is 60 years, but about 5% of patients have an Early Onset PD (EOPD) before 40 years [18]. Although the etiology of PD has not been fully elucidated yet, it is generally considered the result of the interaction between genetic and environmental factors [23]. Several genes involved in familial PD have been identified. In particular, three genes – Parkin (PRKN), DJ1 (locus PARK7), and PINK1 (locus PARK6) – are involved in autosomal recessive Parkinson's disease [7]. In these cases, age at onset tends to be earlier, typically before 40 years.
PARK-7 locus was identified by van Duijn and colleagues [31], in a family-based linkage analysis that led to the identification of two families with mutations in the DJ1 gene [3]. The former was a Dutch family with a large homozygous deletion of exons 1–5. The latter family of Italian origin harbored a homozygous missense mutation, the p.Leu166Pro, in a highly conserved coding position of the protein.
The DJ1 gene encodes a ubiquitous, highly conserved small protein of 189 amino acids, expressed in the brain areas and extra-cerebral tissues [2]. The gene maps to chromosome 1p36.23 and comprises nine exons, the first two (1A and 1B) being alternatively spliced and non-coding. Crystal structure and biochemical data show that DJ1 protein is biological active as a dimer, but its role in PD pathogenesis is still unclear [33]. In model systems, DJ1 protects dopaminergic neurons against various insults, including rotenone, mutant α-synuclein, hydrogen peroxide, and 6-hydroxydopamine [18,19]. Moreover, recent studies indicate DJ1 as a multifunctional protein involved not only in oxidative cellular stress response, maintaining normal mitochondrial function, but also in modulator of transcriptional activity and regulation of RNA stability [21,32].
Since the identification of the DJ1 gene, molecular analyses in PD patients identified homozygous and compound heterozygous nucleotide substitutions, including missense, truncating, splice-site mutations, and large deletions [1,6,11–13,20,21].
Few studies searched DJ1 mutations in Italian population including a relatively low number of early onset PD patients [16,28]. In this context, the aim of our study was to determine the frequency of DJ1 mutations in a large consecutive series of EOPD patients enrolled in a single Italian clinical referral centre.
2. Methods
2.1. Subjects
We studied 263 subjects, of which 163 were affected by PD and 100 were healthy controls. The population consisted of unrelated consecutive patients with EOPD (onset ≤40 years of age), who contributed to the Parkinson Institute Biobank of Milan-Italy (http://www.parkinsonbiobank.com/). The clinical diagnosis of PD was established according to the UK Parkinson's Disease Society Brain Bank Criteria which require the presence of bradykinesia, the absence of atypical features or other causes of parkinsonism, and at least 1 of the following signs: resting tremor, rigidity and postural instability, and a positive response to dopaminergic therapy [14,15]. Neurological examination was performed by neurologists with experience in movement disorders.
Among the 163 EOPD patients, 99 were male (61%), the mean age at onset was 35.4 ± 4.8 years (range 14–40) and the mean disease duration was 19.3 ± 9 years (range 5–36). We considered as age at onset, the age at which the patient noticed the first PD symptom. Forty-three were familial cases (with at least one 1st or 2nd degree relative with a formal diagnosis of PD), while the remaining patients were sporadic. Our cohort of 163 EOPD was also screened for PRKN gene mutations [25] and for the presence of the p.G2019S LRRK2 mutation [9]. The mutation carriers of these 2 genes (11 subjects with 2 mutations in the PRKN gene, 4 with one mutation in the PRKN gene, 4 subjects with the p.G2019S mutation in the LRRK2 gene) were not excluded from this study.
The control group was composed by individuals free from any neurodegenerative disorder, with no family history of PD (usually patients’ partners or caregivers). The mean age of control subjects at the time of blood collection was 64 ± 7.2 years. All patients and controls were Caucasians and of Italian origin with the exception of 8 affected individuals from 7 different Countries: Argentina (2), Albania, Colombia, France, Ireland, Sri Lanka and Spain. In addition, 50 South European healthy subjects were tested for a specific variant identified in a non-Italian patient.
The project was approved by the local Ethics Committee. Each participant signed an informed consent prior to participate to the study.
2.2. Genomic DNA analysis
Genomic DNA from PD patients and control subjects was isolated from peripheral blood using a semiautomatic extractor (QuickGeneDNA Whole Blood Kit; FUJIFILM Europe GmbH Life Science, Düsseldorf). To sequence DJ1 coding region the seven exons and intron-exon boundaries were amplified by PCR (primers sequences and PCR conditions available upon request).
To sequence the genomic region upstream exon 2, a genomic fragment of 1407 bp spanning exon 1, intron 1 and exon 2, was amplified using TaKaRa LA Taq polymerase with GC buffer II (Takara Biomedicals).
PCR products were purified using ExoSAP-IT and directly sequenced using Big Dye Terminator Kit (version 3.1, Applied Biosystems), loaded on an ABI3100 automatic sequencer and analyzed with SeqScape software (Applied Biosystems).
In order to detect the presence of exon rearrangements, patients carriers of mutations were also analyzed using the MLPA (Multiplex Ligation-dependent Probe Amplification) assay Kit “SALSA P051” (MRC-Holland, Amsterdam, Netherlands – http://www.mrc-holland.com). MLPA was performed according to manufacturer instructions. All samples were tested in duplicate.
The MLPA fragments were analyzed on an ABI Prism 3130 XL automatic sequencer (PE Applied Biosystems) with the Genescan software using 500 ROX size standards (PE Applied Biosystems). The individual peak corresponding to each exon was identified based on the difference in migration relative to the size standards. The peak area of each fragment was compared to that of 3 control samples. Raw data were analyzed using Coffalyser software v.9.
2.3. cDNA analysis
RNA was isolated from peripheral blood using RNA-Beads protocols. RT-PCR was performed using iScript™cDNA Synthesis Kit (Bio Rad) and 4 μg of each RNA sample.
A 770 bp fragment of the DJ1 cDNA spanning exons 1a-7, but lacking ex1b, was amplified from the RT-PCR material (Taq polymerase Invitrogen) and the following primers: forward 5′-gcgtgctggcgtgcgttc-3′, and reverse 5′-tgacttccatacttccgcaaa-3′.
The PCR products were loaded on a 2% Agarose gel. After separation, the fragments were isolated and purified using Illustra GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare). Fragments were directly sequenced as previously described. The consequences of the variants identified were predicted at the protein level according to the DJ1 mRNA sequence (accession number NM_007262.3).
2.4. Bioinformatic analysis
To obtain sequences of DJ1 homologues, we performed a BLASTP search against the Swiss-Prot + TrEMBL complete proteome database. The multiple amino acid sequence alignment was performed using the T-COFFIE program, which was manually edited with GeneDoc version 2.6 [8].
The DJ1 structural model was generated by using the Molscript software on the basis of the crystal structure of the human DJ1 protein (Protein Data Bank code 1PS4).
Scores for enhancer/silencer sequence binding sites were predicted by ESE finder 2.0 software http://rulai.cshl.edu/tools/ESE/index.html [4].
Amino acid substitutions were predicted by PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and SIFT [22].
3. Results
The DJ1 gene was fully analyzed in 163 EOPD patients and 100 healthy controls. We found no homozygous or compound heterozygous mutations with an obvious pathogenic effect. A total of twenty-nine variants were identified, 10 of which were novels. All variants had similar frequency in patients and controls (Table 1).
Table 1.
DJ1 genomic variants identified in 163 EOPD patients and 100 healthy controls.
| Exon | Variant | Genotype | PD subjects | Allelic % | CNT subjects | Allelic % | p* |
|---|---|---|---|---|---|---|---|
| 1 | c.-84-52A>G rs17523802 | HET | 0 | 2.5 | 12/100 | 6.0 | 0.039 |
| HOMO | 4/163 | 0 | |||||
| 1 | c.-84-15T>C rs226249 | HET | 71/163 | 55.5 | 58/100 | 56.0 | 0.915 |
| HOMO | 55/163 | 27/100 | |||||
| 1 | c.-70C>T rs11121064 | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 | |||||
| 1 | c.-24+66C>G (Tarantino, 2010) | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 | |||||
| 1 | c.-24+92_111del18bp rs7944515 | HET | 21/163 | 8.9 | 19/100 | 10.5 | 0.543 |
| HOMO | 4/163 | 1/100 | |||||
| 1 | c-24+120G>T rs35675666 | HET | 4/163 | 2.5 | 1/100 | 0.5 | 0.193 |
| HOMO | 2/163 | 0 | |||||
| 2 | c.-22C>T rs12077723 | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 | |||||
| 2 | c.71A>C (p.D24A) | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 | |||||
| 3 | c.91-102A>G | HET | 0 | 0.0 | 1/100 | 0.5 | 0.301 |
| HOMO | 0 | 0 | |||||
| 3 | c.91-109C>T rs7517357 | HET | 28/163 | 11.7 | 16/100 | 8.0 | 0.201 |
| HOMO | 5/163 | 0 | |||||
| 3 | c.192+81A>G rs71653618 | HET | 0 | 0.0 | 1/100 | 0.5 | 0.201 |
| HOMO | 0 | 0 | |||||
| 4 | c.+193-86 A>G rs41278962 | HET | 11/163 | 3.4 | 10 | 5.0 | 0.355 |
| HOMO | 0 | 0 | |||||
| 4 | c.+252+47A>G | HET | 1/163 | 0.3 | 1/100 | 0.5 | 0.727 |
| HOMO | 0 | 0 | |||||
| 4 | c.+252+37_46delG | HET | 7/163 | 2.1 | 2/100 | 1.0 | 0.494 |
| HOMO | 0 | 0 | |||||
| 4 | c.+252+46insG | HET | 5/163 | 2.1 | 7/100 | 4.5 | 0.325 |
| HOMO | 1/163 | 1/100 | |||||
| 4 | c.+252_253GG>AA | HET | 30/163 | 14.1 | 17/100 | 8.5 | 0.054 |
| HOMO | 8/163 | 0 | |||||
| 4 | c.+252+30T>G rs2641116 | HET | 32/163 | 12.9 | 18/100 | 9.0 | 0.174 |
| HOMO | 5/163 | 0 | |||||
| 4 | c.+252+3-8insA (Tarantino, 2010) | HET | 1/163 | 0.3 | 2/100 | 1.0 | 0.305 |
| HOMO | 0 | 0 | |||||
| 5 | c.253-109A>G | HET | 0 | 0.0 | 1/100 | 0.5 | 0.201 |
| HOMO | 0 | 0 | |||||
| 5 | c.253-98G>A rs6703670 | HET | 15/163 | 5.2 | 12/100 | 7.0 | 0.399 |
| HOMO | 1/163 | 1/100 | |||||
| 5 | c.+253-31C>T rs7534132 | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 | |||||
| 5 | c.+322+31C>T rs389298 | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 | |||||
| 5 | c.293G>A (p.R98Q) rs71653619 | HET | 6/163 | 1.8 | 4/100 | 2.0 | 0.897 |
| HOMO | 0 | 0 | |||||
| 6 | c.323-216G>A rs161807 | HET | 61/163 | 34.7 | 58/100 | 46.0 | 0.010 |
| HOMO | 26/163 | 17/100 | |||||
| 6 | c.323-14A>G rs72854882 | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 | |||||
| 6 | c.323-48A>G rs72854880 | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 | |||||
| 6 | c.+409+33 insGTT | HET | 1/163 | 0.3 | 1/100 | 0.5 | 0.727 |
| HOMO | 0 | 0 | |||||
| 7 | c.486C>A (p.F162L) | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 | |||||
| 7 | c.+410-29A>G | HET | 1/163 | 0.3 | 0 | 0.0 | 0.433 |
| HOMO | 0 | 0 |
Novel variants are in bold.
p value were calculated on allelic frequencies with chi-square test. All values were non-significant according to Bonferroni correction (p < 0.002).
Subjects carried only one variant, with the exception of one patient (PD212) who carried six known polymorphisms: c.323-14A->G (rs72854882); c.323-48A>G (rs72854880); c.-84-15T>C (rs226249); c.-70C>T (rs11121064); c.-22C>T (rs12077723); c.+253-31C>T (rs7534132). Except for the c.-84-15T>C, they resulted present only in this patient and not in controls. In silico analysis (Mutation Tester Program) suggested that all six variants, including the two located in the 5′UTR (c.-70C>T and c.-22C>T), have no functional effects.
Two novel substitutions, the p.Asp24Ala and the p.Phe162Leu, were identified in two patients both in heterozygous state (Table 1).
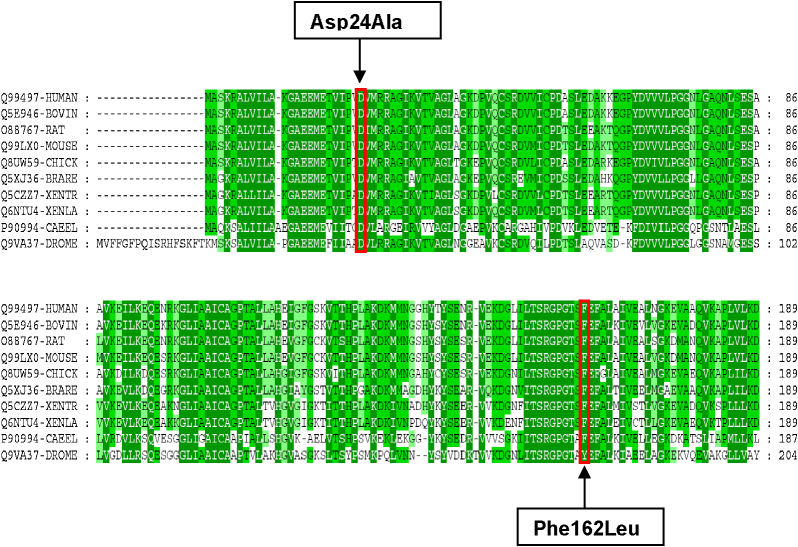
The substitution p.Asp24Ala (c.71A>C) in exon 2 has been found in a sporadic case of Italian origin (PD1219) and is predicted to replace a highly conserved Aspartic acid with Alanine in amino acid position 24 (Fig. 1). The complete sequence and quantitative analysis of DJ1 coding region did not reveal a second mutation. In order to exclude intronic changes that might unbalance the formation of the two mRNA isoforms, we sequenced the genomic region upstream exon 2 (1407 bp encompassing exon 1 and intron 1) in the p.Asp24Ala carrier. No new variants were detected.
Fig. 1.
Alignment and comparison of several DJ-1 protein sequences. [Q99497 – Homo sapiens (Human), Q5E946 – Bos taurus (Bovine), O88767 – Rattus norvegicus (Rat), Q99LX0 – Mus musculus (Mouse), Q8UW59 – Gallus gallus (Chicken), Q5XJ36 – Brachydanio rerio (Zebrafish), Q5CZZ7 – Xenopus tropicalis, Q6NTU4 – Xenopus laevis, P90994 – Caenorhabditis elegans, Q9VA37 – Drosophila melanogaster]. The black arrows indicate the highly conserved positions 24 of aspartic acid and 162 of phenylalanine.
The second newly identified variant is a nucleotide substitution in exon 7, the c.486C>A, leading to a missense mutation in the coding region, the p.Phe162Leu. This change was found in a PD subject resulted previously positive at the PRKN gene analysis: she was carrier of the well-known mutation p.Cys253Tyr in homozygous state. She was not of Italian origin and developed the first signs of PD (resting tremor at the right arm) when she was 20. She slowly developed bradykinesia and rigidity. Diagnosis of PD was confirmed by Dopamine transporter SPECT imaging (FP-CIT) and by good response to levodopa treatment. Similarly, one sister developed PD at 20 years of age: she resulted homozygous for the p.Cys253Tyr PRKN mutation, but was non-carrier of the p.Phe162Leu DJ1 gene variant.
Amino acid Phe162 is well conserved at phylogenic level (Fig. 1). Although this missense variant occurs in a highly conserved region of the protein, the amino acid change is conservative and the mutation is predicted to be benign by PolyPhen-2 (1.06 of score – sensitivity 0.92, specificity 0.59) and by SIFT prediction WEB tools for amino acid substitutions (score of 0.07). This mutation was not observed in 150 healthy controls (100 Italian and 50 from the same South European country of the patient) excluding the chance to be a common polymorphism.
DJ1 quantitative analysis (MLPA) analysis did not detect any exon rearrangements in these two patients.
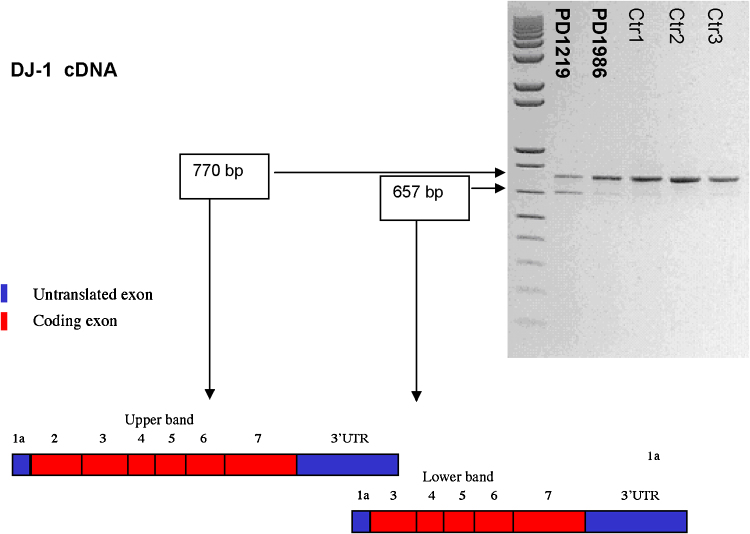
To investigate the presence of a second mutation that might be lost in the screening at DNA level, missense mutations cDNA analysis was performed in both cases. cDNA analysis (from RNA patient's leukocytes and from three control subjects) revealed two cDNA fragments: one of the expected size (770 base pairs) spanning the seven exons of DJ-1 gene, and one smaller band of 657 bp. The sequencing of the larger cDNA fragment confirmed the heterozygosity identified by genomic sequencing. Sequencing of the smaller band showed an mRNA isoform missing the entire exon 2, that contains the protein start codon. For this reason, and also because no other ORFs are predicted within the remaining coding sequence, it is likely that the smaller mRNA is untranslated. It seems that this transcript physiologically exists in the leukocytes, although at very low levels, since it is present in healthy subjects as well as in affected patients. Interestingly, the ratio between these two isoforms (larger vs. smaller) is much lower in the patient carrying the p.Asp24Ala substitutions that in controls (Fig. 2). This experiment was replicated three times starting from three different RT-PCR products obtaining similar results. In order to exclude intronic changes that might unbalance the formation of the two mRNA isoforms we also sequenced the genomic region upstream exon 2 (1407 bp encompassing exon 1 and intron 1) in the p.Asp24Ala carrier. No new variants were detected.
Fig. 2.
RT-PCR experiments on peripheral DNA leukocytes from patients PD1219 and PD1986, and from three control subjects. Two cDNA fragments were amplified: one of the expected size (770 base pairs) spanning the seven exons of DJ-1 gene, and one smaller isoform (657 bp), lacking the entire exon 2 containing the start codon.
4. Discussion
We analyzed 163 EOPD patients with onset before 40 years of age and none was homozygous or compound heterozygous of variants with either a known or a predictable pathological effect on the protein. These findings reveal that DJ1 mutations are very rare in Italian EOPD patients.
Data on DJ1 mutations frequency are limited. Several studies in PD show a very low rate of mutations on the DJ1 gene, ranging from 0% [20,24,30] to 2% [6,10,27]. These studies were mostly carried out using a multiethnic population and results were difficult to compare one another. Sleiman and colleagues screened 190 PD subjects from UK and found 2 positive cases: p.Met26Ile in homozygous state in an Ashkenazi Jewish patient and p.Asp149Ala in heterozygous state in an Afro-Caribbean patient [27]. Clark and colleagues screened 89 EOPD patients of different ethnic background [6] and found the p.Ala104Thr mutation in heterozygous state in an Asian patient and a rare silent variant (the p.Ala167Ala) in an African control. The estimated rate of mutant alleles by these authors is approximately 1%. Pankratz and colleagues screened 93 PD patients from 63 families (95% Caucasian and 5% Hispanic) finding two silent substitutions: c.480C>A (p.Thr160Thr) and c.501A>G (p.Ala167Ala) [24]. Guo and colleagues screened 29 highly selected Chinese families with Autosomal Recessive PD and found one heterozygous subject carrying the p.Leu10Pro mutation [10].
In the Italian population, two studies on relatively small group of patients failed to identify exonic variants working in a clear recessive manner [16,28]. Klein and colleagues did not find any DJ1 mutation in 65 PD patients with onset before 50 years of age [16]. Tarantino and colleagues tested 40 PD patients with onset before 45 years of age and found a single patient carrying two de novo heterozygous nucleotidic substitutions, both located in non-coding regions [28]. We found the same variants in one PD patients as well as in healthy controls (see Table 1). For these and other reasons previously reported [26], we considered these two variants as polymorphisms.
In this study, we have identified two novel missense variants at heterozygous state: the p.Asp24Ale and the p.Phe162Leu. Given that DJ1 works as a dimer, it has been suggested that some mutations may alter the quaternary structure of the protein with a dominant-negative effect against wild-type DJ1. However, the two cases identified did not support this hypothesis. Indeed, neither of our patients had a family history suggestive of dominant inheritance pattern. The patient carrying the p.Asp24Ala mutation did not report any relative with movement disorders, consistent with the diagnosis of PD. While the patient carrier of the p.Phe162Leu mutation had a sister affected by PD not carrying the DJ1 mutation, both sisters were carriers of a PRKN mutation in homozygous state. There were no relevant differences in the clinical course of the two sisters and there were no other PD cases in the family. This suggests that the p.Phe162Leu variant was not the cause of PD in this family. It is also possible that the effect of PRKN mutation was stronger than DJ1 mutation to produce the PD phenotype. The absence of a combined effect between the two genes confirms the skepticism of Thomas and colleagues [29] about the existence of a functional ubiquitin E3 ligase complex (PPD complex) comprising PRKN, PINK1 and DJ1 proteins [34].
However, we cannot exclude that p.Phe162Leu and p.Asp24Ala variants may cause PD when present in homozygous or compound heterozygous state with other mutations.
On the other hand, it might be speculated that p.Phe162Leu and p.Asp24Ala variants are real dominant mutations with a possible restoring mechanism, as already demonstrated for the p.Leu166Pro mutation [5]. The use of computational techniques to simulate molecular dynamics and protein–protein interactions should help to elucidate this hypothesis.
We analyzed the messenger RNA in our heterozygous patients to search for a second mutation at DNA level, but did not find it. In both cases, the sequence of the complete cDNA fragment confirmed the heterozygosity for the previous identified variants.
It is worth noting that in one patient (PD212) we have identified six different variants, which individually have no significant effect on the protein. However, most of these variants are novel and have not been identified so far either in controls or in other patients. In this patient, mRNA analysis was not possible and thus we cannot exclude an overall effect of these variants as the cause of PD, in particular by acting on either gene expression or mRNA stability.
In our analysis, we also found several known and novel variants both in cases and controls. These variants had a similar frequency in the two populations. This suggests to exclude these variants as risk factors for PD, although a larger number of cases and controls need to be studied to fully evaluate their pathogenic effect.
We conclude that DJ1 PD causing mutations are very rare in Italian population. Other genes and risk factors for PD are still to be identified.
Acknowledgments
This work was supported by Italian Telethon Foundation (grant no. GGP11164). The DNA samples were from the “Parkinson Institute Biobank” (www.parkinsonbiobank.com), supported by the Italian Telethon Foundation (grant no. GTB12001) and by the “Fondazione Grigioni per il Morbo di Parkinson”.
We thank Prof. Vincenzo Bonifati, dr A. Di Fonzo and Dr. M. Travi for their help and support.
Dr. Francesca Sironi personally thanks Sophie Lagues, as member of Rotterdam SGI-Holland, for her personal inspiration in her daily life.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Annesi G., Savettieri G., Pugliese P., D’Amelio M., Tarantino P., Ragonese P., La Bella V., Piccoli T., Civitelli D., Annesi F., Fierro B., Piccoli F., Arabia G., Caracciolo M., Cirò Candiano I.C., Quattrone A. DJ1 mutations and parkinsonism-dementia-amyotrophic lateral sclerosis complex. Ann. Neurol. 2005;58:803–807. doi: 10.1002/ana.20666. [DOI] [PubMed] [Google Scholar]
- 2.Bader V., Ran Zhu X., Lubbert H., Stichel C.C. Expression of DJ1 in the adult mouse CNS. Brain Res. 2005;1041:102–111. doi: 10.1016/j.brainres.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Bonifati V., Rizzu P., van Baren M.J., Schaap O., Breedveld G.J., Krieger E., Dekker M.C., Squitieri F., Ibanez P., Joosse M., van Dongen J.W., Vanacore N., van Swieten J.C., Brice A., Meco G., van Duijn C.M., Oostra B.A., Heutink P. Mutations in the DJ1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299(5604):256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 4.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C.Y.C. Mechanism of BAG1 repair on Parkinson's disease linked DJ1 mutation. J. Biomol. Struct. Dyn. 2012;30(1):1–12. doi: 10.1080/07391102.2012.674182. [DOI] [PubMed] [Google Scholar]
- 6.Clark L.N., Afridi S., Mejia-Santana H., Harris J., Louis E.D., Cote L.J., Andrews H., Singleton A., Wavrant De-Vrieze F., Hardy J., Mayeux R., Fahn S., Waters C., Ford B., Frucht S., Ottman R., Marder K. Analysis of an early-onset Parkinson's disease cohort for DJ1 mutations. Mov. Disord. 2004;19(7):796–800. doi: 10.1002/mds.20131. [DOI] [PubMed] [Google Scholar]
- 7.Corti O., Lesage S., Brice A. What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol. Rev. 2011;91(4):1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- 8.Deerfield D.W., 2nd, Holland-Minkley A.M., Geigel J., Jr., Nicholas H.B. Classification of the environment of protein residues. J. Protein Chem. 1997;16(5):441–447. doi: 10.1023/a:1026349124850. [DOI] [PubMed] [Google Scholar]
- 9.Goldwurm S., Zini M., Di Fonzo A., De Gaspari D., Siri C., Simons E.J., van Doeselaar M., Tesei S., Antonini A., Canesi M., Zecchinelli A., Mariani C., Meucci N., Sacilotto G., Cilia R., Isaias I.U., Bonetti A., Sironi F., Ricca S., Oostra B.A., Bonifati V., Pezzoli G. LRRK2 G2019S mutation and Parkinson's disease: a clinical, neuropsychological and neuropsychiatric study in a large Italian sample. Parkinsonism Relat. Disord. 2006;12:410–419. doi: 10.1016/j.parkreldis.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Guo J., Xiao B., Liao B., Zhang X., Nie L., Zhang Y., Shen L., Jiang H., Xia K., Pan Q., Yan X., Tang B. Mutation analysis of Parkin, PINK1, DJ1 and ATP13A2 genes in Chinese patients with autosomal recessive early-onset parkinsonism. Mov. Disord. 2008;23(14):2074–2093. doi: 10.1002/mds.22156. [DOI] [PubMed] [Google Scholar]
- 11.Hague S., Rogaeva E., Hernandez D., Gulick C., Singleton A., Hanson M., Johnson J., Weiser R., Gallardo M., Ravina B., Gwinn-Hardy K., Crawley A., George-Hyslop P.H., Lang A.E., Heutink P., Bonifati V., Hardy J., Singleton A. Early-onset Parkinson's disease caused by a compound heterozygous DJ1 mutation. Ann. Neurol. 2003;54(2):271–274. doi: 10.1002/ana.10663. [DOI] [PubMed] [Google Scholar]
- 12.Hedrich K., Djarmati A., Schäfer N., Hering R., Wellenbrock C., Weiss P.H., Hilker R., Vieregge P., Ozelius L.J., Heutink P., Bonifati V., Schwinger E., Lang A.E., Noth J., Bressman S.B., Pramstaller P.P., Riess O., Klein C. DJ1 (PARK7) mutations are less frequent than Parkin (PARK2) mutations in early-onset Parkinson disease. Neurology. 2004;62(3):389–394. doi: 10.1212/01.wnl.0000113022.51739.88. [DOI] [PubMed] [Google Scholar]
- 13.Hedrich K., Schafer N., Hering R., Hagenah J., Lanthaler A.J., Schwinger E., Kramer P.L., Ozelius L.J., Bressman S.B., Abruzzese G., Martinelli P., Kostic V., Pramstaller P.P., Vieregge P., Riess O., Klein C. The R98Q variation in DJ1 represents a rare polymorphism. Ann. Neurol. 2004;55(1):145. doi: 10.1002/ana.10816. [DOI] [PubMed] [Google Scholar]
- 14.Hughes A.J., Danie S.E., Kilford L., Lees A.J. Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry. 1992;55(3):181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes A.J., Daniel S.E., Lees A.J. Improved accuracy of clinical diagnosis of Lewy body Parkinson's disease. Neurology. 2001;57(8):1497–1499. doi: 10.1212/wnl.57.8.1497. [DOI] [PubMed] [Google Scholar]
- 16.Klein C., Djarmati A., Hedrich K., Schäfer N., Scaglione C., Marchese R., Kock N., Schüle B., Hiller A., Lohnau T., Winkler S., Wiegers K., Hering R., Bauer P., Riess O., Abbruzzese G., Martinelli P., Pramstaller P.P. PINK1, Parkin, and DJ1 mutations in Italian patients with early-onset parkinsonism. Eur. J. Hum. Genet. 2005;13:1086–1093. doi: 10.1038/sj.ejhg.5201455. [DOI] [PubMed] [Google Scholar]
- 17.de Lau L.M., Breteler M.M. Lancet Neurol. 2010;5(6):525–535. doi: 10.1016/S1474-4422(06)70471-9. [DOI] [PubMed] [Google Scholar]
- 18.Lees A.J., Tamas Revesz J.H. Parkinson's disease. Lancet. 2009;373:2055–2066. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 19.Lev N., Ickowicz D., Barhum Y., Lev S., Melamed E., Offen D. DJ1 protects against dopamine toxicity. J. Neural Transm. 2009;116:151–160. doi: 10.1007/s00702-008-0134-4. [DOI] [PubMed] [Google Scholar]
- 20.Lohmann E., Dursun B., Lesagec S., Hanagasia H.A., Sevinc G., Honore A., Bilgic B., Gurvit H., Dogu O., Kaleagası H., Babacan G., Yazici J., Erginel-Unaltuna N., Brice A., Emre M. Genetic bases and phenotypes of autosomal recessive Parkinson disease in a Turkish population. Eur. J. Neurol. 2012;19:769–775. doi: 10.1111/j.1468-1331.2011.03639.x. [DOI] [PubMed] [Google Scholar]
- 21.Nuytemans K., Theuns J., Cruts M., Van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Hum. Mutat. 2010;31(7):763–780. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng P.C., Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2010;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obeso J.A., Rodriguez-Oroz M.C., Goetz C.G., Marin C., Kordower K.H., Rodriguez M., Hirsch E.C., Farrer M., Schapira A.H., Halliday G. Missing pieces in the Parkinson's disease puzzle. Nat. Med. 2010;16(6):653–661. doi: 10.1038/nm.2165. [DOI] [PubMed] [Google Scholar]
- 24.Pankratz N., Pauciulo M.W., Elsaesser V.E., Marek D.K., Halter C.A., Wojcieszek J., Rudolph A., Shults C.W., Foroud T., Nichols W.C., Parkinson Study Group – PROGENI Investigators Mutations in DJ1 are rare in familial Parkinson disease. Neurosci. Lett. 2006;408(3):209–213. doi: 10.1016/j.neulet.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sironi F., Primignani P., Zini M., Tunesi S., Ruffmann C., Ricca S., Brambilla T., Antonini A., Tesei S., Canesi M., Zecchinelli A., Mariani C., Meucci N., Sacilotto G., Cilia R., Isaias I.U., Garavaglia B., Grezzi D., Coviello D.A., Pezzoli G., Goldwurm S. Parkin analysis in early onset Parkinson disease. Parkinsonism Relat. Disord. 2008;14:326–333. doi: 10.1016/j.parkreldis.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Sironi F., Primignani P., Goldwurm S. Comment on compound heterozygosity in DJ1 gene non-coding portion related to Parkinsonism. Parkinsonism Relat. Disord. 2010;16(5):360–361. doi: 10.1016/j.parkreldis.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Sleiman P.M., Healy D.G., Quinn N., Lees A.J., Wood N.W. The role of pathogenic DJ1 mutations in Parkinson's disease. Ann. Neurol. 2003;54(3):283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 28.Tarantino P., Donatella C., Annesi F., De Marco E.V., Rocca F.E., Pugliese P., Nicoletti G., Carrideo S., Provenzano G., Annesi G., Quattrone A. Compound heterozygosity in DJ1 gene non-coding portion related to parkinsonism. Parkinsonism Relat. Disord. 2009;15:324–326. doi: 10.1016/j.parkreldis.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Thomas K.J., McCoy M.K., Blackinton J., Beilina A., vanderBrug M., Sandebring A., Miller D., Maric D., Cedazo-Minguez A., Cookson M.R. DJ1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 2011;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomiyama H., Li Y., Yoshino H., Mizuno Y., Kubo S., Toda T., Hattori N. Mutation analysis for DJ1 in sporadic and familial parkinsonism: screening strategy in parkinsonism. Neurosci. Lett. 2009;455:159–161. doi: 10.1016/j.neulet.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 31.van Duijn C.M., Dekker M.C., Bonifati V., Galjaard R., Houwing-Duistermaat J., Snijders P., Testers L., Breedveld G., Horstink M., Sandkuijl L., van Swieten J., Oostra B., Heutink P. Park7, A novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36. Am. J. Hum. Genet. 2001;69:629–634. doi: 10.1086/322996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilhelmus Micha M.M., Nijland P.G., Drukarch B., deVries H.E., van Horssen J. Involvement and interplay of Parkin, PINK1, and DJ1 in neurodegenerative and neuroinflammatory disorders. Free Radic. Biol. Med. 2012;53:983–992. doi: 10.1016/j.freeradbiomed.2012.05.040. [DOI] [PubMed] [Google Scholar]
- 33.Wilson M.A., Collins J.L., Hod Y., Ringe D., Petsko G.A. The 1.1-A resolution crystal structure of DJ1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong H., Wang D., Chen L., Choo Y.S., Ma H., Tang C., Xia K., Jiang W., Ronai Z., Zhuang X., Zhang Z. Parkin, PINK1, and DJ1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J. Clin. Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]