Abstract
Background
Qualitative and quantitative analysis of small non-coding RNAs by next generation sequencing (smallRNA-Seq) represents a novel technology increasingly used to investigate with high sensitivity and specificity RNA population comprising microRNAs and other regulatory small transcripts. Analysis of smallRNA-Seq data to gather biologically relevant information, i.e. detection and differential expression analysis of known and novel non-coding RNAs, target prediction, etc., requires implementation of multiple statistical and bioinformatics tools from different sources, each focusing on a specific step of the analysis pipeline. As a consequence, the analytical workflow is slowed down by the need for continuous interventions by the operator, a critical factor when large numbers of datasets need to be analyzed at once.
Results
We designed a novel modular pipeline (iMir) for comprehensive analysis of smallRNA-Seq data, comprising specific tools for adapter trimming, quality filtering, differential expression analysis, biological target prediction and other useful options by integrating multiple open source modules and resources in an automated workflow. As statistics is crucial in deep-sequencing data analysis, we devised and integrated in iMir tools based on different statistical approaches to allow the operator to analyze data rigorously. The pipeline created here proved to be efficient and time-saving than currently available methods and, in addition, flexible enough to allow the user to select the preferred combination of analytical steps. We present here the results obtained by applying this pipeline to analyze simultaneously 6 smallRNA-Seq datasets from either exponentially growing or growth-arrested human breast cancer MCF-7 cells, that led to the rapid and accurate identification, quantitation and differential expression analysis of ~450 miRNAs, including several novel miRNAs and isomiRs, as well as identification of the putative mRNA targets of differentially expressed miRNAs. In addition, iMir allowed also the identification of ~70 piRNAs (piwi-interacting RNAs), some of which differentially expressed in proliferating vs growth arrested cells.
Conclusion
The integrated data analysis pipeline described here is based on a reliable, flexible and fully automated workflow, useful to rapidly and efficiently analyze high-throughput smallRNA-Seq data, such as those produced by the most recent high-performance next generation sequencers. iMir is available at http://www.labmedmolge.unisa.it/inglese/research/imir.
Keywords: Next generation sequencing, SmallRNA-Seq, Data analysis pipeline, Breast cancer, Small non-coding RNA, microRNA, Piwi-interacting RNA
Background
Small RNA analysis by massively parallel sequencing (smallRNA-Seq) represents an increasingly popular method to address different questions concerning the biological role of miRNAs and other regulatory small transcripts, such as piwi-interacting (piRNAs), small inhibitory (siRNAs), transcription initiation (tiRNAs), transfer (tRNAs) and other small non-coding (sncRNAs) RNAs, including also extra-cellular small RNAs (exRNAs). Among sncRNAs, miRNAs and piRNAs are emerging as key regulators in multiple cellular functions and for this reason are widely studied by direct sequencing. miRNAs, the best know and studied class of sncRNAs, are interesting to investigate due to their ability to control gene expression in eukaryotes by fine tuning mRNA translation [1-3]. They represent a class of short (~ 22 nucleotides) RNA molecules that play pivotal roles in a variety of molecular processes, such as immune response [4], differentiation [5], development [6-8], infection [9,10] and carcinogenesis [11-13]. miRNA genes are synthesized as long precursor RNA molecules (pri-miRNAs), usually by RNA polymerase II [14], that are rapidly processed in the nucleus by Drosha RNase III to release approximately 70 nucleotides long miRNA precursor stem loop (pre-miRNA) [15] that in turn are exported to the cytoplasm by Exportin 5 [16]. In the cytoplasm, mature miRNAs are produced through the action of Dicer RNase [17]. These small RNAs regulate gene expression by binding to targets sites generally in the 3′ untraslated region (3′ UTR) of target mRNAs, resulting in mRNA degradation or translation inhibition [1,18]. miRNAs recognition of the 3′ UTR of their target mRNA is mediated by hybridization between nucleotides 2–8 at 5′ end of the small RNA (seed sequence) and the complementary sequences present in the 3′ UTR of the mRNA [1,19,20]. On the other hand, small non-coding RNAs that interact with Piwi proteins, called piRNAs, are emerging as regulatory transcripts able to control a broad range of biological processes. The main roles of these molecules has been investigated mainly in germline stem cells, where they are involved in: (i) regulation of transposone activity; (ii) modulation of genome epigenetic state, (iii) development and (iv) spermatogenesis [21]. However piRNAs have been also identified in somatic cells, including human cancer cells [22], suggesting their possible involvement in tumors. This aspect highlights the need for sensitive and efficient bioinformatics tools to study these novel class of sncRNAs in smallRNA-Seq datasets. SmallRNA-Seq allows detection of RNAs with a high dynamic range and reliably measures small differences in RNA concentration between samples, enabling also to discover novel RNA molecules not annotated in databases. Generally, data analysis is performed by combining multiple statistical and bioinformatics tools available from different sources. Many useful programs for processing these data exist nowadays, such as RandA [23], Shortran [24], UEA sRNA Workbench [25], DSAP [26], miRTools 2.0 [27] and miRExpress [28]. Two main issues hamper diffusion and implementation of such programs: (i) web-based tools have some restriction on data upload; (ii) stand-alone programs often lack one or more analysis steps, such as for example prediction of novel sncRNAs. As main consequence, the analytical workflow is slowed down by the need for the continuous interventions by the operator, a critical factor when a large number of samples need to be analyzed at once. A main challenge in bioinformatics is thus to create comprehensive computational tools for handling and analyzing, in an automated manner, the huge amount of data generated by these experiments.
We describe here a modular analysis pipeline, iMir, for comprehensive analyses of smallRNA-Seq data integrating multiple open source modules and resources linked together in automated way. The pipeline allows identification of miRNAs and other sncRNAs, such as piRNAs, to perform differential expression analysis and, for miRNAs, to predict the corresponding mRNA targets. In addition, iMir provides the possibility to perform hierarchical clustering and to apply different statistical approaches to the analysis, improving discrimination of expressed sncRNAs and allows to identify those more likely to be biologically relevant. The pipeline output includes graphics and text files that are useful for a better interpretation of the results. iMir is well suited for the analysis of smallRNA-seq data obtained from animal samples. Moreover, it can be used to investigate the role of sncRNAs in plants adding the appropriate reference tracks in iMir database.
Implementation
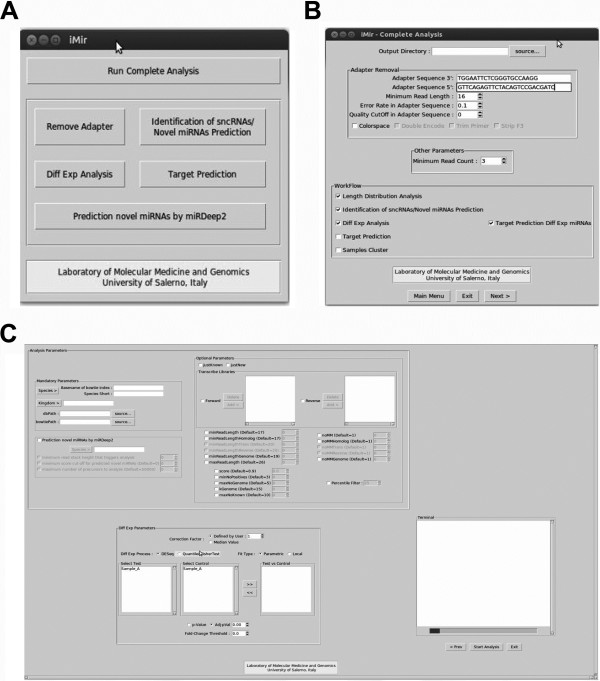
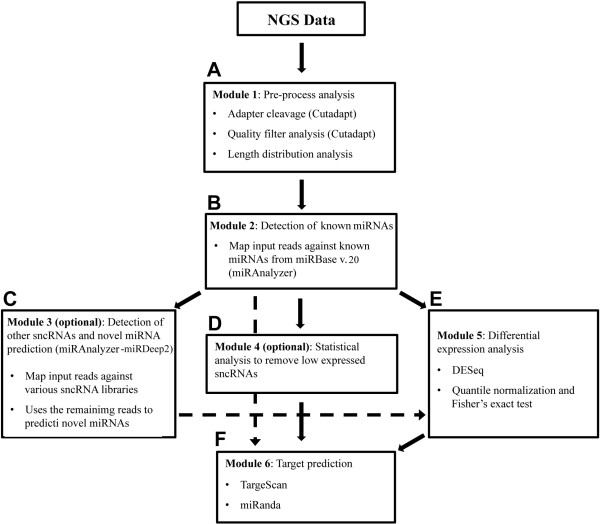
One of the main problems when dealing with the large datasets generated with the currently available Next Generation Sequencing (NGS) technologies are represented by the difficulties in their management and analysis. Analysis of smallRNA-Seq data, for example, requires implementation of different bioinformatics tools and the possibility to perform multiple, subsequent file format conversions that slows-down and makes cumbersome the analytical procedure. The need of a bioinformatics instrument that may help solve these problems in a user-friendly and handy way led us to devise a tool, called iMir, that integrates various open source modules and resources and, in addition, implementing different statistical approaches for sncRNAs expression analyses allows users to select the most performing and relevant method for analysis of their data. The analytical modules included in iMir, selected among the best available, have been made more performant thanks to home-made scripts that allows the user to create self-defined analytical flows. Indeed, the resources integrated in iMir were selected after careful comparison and throughout evaluation of the performance of software widely used in smallRNA-Seq data analysis, according also to what recently reported by Cordero et al. [29] and Williamson et al. [30]. iMir was implemented using an object-oriented programming language, Python, and comprises also a Graphical User Interface (GUI, Figure 1A-C and Additional file 1) that makes it easier the use of command line tools, so that the pipeline is particularly suited for biologist and early stage bioinformaticians, also because it simplify the way how projects are created, parameters are specified for each of several steps of the analysis and the different algorithms are run on project data (Figure 1A). In addition, terminal display window (Figure 1C) can be used to follow the flow of analysis. iMir provides stepwise planning that allows user to select the desired combination of analytical tools in the workflow. Some modules are mandatory while others are, instead, optional. A schematic representation of the iMir workflow is shown in Figure 2A-F. The pipeline takes in input the deeply sequenced reads in FASTQ format. As mentioned above, iMir offers the possibility to run different modules independently and this can be considered one of its main advantages. Indeed, in some cases user can work using pre-analyzed datasets (e.g. reads clipped from adapters sequences, or table with read-counts for each sncRNAs detected in test vs control samples), or may need to perform only a specific analytical step, such as adapter cleavage from input reads, detection of known and/or novel miRNAs, or to map sequence reads against other sncRNA libraries and then to perform differential expression analysis. In all cases, it is possible to start the analytical flow at that step simply by using the input file specific for it. The initial analytical step described in Module 1 (Figure 2A) allows to perform a pre-process analysis of the input files by setting user-defined options for performing adapter cleavage with cutadapt tool [31], as well as quality filtering and analysis of the length distribution of reads. Cutadapt is used for adapter trimming and differs from other adapter trimming tools because it provides several useful option, e.g. error rate assessment in adapter cleavage or search and removal of multiple adapter sequences, essential to get rid of adapter duplications occurring during sequencing library preparation. Module 2 (Figure 2B) allows detection of known miRNAs. To this aim, iMir integrates in its pipeline miRanalyzer stand-alone tool [32], that in its last version (miRanalyzer version 0.3) was improved in speed and features, including a comprehensive analysis of sequences corresponding to isomiR [33]. At this step it is possible to perform also cluster analyses, carrying out PCA analysis and/or applying different hierarchical clustering algorithms. This feature, in fact, is useful when dealing with a very large number of samples to assess similarities and differences among them, such as for example when analyzing results from large cohorts of tumor biopsies. One main advantage of small non-coding RNA sequencing is the possibility to predict novel miRNAs not annotated in databases. This procedure (Module 3, Figure 2C) is performed in iMir with miRanalyzer stand-alone tool [32] and miRDeep2 [34]. With this process it is possible to achieve a dual purpose: (i) to obtain more accurate results on novel miRNAs, that can then be experimentally validated and (ii) to evaluate presence and concentration of reads relative to other sncRNAs in the same datasets. We included in iMir the possibility to implement an intermediate step (Module 4, Figure 2D), before proceeding to differential expression analysis step (Module 5, Figure 2E), to remove the noise and less informative reads, e.g. miRNAs or sncRNAs expressed with very low read counts, that is based on the following statistics: (i) cumulative distribution function and (ii) arbitrary value approaches. Furthermore, low read counts might not reveal a real biological information, being due to sequencing errors or inaccuracy during the procedure of read alignment to the reference genome, such as cross mapping artefacts. To account for this problem, a minimum read count value can be used to filter out reads detected below the cutoff (“Minimum Read Count”, Figure 1B). In addition, after known miRNAs detection (Module 2, Figure 2B), we included statistical approaches to evaluate the cumulative distribution function, such as quartile or percentile values that are computed considering the whole reads-counts to exclude from the list of expressed miRNAs in a given sample those showing read count below that value. This approach can be used also with any sncRNA libraries. The other statistical approach implemented in iMir addresses a common problem encountered when calculating fold-change values (test/control read counts ratios) for RNAs present in the samples at very low levels. Considering a fold-change threshold of ±1.5 and p-value <0.05, when read counts for a given sncRNA are very low this setting can generate biologically irrelevant results. For example, considering a case where 10 tags are assigned to a given RNA in the test sample and 5 in the control, both derived for large cell populations, the resulting fold-change (2.0) may be statistically significant but of doubtful biological relevance [35], contrary to what occurs for sncRNAs showing in the same conditions high read counts. To overcome this problem, we included in iMir the possibility to add in such cases a correction factor. This, computed automatically by the tool as the median of the read count distribution relative to the sncRNA datasets of interest, can be added to the actual read counts of all entries in the datasets. In the case described above, considering a calculated correction factor of 30 the fold-change value calculated for such low-expressed sncRNA will be 1.14, and thus below the threshold set, while for an RNA of the same dataset that shows 4,000 read counts in the sample and 2,000 in the control this adjustment will be irrelevant (see also: Results and discussion). Of note, the user can either disregard this function or use at will an alternative value for this parameter, calculated by any other means desired. These approaches are useful to reduce the number of false-positives detected by differential expression analysis, as these most likely occur among RNAs expressed at a very low level.
Figure 1.
iMir Graphical User Interface. iMir GUI screen-shots. A: Once iMir is launched the user can define which step of the analysis perform and set different parameters for adapter cleavage. B: In the next step, the user can select and rename the samples, while the next windows C: allows to set parameter for detect known and novel miRNAs and differential expression analysis.
Figure 2.
iMir workflow. Graphic summary of iMir workflow: the pipeline accepts NGS data as input and then proceeds automatically to perform several independent analyses, most of which can be selected or excluded according to the user’s needs. Dotted lines represent optional steps of the pipeline. (A) Module 1: Pre-process analyis. (B) Module 2: Detection of known miRNAs. (C) Module 3: Detection of other sncRNAs and novel miRNAs prediction. (D) Module 4: Statistical analysis to remove low expressed sncRNAs. (E) Module 5: Differential expression analysis. (F) Module 6: Target prediction.
Identification and analysis of differentially expressed sncRNAs using digital data is implemented in iMir with two different methods (Module 5, Figures 1C and 2E). The first one, based on the DESeq bioconductor package [36], is particularly suited when biological or technical replicates are available [29]. The second, based on quantile normalization and Fisher’s exact test to assess the statistical relevance, is specially designed for use when no replicates are available [37]. The last iMir module (Module 6, Figure 2F) is designed to perform mRNA targets prediction of expressed, or differentially expressed, miRNAs. mRNA targets are predicted by using miRanda [38,39], that includes current knowledge on target rules and uses a compendium of mammalian miRNAs, and TargetScan [40,41], that computes mRNA targets by searching for the presence of 8mer and 7mer sites matching the seed region of each miRNA. iMir includes in its databases different sncRNAs, such as miRNAs, piRNAs, tRNAs, mRNAs and data from RFam for human, rat and mouse (Additional file 2: Table S1). Performance of iMir was compared with that of the individual bioinformatics tools considered by Williamson et al. [30], selected on the basis of their popularity highlighted by number of citations in the literature. Furthermore, the number of known (available in miRBase) and of novel (absent from the latest release of miRBase) miRNAs detected and the time required to carry to completion the whole analytical flow were evaluated on multiple datasets generated in our laboratory and available from public data repositories and then taken as indicators of iMir performance. The results obtained are in line with what previously reported [30], suggesting reliability of this new tool.
Results and discussion
As an example of the performance of the iMir pipeline, the tool was applied to analyze patterns of sncRNA expression and changes in human breast cancer MCF-7 cells maintained in two different culture conditions affecting cell cycle progression, e.g. growth-arrest and exponential growth [42,43] (see: Additional file 3 for details). For each experimental condition, three sequencing replicates were analyzed to gather a correct estimation also of the technical variability occurring during differential expression analyses. A comprehensive smallRNA-Seq data analysis was performed running all iMir functions with default parameters and the performances of the tool are summarized in Figure 3. To detect classes of sncRNAs other than miRNAs, the raw reads not mapping to known mature miRNAs were aligned against tRNA and mRNA sequences from UCSC Genome Browser, rRNA and piRNA sequences from Nucleotide NCBI database and other sncRNA sequences from RFam [44] (see: Additional file 3 for details). One of the main advantages when applying this procedure is the possibility to reduce false-positive rate in novel miRNAs prediction, while at the same time allowing to search for and analyze other classes of sncRNAs in the datasets. For each sncRNA library sequenced, ~4,0 M raw reads/sample for exponentially growing and ~5.8 M reads/sample for growth arrested cells were obtained (Figure 3 and Table 1). After the pre-process analysis, a small percentage of reads, all <15nt-long, is discarded as the algorithm is unable at present to manage them. The read-length distribution after adapter cleavage in all samples is reported to the right of Module 1 in Figure 3, to show how the majority of reads obtained after this first step are ~22nt long, suggesting that they are mainly due to miRNAs. This observation is further confirmed by the number of reads that actually match known miRNAs (Table 1), computed to account for more than 50% of the entire dataset in each case. The remaining reads, showing a length distribution between 26 and 31nt could include piRNAs, while those 36nt-long could result from longer RNA degradation products. The heatmap reported in Figure 3 to the right of Module 5 highlights the high degree of similarity of miRNA profiles in the three replicates for each sample sequenced (A1-3 for the Case and B1-3 for the Control). A similar results was obtained for piRNAs (data not shown). After that, the iMir module that computes differential expression analysis (Module 5, Figure 2E) was run. miRNA analysis led to the identification of about 460–70 miRNAs per sample (Figure 3 and Table 2), some of which differentially expressed. The pie-charts in Figure 3 summarizes the results of miRNA differential expression analysis performed with DESeq [36], expressed as percentage of detected RNAs showing statistically significant differences in concentration between samples beyond a standard threshold (fold-change ≤ −1.5 or ≥1.5 in exponentially growing vs quiescent cells, p-value ≤ 0.05) or not (70.5%). These results, when compared with previously published data relative to miRNA modulation in the same cell line [13,42,43], confirm that iMir is useful to rapidly and efficiently perform differential analysis for these sncRNAs.
Figure 3.
Graphic representation of iMir pipeline performances. Datasets obtained from smallRNA-Seq analysis in exponentially growing (sample A) or growth-arrested (sample B) MCF-7 cells, performed in triplicate as described in the text, were input in iMir and analyzed with the standard, complete analytical workflow of the tool. The processing time of each module are highlighted in yellow and the graphic outputs of Modules 1 (histograms showing sequence read length distribution in each replicate) and 5 (heat-map visualization of sncRNA profile differences among samples and pie-chart summarizing the results of the differential expression analysis) are shown to their right.
Table 1.
Number of reads before and after adapter cleavage and reads mapped in each sncRNA library included in iMir
| MCF-7 cells | Raw reads | Reads after adapter Cleavage | miRNA reads | tRNA reads | rRNA reads | mRNA reads | piRNA reads | Remaining reads mapping on the genome | Reads not assigned | |
|---|---|---|---|---|---|---|---|---|---|---|
| Exponentially growing |
Replicate 1 |
4,327,501 |
4,068,141 |
2,310,200 |
16,989 |
91,040 |
391,750 |
15,753 |
597,037 |
69,516 |
| |
Replicate 2 |
4,337,535 |
4,075,320 |
2,314,040 |
17,042 |
92,178 |
404,148 |
16,438 |
614,614 |
70,165 |
| |
Replicate 3 |
4,354,046 |
4,091,633 |
2,374,218 |
17,737 |
94,961 |
420,175 |
16,949 |
636,708 |
71,737 |
| Growth-arrested |
Replicate 1 |
6,071,484 |
5,844,875 |
4,626,170 |
13,588 |
72,460 |
181,084 |
14,831 |
234,955 |
40,941 |
| |
Replicate 2 |
6,075,950 |
5,846,690 |
4,621,008 |
12,470 |
75,251 |
185,803 |
15,122 |
242,065 |
40,667 |
| Replicate 3 | 6,085,784 | 5,855,090 | 4,725,975 | 12,705 | 77,842 | 192,161 | 15,582 | 249,638 | 41,494 |
Table 2.
Number of known RNAs and of predicted novel miRNAs identified with iMir in replicate smallRNA-Seq datasets from MCF-7 cells
| |
Exponentially growing cells |
Growth-arrested cells |
||||
|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | Replicate 1 | Replicate 2 | Replicate 3 | |
| miRNA (miRBase v.20) |
473 |
469 |
476 |
461 |
467 |
473 |
| tRNA (UCSC Genome Browser) |
56 |
56 |
54 |
45 |
48 |
47 |
| rRNA (NCBI Nucleotide) |
4 |
4 |
4 |
4 |
4 |
4 |
| mRNA (RefSeq) |
308 |
307 |
307 |
297 |
320 |
325 |
| piRNA (NCBI Nucleotide) |
86 |
85 |
84 |
73 |
70 |
67 |
| Novel miRNA predicted | 46 | 57 | 55 | 38 | 39 | 42 |
Recently, an increasing number of studies highlighted the role of piRNAs in breast cancer [45,46]. Since the average length of these RNAs is ~30nt (see: Figure in Additional file 3: Figure S1), smallRNA-Seq represents an efficient analytical approach to assess also absolute and relative expression of these molecules. Based on this assumption, we searched for and analyzed piRNAs in the datasets selected to test iMir performance. To reduce cross-mapping artifacts, reads corresponding to other RNAs, in particular miRNAs, tRNAs, rRNAs and mRNAs, were first filtered out with iMir mapping them against the selected transcribed RNA libraries. This allowed at once to start from a set of more reliable data and to gather information concerning other small RNAs detected by sequencing (Table 1 and Table 2). This analysis led to the identification of 70 and 85 piRNAs expressed in growth-arrested and exponentially growing MCF-7 cells, respectively. Differential piRNA expression analysis and statistical significance testing performed with iMir revealed 12 downregulated and 25 upregulated piRNAs in growing cells, when compared to quiescent ones (p-value = 0.05, threshold = 1.5; Figure 3). We do not have a ready explanation for these relatively low numbers of piRNAs identified in breast cancer cells, except for the fact that piRNAs know to date have been identified in germ cells [21] and it is thus possible that the majority of them is expressed only in these cell types. Furthermore, most piRNAs identified so far associate with the piRNA biogenesis factor Piwil1 (Hiwi) [21,47], that is not detectable in MCF-7 cells, where only Piwil 2 (Hili) and Piwil4 (Hiwi2) are detected [Hashim et al., manuscript in preparation]. As new validated piRNA datasets will become available, for example those identified by association to Piwil 2 and 4, the possibility built into iMir to customize its database will allow to include these in the analysis. The decision to focus here on individual piRNAs instead of considering their genomic organization in clusters is based on the observation that in somatic cells piRNAs deriving from a given cluster show different levels of steady-state expression, possibly due to a specific mechanism of precursor RNA maturation active in these cells or to differences in their half-life. In addition, recent results suggest that individual piRNAs could play important roles in tumor cells [48-50].
We then tested another function of iMir by performing differential miRNAs expression analysis in two different ways: (i) starting directly from the number of raw read-counts obtained with miRanalyzer [32] or (ii) by adding to each of these counts a correction factor (31), computed as the median of the whole read dataset (see above). Once compared, the results obtained with the two approaches showed slight but substantial differences, since ~10% of the miRNAs identified with the first method (pvalue ≤ 0.05) were excluded by the second one (Additional file 4: Table S2). This is explained by the fact that the RNAs expressed at a very low level under both experimental conditions, and thus of uncertain biological significance, were filtered out when using this correction. iMir offers the possibility to choose this method, when needed, also to other classes of sncRNAs.
With respect to the possibility to perform target prediction for selected miRNAs using miRanda and TargetScan databases, another useful function of the tool, it is worth mentioning the possibility for the user to update when required these and the other databases associated to the pipeline, such as those of miRNAs from miRBase, [51-53], of other sncRNAs from different sources and of mRNA targets from TargetScan [40,41,54] and miRanda [38,39].
Conclusion
We designed, built and describe here iMir, a pipeline that integrates multiple open source modules/resources and implements statistical approaches, combined in an automated flow for high-throughput smallRNA-Seq data analysis. iMir is rapid, accurate and efficient, allowing to examine multiple samples at once and thereby addressing a critical factor for high-throughtput analysis of sncRNA sequencing data, represented by the need for continuous interventions by an operator skilled in informatics and programming. The graphical user interface of iMir, allows a simplified use of the many tools integrated in the pipeline and to customize data analysis according to different needs. In addition, the implementation of different statistical approaches provides the possibility to analyze data according to standard, widely used, as well as to specific needs. Finally, iMir works on Linux and Mac operative systems, user-friendly for biologists with limited skills in informatics. In the future, following the evolution of NGS technologies and recommendations by the scientific community, we plan to keep improving iMir features, including for example tools for sequence variants detection, evolutionary sncRNAs analysis across multiple species and adding specific functions for analysis of emerging classes of small RNAs (pi-, si-, sn-, sno-, ti-RNA, etc.).
Availability and requirements
Project name: iMir.
Project home page:http://www.labmedmolge.unisa.it/inglese/research/imir.
Operating System(s): Unix/Linux based.
Other requirements: Python, Java, Perl, R, DESeq, Bowtie, Vienna RNA Secondary Structure package.
License: GNU GPL v3.
Any restrictions to use by non-academics: specified by GNU GPL v3.
Abbreviations
sncRNA: Small non-coding RNA; miRNA: microRNA; NGS: Next generation sequencing.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GG and MRDF lead in the conception and design of the software, performed the statistical analyses and coordinated manuscript drafting and revision. AR participated in conception and development of the software. AH and FR contributed to software conception and design and to manuscript drafting. GN, MR and RT carried out smallRNA-Seq analyses, manuscript drafting and contributed to software conception. AW coordinated the project, participated in conception of the software and participated in drafting and finalization of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
User Manual.
Summary of annotated small non-coding RNAs included in iMir database.
Additional Materials and Methods and Additional Figure Legend.
List of miRNAs differentially expressed in exponentially growing vs growth-arrested MCF-7 human breast cancer cells (raw and adjusted read-counts, with associated fold-changes and p-values).
Contributor Information
Giorgio Giurato, Email: ggiurato@unisa.it.
Maria Rosaria De Filippo, Email: mdefilippo@unisa.it.
Antonio Rinaldi, Email: arinaldi@unisa.it.
Adnan Hashim, Email: ahashim@unisa.it.
Giovanni Nassa, Email: gnassa@unisa.it.
Maria Ravo, Email: mravo@unisa.it.
Francesca Rizzo, Email: frizzo@unisa.it.
Roberta Tarallo, Email: rtarallo@unisa.it.
Alessandro Weisz, Email: aweisz@unisa.it.
Acknowledgements
Work supported by: Italian Ministry for Education, University and Research (Grant PRIN 2010LC747T_002 to AW and FIRB RBFR12W5V5_003 to RT), Italian Association for Cancer Research (Grant IG-13176), Fondazione Umberto Veronesi’ (Grant 2012–13), EU SeqAhead COST Action (BM1006), CNR Epigen Flagship Project (Grant 2013), University of Salerno (Fondi FARB 2011–2012). GN is supported by a ‘Mario e Valeria Rindi’ fellowship of the Italian Foundation for Cancer Research, MR is supported by a ‘Vladimir Ashkenazy’ fellowship of Italian Association for Cancer Research, FR is supported by a “Young Investigator Programme” fellowship of the Fondazione ‘Umberto Veronesi’, University of Napoli ‘Federico II’ and AH is a PhD student in “Experimental Physiopathology and Neurosciences”, Second University of Napoli.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;14(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;14(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;14(11):831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA. et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;14(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, Wang W, Liu R, George J, Ng HH, Perera RJ. et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;14(1):17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;14(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;14(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;14(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;14(7098):82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;14(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ. et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;14(2):202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF. et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;14:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris O, Ferraro L, Grober OMV, Ravo M, De Filippo MR, Giurato G, Nassa G, Tarallo R, Cantarella C, Rizzo F. et al. Direct regulation of microRNA biogenesis and expression by estrogen receptor beta in hormone-responsive breast cancer. Oncogene. 2012;14(38):4196–4206. doi: 10.1038/onc.2011.583. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;14(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S. et al. The nuclear Rnase III Drosha initiates microRNA processing. Nature. 2003;14(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;14(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;14(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;14(5):243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;14(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;14(5740):1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14(8):523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;14(17–18):1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Isakov O, Ronen R, Kovarsky J, Gabay A, Gan I, Modai S, Shomron N. Novel nsight into the non-coding reperto ire through deep sequencing analysis. Nucleic Acids Res. 2012;14:1093. doi: 10.1093/nar/gks228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Markmann K, Pedersen CN, Stougaard J, Andersen SU. shortran: a pipeline for small RNA-seq data analysis. Bioinformatics. 2012;14(20):2698–2700. doi: 10.1093/bioinformatics/bts496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics. 2012;14(15):2059–2061. doi: 10.1093/bioinformatics/bts311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PJ, Liu YC, Lee CC, Lin WC, Gan RR, Lyu PC, Tang P. DSAP: deep-sequencing small RNA analysis pipeline. Nucleic Acids Res. 2010;14:W385–W391. doi: 10.1093/nar/gkq392. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liu Q, Wang X, Zheng J, Wang T, You M, Sheng Sun Z, Shi Q. mirTools 2.0 for non-coding RNA discovery, profiling, and functional annotation based on high-throughput sequencing. RNA Biol. 2013;14(7):1087–1092. doi: 10.4161/rna.25193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC bioinformatics. 2009;14:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero F, Beccuti M, Arigoni M, Donatelli S, Calogero RA. Optimizing a massive parallel sequencing workflow for quantitative miRNA expression analysis. PLoS One. 2012;14(2):e31630. doi: 10.1371/journal.pone.0031630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V, Kim A, Xie B, McMichael GO, Gao Y, Vladimirov V. Detecting miRNAs in deep-sequencing data: a software performance comparison and evaluation. Brief Bioinform. 2013;14(1):36–45. doi: 10.1093/bib/bbs010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;14:1. [Google Scholar]
- Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 2011;14:W132–W138. doi: 10.1093/nar/gkr247. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M. et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;14(4):610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;14(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;14(5):1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;14(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmire LX, Subramaniam S. Evaluation of normalization methods in mammalian microRNA-Seq data. RNA. 2012;14(6):1279–1288. doi: 10.1261/rna.030916.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;14(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;14:D149–D153. doi: 10.1093/nar/gkm995. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;14(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;14(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicatiello L, Mutarelli M, Grober OM, Paris O, Ferraro L, Ravo M, Tarallo R, Luo S, Schroth GP. et al. Estrogen receptor alpha controls a gene network in luminal-like breast cancer cells comprising multiple transcription factors and microRNAs. Am J Pathol. 2010;14(5):2113–2130. doi: 10.2353/ajpath.2010.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro L, Ravo M, Nassa G, Tarallo R, De Filippo MR, Giurato G, Cirillo F, Stellato C, Silvestro S, Cantarella C. et al. Effects of estrogen on microRNA expression in hormone-responsive breast cancer cells. Horm Cancer. 2011;14(5):610–621. doi: 10.1007/s12672-012-0102-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;14:D226–D232. doi: 10.1093/nar/gks1005. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito T, Magliocca S, Formicola D, Gianfrancesco F. piR_015520 belongs to Piwi-associated RNAs regulates expression of the human melatonin receptor 1A gene. PLoS One. 2011;14(7):e22727. doi: 10.1371/journal.pone.0022727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2012. [Epub ahead of print] [DOI] [PubMed]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;14(7099):203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;14(1):12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;14(7):563–568. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;14(6):1165–1173. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;14:D152–D157. doi: 10.1093/nar/gkq1027. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;14:D140–D144. doi: 10.1093/nar/gkj112. Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;14:D154–D158. doi: 10.1093/nar/gkn221. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;14(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
User Manual.
Summary of annotated small non-coding RNAs included in iMir database.
Additional Materials and Methods and Additional Figure Legend.
List of miRNAs differentially expressed in exponentially growing vs growth-arrested MCF-7 human breast cancer cells (raw and adjusted read-counts, with associated fold-changes and p-values).